Abstract
Phosphatidylcholine (PtdCho), the major phospholipid of animal membranes, is generated by its remodeling and de novo synthesis. Overexpression of the remodeling enzyme, LPCAT1 (acyl-CoA:lysophosphatidylcholine acyltransferase) in epithelia decreased de novo PtdCho synthesis without significantly altering cellular PtdCho mass. Overexpression of LPCAT1 increased degradation of CPT1 (cholinephosphotransferase), a resident Golgi enzyme that catalyzes the terminal step for de novo PtdCho synthesis. CPT1 degradation involved its multiubiquitination and processing via the lysosomal pathway. CPT1 mutants harboring arginine substitutions at multiple carboxyl-terminal lysines exhibited proteolytic resistance to effects of LPCAT1 overexpression in cells and restored de novo PtdCho synthesis. Thus, cross-talk between phospholipid remodeling and de novo pathways involves ubiquitin-lysosomal processing of a key molecular target that mechanistically provides homeostatic control of cellular PtdCho content.
Keywords: CELL/Epithelial, Development Differentiation, Proteases/Ubiquitination, Protein/Degradation, Protein/Stability, Pulmonary Surfactant Apoproteins, Tissue/Organ Systems/Lung
Introduction
Mammalian membranes are enriched with phosphatidylcholine (PtdCho),2 a zwitterionic phospholipid that serves as a major component of various secretory products, including bile, high density lipoproteins, and pulmonary surfactant. With the exception of hepatic tissues, where PtdCho synthesis may involve sequential N-methylation of phosphatidylethanolamine, PtdCho biosynthesis in mammalian cells occurs primarily via the CDP-choline or de novo pathway. This pathway requires three enzymes: choline kinase (CK) (EC 2.7.1.32), which catalyzes the first committed step; CTP:phosphocholine cytidylyltransferase (CCT) (EC 2.7.7.15), the penultimate enzyme; and cholinephosphotransferase (CPT) (EC 2.7.8.2), which catalyzes the terminal reaction within the CDP-choline pathway generating PtdCho (1). Of these enzymes, CCT is rate-limiting and rate-regulatory. CCT is an amphitrophic enzyme and thus can switch between an inactive soluble or cytoplasmic form and an active, membrane-bound species within the nucleus. There are four isoforms of CCT, and deficiency of a major species, CCTα, is associated with impaired cell growth, apoptosis, and embryonic lethality (1). CK may also be critical for embryonic development, and genomic deletion of the CK gene results in bone deformities and muscular dystrophy (2, 3). CPT (CPT1) catalyzes the transfer of the phosphocholine moiety from CDP-choline to diacylglycerol. A second human choline/ethanolaminephosphotransferase (CEPT1) that displays dual specificity using either CDP-choline or CDP-ethanolamine as substrates with significant primary sequence identity to CPT1 may also regulate PtdCho availability (4). CPT1 exists in the Golgi, whereas CEPT1 is detected within the endoplasmic reticulum (5). The substrate requirements of CPT1 and CEPT1 for diacylglycerol would impact the molecular species profile of the newly synthesized PtdCho. Genetic inactivation of CPT genes results in reduced phospholipid synthesis, and its inhibition by isoprenoids and ceramides triggers apoptosis, underscoring CPT as a key regulator of PtdCho synthesis (6–8). Unlike CCT, there is limited information on the molecular control of CK, CPT1, and CEPT1.
In addition to the CDP-choline pathway, the generation of PtdCho in tissues involves its remodeling. In the lung, this remodeling mechanism constitutes a major route for generation of dipalmitoylphosphatidylcholine (DPPtdCho), the major component of surfactant that lowers alveolar surface tension (9). In this pathway, a phospholipase A2 deacylates PtdCho at the sn-2-position of the glycerol backbone, typically releasing an unsaturated fatty acid. The resultant lysophosphatidylcholine (LysoPtdCho) is then reacylated with a saturated fatty acid (typically 16:0 or 14:0), generating surfactant DPPtdCho. The identity of the deacylating phospholipase A2 is a calcium-independent, acid pH-optimal lysosomal enzyme (aiPLA2) (10). The identity of the enzyme that catalyzes the reacylation step remained elusive, but it recently was shown to be an LPCAT1 (acyl-CoA:lysophosphatidylcholine acyltransferase) (11, 12). A conserved HX4D motif is necessary for enzymatic activity (13), and LPCAT1 expression increases with lung development and in response to glucocorticoids (11). LPCAT1 mRNA levels have also been shown to be regulated by keratinocyte growth factor (KGF) in cultured rat type II cells (11). The biochemical and molecular regulation of LPCAT1 and its role in PtdCho synthesis in vivo are largely unknown.
Lung epithelia must tightly balance levels of surfactant (DPPtdCho) lipid secreted into airways versus nonsurfactant PtdCho destined for membranes. Although the de novo pathway generates both nonsurfactant PtdCho and DPPtdCho, the remodeling pathway would be predicted to synthesize primarily surfactant. Thus, it is possible that cross-talk between these pathways exists to modify pulmonary lipid composition, depending on cellular needs. In other systems, such interdependency exists between lipogenic pathways. For example, rates of PtdCho synthesis are coupled to its degradation (14–16). Overexpression of CCT in COS cells increases PtdCho mass only modestly yet triggers a 3-fold increase in its degradation rate (14). This coupling between biosynthetic and degradative pathways indicates close regulation of PtdCho metabolism, perhaps as a means to avoid cellular lipotoxicity. Further, overexpression of PEMT2 (phosphatidylethanolamine N-methyltransferase-2) in hepatoma cells results in feedback inhibition of CCT by reducing its transcriptional rate without altering PtdCho content (17). These latter data suggest interdependency between the N-methylation and CDP-choline pathways for PtdCho biosynthesis. The relationship between the remodeling and de novo pathways for PtdCho synthesis has not been investigated.
Herein, we hypothesized that the remodeling pathway regulates the CDP-choline pathway to maintain PtdCho balance. Overexpression of the remodeling enzyme, LPCAT1, in lung epithelia significantly decreased de novo PtdCho synthesis without altering cellular PtdCho levels. LPCAT1 expression increased degradation of the final enzyme within the CDP-choline pathway, CPT1, a multiubiquitinated enzyme, through its lysosomal elimination. CPT mutants harboring arginine substitutions at multiple putative ubiquitination acceptor sites conferred proteolytic resistance to inhibitory effects of LPCAT1 expression in cells. The data provide new insight into the molecular processing of a key regulatory enzyme involved in PtdCho biosynthesis and are the first indicating interdependence between remodeling and de novo pathways to preserve lipid homeostasis.
EXPERIMENTAL PROCEDURES
The sources of murine lung epithelial (MLE) cells, culture medium, immunoblotting materials, and radiochemicals were described previously (18). Rat primary alveolar type II epithelia were isolated as described (19). Mouse monoclonal ubiquitin antibody was purchased from Cell Signaling (Danvers, MA). The pAmCyan1-C1 and pZsYellow-C1 vector was purchased from Clontech. LysoTracker Red; mouse monoclonal V5 antibody; the To-Pro-3 nuclear staining kit; the PCRTOPO4.1 cloning kit; pcDNA-DEST40, pcDNA3.1/nV5-DEST, and pLenti6/V5-Dest cloning vectors; Escherichia coli One Shot competent cells; the pENTR Directional TOPO cloning kits; LR Clonase II recombinase; the Superscript III RT kit; and the Gateway mammalian expression system were purchased from Invitrogen. Ni2+ resin, HIS-select nickel affinity gel, Tri Reagent®, human KGF, and β-actin primary mouse monoclonal antibody were obtained from Sigma. LPCAT antibody was generated by Covance (Princeton, NJ). Amicon Ultra-4 centrifugal filter devices were from Millipore (Billerica, MA). The QuikChangeTM site-directed mutagenesis kit, XL-gold cells, and pCMV-Tag1 vector were from Stratagene (La Jolla, CA). A ubiquitin plasmid was constructed as described (20). The gel extraction kit and QIAprep Spin Miniprep kits were from Qiagen (Valencia, CA). NucleoBond Xtra Maxi prep kits were obtained from Macherey-Nagel (Bethlehem, PA). Cyclohexamide (CHM) and UbiQapture-Q matrix were from Biomol (Plymouth Meeting, PA). HA-tagged ubiquitin was a gift from Dr. Peter Snyder (University of Iowa). β-1,3-Galactosyltransferase 2 goat polyclonal primary antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Power CYBR Green PCR master mix was from Applied Biosystems (Carlsbad, CA). All restriction enzymes and ligases were purchased from New England Biolabs (Ipswich, MA). The TNT coupled reticulocyte lysate system and RQ1 DNase kit were purchased from Promega (Madison, WI). All DNA sequencing was performed by the University of Iowa DNA Core Facility. Cloning primers were purchased from IDT (Coralville, IA). The Zeiss LSM 510 confocal microscope is part of the University of Iowa Central Microscopy Research Facility.
Quantitative Real-time PCR (qRT-PCR)
RNA was purified from primary murine lung cells (21). Total cellular RNA was isolated using Tri Reagent, and cDNA was obtained by reverse transcription followed by DNase I digestion, amplification, and detection by a Chromo 4 real-time PCR detector (22). Levels of transcripts were measured relative to GAPDH or 18 S mRNA.
Expression Plasmids
The coding sequence available for LPCAT1, CPT1, and CEPT1 on the NCBI Web site (NM_145376, NM_144807, NM_133869) were used to construct primers for cloning of genes from cDNA from mRNA via reverse transcriptase PCR from murine liver and kidney tissues. Amplified fragments were subcloned into PCRTOPO4.1, sequenced, and were found to be identical to the deposited NCBI LPCAT1 sequence. The resulting PCRTOPO4.1 vector served as a source for cloning into pacAd5 CMV internal ribosome entry site enhanced green fluorescent protein pA (University of Iowa DNA Core) using ClaI and BamHI restriction sites. For CPT1 and CEPT1, the Invitrogen Gateway system was used. Primers constructed containing CACC overhangs upstream of the 5′ ATG and antisense sequence with (V5-CEPT1) or without (CPT-V5his) a stop codon were used for amplification using a blunt end polymerase and pENTR/D-TOPO per the manufacturer's instructions. LR Clonase II recombinase was used for cloning of CPT1 and CEPT1 sequences into pcDNA-DEST40 or pcDNA3.1/nV5-DEST gateway vectors, respectively (CPT1-V5his, V5-CEPT1). CPT1K4R and CPT1K6R were constructed by performing site-directed mutagenesis on CPT1-V5his at Lys254, Lys282, Lys283, and Lys292 (CPT1K4R) and additionally Lys307 and Lys311 for CPT1K6R using the QuikChange II XL site-directed mutagenesis (Stratagene) kit. CFP-CPT1 was generated, similar to CFP-CCT (22). The CPT1 construct CPT1K6R was digested with BglII and SalI, purified, and ligated into pAmCyan-C1 as described, generating CFP-CPT1K6R (20). FLAG-CPT1 was constructed by amplifying CPT1 and ligation into pCMV-Tag1. A βGT-YFP (carboxyl-terminal YFP tag) plasmid was constructed by amplifying or digesting three separate fragments for ligation. First, a human cDNA was used as a template to amplify the first 249 base pairs of the UDP-Gal:βGlcNAcβ1,4-galactosyltransferase gene (B4GALT1) (GenBankTM number BC045773) with flanking ClaI/EcoRV restriction sites. Fragment 2 was an EcoRV/EcoRI YFP fragment amplified from pZsYellow-C1. Finally a pacAd5 CMV internal ribosome entry site enhanced green fluorescent protein pA vector, and the two amplified fragments were digested with appropriate restriction enzymes and gel-extracted. The B4GALT1 and YFP fragments were ligated into the pacAd5 CMV internal ribosome entry site enhanced green fluorescent protein pA to generate βGT-YFP. Tandem Ubi-CFP-CPT fusion constructs were generated by cloning the ubiquitin coding sequence at the carboxyl terminus of CFP-CPT1 using SalI and ApaI sites to generate CFP-CPT1×Ub, CFP-CPT3×Ub, and CFP-CPT4×Ub.
Cell Isolation, Culture, and Transient Transfection
Alveolar macrophages, primary type II cells, and fibroblasts were isolated as described previously (23). MLE cells were maintained in Hite's medium (Dulbecco's modified Eagle's medium/F-12 medium) with 2% fetal bovine serum at 37 °C in 5% CO2. After reaching 80% confluence, the cells were harvested using 0.25% trypsin and 0.1% EDTA, resuspended in medium, and plated onto appropriate culture dishes containing a 3 μl/1 μg DNA ratio of FuGENE6 lipofection reagent and appropriate expression vector. After incubation overnight, the medium was replaced with Hite's medium with 2% fetal bovine serum for 8 h before cell harvesting. In some studies, an Amaxa electroporation device with program T-013 and solution L was used for plasmid transfection of cells. Cells were maintained as described above, except after trypsinization, cells were resuspended in a small volume of solution L per the manufacturer's directions. Plasmids were transfected into cells, or cells were left untransfected. Medium was aspirated 24 h post-transfection, and cells were treated an additional 24 h with 0% fetal bovine serum Dulbecco's modified Eagle's medium/F-12 medium supplemented with or without NH4Cl (25 mm). Cell lysates were harvested by brief sonication in 150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA (no EDTA if Ni2+ purification was performed), 2 mm dithiothreitol, 0.025% sodium azide, and 1 mm phenylmethylsulfonyl fluoride (pH 7.4), at 4 °C. Alternatively, cells were switched to Dulbecco's modified Eagle's medium/F-12 medium without fetal bovine serum and treated for 6 h with CHM (18 μg/ml) with either NH4Cl (25 mm) or lactacystin (25 μm). Cells were collected in Buffer A plus 0.5% Triton X-100 plus 0.5% Nonidet P-40 and sonicated for further analysis. MLE cells were also exposed to human KGF (20 ng/ml) for 24 h prior to harvest. Rat type II cells were transduced for 48 h with lentivirus, constructed at the University of Iowa gene transfer vector core, containing LPCAT1 in a pLenti6/V5-Dest vector.
Isolation of Microsome Fractions
Cells were resuspended in Buffer A, and samples were first centrifuged at 16,000 × g for 10 min at 4 °C. The resulting supernatant was centrifuged at 100,000 × g for 60 min at 4 °C. The resulting microsomal pellet was resuspended in Buffer A using a 25-gauge needle.
Phospholipid Analysis
Lipids were extracted from equal amounts of membrane protein, and levels of the individual phospholipids were quantitated with a phosphorus assay (24). DPPtdCho was assayed as before (25). For PtdCho de novo synthesis, cells were pulsed with 1 μCi of [methyl-3H]choline chloride during the final 2 h of incubation with choline-depleted medium. For remodeling activity, cells were labeled with 1.75 nm [14C]LysoPtdCho (55 mCi/mmol) for 3 h. Total cellular lipids were extracted, resolved using TLC, and processed for scintillation counting (18).
LPCAT Activity
Cells were harvested in lysis buffer (250 mm sucrose, 10 mm Tris-HCl, pH 7.4), and equal amounts of microsomal cellular protein were used in the assay. 1 nmol of 1-palmitoyl-sn-glycero-3-phosphocholine (50.44 mm solution in 1:1 chloroform/methanol)/μl of 5× assay buffer was added and sonicated with 5× assay buffer (final concentration 65 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 12.5 mm fatty acid-free bovine serum albumin, 2 mm EDTA). 35 μl of H2O, and cellular protein was added to 10 μl of sonicated assay buffer and 5 μl of [1-14C]acyl-CoA (0.1 μCi, 1.8 nmol) for a total assay volume of 50 μl. Upon the addition of [1-14C]acyl-CoA, samples were incubated at 30 °C for 10 min, after which the reaction was terminated by the addition of chloroform/methanol/H2O (1:2:0.70, v/v/v). Total cellular lipids from reaction mixtures were extracted by the method of Bligh and Dyer (26) and spotted onto LK5D plates, and PtdCho was resolved using TLC and detected using a plate reader (18, 27).
De Novo Enzyme Activities
The activity of CK was assayed as described (18). CCT activity was determined by using a charcoal extraction method (28). CPT activity was assayed as described (29). Each reaction mixture contained 50 mm Tris·HCl buffer (pH 8.2), 0.1 mg/ml Tween 20, 1 mm 1,2- dioleoylglycerol, 0.8 mm phosphatidylglycerol, 0.5 mm [14C]CDP-choline (specific activity 1,110 dpm/nmol), 5 mm dithiothreitol, 5 mm EDTA, 10 mm MgCl2, and 30–40 μg of protein. The lipid substrate was prepared by combining appropriate amounts of 1,2-dioleoylglycerol (1 mm) and phosphatidylglycerol (0.8 mm) in a test tube, drying under nitrogen gas, and brief sonication before the addition to the assay mixture to achieve the final desired concentration. The reaction proceeded for 1 h at 37 °C and terminated with 4 ml of methanol/chloroform/water (2:1:7, v/v/v). The remainder of the assay was performed exactly as described (29). A Bioscan AR-2000 plate reader was used for detection of radiolabeled PtdCho on LK5D TLC plates with data quantified using WinScan software. PtdCho bands on the LK5D silica plates were also scraped and quantified by liquid scintillation counting.
Immunoblot Analysis
Immunoblotting was performed as described (22, 30). The dilution factors for the LPCAT1, V5, FLAG, and β-actin antibodies were 1:2000, 1:2000, 1:5000, and 1:10,000, respectively.
Immunoprecipitation
Cell lysates were also incubated with 3 μl of HA antibody (Sigma) at 4 °C overnight, incubated with 20 μl of Pierce protein A/G matrix for 2 h, and eluted with protein sample buffer/lysis buffer prior to immunoblot analysis (22, 30).
Immunofluorescence Microscopy
MLE cells were plated at 30% confluence on 35-mm MetTek glass bottom culture dishes and transfected with individual CFP plasmids. Cells were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde for 20 min and then exposed to 15% bovine serum albumin, 1:200 β1,3-galactosyltransferase 2 primary goat antibody, and 1:200 fluorescein isothiocyanate-conjugated AffiniPure Donkey anti-goat IgG (H + L) (Jackson Immunoresearch) to visualize the trans-Golgi; cells were also incubated with an Alexa568-labeled goat anti-rabbit secondary antibody. Nuclei were visualized using To-Pro-3 (1:2000 dilution). Immunofluorescent cell imaging was performed on a Zeiss LSM 510 confocal microscope using the 458-, 568-, or 615-nm wavelength. All experiments were done with a Zeiss ×63 or ×100 oil differential interference contrast objective lens. The 458-nm wavelength was used to excite the CFP-CPT1 fusion proteins, with fluorescence emission collected through a 475-nm filter. A 488-nm wavelength was used to excite fluorescein dye, with fluorescence emission collected through a 505–530-nm filter. A 488-nm wavelength was used to excite βGT-YFP, with fluorescence emission collected through a 530–600-nm filter. A 613-nm wavelength was used to excite To-Pro-3 dye, with fluorescence emission collected through a 633-nm filter. Scanning was bidirectional at the highest possible rate measurement using a digital 1× zoom.
UbiQapture-Q Matrix Pull-down
Cell lysates were incubated with 40 μl of agarose beads complexed to ubiquitin-binding domain peptide overnight at 4 °C. An aliquot of prebound lysate was also resolved by 10% SDS-PAGE to determine loading onto the resin. The matrix resin was washed with 5 × 1 volume of 9:1 phosphate-buffered saline to lysis buffer as described in the Biomol protocol, and protein sample buffer was used to elute bound protein.
Ni2+ Resin Purification
Cell lysates were incubated with 40 ml of His-select nickel affinity gel resin overnight at 4 °C. The matrix resin was washed sequentially with a 3 × 3 bead volume of Buffer A without EDTA, 0.05% Triton X-100, and 0.05% Nonidet P-40; a 2 × 3 bead volume of Buffer A without EDTA, 0.005% Triton X-100, 0.005% Nonidet P-40, 500 mm NaCl, and 6 mm imidazole; and finally 1 volume of Buffer A without EDTA, 0.005% Triton X-100, 0.005% Nonidet P-40, 500 mm NaCl, and 200 mm imidazole. After 30 min, protein sample buffer was added to the resin/buffer mixture, samples were incubated at 25 °C overnight, and proteins were visualized by immunoblot analysis.
Statistical Analysis
Statistical analysis was performed by two-way analysis of variance or Student's t test. Data are presented as mean ± S.E.
RESULTS
LPCAT1 Is Highly Expressed in Distal Lung Epithelia
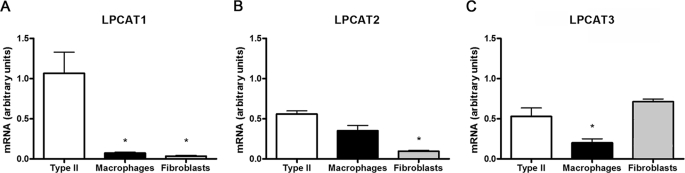
Lung epithelial type II cells synthesize and secrete surfactant composed of the major surface-active species, DPPtdCho. At least three acytransferases might participate in remodeling of PtdCho within lung epithelia generating DPPtdCho. These proteins include LPCAT1 and two variants harboring similar catalytic domains, termed LPCAT2 (31) and LPCAT3 (32) (AGPAT7 and LPEAT2); these enzymes exhibit similar substrate requirements with regard to saturated (16:0) fatty acyl-CoA donors (33). To evaluate which species may represent the primary acyltransferase involved in surfactant remodeling, we isolated mouse epithelial type II cells, alveolar macrophages, and lung fibroblasts. qRT-PCR shows that LPCAT1 mRNA is highly expressed in surfactant-producing mouse primary type II cells, with markedly reduced expression in macrophages or fibroblasts. Lung epithelia also expressed mRNAs encoding LPCAT2 and LPCAT3, albeit at lower levels (Fig. 1, B and C). Of the three homologues, LPCAT2 expression was highly expressed within macrophages, and LPCAT3 was most predominant within fibroblasts (Fig. 1, B and C). Thus, LPCAT1 appears to be a major regulator of surfactant remodeling, but the data do not exclude redundancy with other related acyltransferases.
FIGURE 1.
Pulmonary expression of LPCAT1 (A), LPCAT2 (B), and LPCAT3 (C). Primary murine lung type II cells, alveolar macrophages, and fibroblasts were isolated, and total cellular RNA was reverse transcribed to cDNA for subsequent qRT-PCR. The mRNA levels of each LPCAT were measured and normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. The data represent cells isolated from at least 5 mice/group. *, p < 0.05 versus type II cells.
Overexpression of LPCAT1 Reduces PtdCho de Novo Synthesis
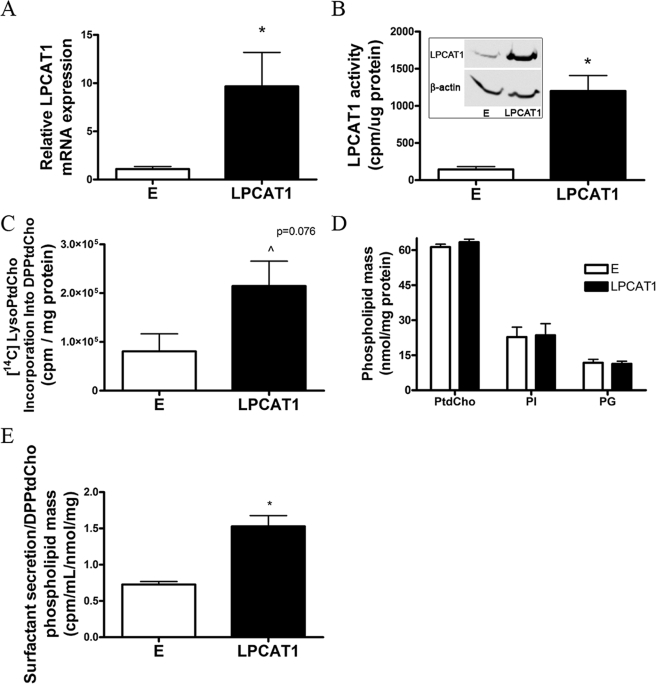
Because LPCAT1 was highly expressed in surfactant-producing lung cells, this enzyme was cloned and expressed in murine lung epithelia to test its regulatory activity on PtdCho metabolism. Using nucleofection, we were able to achieve high level (>90%) transfection efficiency in mammalian epithelia within a strong CMV-driven mammalian expression vector (Adv-CMV-LPCAT1). Following expression of this plasmid, we detected a ∼10-fold increase in LPCAT1 mRNA (Fig. 2A), a 28-fold increase in immunoreactive LPCAT1 (Fig. 2B, inset), a ∼7-fold increase in LPCAT activity (Fig. 2B), and a nearly 3-fold increase in [3H]LysoPtdCho incorporation into DPPtdCho (Fig. 2C). Importantly, despite up-regulating LPCAT1 expression and remodeling activity, total levels of cellular PtdCho (Fig. 2D) and DPPtdCho (data not shown) remained unchanged. This suggests the existence of a compensatory feedback mechanism(s) to preserve PtdCho balance. LPCAT1 overexpression also did not alter cellular mass of phosphatidylinositol or phosphatidylglycerol. Although PtdCho and DPPtdCho remained constant in cellular samples, surfactant secretion increased 2-fold in cells overexpressing LPCAT1 (Fig. 2E). These data are consistent with the ability of type II epithelia to rapidly mobilize intracellular DPPtdCho for export (34) and suggest a novel mechanism for increased surfactant secretion without alteration of cellular PtdCho mass. Thus, type II cells are readily able to adapt to modest surges in surfactant secretion by multiple homeostatic control mechanisms to maintain steady-state levels of lipid mass.
FIGURE 2.
LPCAT1 expression regulates surfactant metabolism. MLE cells were nucleofected with an LPCAT1 plasmid or an empty (E) vector. After 24 h, cells were collected and processed for LPCAT1 mRNA (measured relative to 18 S mRNA) by qRT-PCR (A), LPCAT1 and β-actin immunoblotting (B, inset), or enzyme activity (B), [14C]LysoPtdCho incorporation into surfactant DPPtdCho (C), or cellular mass of PtdCho, phosphatidylinositol (PI), and phosphatidylglycerol (PG) (D). B–D were normalized for cellular protein. E, surfactant secretion was measured by [3H]palmitic acid labeling of cells and activities retrieved in medium. Data are expressed as cpm medium/ml/total cellular DPPtdCho phospholipid phosphorus/mg of protein. Results are mean ± S.E. from at least three individual experiments except for PtdCho mass (two experiments) and phosphatidylinositol and phosphatidylglycerol mass (n = 1). *, p < 0.05 versus empty construct.
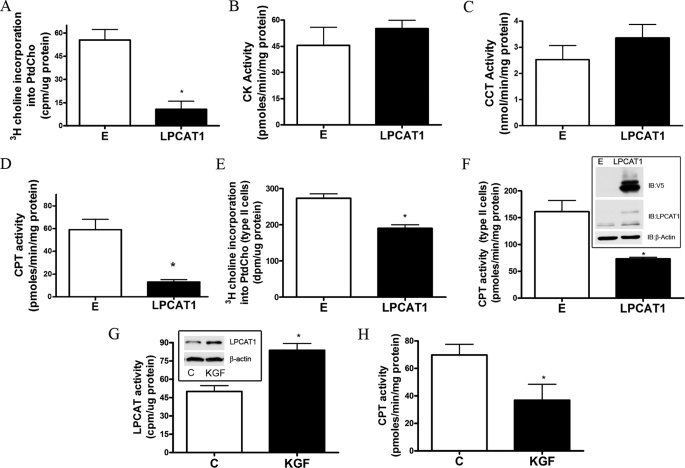
Next, we tested the hypothesis that LPCAT1 overexpression coordinately down-regulates de novo synthesis of PtdCho to maintain steady-state levels of phospholipid mass. This might occur by inhibiting the CDP-choline pathway. LPCAT1 overexpression decreased cellular [3H]choline incorporation into PtdCho, a marker of de novo synthesis, by nearly 80% versus empty vector-transfected cells (Fig. 3A). LPCAT1 overexpression reduced PtdCho synthesis within the CDP-choline pathway in MLE cells by significantly reducing CPT activity, without altering activities of either CK or CCT (Fig. 3, B–D). These effects were also seen in primary type II cells transduced with lentivirus encoding LPCAT1-V5 (Fig. 3, E and F).
FIGURE 3.
LPCAT1 expression inhibits the denovo (CDP-choline) pathway. LPCAT1 was expressed in cells and processed for [3H]choline incorporation into PtdCho as a measure of de novo synthesis (A), CK activity (B), CCT activity (C), and CPT activity (D). E and F, primary rat type II cells were isolated, cultured, and infected with LPCAT1-V5 packaged in lentivirus prior to assays for [3H]choline incorporation into PtdCho (E), V5 and LPCAT1 immunoblotting (F, inset), and CPT1 activity (F). G and H, cells were left untreated or treated with KGF (20 ng/ml), and 24 h post-treatment, cells were harvested and processed for LPCAT immunoblotting and activity (G) and CPT activities (H). Results are mean ± S.E. from at least three individual experiments except for LPCAT activity (G) (two experiments). *, p < 0.05 versus empty construct.
We next modulated LPCAT1 expression using nontransfectional approaches to assess physiological relevance. KGF increases type II cell surfactant synthesis by increasing LPCAT1 content (11). KGF increased LPCAT1 protein levels and activity and also decreased CPT activity in lung epithelia (Fig. 3, G and H). In preliminary studies, mechanical ventilation of mice using high tidal volumes also increases immunoreactive LPCAT1 levels concomitant with decreased CPT activity in mouse lung (data not shown). Hyperventilation also increases surfactant production (11, 35, 36). In the primary type II cell, KGF, and ventilated models, LPCAT1 protein levels increase more modestly: 3.5–5.7 and 4.4-fold, respectively (and not 28-fold as with MLE cells). This demonstrates that the cross-talk between the remodeling and de novo pathways occurs at more modest levels of LPCAT1 overexpression, consistent with what has been shown for other lipogenic enzymes during development and malignant transformation (37, 38). Collectively, these observations strongly suggest tight control of PtdCho metabolism in lung epithelia; the data also suggest that remodeling and de novo pathways in lung epithelia are physiologically linked and biologically relevant.
LPCAT1 Overexpression Increases CPT Degradation via Endosomal/Lysosomal Processing
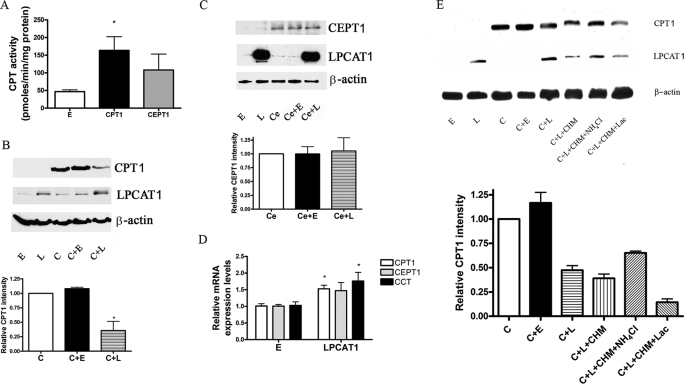
To evaluate the mechanism whereby LPCAT1 overexpression reduces CPT activity, we expressed recombinant CPT1 and CEPT1 proteins harboring V5 and/or His tags individually or in combination with LPCAT1 plasmid in epithelia. CPT1-V5his and V5-CEPT1 cellular expression resulted in increased CPT activity (Fig. 4A). Co-expression of the CPT1 or CEPT1 recombinant plasmids in cells with LPCAT1 resulted in decreased immunoreactive CPT1 protein levels and no significant difference in immunoreactive CEPT1 levels (Fig. 4, B and C). In contrast, expression of LPCAT2-V5 or LPCAT3-V5 did not alter CPT1 protein levels or affect [3H]choline incorporation into PtdCho (data not shown). LPCAT1 overexpression significantly increased CPT1 and CCT mRNA by qRT-PCR (p < 0.05 versus control), but effects on CEPT1 mRNA did not reach significance (Fig. 4D). These data suggest that LPCAT1 overexpression selectively enhances CPT1 protein degradation or perhaps regulates translational efficiency of the de novo biosynthetic enzymes.
FIGURE 4.
LPCAT1 expression promotes CPT degradation. A, MLE cells were nucleofected with empty, CPT1, or CEPT1 cDNA vectors, and CPT activity was assayed. B, cells were transfected with empty (E), LPCAT1 (L), or CPT1 (C) or in combination with empty or LPCAT1 plasmids, and after 16 h, CPT1, LPCAT1, and β-actin levels were detected by immunoblotting. C, experiments similar to B were performed, except CEPT1 was overexpressed instead of CPT1. D, cells were transfected with empty (E) or LPCAT1 expression vectors, and relative mRNA levels of CPT1, CEPT1, and CCT in cells were assayed using qRT-PCR. E, empty (E), LPCAT1 (L), or CPT1 (C) plasmids were transfected in cells alone or in combination, and after 12 h, cells were exposed to CHM with or without either NH4Cl or lactacystin (Lac). After 18 h, cells were harvested and processed for immunoblotting for V5-CPT1, LPCAT1, and β-actin protein levels. The lower graphs in B, C, and E show densitometric values of bands from respective immunoblots. Data in each panel (A–E) represent three individual experiments (*, p < 0.05 versus empty group).
Two major avenues for cellular protein disposal include proteasomal degradation and turnover within the endosomal-lysosomal compartment. To identify a role for these pathways in CPT1 degradation, we impaired acidification of endocytic vesicles with NH4Cl or inhibited proteasomal degradation with lactacystin after cells were transfected with relevant plasmids. In addition, cells were treated with the protein synthesis inhibitor, cyclohexamide. Fig. 4E demonstrates that co-transfection of CPT1 with LPCAT1 or inclusion of cyclohexamide in the medium reduced immunoreactive levels of CPT1. Compared with cells transfected with CPT1 (control), cells transfected with LPCAT1 alone or in combination with either cyclohexamide or lactacystin resulted in lower levels of CPT1. Unlike the effects of lactacystin, inclusion of NH4Cl in the medium significantly blocked LPCAT1 reduction of CPT1 levels (Fig. 4E). Plasmid co-transfection or NH4Cl was not toxic to cells (data not shown).
CPT Accumulates within Lysosomes
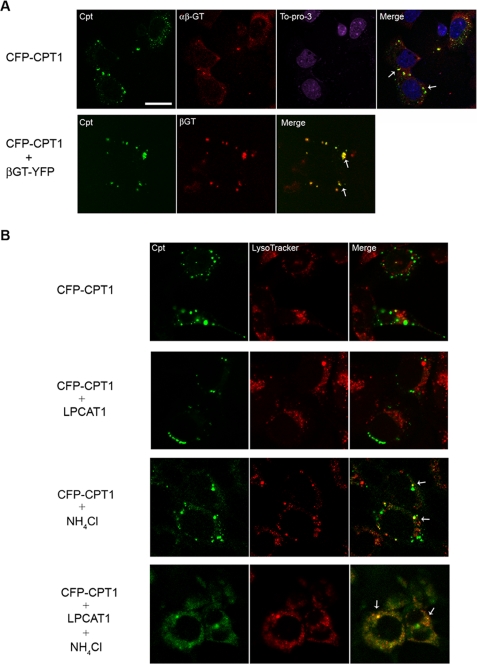
To further assess CPT1 in lysosomal trafficking, we examined CFP-CPT1 subcellular localization in epithelia. Immunofluorescent studies revealed co-localization of CFP-CPT1 with β1,3-galactosyltransferase 2 (αβ-GT), indicative of CPT1 localization within the Golgi apparatus (Fig. 5A, top row). Because MLE cells are a transformed cell line, cells contained Golgi, as represented by larger inclusions rather than the more classic perinuclear labeling as seen with other mammalian cells. When CFP-CPT1 was cotransfected with a plasmid encoding a fragment of another Golgi-resident enzyme, βGT-YFP, the fusion proteins also co-localized (Fig. 5A, bottom row) (39). We also used LysoTracker Red, which fluoresces within the acidic pH compartments to examine CPT1 trafficking within lysosomes (Fig. 5B). Unlike data from Fig. 5A, where CFP-CPT1 co-localizes with β-GT, CFP-CPT1 under native conditions does not appear to significantly co-localize with LysoTracker Red. Co-transfection of CFP-CPT1 with LPCAT1 also results in limited CFP-CPT1 immunofluorescence within cells (Fig. 5B, second row). Importantly, only when NH4Cl was used to impair late endosome/lysosomal function (with or without LPCAT1 transfection) did we detect increased CFP-CPT1 immunofluorescence that co-localized with LysoTracker Red (Fig. 5B, lower rows) and the endosomal marker, Rab5 (data not shown). CFP-CPT1 co- localization with LysoTracker was apparent by small punctate yellow cytosolic signals (arrows). Together, these data are consistent with LPCAT1-induced CPT1 sorting through the endosomal/lysosomal pathway for its degradation.
FIGURE 5.
CPT1 is localized to Golgi but degraded within the lysosomes. MLE cells were transfected with CFP-CPT1, βGT-YFP, and/or LPCAT1. A (top), cells were fixed and stained using a β-1,3-Gal-T2 (αβ-GT) primary antibody and an Alexa568-labeled goat anti-rabbit secondary antibody. To-Pro-3 was used to visualize nuclei. Bottom, cells were co-transfected with CFP-CPT1 and βGT-YFP and processed for imaging. White bar, 20 nm. B, transfected cells were exposed to NH4Cl for 24 h as indicated. Cells were allowed to recover (without NH4Cl) for 2 h prior to incubation with LysoTracker dye for 1 h. Cells were then fixed and visualized using a confocal microscope. The arrows in merged panels represent co-localization with LysoTracker.
Multiple CPT1 Ubiquitination Sites Responsible for Trafficking through the Endosomal Pathway
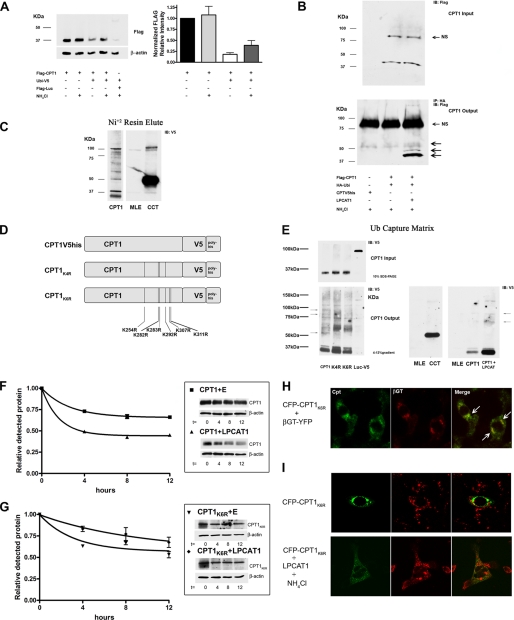
Generally, monoubiquitinated or multiubiquitinated proteins are sorted and degraded in the lysosome, whereas polyubiquitinated proteins are degraded in the 26 S proteasome (40, 41). To determine if CPT1 is ubiquitinated, we first co-transfected a ubiquitin plasmid with FLAG-CPT1 and observed that levels of CPT1 decreased compared with control, an effect lessened after exposing cells to NH4Cl (Fig. 6A). In separate studies, cells were co-transfected with HA-ubiquitin and FLAG-CPT1 with NH4Cl (Fig. 6B). Ubiquitin was immunoprecipitated using HA antibody, and samples were processed for FLAG immunoblotting. Here, LPCAT1 increased intensity of higher molecular weight species (Fig. 6B, bottom, arrows). We next expressed CPT1-V5his or CCT-V5his in MLE cells and partially purified the proteins using Ni2+ chromatography prior to V5 immunoblotting (Fig. 6C). We observed several slower migrating immunoreactive bands after CPT1-V5his expression; these results differ from CCT, where a single monoubiquitinated band at ∼50 kDa and dimeric species were recently detected (20). As a negative control, no major bands were detected after probing eluants from untransfected cell lysates alone.
FIGURE 6.
CPT1 is ubiquitinated. A, cells were co-transfected with ubiquitin plasmid, FLAG-luciferase (Luc), and FLAG-CPT1 with or without NH4Cl (25 mm) for 24 h. Total cell lysates were processed for FLAG and β-actin immunoblotting. The graph on the right represents densitometric analysis of immunoblots. B, cells were nucleofected with FLAG-CPT1, HA-ubiquitin, or CPT1-V5his, with or without or LPCAT1, incubated for 18 h, and treated with NH4Cl for 24 h using methods described previously. Cell lysates were loaded onto SDS-PAGE (Input) or incubated with HA antibody/protein A/G matrix, eluted, and visualized by FLAG immunoblotting (top and bottom). NS, nonspecific bands; arrows denote higher migrating CPT1-UB conjugates. C, cells were nucleofected with CPT1-V5his or CCT-V5, and 18 h after transfection, cells were treated with NH4Cl for 24 h. Cells were harvested and incubated with Ni2+ resin at 4 °C for 4 h, and protein was eluted and visualized using SDS-PAGE and V5 immunoblotting. D, schematic of candidate ubiquitin acceptor sites targeted for generation of multiple Lys → Arg CPT1-V5his mutants. The CPT1K4R mutant harbors mutations at Lys254, Lys282, Lys283, and Lys292, and the CPT1K6R construct also harbors substitutions at Lys307 and Lys311. E (left), CPT1, CPT1K4R, CPT1K6R, and luciferase-V5 (Luc) plasmids were transfected into cells; after 18 h, cells were exposed to NH4Cl for 24 h and harvested. Cell lysates were loaded onto SDS-PAGE (Input) or incubated with UbiQapture-Q kit matrix (4 °C for 4 h), eluted, and visualized by V5 immunoblotting (bottom). The arrows indicate slower migrating bands that exhibit differential levels of intensities between recombinant CPT1 and mutant constructs. Right, untransfected cells, CCT, and CPT1 alone or co-transfected with LPCAT1 lysates were processed using UbiQapture-Q matrix and visualized. All panels were exposed extensively to detect ubiquitinated products except for the far right panel. Results are representative of at least three individual experiments. F and G, cells were co-transfected with CPT1 or CPT1K6R plasmid with either an empty (E) or LPCAT1 plasmid and exposed to CHM. Cells from each group were harvested at times after CHM treatment and processed for V5 immunoblotting (left) and densitometric analysis (right) of autoradiograms. The starting (0 h) time point was normalized to equal 1 for each test group. Best fit lines are a result of one-phase exponential decay line fitting performed by Prism graphing software. Data in F and G represent two individual experiments. H and I, cells were transfected with CFP-CPT1K6R alone or in combination with βGT-YFP or LPCAT1. F, cells were co-transfected with CFP-CPT1K6R and βGT-YFP, fixed, and visualized. The arrows indicate co-localization of CFP-CPT1K6R with βGT-YFP. G, transfected cells were exposed to NH4Cl treatment for 24 h. Cells were allowed to recover (no NH4Cl) for 2 h prior to incubation with LysoTracker dye for 1 h. Cells were then fixed and visualized by confocal microscopy.
To further study ubiquitination, we constructed and expressed CPT1 mutants containing either four or six Lys → Arg mutations, termed CPT1K4R and CPT1K6R, respectively (Fig. 6D). Specific candidate lysines were mutated based on the enzyme's proposed secondary structure where these residues are probably cytosolically exposed and conserved between mouse and human sequences (42). Cells were first transfected with CPT1-V5his, CPT1K4R, CPT1K6R, and a luciferase-V5 negative control. Transfectants were then applied to ubiquitin capture columns, where agarose was complexed to ubiquitin-binding domain peptide, and reaction mixtures were processed for V5 immunoblotting (Fig. 6E) (43). Comparable levels of the ∼37-kDa CPT1 were detected before resin loading (Fig. 6E, Input). Multiple high molecular weight bands were seen in cells transfected with CPT1-V5his in a pattern similar to results using Ni2+ chromatography. These bands were only visualized after prolonged exposure of autoradiograms. Interestingly, bands at ∼35 and 37 kDa were also visualized, suggesting that full-length CPT1 might aggregate with ubiquitinated CPT1 during incubation with ubiquitin-binding domain peptide and that these products are dissociated during SDS-PAGE. Importantly, the intensity of several higher molecular weight immunoreactive V5 bands was reduced, some to undetectable levels, in samples that were analyzed from preparations using CPT1K4R and CPT1K6R mutants (Fig. 6E, bottom, arrows). Cells transfected with CPT1-V5his with or without LPCAT1 were also processed similarly (Fig. 6E, far right panel). After a short exposure of films, LPCAT1 increased the appearance of slower migrating bands.
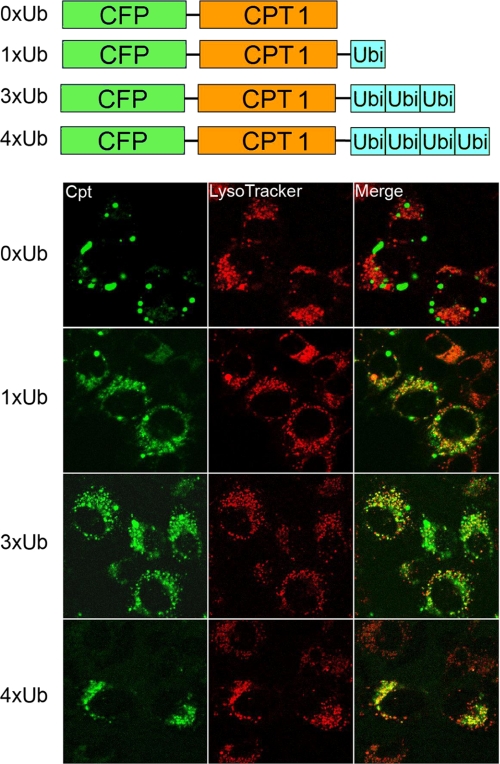
To address the functionality of ubiquitin acceptor sites, cells were transfected with CPT1-V5his, CPT1K6R, or an empty vector with or without LPCAT1, and decay studies were performed using CHM (Fig. 6, F and G). Phase exponential decay line fitting was performed by graphing software based on densitometric data (Fig. 6, F and G, left) from immunoblots (Fig. 6, F and G, right). Cells transfected with CPT1 and an empty vector displayed an extended half-life (>12 h), whereas CPT1 t½ was reduced to ∼4 h in the presence of LPCAT1 (Fig. 6F). Further, overall stability of CPT1K6R was similar to that of CPT1-V5his despite co-transfection of LPCAT1 (Fig. 6G). The results suggest that specific ubiquitination acceptor sites within CPT1 regulate its stability. Finally, a CFP-CPT1K6R construct was used for subcellular localization. Unlike CFP-CPT1, the CFP- CPT1K6R construct failed to co-localize with LysoTracker Red despite overexpression of LPCAT1 or treatment with NH4Cl (Fig. 6, H and I). CFP-CPT1K6R does, however, co-localize with βGT-YFP, similar to CFP-CPT1 (Fig. 6H, white arrows). Because CPT1 appears to be multiubiquitinated, CFP-CPT1 was fused to tandem repeats of ubiquitin, and localization was also examined (Fig. 7). Cells transfected with increasing copies of carboxyl-terminal ubiquitin linked to CFP-CPT1 displayed a progressive increase in the levels of co-localization to lysosomes (evidenced by increases in punctate yellow signals/cell).
FIGURE 7.
CPT1 ubiquitin fusion proteins target the lysosome. MLE cells were transfected with CFP-CPT1, CFP-CPT1–1×Ub, CFP-CPT1–2×Ub, CFP-CPT1–3×Ub, or CFP-CPT1–4×Ub. After 18 h, cells were incubated with LysoTracker dye for 1 h. Cells were then fixed and visualized using confocal microscopy.
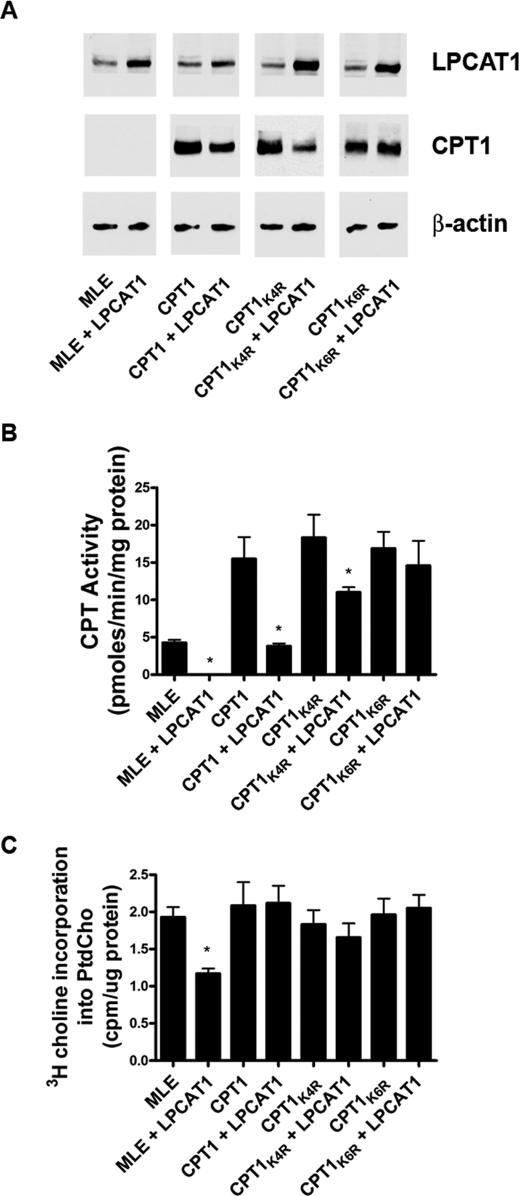
Last, cells were co-transfected with CPT1-V5his, CPT1-V5his mutants, and LPCAT1, and enzyme activity and de novo PtdCho synthesis were measured (Fig. 8). Although LPCAT1 decreased immunoreactive levels of CPT1-V5his and CPT1K4R, levels were comparable with control in cells co-expressing the CPT1K6R mutant with LPCAT1 (Fig. 8A). Overexpression of each construct led to a robust increase in CPT activity, indicating that constructs were functional (Fig. 8B). Cells transfected with CPT1K4R and LPCAT1 exhibited a ∼45% reduction in CPT1 activity versus cells transfected with CPT1K4R alone; nevertheless, cells from the former group exhibited greater CPT activity than untransfected cells or cells co-transfected with wild-type CPT1 and LPCAT that were also sufficient to restore PtdCho synthesis (Fig. 8C). Compared with CPT1-V5his, overexpression of CPT1K6R restored both CPT activity and PtdCho synthesis despite LPCAT1 expression (Fig. 8, B and C). As a whole, the data support CPT1 multiubiquitination at highly conserved lysines and indicate that expression of CPT1 lysine mutants confers resistance to inhibitory actions of LPCAT1 within the phospholipid pathway.
FIGURE 8.
CPT1 Lys → Arg mutants are resistant to LPCAT1-induced degradation. A, MLE cells were untransfected or transfected with CPT1-V5his, CPT1K4R, or CPT1K6R constructs alone or in combination with LPCAT1 and processed for LPCAT1, V5 (CPT1), and β-actin immunoblotting (A), CPT activities (B), or [3H]choline incorporation into PtdCho (C). Results are means ± S.E. from at least three individual experiments. *, p < 0.05 versus paired controls.
DISCUSSION
Surfactant PtdCho is relatively unique because it is selectively enriched with 16:0/16:0 molecular species (DPPtdCho) on the phospholipid glycerol backbone. This molecular species specificity is essential to optimize surfactant function and is conferred, in part, by a constitutively active LPCAT1 enzyme within lung epithelia. Indeed, perhaps ∼55–75% of surfactant PtdCho is synthesized by the remodeling pathway with the remainder generated by de novo synthesis (9, 44, 45). These results suggest that when LPCAT1 levels are increased, as occurs during alveolar stress, when demands for surfactant (DPPtdCho) are high, lung epithelia might coordinately reduce de novo synthesis of PtdCho by increasing CPT1 degradation through its ubiquitination. This cellular mechanism may be favorable to shift the balance toward DPPtdCho enrichment for surfactant secretion into the airways via LPCAT1 remodeling, given that the CDP-choline pathway is also active in membrane (nonsurfactant) phospholipid biosynthesis. Thus, our results unveil a previously unrecognized molecular mechanism for feedback control for PtdCho homeostasis.
Comparative analysis of LPCAT mRNAs revealed differential expression of three related acyl-CoA LysoPtdCho acyltransferases that may exhibit synthetic capacity for DPPtdCho. LPCAT1 was a primary candidate based on prior data showing high level pulmonary expression and preference for saturated (16:0) fatty acids as a donor and 1-myristoyl or 1-palmitoyl-LPtdCho as an acceptor (11, 12). LPCAT2 and LPCAT3 (AGPAT7 and LPEAT2) also contain similar conserved motifs, including acyl-transferase, transmembrane, and EF-hand domains (46). Our observations that LPCAT1 mRNA was highest in type II epithelia versus macrophages or fibroblasts underscore its role as a primary regulator of PtdCho remodeling, although confirmation awaits analysis of individual endogenous LPCAT proteins. There may be redundancy in this system because type II epithelia also express modest levels of LPCAT2/LPCAT3 (Fig. 1); their contribution to surfactant remodeling will require more sophisticated in vivo genetic ablation systems.
Multiple mechanisms exist that maintain stable steady-state total cellular PtdCho mass despite its reduced de novo synthesis and increased surfactant secretion after LPCAT expression (Fig. 2, C–E, and 3E). We observed a robust increase (∼3-fold) in remodeling activity by LPCAT1 (Fig. 2C). Although LPCAT1 is involved surfactant synthesis, the enzyme also exhibits some promiscuity, with significant substrate activity for other unsaturated fatty acyl species that may be destined for incorporation into the cell membrane (12). This would explain, in part, our observation that total cell PtdCho levels are relatively unchanged after LPCAT1 overexpression. Second, type II cells may exploit other compensatory mechanisms to maintain constant levels of cellular PtdCho, including reduced phospholipid degradation rates or increased PC uptake. Overexpression of some biosynthetic lipogenic enzymes reduces degradation rates (14). Up to 50–85% of secreted PC can be taken up and reutilized (47). Because the actual mass of lipid secreted by type II epithelia is only 1–2% of total cellular phospholipid per hour, type II cells have significant reserves of intracellular lipid that can offset abrupt alterations in synthetic or secretory rates (34). LPCAT1 inhibition of de novo PtdCho synthesis appears unidirectional because RNA silencing of LPCAT1 did not coordinately increase CPT activity (data not shown), suggesting a more complex mode of regulation. Nonetheless, transfection of LPCAT1 plasmid in lung epithelia resulted in its robust expression coupled with increases in DPPtdCho synthesis yet did not alter total cellular phospholipid mass, suggestive of feedback inhibition within the de novo pathway.
Genetic inactivation CPT1 and CEPT1 impairs de novo PtdCho synthesis in Saccharomyces cerevisiae, but their biochemical and molecular characterization has been hampered because these mammalian enzymes have not been purified to homogeneity (6). Thus, we expressed plasmids encoding wild-type and mutant CPT1 in cells where tagged proteins were functional and detected within the Golgi (5). The Golgi pattern in MLE cells was somewhat atypical, but CFP-CPT1 was observed to co-localize with known markers. Exogenously expressed CPT1 also appears to have an extended t½ that exceeds other lipogenic enzymes, including hydroxymethylglutaryl-CoA reductase or CCT, suggestive of stabilizing ligands or post-translational modifications within its primary structure (data not shown) (22, 48). LPCAT1 overexpression selectively reduced CPT activity and immunoreactive levels by shortening CPT1 t½ in lung epithelia via sorting from the Golgi to endosomal/lysosomal organelles. Co-localization of CPT1 with LysoTracker Red was limited and only observed after simultaneous treatment with NH4Cl because acidic compartments limit fluorescence of CFP-tagged constructs (49).
Ubiquitin can divert proteins from either the cell surface, endosomes, or trans-Golgi network to lysosomes, thus serving as an important sorting signal for degradation of transmembrane proteins (50). In addition, several proteins containing ubiquitin-binding domains comprise the machinery to assist in the trafficking of ubiquitinated cargoes to lysosomes. These cargoes may be monoubiquitinated, multiubiquitinated, and less often polyubiquitinated. Evidence in support of CPT1 ubiquitination includes our observations that (i) CPT1 cellular levels accumulated after treatment with NH4Cl, which impairs the clearance of ubiquitinated substrates, (ii) overexpression of ubiquitin decreased CPT1 levels, (iii) CPT1 effectively bound ubiquitin-binding domain peptide, (iv) LPCAT1-dependent reduction in CPT1K6R t½ is not observed, and (v) tandem fusions of ubiquitin to CPT1 were sufficient for lysosomal targeting. Last, expression of CPT1 mutants harboring multiple Lys-Arg substitutions at candidate ubiquitin acceptor sites led to reduced binding to ubiquitin-binding domain peptide and resulted in proteolytic resistance to actions of LPCAT1. However, LPCAT1 might still act through non-ubiquitin-dependent mechanisms or even indirectly to alter CPT1 stability. CPT1 mutants still, however, show some degree of residual binding to ubiquitin-binding domain peptide, suggesting that several yet unidentified lysines partake as ubiquitin acceptor sites when some sites are mutated out (Fig. 6E). Although LPCAT1 might itself act as a novel E3 ligase, it is more likely that other downstream events are activated (e.g. E3 ligases) or inhibited (deubiquitinating enzymes) after LPCAT1 overexpression to modulate CPT1 protein stability. It is unlikely that LPCAT1 itself exhibits ubiquitin E3 ligase activity because it is not bound to CPT1 (data not shown). Further, expression of a catalytically inactive LPCAT1 protein containing an amino acid substitution at His135 does not alter CPT1 levels or significantly reduce de novo PtdCho synthesis (data not shown). This suggests a mechanism involving lipid-dependent signaling. Ongoing studies suggest that β-transducin repeat-containing protein might serve as a putative E3 ligase targeting CPT1 for its degradation (data not shown).
CPT1 targeting to the endosome/lysosome pathway may also involve GGA (Golgi-localized, γ-ear-containing, Arf-binding) proteins that specifically target Golgi-localized ubiquitinated proteins to lysosomes through the GGA protein GAT domain (51). This interaction has been primarily described in yeast but may also occur in mammalian cells requiring phosphatidylinositol 4-phosphate kinase IIa for optimal sorting (52). Thus, it is possible that LPCAT1-directed CPT1 ubiquitination and lysosomal targeting require these GGA-like adaptor molecules for its cellular trafficking/elimination.
These results do not exclude the possibilities that LPCAT1 triggers polyubiquitination or even monoubiquitination of CPT1 as a means to regulate phospholipid metabolism. In this manner, CPT1 would be modified and processed similarly to receptor tyrosine kinases that appear covalently attached at multiple or single sites to ubiquitin (53). However, it is very difficult to detect monoubiquitinated proteins in cells because the subpopulation of select proteins that are ubiquitinated at any given time is extremely small (54). Multiubiquitinated CPT1 was also very difficult to detect despite co-expression of various tagged CPT1 and ubiquitin constructs and co-immunoprecipitation (data not shown). Detection also depended on optimal exposure of our autoradiograms. Levels of these ubiquitinated cargoes are also regulated by activities of deubiquitinating enzymes, making detection difficult. The use of new approaches such as ubiquitin-mediated fluorescence complementation may aid in resolving detection difficulties in future studies (54). One intriguing possibility that deserves further study is the hypothesis that a subpopulation of ubiquitinated CTP1 may traffic to lysosomes independently of Golgi-localized ubiquitin-binding proteins and through autophagy. This mechanism has recently been described for lysosomal degradation of long lived monoubiquitinated proteins during cell stress requiring the ubiquitin-binding protein, p62 (55).
Acknowledgments
We thank Dr. Bill Chen for assistance with generation of CPT1 ubiquitin fusion plasmids. We also thank Dr. Peter Snyder and Dr. Ernesto Fuentes for suggestions and critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health R01 Grants HL097376, HL096376, HL098174, HL081784, and HL068135 (to R. K. M.). This work was also supported by a Merit Review Award from the Department of Veterans Affairs.
- PtdCho
- phosphatidylcholine
- CCT
- cytidylyltransferase
- CDP-choline
- cytidine diphosphocholine
- CFP
- cyan fluorescent protein
- CHM
- cycloheximide
- CK
- choline kinase
- CPT
- cholinephosphotransferase
- DPPtdCho
- dipalmitoylphosphatidylcholine
- KGF
- keratinocyte growth factor
- LPCAT
- acyl-CoA:lysophosphatidylcholine acyltransferase
- MLE
- murine lung epithelial
- YFP
- yellow fluorescent protein
- βGT
- UDP-Gal:β-GlcNAcβ1,4-galactosyltransferase
- LysoPtdCho
- lysophosphatidylcholine
- HA
- hemagglutinin
- CMV
- cytomegalovirus
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1.Jackowski S., Fagone P. (2005) J. Biol. Chem. 280, 853–856 [DOI] [PubMed] [Google Scholar]
- 2.Sher R. B., Aoyama C., Huebsch K. A., Ji S., Kerner J., Yang Y., Frankel W. N., Hoppel C. L., Wood P. A., Vance D. E., Cox G. A. (2006) J. Biol. Chem. 281, 4938–4948 [DOI] [PubMed] [Google Scholar]
- 3.Wu G., Aoyama C., Young S. G., Vance D. E. (2008) J. Biol. Chem. 283, 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henneberry A. L., Wistow G., McMaster C. R. (2000) J. Biol. Chem. 275, 29808–29815 [DOI] [PubMed] [Google Scholar]
- 5.Henneberry A. L., Wright M. M., McMaster C. R. (2002) Mol. Biol. Cell 13, 3148–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMaster C. R., Bell R. M. (1994) J. Biol. Chem. 269, 28010–28016 [PubMed] [Google Scholar]
- 7.Wright M. M., Henneberry A. L., Lagace T. A., Ridgway N. D., McMaster C. R. (2001) J. Biol. Chem. 276, 25254–25261 [DOI] [PubMed] [Google Scholar]
- 8.Bladergroen B. A., Bussière M., Klein W., Geelen M. J., Van Golde L. M., Houweling M. (1999) Eur. J. Biochem. 264, 152–160 [DOI] [PubMed] [Google Scholar]
- 9.den Breejen J. N., Batenburg J. J., van Golde L. M. (1989) Biochim. Biophys. Acta 1002, 277–282 [DOI] [PubMed] [Google Scholar]
- 10.Fisher A. B., Dodia C. (1997) Am. J. Physiol. 272, L238–L243 [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 13.Heath R. J., Rock C. O. (1998) J. Bacteriol. 180, 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walkey C. J., Kalmar G. B., Cornell R. B. (1994) J. Biol. Chem. 269, 5742–5749 [PubMed] [Google Scholar]
- 15.Tijburg L. B., Nishimaki-Mogami T., Vance D. E. (1991) Biochim. Biophys. Acta 1085, 167–177 [DOI] [PubMed] [Google Scholar]
- 16.Tercé F., Record M., Tronchère H., Ribbes G., Chap H. (1991) Biochim. Biophys. Acta 1084, 69–77 [DOI] [PubMed] [Google Scholar]
- 17.Cui Z., Houweling M., Vance D. E. (1995) Biochem. J. 312, 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallampalli R. K., Ryan A. J., Salome R. G., Jackowski S. (2000) J. Biol. Chem. 275, 9699–9708 [DOI] [PubMed] [Google Scholar]
- 19.Salome R. G., McCoy D. M., Ryan A. J., Mallampalli R. K. (2000) J. Appl. Physiol. 88, 10–16 [DOI] [PubMed] [Google Scholar]
- 20.Chen B. B., Mallampalli R. K. (2009) Mol. Cell. Biol. 29, 3062–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J., Wu Y., Henderson F., McCoy D. M., Salome R. G., McGowan S. E., Mallampalli R. K. (2006) Gene Ther. 13, 974–985 [DOI] [PubMed] [Google Scholar]
- 22.Chen B. B., Mallampalli R. K. (2007) J. Biol. Chem. 282, 33494–33506 [DOI] [PubMed] [Google Scholar]
- 23.McCoy D. M., Fisher K., Robichaud J., Ryan A. J., Mallampalli R. K. (2006) Am. J. Respir. Cell Mol. Biol. 35, 394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallampalli R. K., Walter M. E., Peterson M. W., Hunninghake G. W. (1994) Am. J. Respir. Cell Mol. Biol. 10, 48–57 [DOI] [PubMed] [Google Scholar]
- 25.Agassandian M., Miakotina O. L., Andrews M., Mathur S. N., Mallampalli R. K. (2007) Biochem. J. 403, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 27.Mallampalli R. K., Salome R. G., Spector A. A. (1994) Am. J. Physiol. 267, L641–L648 [DOI] [PubMed] [Google Scholar]
- 28.Mallampalli R. K., Mathur S. N., Warnock L. J., Salome R. G., Hunninghake G. W., Field F. J. (1996) Biochem. J. 318, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J. C., Weinhold P. A. (1981) J. Biol. Chem. 256, 12662–12665 [PubMed] [Google Scholar]
- 30.Agassandian M., Zhou J., Tephly L. A., Ryan A. J., Carter A. B., Mallampalli R. K. (2005) J. Biol. Chem. 280, 21577–21587 [DOI] [PubMed] [Google Scholar]
- 31.Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. (2007) J. Biol. Chem. 282, 6532–6539 [DOI] [PubMed] [Google Scholar]
- 32.Ye G. M., Chen C., Huang S., Han D. D., Guo J. H., Wan B., Yu L. (2005) DNA Seq. 16, 386–390 [DOI] [PubMed] [Google Scholar]
- 33.Soupene E., Fyrst H., Kuypers F. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M., Brown L. A. (1990) Am. J. Physiol. 258, L195–L200 [DOI] [PubMed] [Google Scholar]
- 35.Nicholas T. E., Power J. H., Barr H. A. (1982) Respir. Physiol. 49, 315–324 [DOI] [PubMed] [Google Scholar]
- 36.Dietl P., Frick M., Mair N., Bertocchi C., Haller T. (2004) Biol. Neonate 85, 299–304 [DOI] [PubMed] [Google Scholar]
- 37.Mallampalli R. K., Hunninghake G. W. (1993) Pediatr. Res. 34, 502–511 [DOI] [PubMed] [Google Scholar]
- 38.Piyathilake C. J., Frost A. R., Manne U., Bell W. C., Weiss H., Heimburger D. C., Grizzle W. E. (2000) Hum. Pathol. 31, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 39.Hu W., Xu R., Zhang G., Jin J., Szulc Z. M., Bielawski J., Hannun Y. A., Obeid L. M., Mao C. (2005) Mol. Biol. Cell 16, 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carter S., Bischof O., Dejean A., Vousden K. H. (2007) Nat. Cell Biol. 9, 428–435 [DOI] [PubMed] [Google Scholar]
- 41.Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 42.McMaster C. R., Bell R. M. (1997) Biochim. Biophys. Acta 1348, 100–110 [DOI] [PubMed] [Google Scholar]
- 43.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 44.Lands W. E. (1960) J. Biol. Chem. 235, 2233–2237 [PubMed] [Google Scholar]
- 45.Mason R. J., Nellenbogen J. (1984) Biochim. Biophys. Acta 794, 392–402 [DOI] [PubMed] [Google Scholar]
- 46.Harayama T., Shindou H., Ogasawara R., Suwabe A., Shimizu T. (2008) J. Biol. Chem. 283, 11097–11106 [DOI] [PubMed] [Google Scholar]
- 47.Wright J. R. (1990) Am. J. Physiol. 259, L1–L12 [DOI] [PubMed] [Google Scholar]
- 48.Dice J. F. (1987) FASEB J. 1, 349–357 [DOI] [PubMed] [Google Scholar]
- 49.Tsien R. Y. (1998) Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 50.Piper R. C., Luzio J. P. (2007) Curr. Opin. Cell Biol. 19, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelham H. R. (2004) Curr. Biol. 14, R357–R359 [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. (2007) Mol. Biol. Cell 18, 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 54.Ikeda H., Kerppola T. K. (2008) Mol. Biol. Cell 19, 4588–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]