Abstract
Mono- and polyubiquitylation of proteins are key steps in a wide range of biological processes. However, the molecular mechanisms that mediate these different events are poorly understood. Here, we employed NMR spectroscopy to map a non-covalent ubiquitin binding surface (UBS) on the Smurf ubiquitin ligase HECT domain. Analysis of mutants of the HECT UBS reveal that interfering with the UBS surface blocked Smurf-dependent degradation of its substrate RhoA in cells. In vitro analysis revealed that the UBS was not required for UbcH7-dependent charging of the HECT catalytic cysteine. Surprisingly, although the UBS was required for polyubiquitylation of both Smurf itself and the Smurf substrate RhoA, it was not required for monoubiquitylation. Furthermore, we show that mutating the UBS interfered with efficient binding of a monoubiquitylated form of RhoA to the Smurf HECT domain. Our findings suggest the UBS promotes polyubiquitylation by stabilizing ubiquitylated substrate binding to the HECT domain.
Keywords: Cytokines/TGFβ, Enzymes/Catalysis, Proteases/Ubiquitination, Protein/Degradation, Protein/Structure, HECT-type Ubiquitin Ligase, NMR
Introduction
Ubiquitylation of protein substrates via an E1-E2-E3 enzymatic cascade is important for many biological processes. There is a panoply of ubiquitin modifications that can regulate the outcome of ubiquitylation, in particular, chain length. For example, monoubiquitylation is critical for directing trafficking of proteins through the endosomal system, whereas polyubiquitylation plays a key role in directing substrates to the proteasome for degradation (1). One class of E3 ubiquitin ligases is HECT domain ubiquitin ligases that can both mono- and polyubiquitylate substrates. How mono- versus polyubiquitylation of substrates is mediated is unknown (1–4). Smurf1 and Smurf2 are HECT domain ubiquitin ligases that regulate transforming growth factor-β signaling as well as cell motility and polarity in part through targeting the GTPases RhoA and Rap1 as well as talin and core planar cell polarity components for polyubiquitin-dependent degradation (5–7).
Recently, non-covalent ubiquitin binding to the HECT domain of Rsp5 was characterized and proposed to play a role in regulating polyubiquitylation (8). Here we employ NMR spectroscopy to map the non-covalent ubiquitin binding surface (UBS)5 on the HECT domain of Smurf2. We show that mutation of a conserved surface tyrosine residue Tyr-459 on the UBS interferes with Smurf-dependent degradation of RhoA and blocks polyubiquitylation but not monoubiquitylation by the Smurf HECT domain. Furthermore, we show that efficient binding of a monoubiquitylated version of RhoA to the HECT domain is dependent on the UBS. Our results point to a model in which non-covalent binding of ubiquitin by HECT domains promotes polyubiquitylation by stabilizing interaction with monoubiquitylated substrates.
EXPERIMENTAL PROCEDURES
NMR Analysis
For NMR structure studies, ubiquitin (aa 1–76), the Smurf2 HECT domain (aa 366–748), and its N2 (aa 519–590) and C-lobe (aa 630–748) subdomains were expressed in Escherichia coli BL21(DE3) CodonPlus cells upon induction with isopropyl 1-thio-β-d-galactopyranoside. Cells expressing ubiquitin were grown in LB or M9 minimal medium containing 15NH4Cl as the sole sources of nitrogen. The N2 and C-lobe subdomains were expressed in 100% H2O M9 minimal medium containing 15NH4Cl and d-[13C]glucose as the sole sources of nitrogen and carbon. [U-methyl-δ1-13C,1H]Ile-labeled Smurf2 HECT domain was obtained by overexpression in 100% D2O M9 minimal medium as described (9, 10). The His-tagged recombinant proteins were purified by nickel affinity chromatography followed by tobacco etch virus protease cleavage. Untagged cleavage products were separated from the His tag and His-tagged tobacco etch virus protease by nickel affinity chromatography followed by size-exclusion gel filtration.
For resonance assignment, 0.6 mm 13C,15N-labeled N2 subdomain and 1 mm 13C,15N-labeled C-lobe samples were used in NMR buffer (20 mm HEPES buffer, pH 6.8, 200 mm NaCl, 0.03% (w/v) NaN3, and 3 mm dithiothreitol) in 90% H2O/10% D2O. 13C,1H and 15N chemical shifts were assigned using standard experiments (11) recorded at 30 °C on Varian Inova 500 MHz spectrometers. All spectra were processed with the NMRPipe/NMR Draw package (12) and analyzed with XEASY (13). All structure representations were prepared with PyMOL.
Chemical shift perturbation studies were performed using 0.1 mm 15N-labeled ubiquitin dissolved in NMR buffer, pH 6.8, for the interaction with the Smurf2 HECT domain, whereas an 80 μm [δ1-13C,1H]Ile-labeled Smurf2 HECT domain sample in NMR buffer, pH 7.0, was used for titration with ubiquitin. 1H,15N two-dimensional heteronuclear single quantum correlation spectra for chemical shift mapping were recorded at 30 °C on a Varian Inova 800 MHz spectrometer for the HECT domain titration with 15N-labeled ubiquitin. All 13C,1H HMQC spectra of [U-δ1-13C,1H]Ile-labeled Smurf2 HECT domains in 100% D2O NMR buffer were recorded at 30 °C on a Varian Inova 500 MHz spectrometer.
Cell and Biochemical Analyses
Full-length Smurf2, wild type, mutants (F29A/F30A), and UBS mutants (E404K and Y459A) used in ubiquitylation assays were generated using standard approaches in pCMV5. The Smurf2 WW-HECT construct for wild-type Smurf2 was previously described (14), and UBS mutants were mutagenized by PCR and cloned into pGEX-4T-1 and pProEx-HTa vectors. For mammalian cell assays, FLAG-tagged RhoA, wild-type Smurf2, or the corresponding Myc-tagged Smurf1 wild-type and the UBS mutants (E384K and Y439A) were subcloned in pCMV5b vector and transiently expressed as described previously (15). Chimeric UbG76V-RhoA was amplified by PCR and designed such that Ub is fused to the N terminus of RhoA and cloned into modified pGEX4T-1 with the tobacco etch virus protease site. The Gly-76 in Ub was mutated to valine to prevent Ub protease cleavage. This chimera was expressed in BL21 (DE3) codon-plus cells, and GST was affinity-purified from bacterial cell lysates as previously described (16). For in vitro Ub thioester assays, removal of the last four residues of the HECT domain is required to block autoubiquitylation (17) and stabilize thioester formation on the catalytic cysteine of HECT domains. Thus, wild-type Smurf2-HECT (−4 aa), Smurf2-HECT Y459A (−4 aa), and His-tagged Smurf2 HECT C716A were subcloned into pProExHTb vectors for expression in E. coli BL21(DE3) Codon Plus cells.
Immunoprecipitation and Blotting
For immunoprecipitation and immunoblotting, antibodies and their suppliers were α-Ub (P4D1; Santa Cruz), α-His5 (Qiagen), α-FLAG (M2; Sigma), and α-RhoA (Santa Cruz). α-Smurf2 antibodies were generated as described (15). For in vivo expression and assays, FLAG-tagged RhoA and/or wild-type and mutant Smurf2 were transfected in HEK293T cells as described (15, 18). Smurf2 and RhoA steady-state levels were analyzed by SDS-PAGE and immunoblotting using α-FLAG and α-Smurf2 antibody, respectively. All in vitro ubiquitylation and Ub thioester assays were performed in 15-μl reactions as previously described (16). Ubiquitylated species were detected using α-His5, α-RhoA, and/or α-Ub antibody for autoubiquitylation assays. GST pulldown experiments were performed as described previously (16). Purified and tobacco etch virus protease cleaved wild-type RhoA, or a Ub-RhoA fusion was incubated at 4 °C for 1 h with GST or GST-tagged Smurf2 ww-HECT bound to GST-beads in TNT (0.1% Triton X-100, 150 mm NaCl and 50 mm Tris·Cl, pH 7.5). GST beads were washed 4 times in 50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 5 mm β-mercaptoethanol. Bound proteins were analyzed by SDS-PAGE and immunoblotted using α-RhoA and α-Ub antibodies. For analysis of RhoA multi-monoubiquitylated species, RhoA was immunoprecipitated from ubiquitylation reactions and analyzed by immunoblotting.
RESULTS
To begin to understand the catalytic process of HECT domains, we explored by NMR spectroscopy whether ubiquitin binds to the Smurf2 HECT domain non-covalently. Using two-dimensional 1H,15N correlation experiments, we observed numerous chemical shift changes when the HECT domain was titrated into 15N-labeled ubiquitin (Fig. 1a). Mapping these chemical shift perturbations onto the ubiquitin structure, we observed that the interaction occurred with a mostly hydrophobic and positively charged surface centered around Arg-42 and Val-70 (Fig. 1, b and c). This is in agreement with the recently published Rsp5p HECT domain interaction surface of ubiquitin (8) and corresponds closely to those of most other ubiquitin binding proteins (19) characterized to date, such as the UBA domain of Dsk2p (Fig. 1d). However, our observations show that in addition to the surface centered on Arg-42 and Val-70, the Smurf2 HECT domain binding surface also includes the very C terminus of ubiquitin that is used for isopeptide linkage with a substrate lysine residue.
FIGURE 1.
Ubiquitin interacts non-covalently with the HECT domain of Smurf2. a, shown is an overlay of representative regions of 1H,15N two-dimensional correlation spectra of 15N-labeled ubiquitin in the absence (black; reference spectrum) and presence of increasing concentrations of Smurf2 HECT domain. b, residues experiencing significant chemical shift changes were mapped onto the structure of ubiquitin (PDB ID 1UBQ) and colored with a linear gradient from white (ΔδAv ≤ 0.03 ppm) to green (ΔδAv = 0.2 ppm). Spheres represent the nitrogen atoms of affected residues. Average chemical shift changes in proton and nitrogen were calculated as ΔδAv = ((Δδ1H)2 + (Δδ15N)2/5)1/2. c, shown is an electrostatic surface potential representation of ubiquitin in the same orientation as in b. Residues constituting the HECT domain binding site are highlighted on the ubiquitin surface. d, shown is structural superposition of ubiquitin color-coded as in b with ubiquitin in complex with the UBA domain of Dsk2p (PDB ID 1WR1).
We next sought to map the UBS on the Smurf2 HECT domain. Because the poor stability of this domain precludes traditional backbone-directed resonance assignment experiments, we made use of an isotope-labeling scheme where the Smurf2 HECT domain is 13C,1H-labeled exclusively at isoleucine δ1-methyl positions (16, 20). Upon stepwise addition of unlabeled Ub, we observed that a subset of Smurf2 HECT domain resonances changed in the two-dimensional 13C,1H correlation (HMQC) spectra (Fig. 2a). To obtain resonance assignments for the 25 Ile δ1-methyl groups in the Smurf2 HECT domain, we compared 13C,1H correlation spectra of the wild-type HECT domain to a set of 15 Ile mutants (Fig. 2b, right side), which allowed us to assign peaks for these residues (examples in Fig. 2b, left side). Next, we dissected the HECT domain into its subdomains. First, we expressed and purified a subdomain of the N-lobe (N2; Fig. 2c), which comprises the E2 binding pocket, and obtained the peak assignments by traditional backbone-directed NMR experiments (Fig. 2c). We also performed this with the C lobe (Fig. 2d). Altogether this allowed us to unambiguously assign 22 of the 25 Ile δ1-methyl groups in the Smurf2 HECT domain. With these assignments in hand, we then mapped the Smurf2 HECT domain residues affected by ubiquitin binding. This revealed a mostly acidic and hydrophobic region on the N-lobe of the Smurf2 HECT domain centered on Ile-594 (Fig. 3, a and b). Interestingly, this encompasses the region proposed for the Rsp5p HECT ubiquitin binding surface (8) but is more extensive. In those studies GST pulldown experiments with the Rsp5p HECT domain revealed seven alanine mutants (N513A, Y516A, Y521A, N534A, F618A, V621A/V622A, and R651A) that significantly affected Ub binding. These mutants correspond to residues Asn-451, Tyr-454, Tyr-459, Asp-472, Tyr-556, Ile-559, Ile-560, and Arg-589 in the Smurf2 HECT domain, of which Ile-559 and Ile-560 showed substantial chemical shift perturbations in our NMR titrations.
FIGURE 2.
Resonance assignment strategy for the Smurf2 HECT domain Ile δ1 positions. a, shown is an overlay of the representative regions of 13C,1H HMQC spectra of [δ1-methyl-13C,1H]Ile-labeled Smurf2 HECT domain in the absence (black; reference spectrum) and presence of increasing concentrations of ubiquitin. b, left and center panel, shown is an overlay of 13C,1H HMQC spectra of WT and mutant Smurf2 HECT domains. Right panel, shown are Ile residues (blue) that were mutated in the N1 domain of the Smurf2 HECT domain to enable Ile assignment. c, left panel, shown is an overlay of 13C,1H HMQC spectra of Smurf2 HECT domain (black) and its N2 subdomain (green). Right panel, shown are Ile residues in the N2 subdomain (green) of the Smurf2 HECT domain. Center panel, shown is a 1H,15N heteronuclear single quantum correlation spectrum of the N2 subdomain. d, left panel, shown is an overlay of 13C,1H HMQC spectra of Smurf2 HECT domain (black) and its C-lobe (red). Right panel, shown are Ile residues in the C-lobe (red) of the Smurf2 HECT domain. Center panel, shown is the 1H,15N heteronuclear single quantum correlation spectrum of the C-lobe.
FIGURE 3.
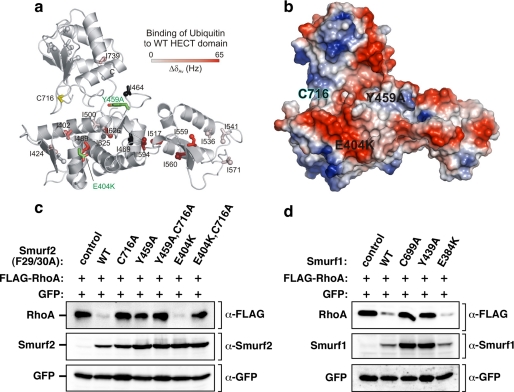
The ubiquitin binding surface of Smurfs is required for targeting RhoA in cells. a, residues experiencing significant chemical shift changes were mapped onto the structure of the Smurf2 HECT domain (PDB ID 1ZVD) and colored with a linear gradient from white (ΔδAv ≤ 0.0 Hz) to red (ΔδAv = 65 Hz). Spheres represent the δ1 carbon atoms of affected Ile residues. Ile residues with missing resonance assignments (I464, I469, and I625) are shown in black. The active site cysteine (Cys-716) and the mutations interfering with ubiquitin binding (E404K and Y459A) are highlighted in yellow and green. b, shown is an electrostatic surface potential representation of the Smurf2 HECT domain in the same orientation as in Fig. 4a, with the positions of the E404K and Y459A mutations labeled on the HECT domain surface. c, mutation in the Smurf UBS interferes with RhoA targeting in cells. HEK293T cells were transiently transfected with FLAG-tagged RhoA in combination with empty vector, F29A/F30A, or F29A/F30A catalytically inactive Myc-tagged Smurf2 with an otherwise WT HECT domain or a HECT domain harboring the indicated UBS mutants. RhoA and Smurf2 steady-state levels in total cell lysates were determined by immunoblotting with α-FLAG and α-Smurf2 antibody (top panel and bottom panel), respectively. d, mutation in the Smurf1 UBS interferes with RhoA targeting in cells. HEK293T cells were transiently transfected with FLAG-tagged RhoA in combination with empty vector or Myc-tagged Smurf1 that was WT, C699A, and Y439A, or E384K (which correspond, respectively, to Tyr-459 and Glu-404 in Smurf2). RhoA and Smurf1 steady-state levels in total cell lysates were determined by immunoblotting with α-FLAG and α-Smurf1 antibody (top panel and bottom panel), respectively.
To explore the function of the Smurf2 UBS, we used our NMR analysis to design mutants on the surface of the UBS (Fig. 3b), specifically a highly conserved glutamate (Glyu-404 to Lys) at the edge of the UBS close to the catalytic Cys and a highly conserved tyrosine (Tyr-459 to Ala) that is equivalent to the Y521A mutant in Rsp5p shown to inhibit but not abolish ubiquitin binding to the HECT domain. Next we examined in vivo whether these mutants of the HECT domain affected targeting of the GTPase RhoA, which is a Smurf target involved in the regulation of cell motility and polarity. For this, we introduced the WT or UBS HECT domain mutants into an activated form of Smurf2 in which two phenylalanine residues that mediate autoinhibition of the Smurf2 C2 domain are mutated to alanine residues (Smurf2 (F29A/F30A) (16). These mutants were then co-expressed with RhoA in HEK293T cells, and steady-state levels of RhoA were assessed by Western blotting. Under these conditions Smurfs actively target RhoA for polyubiquitylation and proteasome-mediated degradation (3, 5, 16, 21). Both activated WT Smurf2 and the E404K mutant strongly decreased RhoA steady-state levels but not the control GFP protein (Fig. 3c). This loss was abrogated by a mutation in the HECT catalytic cysteine residue (C716A). This was confirmed using an E404K mutant harboring a concomitant mutation in the catalytic cysteine (E404K, C716A) that prevented loss of RhoA. In contrast, the Y459A mutant of Smurf2 displayed only marginal activity toward RhoA, and when in combination with the C716A mutation we observed only a small increase in RhoA steady-state levels (Fig. 3c). Similar results were obtained for Smurf1, which is closely related to Smurf2 (Fig. 3d). These results indicate that the surface-exposed tyrosine 459 within the UBS of the HECT domain is critical for targeting RhoA for degradation.
To examine how mutation of the UBS affected the catalytic activity of the Smurf2 HECT domain, we performed in vitro autoubiquitylation assays. Under these conditions both WT and the E404K mutant of Smurf2 underwent efficient autoubiquitylation, although the E404K mutant displayed less activity compared with WT Smurf2 (Fig. 4a). Surprisingly, when we examined the Y459A mutant, most ubiquitylation was monoubiquitylation, with little polyubiquitylated species apparent. This suggested that the Smurf2 (Y459A) mutant interferes primarily with polyubiquitylation. To directly test this, we performed autoubiquitylation assays using ubiquitin in which all lysines are mutated (UbNOK), thus preventing ubiquitin chain elongation. Under these conditions, the WT HECT domain revealed multiple isoforms corresponding to multi-monoubiquitylation (Fig. 4b). The Y459A mutant also displayed similar levels of monoubiquitylation, albeit the pattern was different, with single and di-ubiquitylated species more prominent. Because the polyubiquitylation activity of the Y459A mutant was crippled, we next examined whether ubiquitin thioester formation was affected. For this we employed a truncation of the last four residues of the HECT domain, which was previously shown to block ubiquitylation and stabilize thioester formation on the catalytic cysteine of HECT domains (17). This revealed that dithiothreitol-sensitive ubiquitin conjugation to cysteine 716 was comparable in both WT and the Y459A mutant (Fig. 4c). Thus, the Y459A mutant does not interfere with ubiquitylation by affecting ubiquitin charging of the catalytic cysteine by the E2.
FIGURE 4.
Disruption of the UBS inhibits autopolyubiquitylation by the Smurf HECT domain. a, shown is Smurf2 UBS mutant autoubiquitylation in vitro. In vitro autoubiquitylation assays were performed for the indicated times using purified wild-type Smurf2 and Smurf2 E404K and Y459A mutants. Autoubiquitylated Smurf2 (top panel) or total Smurf2 in the reaction (bottom panel) was analyzed by immunoblotting using anti-Ub and anti-His antibody, respectively. b, WT Smurf2 and mutants form multimonoubiquitylated adducts. In vitro autoubiquitylation assays were carried out for the indicated times using Smurf2 WT and E404K and Y459A mutants in the presence of lysine-null ubiquitin (UbNOK) and analyzed by immunoblotting with an anti-His antibody. c, WT-Smurf2 HECT domain (−4 aa) and Y459A HECT (−4 aa) mutants efficiently form Ub thioester intermediates. Wild-type and catalytically inactive and Smurf2 Y459A HECT domain missing the last four amino acids (−4 aa) were charged for 10 min at room temperature and analyzed by immunoblotting under non-reducing (−dithiothreitol (−DTT, left panel) or reducing conditions (+DTT, right panel) using an α-Ub antibody.
To confirm these observations we performed a kinetic analysis of poly- and monoubiquitylation of WT and the Y459A mutant. In the presence of WT ubiquitin, the Smurf2 HECT domain achieved maximal levels of polyubiquitylation by ∼90 min, whereas polyubiquitylation of the Y459A mutant was almost undetectable despite the appearance of monoubiquitylated species (Fig. 5a). In parallel, we performed assays using UbNOK, which revealed that the WT HECT domain efficiently catalyzed monoubiquitylation with maximal levels achieved by ∼20 min (Fig. 5b). Unlike polyubiquitylation, the Y459A mutant achieved similar overall levels of monoubiquitylation as WT (Fig. 5b), although the Y459A mutant caused a lag in the addition of monoubiquitylation such that half-maximal levels of monoubiquitylation were achieved by 20 min compared with ∼9 min for WT Smurf. These results show that mutation of the UBS leads to a strong reduction in polyubiquitylation activity of the Smurf HECT domain.
FIGURE 5.
Disruption of the Smuf2 HECT domain UBS inhibits polyubiquitylation but not monoubiquitylation. In vitro autoubiquitylation assays were carried out for the indicated time points using purified wild-type Smurf2 or the Smurf2 Y459A mutant as indicated and either wild-type Ub to assess polyubiquitylation (a) or lysine-null Ub (UbNOK) (b) to assess monoubiquitylation. The autoubiquitylation profiles were analyzed by immunoblotting with anti-His antibody. To assess the kinetics of autoubiquitylation, the levels of polyubiquitin or monoubiquitin were quantified at each time point and are plotted as the percent of the input HECT domain. The results are the mean of three independent experiments (±S.E.).
We next sought to confirm that ubiquitylation of the Smurf substrate, RhoA, was similarly affected by the Y459A mutant in vitro. Although polyubiquitylation of RhoA was strongly inhibited by the Y459A mutant, monoubiquitylated RhoA was readily apparent (Fig. 6a). We confirmed this employing UbNOK, which revealed that both the WT and the Y459A HECT domains catalyzed monoubiquitylation of RhoA (Fig. 6b). Together with our analysis of autoubiquitylation, these results indicate that the UBS is critical for polyubiquitylation but is not essential for monoubiquitylation. One possibility is that binding of free ubiquitin to the HECT plays a role in catalyzing chain elongation. However, in Rsp5p, mutation of Tyr-521 (that corresponds to Tyr-459 in Smurf2) retains ubiquitin binding activity (8), albeit reduced compared with WT, and our analysis of the Smurf2 Y459A HECT mutant by NMR also revealed ubiquitin binding (data not shown). We, therefore, considered the possibility that the UBS may be primarily functioning in a different context; that is, to stabilize the association of a ubiquitin adduct to the HECT domain, in particular monoubiquitylated substrates. To test this we took advantage of the fact that the lysines in RhoA that are targeted for ubiquitylation by the Smurf HECT domain reside near the N terminus of RhoA at lysine residues 6 and 7 (22). Therefore, to mimic monoubiquitylated RhoA, we constructed a chimeric protein (Ub-RhoA) in which Ub was fused to the N terminus of RhoA. Analysis of RhoA binding to the WT or the Y459A mutant HECT domains revealed equivalent RhoA binding (Fig. 6c). Interestingly, binding of Ub-RhoA to the WT HECT domain was greater than that observed for RhoA alone (Fig. 6c). In contrast, Ub-RhoA binding to the Y459A mutant was impaired and was comparable with the association of RhoA alone. These results demonstrate that the region of the UBS that encompasses Tyr-459 is required for efficient polyubiquitylation of substrates and functions to promote binding of monoubiquitylated substrates to the HECT domain.
FIGURE 6.
Mutation of the UBS prevents RhoA targeting in vitro and interferes with binding of a monoubiquitylated form of RhoA. a, Smurf2 Y459A mutant monoubiquitylates, but does not polyubiquitylate RhoA. Constitutively active Smurf2 F-29/30-A with either wild type or the Y459A mutant UBS were used in an in vitro ubiquitylation assay with RhoA substrate. RhoA was immunoprecipitated (IP) from ubiquitylation reactions and analyzed by immunoblotting (top panel), whereas polyubiquitylated RhoA was analyzed by immunoblotting ubiquitylation reactions as indicated (as indicated, the top and bottom panels are an analysis of the lower molecular weight and higher molecular weight regions, respectively). RhoA and Smurf2 input were detected by immunoblotting (IP) as indicated. b, ubiquitylation of RhoA using UbNOK. RhoA was subject to an in vitro ubiquitylation reaction as in a, except that UbNOK was employed in the reaction. Ubiquitylated RhoA was detected by subjecting total reactions to immunoblotting with an anti-RhoA antibody as indicated. c, the UBS promotes Smurf2 interaction with a Ub-RhoA fusion protein. Bacterially expressed GST or GST-Smurf2 WW-HECT domain were bound to GST beads and incubated with purified RhoA or Ub-RhoA. Smurf2-bound RhoA or Ub-RhoA (top panel) or totals (bottom panel) were analyzed by immunoblotting with α-RhoA antibodies.
DISCUSSION
We have mapped the UBS onto the HECT domain of Smurf2 by NMR spectroscopy. Previous analysis of determinants on ubiquitin that regulate linkage selectivity by the HECT domain ligase, KIAA10, led to the suggestion that ubiquitin might interact non-covalently with the HECT domain (23). Subsequent analyses of the Rsp5 HECT domain indeed revealed non-covalent ubiquitin binding. Furthermore, using systematic mutagenesis, the Rsp5 ubiquitin binding region was proposed to comprise the same surface as that revealed by our NMR studies of Smurf2 (8). This surface resides on the N1 subdomain of the N lobe of the HECT domain, sitting between the E2 binding N2 domain and the catalytic cysteine in the C lobe. The positioning of the ubiquitin binding surface could, thus, contribute to the catalytic mechanism of ubiquitin conjugation to HECT substrates. However, mutagenesis of this region revealed that interference with free ubiquitin binding did not inhibit in vitro autoubiquitylation (8). Similarly, extensive mutagenesis of ubiquitin led to the proposal that ubiquitin binding might serve to control the type of polyubiquitin linkage catalyzed by KIAA10. Collectively, these studies indicate that noncovalent binding of ubiquitin to the HECT domain is not absolutely required to catalyze the ubiquitin conjugation reaction.
This raises the question of what role ubiquitin binding to the HECT domain might play in ubiquitylation of target proteins. We found that mutation of the surface residues Glu-404 and Tyr-459 had only modest effects on ubiquitin binding, similar to the findings in Rsp5. However, when we analyzed the activity of these Smurf2 HECT mutants, we observed that mutation of Tyr-459 strongly suppressed autopolyubiquitylation as well as polyubiquitylation of the RhoA substrate. This correlated with the inability of Y459A mutants of either Smurf1 or Smurf2 to catalyze efficient degradation of RhoA in cells. Furthermore, when we performed in vitro autoubiquitylation assays using the Smurf2 Y459A mutant, analysis of monoubiquitylated species clearly showed the presence of conjugated species in the presence of the mutant HECT domain, and analysis of kinetics showed a reduced rate, with comparable total levels of monoubiquitylated species observed by the end of the reaction. These studies demonstrate that mutation of the UBS on Smurf HECT domains strongly interferes with polyubiquitylation.
Studies on Rsp5 ubiquitin binding mutants also led to the conclusion that the region functioned predominantly in the context of polyubiquitin chain elongation. However, unlike our studies, they found that chain length might be increased by loss of ubiquitin binding. Although our data clearly show that mutation of the Smurf UBS blocks polyubiquitylation, these studies together suggest that noncovalent ubiquitin binding serves to control chain elongation. How might the UBS modulate chain length? Our analysis of RhoA binding to the Smurf HECT domain showed weak binding to both WT and Y459A mutant HECT domains. However, when we examined a Ub-RhoA fusion protein, we observed enhanced binding to the WT HECT domain that was lost in the Y459A mutant. Thus, we propose that the UBS functions not as a free ubiquitin binding surface but, rather, acts to stabilize binding of ubiquitin-conjugated substrates. This in turn may promote polyubiquitylation by the HECT domain by properly positioning the end of the growing ubiquitin chain for subsequent ubiquitin additions.
We also mapped the interaction surface on ubiquitin and showed that it is composed of the hydrophobic and positively charged surface that is centered around Arg-42 and Val-70. Interestingly, we also observed that interactions extended to the C terminus. This might be consistent with the notion that in the context of catalytic reactions, ubiquitin binding is manifested in the context of conjugated substrates, with the interaction surface extending to the site of conjugation. Furthermore, the surface of ubiquitin that binds the Smurf HECT domain is a common interaction surface for many ubiquitin binding domains and is also used by Rsp5. Given that ubiquitin binding to HECT domains via this surface is important for controlling polyubiquitylation, it will be interesting in the future to define how ubiquitin-binding proteins might affect polyubiquitin chain addition to target substrates in specific cellular contexts.
This work was supported by Canadian Cancer Society Research Institute grants (to J. F.-K.) and Canadian Institutes of Health Research Grants 57795 (to F. S.) and 14339 (to J. L. W.).
- UBS
- ubiquitin binding surface
- aa
- amino acids
- HMQC
- heteronuclear multiple quantum coherence
- Ub
- ubiquitin
- GST
- glutathione S-transferase
- WT
- wild type.
REFERENCES
- 1.Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 2.Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 3.Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 4.Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Nat. Cell Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 5.Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. (2003) Science 302, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 6.Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., Wrana J. L. (2009) Cell 137, 295–307 [DOI] [PubMed] [Google Scholar]
- 7.Huang C., Rajfur Z., Yousefi N., Chen Z., Jacobson K., Ginsberg M. H. (2009) Nat. Cell Biol. 11, 624–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French M. E., Kretzmann B. R., Hicke L. (2009) J. Biol. Chem. 284, 12071–12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner K. H., Kay L. E. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 357–406 [DOI] [PubMed] [Google Scholar]
- 10.Kay L. E., Gardner K. H. (1997) Curr. Opin. Struct. Biol. 7, 722–731 [DOI] [PubMed] [Google Scholar]
- 11.Bottomley M. J., Macias M. J., Liu Z., Sattler M. (1999) J. Biomol. NMR 13, 381–385 [DOI] [PubMed] [Google Scholar]
- 12.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 13.Bartels H., Bennett W. S., Hansen H. A., Eisenstein M., Weinstein S., Müssig J., Volkmann N., Schlünzen F., Agmon I., Franceschi F. (1995) Biopolymers 37, 411–419 [DOI] [PubMed] [Google Scholar]
- 14.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 15.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 16.Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 17.Salvat C., Wang G., Dastur A., Lyon N., Huibregtse J. M. (2004) J. Biol. Chem. 279, 18935–18943 [DOI] [PubMed] [Google Scholar]
- 18.Wang H. R., Ogunjimi A. A., Zhang Y., Ozdamar B., Bose R., Wrana J. L. (2006) Methods Enzymol. 406, 437–447 [DOI] [PubMed] [Google Scholar]
- 19.Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. (2005) Structure 13, 521–532 [DOI] [PubMed] [Google Scholar]
- 20.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E. (2003) J. Am. Chem. Soc. 125, 10420–10428 [DOI] [PubMed] [Google Scholar]
- 21.Doye A., Mettouchi A., Bossis G., Clément R., Buisson-Touati C., Flatau G., Gagnoux L., Piechaczyk M., Boquet P., Lemichez E. (2002) Cell 111, 553–564 [DOI] [PubMed] [Google Scholar]
- 22.Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., Wrana J. L. (2005) Science 307, 1603–1609 [DOI] [PubMed] [Google Scholar]
- 23.Wang M., Cheng D., Peng J., Pickart C. M. (2006) EMBO J. 25, 1710–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]