Abstract
Regulation of the E2F family of transcription factors is important in control of cellular proliferation; dysregulation of the E2Fs is a hallmark of many cancers. One member of the E2F family, E2F1, also has the paradoxical ability to induce apoptosis; however, the mechanisms underlying this selectivity are not fully understood. We now identify a nucleolar protein, RRP1B, as an E2F1-specific transcriptional target. We characterize the RRP1B promoter and demonstrate its selective response to E2F1. Consistent with the activation of E2F1 activity upon DNA damage, RRP1B is induced by several DNA-damaging agents. Importantly, RRP1B is required for the expression of certain E2F1 proapoptotic target genes and the induction of apoptosis by DNA-damaging agents. This activity is mediated in part by complex formation between RRP1B and E2F1 on selective E2F1 target gene promoters. Interaction between RRP1B and E2F1 can be found inside the nucleolus and diffuse nucleoplasmic punctates. Thus, E2F1 makes use of its transcriptional target RRP1B to activate other genes directly involved in apoptosis. Our data also suggest an underappreciated role for nucleolar proteins in transcriptional regulation.
Keywords: Apoptosis, DNA/Damage, DNA/Transcription, Transcription/E2F, Nucleolus, RRP1B
Introduction
E2F1 is a critical regulator of DNA damage response and apoptosis. As part of the E2F family of transcription factors, E2F1 is also involved in regulation of a wide array of genes important for cell cycle progression and other functions (1). Paradoxically, E2F1 has the unique ability to induce apoptosis (2). Overexpression of E2F1 ex vivo leads to apoptosis of breast cancer and other cells (3–5). Deletion of E2F1 in vivo shows a defect in thymocyte apoptosis and increased tumor incidence (6, 7). An endogenous role for E2F1 apoptosis is illustrated by its activation and stabilization by genotoxic stimuli. Overexpression of E2F1 sensitizes cells to radiation and chemotherapy (8, 9). DNA damage activates E2F1 expression and induces E2F1 stabilization through phosphorylation by DNA damage-responsive kinases ATM (ataxia telangiectasia mutated) (10) and Chk2 (checkpoint kinase 2) (11) and through acetylation (12, 13). E2F1 transactivates proapopotic genes, such as p73 (14, 15), Apaf-1 (16), and caspases (16) independently of p53 and cooperates with p53 through transactivation of p19ARF (17). Investigation of how E2F1 specifically regulates apoptosis through selective transcriptional regulation vis-à-vis other E2F family members may reveal targets for future study that might improve the sensitivity of cancer to radiotherapy and chemotherapy.
We therefore attempted to identify genes specifically regulated by E2F1 that potentially regulate E2F1-induced apoptosis. Previously, the Helin group published a microarray data set in which expression profiles were compared between cells that overexpressed E2F1, E2F2, and E2F3 (18). We screened their data set to include only those genes that were significantly induced by E2F1 but whose expression did not change more than 1-fold either positively or negatively upon E2F2 or E2F3 overexpression. The list of genes screened from this study is shown in Table 1. Among them was the gene RRP1B (ribosomal RNA processing 1 homolog B), also known as KIAA0179 or NNP-1B (novel nucleolar protein 1 homolog B). RRP1B is related to RRP1 (ribosomal RNA processing 1), a protein involved in ribosomal biogenesis localized to the nucleolus (19–22). Recent data have shown that RRP1B is involved in suppression of metastasis, and a gene expression profile obtained following its overexpression predicted survival in breast cancers (23). However, the mechanism of how RRP1B reduces tumor burden remains unclear.
TABLE 1.
Post hoc analysis of Muller et al. (18) for genes specifically up-regulated by E2F1 but not other E2Fs
Values indicate -fold induction or repression.
| Gene | E2F1 | E2F2 | E2F3 | Description |
|---|---|---|---|---|
| -fold | -fold | -fold | ||
| EPAS1 | 4.1 | −0.6 | −0.8 | Endothelial PAS domain protein 1 |
| 3.8 | −0.7 | −0.7 | Homo sapiens mRNA; cDNA DKFZp434E1515 | |
| 3.4 | 0.5 | −0.5 | H. sapiens mRNA; cDNA DKFZp564E1363 | |
| ARHH | 2.1 | 0.8 | 0.2 | Ras homolog gene family, member H |
| CHML | 4.8 | 0.2 | 0.3 | Choroideremia-like (Rab escort protein 2) |
| NFRKB | 2.4 | 0.5 | 0.4 | Nuclear factor related to κB-binding protein |
| KIAA0179 | 2.5 | 0.7 | 0.5 | KIAA0179 protein (RRP1B) |
| ABCB2 | 8.5 | −0.3 | 0.6 | ATP-binding cassette, subfamily B (MDR/TAP) |
| CAMKK2 | 3.1 | 0.6 | 0.6 | Calcium/calmodulin-dependent protein kinase kinase |
| NCOA1 | 2.9 | −0.7 | 0.6 | Nuclear receptor coactivator 1 |
| C3 | 2.5 | −0.3 | 0.6 | Complement component 3 |
| MAOA | 2.7 | 0.5 | 0.7 | Monoamine oxidase A |
| OSTF1 | 3.5 | −0.6 | 0.8 | Osteoclast-stimulating factor 1 |
| FBLN5 | 3.2 | −0.5 | 0.8 | Fibulin 5 |
We now provide evidence that RRP1B is specifically regulated by E2F1 and not other E2F family members. RRP1B is important for regulation of apoptosis induced by both DNA damage and E2F1 overexpression. Consistent with its proapoptotic function, RRP1B selectively regulates the expression of several proapoptotic E2F1 target genes through chromatin binding. We also demonstrate a direct interaction between RRP1B and E2F1 in vitro and in vivo in nucleoli and in punctate nucleoplasmic foci. Together, these data suggest that RRP1B is both a novel E2F1 target and a novel coactivator and may prime cells for E2F1-dependent apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293, HEK293T, T98G, NIH3T3, H1299, human foreskin fibroblasts, and Ref52 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS),2 penicillin (50 IU/ml), and streptomycin (50 μg/ml). HCT116 and U2OS cells were grown in McCoy's 5A medium supplemented with 10% FBS, penicillin, and streptomycin. All cells were grown in a humidified incubator at 37 °C with 5% CO2 and 95% air. A standard calcium phosphate method was used for transfection of HEK293, HEK293T, and H1299 cells. NIH3T3 and Ref52 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After transfection, cells were incubated for 48 h before analysis.
Plasmid Construction
The RRP1B promoter was cloned using PCR of genomic DNA, constituting genomic DNA from −2354 to +259 surrounding the cDNA start site. PCR primers contain a XhoI site 5′ to the forward cloning primer and a HindIII site 5′ to the reverse cloning primer. The primers used were as follows: forward promoter, 5′-CGCCTCGAGCAGGGTTGGAGGCTGCA-3′; reverse promoter, 5′-CGCAAGCTTACTGAGAATGTCAGTGATGGGGGA-3′. PCR product was digested with XhoI and HindIII and then ligated together with pGL3-Basic digested with XhoI and HindIII.
A mutation at the putative E2F binding site at +150 was generated in the pGL3-RRP1B promoter by changing two nucleotides (5′-GCGGTCAGCCGCTACACATGGCGGGC-3′) using the QuikChange site-directed mutagenesis kit (Stratagene). To construct pGL3-RRP1B with a mutation at −505 and −400, four nucleotides were changed in two consecutive cycles of a standard megaprimer mutagenesis protocol (24). For pGL3-RRP1B −505, the mutagenic primers used were 5′-AGTGGGGCGTGATGATGCGCGCCTGTAGTC-3′ and GACTACAGGCGCGCATCATCACGCCCCACT-3′ and then 5′-GGGGCGTGATGATGCATGCCTGTAGTCTCAGC-3′ and 5′-GCTGAGACTACAGGCATGCATCATCACGCCCC-3′. For pGL3-RRP1B −400, the mutagenic primers used were 5′-AGCCAGGATCACCGCCAAGATATCGCCACTGCAT-3′ and 5′-ATGCAGTGGCGATATCTTGGCGGTGATCCTGGCT-3′ and then 5′-TCACCGCCAAGATATCGATACTGCATTCCAGCCTGG-3′ and 5′-CCAGGCTGGAATGCAGTATCGATATCTTGGCGGTGA-3′.
To construct a tagged mammalian expression vector for RRP1B, RRP1B cDNA was obtained from ATCC in pBluescript II SK(+). A FLAG tag was inserted 5′ to the transcriptional start site using a PCR primer; a KpnI site, Kozak sequence, and methionine are 5′ to the FLAG tag, and a BglII site was inserted in between the FLAG tag and RRP1B cDNA. The following primers were used: forward, 5′-GCGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGAGATCTATGGCCCCCGCCATGCAGCCGG-3′; reverse, 5′-AGCTTCGAAGACACCCCGAGCTAT-3′. Amplified PCR product was digested with KpnI and BstBI and cloned into pBluescript II SK(+)-RRP1B digested with KpnI and BstBI. pBluescript II SK(+)-FLAG-RRP1B was then digested with KpnI and NotI, and the cDNA insert was ligated with pcDNA3 digested with KpnI and NotI.
pcDNA3-FLAG-RRP1B-(1–473), FLAG-RRP1B-(474–589), or FLAG-RRP1B-(590–758) was cloned from full-length RRP1B with the addition of a BglII site at the 5′-end of the forward primer and a NotI site at the 5′-end of the reverse primer flanking the 3′-end of the coding sequence. PCR products were then digested with BglII and NotI and ligated with the vector sequence from modified pcDNA3-FLAG-RRP1B digested with BglII and NotI. The BglII site in the backbone of pcDNA3 vector was first destroyed by Klenow enzyme. The following primer sets were used: FLAG-RRP1B-(1–473), forward (same as full-length forward sequence) and reverse (5′-CGCGCGGCCGCTCATTTCCTTTTATTGTGCATGGG-3′); FLAG-RRP1B-(474–589), forward (5′-GCGAGATCTCGGCCACGGAAGAAGAGCCCG-3′) and reverse (5′-CGCGCGGCCGCTCATGTTTTCTGGCTGGGCAGGCC-3′); FLAG-RRP1B-(590–758), forward (5′-GCGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGAGATCTGCAAGTTTGAAAAAGAGGAAG-3′) and 5′-CGCGCGGCCGCTCAGAAGAAATCCATAGC-3′ (reverse).
E2F1 domain mutants were constructed into the pGEX-6P1 system (GE Healthcare). To construct pGEX-6P1-E2F1-(1–109), pAS2–1-E2F1-(1–109) (25) was digested with EcoRI and SalI, and the insert was ligated with pGEX-6P1 vector, which was digested with EcoRI and SalI. pGEX-6P1-E2F1-(110–284), pGEX-6P1-E2F1-(285–358), and pGEX-6P1-E2F1-(359–437) were cloned by PCR, using full-length E2F1 cDNA as a template, with the addition of a 5′ BamHI site and a 3′ EcoRI site flanking the primer sequences. PCR products were then digested with BamHI and EcoRI and ligated with pGEX-6P1 digested with BamHI and EcoRI. pGEX-6P1-E2F1-(110–282) was cloned using 5′-GCGGGATCCGGCAGAGGCCGCCATCCA-3′ and 5′-AGCGAATTCTCAAAAGTTCTCCAAGAGTC-3′; pGEX-6P1-E2F1-(283–358) was cloned using 5′-GCGGGATCCCAGATCTCCCTTAAGAGC-3′ and 5′-AGCGAATTCTCACAACAGCGGTTCTGCTC-3′; pGEX-6P1-E2F1-(359–437) was cloned using 5′-GCGGGATCCTCCCGGATGGGCAGCCTG-3′ and 5′-AGCGAATTCTCAGAAATCCAGGGGGGT-3′.
For bimolecular complementation assays, RRP1B was first shuttled from pcDNA3-FLAG-RRP1B by digestion with BglII and BamHI and ligated with pEGFP-C1 digested with BamHI; orientation was checked by digestion with BglII and BamHI. RRP1B was then excised from pEGFP-C1-RRP1B by BspEI and NheI and inserted into pcDNA3.1 yellow fluorescent protein 1-zipper (YFP1, containing enhanced YFP aa 1–158) (26) digested by BspEI and NheI. YFP2-E2F2 was constructed by digestion of pEGFP-E2F2 with BamHI, followed by Klenow digestion, purification, and then sequential digest with NheI and BspEI. The insert was ligated with pcDNA3.1 YFP2-zipper digested with XbaI followed by Klenow digestion, purification, and then digestion with BspEI.
Immunoprecipitation and Western Blot Analysis
Cells prepared for endogenous immunoprecipitation were washed and scraped in phosphate-buffered saline (PBS); nuclei were then extracted twice by incubation on ice for 10 min with nuclear extraction buffer (10 mm Tris, 85 mm KCl, 5 mm EGTA, 0.5% Nonidet P-40) supplemented with protease inhibitors (1 mm dithiothreitol, 1 mm NaF, 1 mm sodium orthovanadate, 20 nm microcystin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml antipain, and 1 μg/ml chymostatin). Nuclei were then lysed in TNN buffer (50 mm Tris, 0.25 m NaCl, 5 mm EDTA, 0.5% Nonidet P-40) with protease inhibitors, sonicated, precleared by nutation at 4 °C for 1 h with protein G-agarose beads (Pierce), and then nutated at 4 °C overnight with 2 μg of E2F1 antibody (KH95, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) or nonspecific mouse IgG (Pierce). Protein G beads were then added, and the sample was nutated at 4 °C for 2 h and then washed five times with ice-cold TNN buffer. Beads were eluted with SDS sample buffer, subjected to SDS-PAGE, and electrotransferred to Immobilon-P membrane (Millipore).
Cells prepared for immunoprecipitation of overexpressed proteins were washed and directly lysed in TNN with protease inhibitors and nutated at 4 °C overnight with anti-FLAG-agarose beads (M2, Sigma). An aliquot of lysate was saved for protein input control. Beads were washed five times with ice-cold TNN buffer, eluted, electrophoresed, and blotted as above.
Cells prepared for direct protein analysis were lysed in SDS lysis buffer (1% SDS, 60 mm Tris). Equal protein amounts were electrophoresed and blotted as above. Equal loading was confirmed by Ponceau S staining. DNA damage was induced by the addition of 1 μm doxorubicin, 20 μm cisplatin, or 0.05, 0.3, or 1.0 μg/ml neocarzinostatin. Densitometric analysis was performed using ImageJ (National Institutes of Health); measurement of RRP1B protein level was normalized against corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein level. For all experiments, specific proteins were detected with the appropriate antibodies. An RRP1B antibody was raised in rabbits against a peptide (ATHPPGPAVQLNKTPSSSKK) by Open Biosystems. Crude rabbit sera were affinity-purified using peptide-conjugated N-hydroxysuccinimide-activated Sepharose (GE Healthcare). Antibodies against E2F1 (KH95 and C20), E2F2 (C20), E2F3 (C18), E2F4 (WUF11), E2F5 (MH5), HA (Y11), and glyceraldehyde-3-phosphate dehydrogenase (0411) were purchased from Santa Cruz Biotechnology. FLAG antibody (F7425) was purchased from Sigma.
Lentivirus Production and Transduction
Knockdown of RRP1B was achieved by infection of cells with lentiviruses expressing RRP1B small interfering RNA (siRNA). pLKO.1 plasmids expressing siRNA sequences (27) were obtained from the RNAi Consortium (Open Biosystems) and screened for knockdown of RRP1B by transient transfection of HEK293T cells, followed by Western blotting. A control nonspecific siScramble pLKO.1 plasmid (28) and pMDG and pCMV ΔR8.2 packaging vectors were obtained from Addgene. Two plasmids containing the following siRNA sequences achieved high knockdown: A, 5′-GATGACCAAATCCTCAGTCAA-3′; B, 5′-GCACATTTGTTCTGCAGACTA-3′. Plasmids achieving high knockdown were used for lentivirus production by cotransfection of pLKO.1 containing siRNA sequences, pMDG, and pCMV ΔR8.2 in HEK293T cells; supernatants containing virus were collected every 24 h, filtered using a 0.3-μm filter, added to target cells, incubated for 48 h, and then selected for stable transduction by the addition of puromycin for 96 h. Knockdown was confirmed by Western blotting.
Luciferase Assays
The expression constructs (5 μg for pcDNA3-E2F1, pcDNA3-E2F2, or pcDNA3-E2F3 or empty vector), the promoter plasmids (1 μg for pGL3-RRP1B and point mutants, pGL3-rRNA promoter, and proximal mutant (29), caspase-7 promoter (30), E2F1 promoter (31), and thymidine kinase (TK) promoter (32)), and 1 μg of pCMV-β-galactosidase plasmids were cotransfected in HEK293T or stably transduced siScramble or siRRP1B H1299 cells. Cells were harvested 48 h later in PBS; an aliquot was lysed in SDS lysis buffer for Western blotting, whereas the rest of the sample was lysed in reporter lysis buffer (Promega). Luciferase activity and β-galactosidase activity were measured according to the manufacturer's instructions. Luciferase activity was normalized against β-galactosidase activity. All transient expression assays were performed in triplicate.
Apoptosis Assays
DNA damage-induced apoptosis was assayed in stably transduced siScramble or siRRP1B U2OS cells, which were untreated or treated with 20 μm cisplatin for 30 h before harvest. Cells were then stained with annexin V-APC or annexin V-PE (BD Biosciences) and 7-amino-actinomycin (BD Biosciences). At least 10,000 cells were profiled for surface annexin-V/7-AAD positivity by flow cytometry. Annexin V+/7-AAD− and annexin V+/7-AAD+ cells were scored as apoptotic. Cell death was also assayed in the same cells untreated or treated with 20 μm cisplatin for 28 h before harvest, followed by staining with propidium iodide (Roche Applied Science) and profiling for DNA content by flow cytometry. The sub-G0/G1 population were scored as dead cells. Alternatively, stably transduced siScramble or siRRP1B U2OS cells were untreated or treated with 1 μm doxorubicin for 8 h, harvested, and then assayed for caspase-3/7 cleavage according to the manufacturer's instructions (Caspase-Glo 3/7, Promega).
E2F1-induced apoptosis was assayed in stably transduced siScramble or siRRP1B U2OS cells infected by adenoviruses expressing E2F1 or empty vector. Adenoviruses were produced in the AdEasy system as previously described (33). Cells were starved in 0.25% fetal bovine serum for 48 h, followed by adenovirus infection (multiplicity of infection of 100) for 28 h. Cells were then harvested and analyzed for surface annexin-V/7-AAD positivity by flow cytometry as above. All apoptosis assays were performed in triplicate.
Cellular Proliferation Assay
1 × 105 stably transduced siScramble and siRRP1B U2OS cells were each plated in six replicates in 3.5-cm diameter 6-well plates, grown for 72 h prior to confluence, trypsinized, and collected. Two aliquots from each well were counted using a hemacytometer. One-quarter of the remaining cells were replated. Assay was repeated on days 6 and 9. Cells were harvested at day 9 in SDS sample buffer for Western blotting. A similar assay was performed identically except that cells were grown in medium containing 2% fetal bovine serum.
Real-time and Semiquantitative PCR
For analysis of RRP1B dependence on E2F family member expression, T98G cells were starved in Dulbecco's modified Eagle's medium containing 0.25% FBS for 48 h and then infected with adenoviruses expressing E2F1, E2F2, E2F3, E2F4, E2F5, or empty vector for 24 h. RNA was then extracted using TRIzol (Invitrogen); 1 μg of RNA was used to produce cDNA using Moloney murine leukemia virus reverse transcriptase (Promega), and then expression of specific targets was assayed by PCR. For analysis of RRP1B expression after E2F family knockdown, U2OS cells were stably transfected with pSuperior.puro containing siGFP, siE2F1, or siE2F3, and puromycin was selected. Parallel aliquots of cells were prepared for RT-PCR and SDS-PAGE as above. Construction and sequences were previously described (34).
For analysis of cell cycle dependent RRP1B RNA levels, human foreskin fibroblast cells were starved in Dulbecco's modified Eagle's medium containing 0.25% FBS for 48 h and then stimulated with 20% FBS at various time points. Harvesting of RNA and semiquantitative PCR was then performed as above. A parallel set of cells was treated identically, harvested, and analyzed for DNA content by propidium iodide staining followed by flow cytometry as previously described (33).
For analysis of RRP1B knockdown and E2F1 target expression, stably transduced siScramble and siRRP1B U2OS cells were harvested in TRIzol and RNA extracted and semiquantitatively analyzed as above. Quantitative PCR was performed in triplicate on an MX3005p thermal cycler (Stratagene) using SYBR Green dye to measure amplification and ROX as a reference dye (Brilliant II SYBR Green QPCR Master Mix, Stratagene). Transcript levels were normalized with GAPDH levels, which were assayed in parallel with test genes. Results were analyzed with MxPro 4.1 QPCR software (Stratagene).
For all experiments, PCR was performed using the following primer sets: RRP1B, 5′-CCCGTCCCTGGAACAGAAC- 3′ and 5′-CTCGGGCCACTCTGAGACA-3′, size 249 bp; p73, 5′-CATGGTCTCGGGGTCCCACT-3′ and 5′-CGTGAACTCCTCCTTGATGG-3′, size 471 bp; Apaf-1, 5′-AATGGACACCTTCTTGGACG-3′ and 5′-GCACTTCATCCTCATGAGCC-3′, size 331 bp; caspase-3, 5′-TCGGTCTGGTACAGATGTCG-3′ and 5′-CATACAAGAAGTCGGCCTCC-3′, size 398 bp; caspase-7, 5′-CAAAGCCACTGACTGAGATG-3′ and 5′-CAACCCAATGAATAAATGAT-3′, size 259 bp; E2F1, 5′-CCGCCATCCAGGAAAAGG-3′ and 5′-GCCCTCAAGGACGTTGGT-3′, size 193 bp; cyclin E, 5′-CTCCAGGAAGAGGAAGGCAA-3′, 5′-GTAAAAGGTCTCCCTGTGAAG-3′, size 421 bp; TK, 5′-ATGAGCTGCATTAACCTGCCCACT-3′, 5′-ATGTGTGCAGAAGCTGCTGC-3′, size 204 bp; GAPDH, 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-AAATGAGCCCCAGCCTTCTCCATG-3′, size 325 bp. We ensured linear amplification in all cases.
Chromatin Immunoprecipitation (ChIP) Assay
U2OS cells were grown in 15-cm diameter dishes, cross-linked with 1% formaldehyde, washed, and scraped with PBS, and nuclei were extracted on ice twice with nuclear extraction buffer with protease inhibitors. Cells were then resuspended in chromatin extraction buffer (1% SDS, 10 mm EDTA, 20 mm Tris) with protease inhibitors and sonicated to an average fragment size of 1000 bp; 0.5% of supernatants were used for control input PCR. All other chromatin was diluted in dilution buffer (0.01% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, 150 mm NaCl) and precleared with salmon sperm DNA/bovine serum albumin-blocked protein G plus protein A-agarose beads (Pierce) for 3 h and then immunoprecipitated with 4 μg of each antibody (E2F1 (C20), E2F2 (C18), E2F3 (C20), E2F4 (C20), RRP1B, and normal rabbit IgG (Pierce)) by nutation at 4 °C overnight. Blocked protein G + A-agarose beads were added for 2 h, and then beads were washed and nutated for 5 min at 4 °C consecutively with ice-cold low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, 150 mm NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, 500 mm NaCl), LiCl buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholic acid, 1 mm EDTA, 10 mm Tris), and twice with TE (10 mm Tris, 1 mm EDTA). Chromatin was eluted in fresh elution buffer (0.1 m NaHCO3, 1% SDS); cross-links were then reversed by incubating samples in high salt conditions for >4 h at 65 °C, followed by digestion of RNA by RNase A and protein by proteinase K. DNA was then purified by dilution in buffer PB (Qiagen) and then purification using a silica column (Qiaquick gel extraction kit, Qiagen).
For re-ChIP assays, cells and chromatin were treated as before; chromatin was immunoprecipitated using 4 μg of antibodies (E2F1 (KH95, Santa Cruz Biotechnology, Inc.) and normal mouse IgG (Pierce)); prior to chromatin elution, antibody-chromatin complexes were eluted in 10 mm dithiothreitol and incubated at 37 °C for 30 min. Supernatants were then diluted 20:1 in re-ChIP buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris) and nutated at 4 °C overnight with 4 μg of antibodies (RRP1B and rabbit IgG (Pierce)). Blocked protein G + A-agarose beads were added for 2 h, and then beads were washed, eluted, and DNA-purified as above.
For all experiments, PCR was performed using the primer sets that flank putative E2F-binding sites within the promoters of the following genes: E2F1, 5′-AGGAACCGCCGCCGTTGTTCCCGT-3′ and 5′-CTGCCTGCAAAGTCCCGGCCACTT-3′, size 124 bp; p73, 5′-CTCTGCCGAAGATCGCGGTCGG-3′ and 5′-GGCCGCGTCCAAGTCGGGGTCC-3′, size 170 bp; β-actin, 5′-ACGCCAAAACTCTCCCTCCTCCTC-3′ and 5′-CATAAAAGGCAACTTTCGGAACGGC-3′, size 166 bp; caspase-7, 5′-TTTGGGCACTTGGAGCGCG-3′ and 5′- AAGAGCCCAAAGCGACCCGT-3′, size 220 bp; GAPDH, 5′-AAAAGCGGGGAGAAAGTAGG-3′ and 5′-CTAGCCTCCCGGGTTTCTCT-3′, size 270 bp; p107, 5′-TCTTTCAGAATCTGAGGTAC-3′ and 5′-CCGACTTCTTTCTCCCTCC-3′, size 198 bp; rRNA, 5′-GTTTTTGGGGACAGGTGT-3′ and 5′-CCAGAGGACAGCGTGTCAGCA-3′, size 146 bp; TK, 5′-TCCCGGATTCCTCCCACGAG-3 and 5′-TGCGCCTCCGGGAAGTTCAC-3′, size 200 bp; RRP1B, 5′-CGGTGAAGAGCTGCGCCAGT-3′ and 5′-CGCAAGCTTACTGAGAATGTCAGTGATGGGGGA-3′, size 180 bp. We ensured linear amplification in all cases. For caspase-3, putative E2F sites in the mouse caspase-3 promoter (30) were compared against the human caspase-3 promoter, and a conserved site was found within the first intron. Primers used to assay this E2F site were 5′-TACTCGCCCTGGGGGCTGAT-3′ and 5′-TGAGCTGCGAGCACTCACGA.
GST Pull-down Assay
Escherichia coli strain BL21 transformed with pGEX or pGEX-E2F1 was cultured in LB medium containing ampicillin at 37 °C to an A600 value of 0.5. GST fusion proteins were induced by 0.02 mm isopropyl- β-d-thiogalactopyranoside at 25 °C for 3 h; cells were then lysed by sonication in PBS containing protease inhibitors and then purified using glutathione-Sepharose 4B (GE Healthcare) (10). 35S-Tagged RRP1B was produced from rabbit reticulocyte lysates according to the manufacturer's instruction (TNT Quick Coupled Transcription/Translation System, Promega). 1 μg of GST or GST-E2F1 on Sepharose beads was combined with 35S-tagged RRP1B in NETN-A buffer (50 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, 0.5% Nonidet P-40) with protease inhibitors and nutated overnight at 4 °C. Sepharose beads were washed four times with NETN-B buffer (100 mm NaCl, 1 mm EDTA), eluted in SDS sample buffer, and then subjected to SDS-PAGE, fixed, enhanced for autoradiography (Enlightening, Dupont), dried, and exposed to film for 1 h at −80 °C. Equal loading of GST proteins was assessed in parallel by SDS-PAGE followed by Coomassie staining.
Alternatively, GST fusion proteins were induced, lysed, and purified by the above method. 2 μg of GST-NHERF-PDZ2 (35) (as a control irrelevant protein), E2F1, or E2F1 mutants on Sepharose beads were nutated overnight at 4 °C with cellular lysates prepared from HEK293T cells that had been transfected with pcDNA3 or pcDNA3-FLAG-RRP1B, FLAG-RRP1B-(1–473), FLAG-RRP1B-(474–589), or FLAG-RRP1B-(590–758), incubated for 48 h, and lysed with NETN-A buffer with protease inhibitors. Sepharose beads were washed five times with NETN-B buffer, eluted in SDS sample buffer, and subjected to SDS-PAGE and immunoblotting. Equal loading of GST proteins was assessed in parallel by SDS-PAGE followed by Coomassie staining.
Immunofluorescence Studies
HEK293, NIH3T3, U2OS, or Ref52 cells were plated on collagen-coated coverslips in 6-well plates and then transfected with pcDNA3-FLAG-RRP1B using the appropriate transfection protocol and incubated for 48 h. Cells were then fixed in 3% paraformaldehyde for 20 min, followed by permeabilization in 0.5% Triton X-100 in PBS for 10 min. Cells were then blocked in 50% calf serum, 50% PBS at room temperature for 30 min and then incubated with primary antibody in blocking solution for 60 min, washed, blocked again, and then incubated with fluorescein-conjugated goat anti-rabbit IgG or Texas Red X goat anti-mouse IgG (Molecular Probes; 1:400 dilution) for 1 h. Cells were then washed, and nuclei were stained using Hoescht 33258 and then mounted. For immunostaining, RRP1B antibody (1:50 dilution, 0.4 μg/μl), FLAG antibody (F7425, Sigma; 1:250 dilution), and nucleolin antibody (MS-3, Santa Cruz Biotechnology; 1:100 dilution) were used. Neutralization of RRP1B antibody was performed by preincubation of RRP1B antibody with a 4 μg/μl peptide antigen solution in PBS overnight at 4 °C. Images were captured on a Zeiss fluorescent microscope (Axioplan 2 imaging system).
For bimolecular fluorescence complementation assays (36), YFP1-RRP1B, YFP2-E2F1 (37), YFP-E2F2, or nonspecific YFP1-zipper and YFP2-zipper (26) were transfected in HEK293 or NIH3T3 cells by an appropriate transfection protocol, incubated for 48 h, fixed, nuclei-stained, and mounted as above.
RESULTS
Expression of RRP1B Is Specifically Controlled by E2F1
We first investigated the potential role and specificity of E2F1 on RRP1B expression. We overexpressed E2F1 through E2F5 using adenoviruses encoding E2F1 to -5 cDNAs or no cDNA (pCMV) in serum-starved T98G cells and then checked for expression of RRP1B by semiquantitative RT-PCR and immunoblotting. RRP1B expression in transcript and protein was induced upon overexpression of E2F1 but not the other E2F family members, E2F2 to -5 (Fig. 1A). We also tested the expression of RRP1B upon knockdown of E2F1, E2F3, or a nonspecific GFP using U2OS cells in which siRNAs against each target were stably transfected. RRP1B transcripts were decreased after knockdown of E2F1 by quantitative RT-PCR but not upon knockdown of E2F3 or nonspecific GFP (Fig. 1B).
FIGURE 1.
Regulation of RRP1B expression by E2F1. A, serum-starved T98G cells were infected with adenoviruses containing either E2F1, E2F2, E2F3, E2F4, E2F5, or the CMV promoter alone. RNA was extracted and subjected to semiquantitative RT-PCR for RRP1B and GAPDH. Cell lysates were also collected for each infection and probed with the indicated antibodies. B, RNA was extracted from U2OS cells that were stably transfected with pSuperior, encoding siRNAs against GFP, E2F1, or E2F3, and subjected to quantitative PCR for RRP1B, levels of which were normalized against GAPDH. Cell lysates for independent experiments were collected for siGFP, siE2F1, and siE2F3 cell lines and probed with the indicated antibodies. *, p < 0.05 compared with both siGFP and siE2F3. C, U2OS or HCT116 cells were treated with 10 μm doxorubicin, neocarzinostatin (NCS), or 20 μm cisplatin for the indicated times and dosages, lysed, electrophoresed, and immunoblotted with the indicated antibodies. The numbers below each lane indicate densitometry of RRP1B levels normalized to GAPDH levels. D, RNA was extracted from U2OS cells treated with 1 μm doxorubicin for the indicated time points and subjected to quantitative PCR for RRP1B, levels of which were normalized against GAPDH. *, p < 0.01 for all treated time points compared with untreated. E, human foreskin fibroblasts were brought to quiescence by serum starvation (0.25% FBS) for 48 h and then reinduced with 20% serum at the indicated time points. Cells were lysed, and RNA and protein were extracted and subjected to semiquantitative RT-PCR or blotting with the indicated primer sets or antibodies. The numbers below each lane indicate percentage of cells in G0/G1, S, and G2 phases of the cell cycle as assayed by propidium iodide DNA histogram analysis. F, representative DNA histogram analysis by propidium iodide flow cytometry.
To further support a role for E2F1 in the control of expression of RRP1B, we investigated the expression of RRP1B during cellular states where E2F1 expression is endogenously induced. E2F1 transcriptional activity is induced following DNA damage (13); if RRP1B is an E2F1 target, RRP1B expression will be increased following DNA damage. Using U2OS cells in which DNA damage was induced by neocarzinostatin or cisplatin for varying times and dosages, we observed that RRP1B expression was induced by genotoxic agents as soon as 15 min following administration, peaking 60 min after administration (Fig. 1C), with a decrease afterward. We also observed similar induction in HCT116 cells after doxorubicin treatment (Fig. 1C). To determine whether RRP1B transcripts were induced following DNA damage, we performed quantitative RT-PCR on U2OS cells that were treated with doxorubicin on a time course. RRP1B transcripts were significantly induced after 15 min of doxorubicin treatment (Fig. 1D).
E2F1 expression is controlled during the cell cycle, where expression peaks at the G1/S transition. We therefore investigated whether RRP1B expression also peaks at the G1/S transition, consistent with E2F1 expression, using semiquantitative RT-PCR and protein blotting for RRP1B expression in primary foreskin fibroblasts that had been serum-starved to quiescence for cell cycle phase synchronization and then stimulated with serum to reinduce cycling. RRP1B transcripts were observed to be induced, peaking at 18 h after cell cycle induction (Fig. 1E), with levels falling afterward, suggesting that RRP1B expression peaks at the G1/S transition (Fig. 1F). This is further supported by the observation of RRP1B protein levels that peaked at 20 h after cell cycle induction, with levels falling afterward (Fig. 1E).
To further test the role of E2F1 in control of RRP1B expression, we cloned the endogenous RRP1B promoter into a reporter luciferase plasmid and assayed the ability of E2F1 to induce RRP1B promoter-driven luciferase activity. A schema of the endogenous RRP1B promoter with putative E2F sites as determined by computer screening (38) is shown in Fig. 2A. We also tested the ability of E2F1 to induce RRP1B promoter reporter activity where putative E2F sites were inactivated by point mutation. E2F1 induced luciferase activity of the wild type promoter, but mutation of the putative E2F site at position +150 from the RRP1B ATG completely abolished induction by E2F1 (Fig. 2B). Two other E2F sites at positions −505 and −400 were also mutated, but the ability of E2F1 to induce luciferase activity was unaffected when compared with the wild type, indicating that these two sites are not relevant to E2F1 induction of RRP1B (Fig. 2C). We finally tested the specific ability of E2F1 to induce RRP1B promoter-driven luciferase activity. Consistent with Fig. 1A, overexpression of E2F1, but not E2F2 or E2F3, was able to significantly induce luciferase activity (Fig. 2D).
FIGURE 2.
E2F1 specifically drives RRP1B expression and binds to the RRP1B promoter. A, schema of the wild type RRP1B promoter. B, HEK293T cells were transfected with either empty vector, wild type RRP1B promoter reporter luciferase vector, or RRP1B promoter vector in which a single E2F site is mutated at +150, with either E2F1 or empty vector and β-galactosidase. 48 h later, cells were lysed for determination of luciferase activity. β-Galactosidase activity was used as a control for transfection efficiency. A protein aliquot from each experimental arm was blotted and probed with the indicated antibodies. *, p < 0.02 between E2F1 transfected arms. C, HEK293T cells were transfected as before but with empty vector, wild type RRP1B promoter, or with RRP1B promoter in which a single E2F site is mutated at −505 or −400 and either E2F1 or empty vector. Luciferase analysis and protein blotting were done as before. D, HEK293T cells were transfected with a RRP1B promoter reporter and either E2F1, E2F2, or E2F3 or empty vector. Luciferase analysis and protein blotting was done as before. *, p < 0.01 between E2F1 arm and all other arms. E, U2OS cells were cross-linked; nuclei were then extracted, sonicated, and incubated with the indicated antibodies, followed by washes and decross-linking. Chromatin was then used for PCR amplification using the indicated primer sets.
Finally, we determined whether E2F1 protein binds to the RRP1B promoter in an endogenous ChIP assay. Using a primer set that encompasses the RRP1B promoter from position +79 to +259, containing the E2F site at +150, we observed binding of E2F1 to the RRP1B promoter (Fig. 2E). The specificity of E2F binding was further shown by immunoprecipitation with E2F2 to -4; although binding of all E2Fs was seen when the p107 promoter was assayed, little binding was seen between E2F2 and -4 on the RRP1B promoter, indicating that E2F1 specifically binds to the RRP1B promoter.
Knockdown of RRP1B Decreases Apoptosis Induced by Genotoxic Agents and E2F1
Because the data above suggest proapoptotic E2F1, and not the other E2Fs, specifically regulates the expression of RRP1B, we investigated what effect RRP1B would have on apoptosis induced by both DNA-damaging genotoxic agents and by overexpression of E2F1 during serum starvation. The effect of RRP1B was tested in U2OS cells that were stably transduced with siRNAs against RRP1B by means of a lentiviral system. Two independent siRNAs against RRP1B were used. We first determined the effect of RRP1B knockdown on apoptosis as induced by cisplatin. Compared with control cells expressing a nonspecific siRNA (siScr), RRP1B knockdown significantly decreased the ability of cisplatin to induce apoptosis as measured by two independent assays: propidium iodide staining/flow cytometry (for detection of the population with sub-2 n DNA contents) and surface annexin V staining/flow cytometry (Fig. 3A). Knockdown of RRP1B protein levels reached nearly 100% in this assay (Fig. 3A, inset). We further tested the role of RRP1B in apoptosis in a caspase cleavage assay in stably transduced U2OS cells expressing siRNA against RRP1B. After treatment with the genotoxic agent doxorubicin, which induces E2F1-dependent apoptosis in HEK293 cells (39), RRP1B knockdown cells had significantly reduced activated caspase activity as compared with control siScr cells (Fig. 3B). Finally, we tested the ability of E2F1 to induce apoptosis in serum-starved U2OS cells expressing siRNAs against RRP1B. Knockdown of RRP1B significantly reduced the ability of E2F1 to induce surface annexin V positivity as compared with control siScr cells (Fig. 3C).
FIGURE 3.
Knockdown of RRP1B reduces DNA damage and E2F1-induced apoptosis but does not affect cellular proliferation. A, stably transduced U2OS cells expressing either nonspecific siScr sequence or two siRNAs against RRP1B (siRRP1B A and siRRP1B B) were seeded equally and induced for apoptosis with 20 μm cisplatin for 28–30 h and then stained by propidium iodide and analyzed by fluorescence-activated cell sorting for sub-G0/G1 population or stained for surface annexin V and analyzed by fluorescence-activated cell sorting. Experiments were done in triplicate. An aliquot of protein from each experimental arm was blotted and probed with the indicated antibodies. **, p < 0.001 between treated siScr and siRRP1B arms. *, p < 0.02 between treated siScr and siRRP1B arms. B, stably transduced siScr or siRRP1B A or B U2OS cells were seeded equally and induced for caspase cleavage with 1 μm doxorubicin for 8 h. *, p < 0.01 between treated siScr and siRRP1B arms. C, stably transduced siScr or siRRP1B A or B U2OS cells were seeded equally, starved for 48 h, and then infected with a multiplicity of infection of 200 of either CMV adenovirus (empty) or E2F1 adenovirus for 36 h and then analyzed by fluorescence-activated cell sorting for surface annexin V staining. Experiments were done in triplicate. An aliquot of protein from each experimental arm was electrophoresed, blotted, and probed with the indicated antibodies. *, p < 0.01 between E2F1 induced siScr and siRRP1B arms. D, stably transduced siScr or siRRP1B A or B U2OS cells were seeded equally on 6-well plates and counted by a hemacytometer at 3 days prior to confluence. Cells were either grown in 10% serum (top) or in 2% serum (middle). Cells were diluted 1:4 and replated and then counted at 6 and 9 days. Cells were lysed at the end of the experiment, electrophoresed, blotted, and probed with the indicated antibodies (bottom). IB, immunoblot.
RRP1B Does Not Affect Cellular Proliferation
Because E2F1 also regulates genes important for cellular proliferation, and RRP1B belongs to the Nop52 family, which is known to regulate ribosomal RNA production, a limiting factor for cellular growth, we assayed the role of RRP1B in cellular proliferation. U2OS cells stably transduced with siRNAs against RRP1B or control nonspecific siScr siRNAs were plated equally, grown, trypsinized, and harvested and counted using a hemacytometer. Knockdown of RRP1B did not appear to change the rate of proliferation of U2OS cells (Fig. 3D, top). Differences in proliferative capacity were not detected between control and RRP1B knockdown cells when cells were grown in low serum conditions (Fig. 3D, middle). This result suggests that RRP1B is not required for cellular proliferation. However, it is possible that Nop52, a homolog of RRP1B, compensates for loss of RRP1B in rRNA production.
RRP1B Selectively Regulates Transcription of E2F1 Target Genes
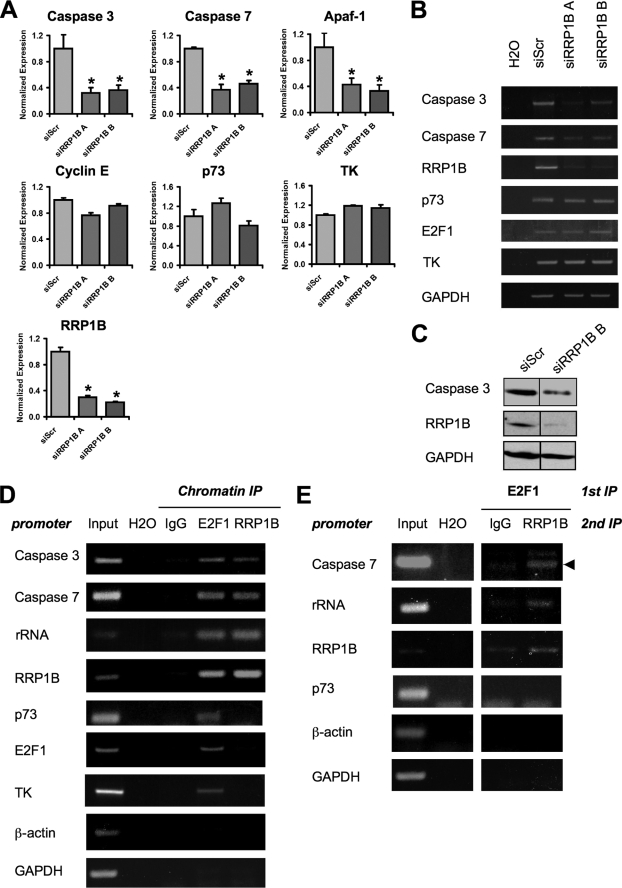
Based on results above showing decrease of the ability of E2F1 to induce apoptosis after knockdown of RRP1B, we investigated whether knockdown of RRP1B could affect the transcription of E2F1 target genes by examining expression in stably transduced U2OS cells expressing siRNAs against RRP1B. E2F1 target genes related to apoptosis, such as p73, Apaf-1, caspase-3, and caspase-7, as well as target genes related to the cell cycle, such as cyclin E and TK, were examined. Transcripts of specific genes were analyzed by quantitative (Fig. 4A) and semiquantitative (Fig. 4B) RT-PCR assays. The effectiveness of RRP1B siRNAs was confirmed, where a 75–80% knockdown of transcripts was observed in both siRNAs tested. Knockdown of RRP1B expression appeared to reduce the expression of caspase-3 and caspase-7 (Fig. 4A), consistent with the caspase cleavage assay above (Fig. 3B), and also reduced the expression of proapoptotic Apaf-1. Furthermore, knockdown of RRP1B correlated with a decrease of procaspase-3 protein level, consistent with the reduction in procaspase-3 transcripts (Fig. 4C). Interestingly, p73, an E2F1 target gene known to be important for apoptosis, and other target genes involved in proliferation, such as TK and cyclin E, were not significantly affected by RRP1B knockdown (Fig. 4A). These results suggest a selective role for RRP1B in regulation of E2F1 target genes.
FIGURE 4.
Knockdown of RRP1B selectively affects E2F1 target levels by binding with E2F1 on E2F-responsive promoters. A, RNA extracted from U2OS cells stably transduced with siScr or siRRP1B A or B was subjected to quantitative PCR for the indicated targets. Expression level was normalized to GAPDH. *, p < 0.02 between siScr and siRRP1B arms. B, in an independent experiment, semiquantitative RT-PCR was performed on RNA extracted from U2OS siScr or siRRP1B cells for the indicated targets. H2O indicates no template control. C, in an independent experiment, U2OS siScr or siRRP1B B cells were lysed, electrophoresed, and blotted and probed with the indicated antibodies. D, U2OS cells were cross-linked, nucleus-extracted, sonicated, precleared, and immunoprecipitated with 4 μg of the indicated antibodies overnight, followed by incubation with protein A + G beads for 3 h and stringent washes. Chromatin was eluted from beads, decross-linked, incubated with RNase A and proteinase K, purified, and subjected to PCR for the indicated E2F-responsive promoters. H2O indicates no template control. E, U2OS cells were cross-linked, nucleus-extracted, sonicated, precleared, and immunoprecipitated with 4 μg of the indicated antibodies overnight followed by incubation with protein A + G beads for 3 h and stringent washes. Chromatin-protein complexes were eluted with 1 mm dithiothreitol, followed by a second immunoprecipitation with the indicated antibodies. Binding to beads, washes, elution, purification, and PCR were done as in D. The arrowheads indicate the expected size of PCR products. IP, immunoprecipitation.
Recently, several nucleolar proteins have been shown to regulate transcription through binding to chromatin (40, 41). We therefore examined a role for RRP1B in E2F1 regulation by assaying the presence of RRP1B on E2F1 target gene promoters through ChIP assays. E2F1 was seen on the promoters of all E2F1 target genes assayed. E2F1 was also seen on the rRNA promoter (29) and the RRP1B promoter (Fig. 2E). Interestingly, RRP1B antibodies precipitated chromatin from the caspase-3 promoter, the caspase-7 promoter, the rRNA promoter, and the RRP1B promoter but not from promoters of other E2F1 target genes assayed, including p73, TK, and E2F1 (Fig. 4D). These data suggest that RRP1B binds only to the promoters of E2F1 target genes that were affected by RRP1B knockdown and not to the promoters of E2F1 target genes not affected by RRP1B knockdown. This suggests that RRP1B binding to specific promoters is important for regulation of E2F1 target genes. We then investigated whether RRP1B and E2F1 were bound together on E2F1 target gene promoters in a ChIP-re-ChIP assay, where two consecutive immunoprecipitations using E2F1 and RRP1B antibodies were performed. RRP1B and E2F1 were shown to interact together on the caspase-7, rRNA, and RRP1B promoters but not on the p73 promoter, suggesting that RRP1B regulation of E2F1 target genes occurs through interaction with E2F1 (Fig. 4E).
We further tested the ability of RRP1B to regulate E2F1 target genes in promoter reporter luciferase assays. We used H1299 cells that were stably transduced with lentiviruses encoding siRNAs against RRP1B. First, we tested the ability of E2F1 to induce the caspase-7, TK, and E2F1 promoters. Consistent with Fig. 4A, RRP1B knockdown inhibited the ability of E2F1 to induce luciferase activity of the caspase-7 promoter reporter (Fig. 5A) but not the E2F1 (Fig. 5B) and TK (Fig. 5C) promoter reporters, further supporting specificity in RRP1B regulation of E2F1 target genes.
FIGURE 5.
Knockdown of RRP1B selectively affects E2F-induced promoter reporter luciferase activity. A, H1299 siScr or siRRP1B A or B cells were transfected with caspase-7 promoter reporter, E2F1 or empty vector, and β-galactosidase; incubated for 48 h; and subjected to a luciferase assay. β-Galactosidase activity was used as a control for transfection efficiency. A protein aliquot from each experimental arm was blotted with the indicated antibodies. *, p < 0.005 between E2F1-transfected siScr cells and both E2F1-transfected siRRP1B cells. B, H1299 siScr or siRRP1B B cells were transfected with E2F1 promoter reporter, E2F1 or empty vector, and β-galactosidase. Luciferase analysis and protein blotting was done as before. C, H1299 siScr or siRRP1B cells were transfected with TK promoter reporter, E2F1 or empty vector, and β-galactosidase. Luciferase analysis and protein blotting was done as before. D, H1299 cells transduced with siRRP1B A or B or no virus were seeded equally and transfected with rRNA promoter reporter, E2F1 or empty vector, and β-galactosidase. Luciferase analysis and protein blotting was done as before. *, p < 0.01 between pcDNA3-transfected arms and between E2F1-transfected arms. E, H1299 cells stably transduced with siScr or siRRP1B B were seeded equally and transfected with an intact rRNA promoter reporter or rRNA promoter, in which the E2F binding site for activation has been mutated, and β-galactosidase. Luciferase analysis and protein blotting was done as before. *, p < 0.05.
Because E2F1 has been reported to bind rRNA promoter and up-regulate its promoter activity (29), we assayed the ability of E2F1 to induce the rRNA promoter in H1299 cells or stably transduced siRRP1B cells. RRP1B knockdown significantly reduced both endogenous and E2F1-induced reporter luciferase activity (Fig. 5D). Similar results were seen in stably transduced U2OS cells expressing RRP1B siRNAs (data not shown). Because the previous assay does not rule out a nonspecific RRP1B effect on transcription, we tested the effect of RRP1B knockdown on reporter luciferase activity of an rRNA promoter containing a mutation through which induction by E2F1 is lost. Consistent with Fig. 5D, RRP1B knockdown significantly reduced the endogenous reporter activity of the wild type promoter (Fig. 5E). However, RRP1B knockdown was not observed to decrease endogenous promoter reporter activity in cells transfected with the mutant rRNA promoter, suggesting that an intact E2F site is required for knockdown of RRP1B to regulate transcriptional activity (Fig. 5B).
E2F1 Interacts Directly with RRP1B
Based on the results above showing coimmunoprecipitation of E2F1 and RRP1B on the chromatin of E2F target gene promoters (Fig. 4E), we tested whether there was a physical interaction between E2F1 and RRP1B in biochemical assays. We examined in vitro binding between purified GST-E2F1 and RRP1B produced in an in vitro transcription/translation system. When 35S-labeled RRP1B was incubated with either GST or GST-E2F1 and pulled down by glutathione-Sepharose, GST-E2F1, but not GST, pulled down RRP1B, demonstrating a direct interaction between E2F1 and RRP1B (Fig. 6A).
FIGURE 6.
Physical interaction between RRP1B and E2F1. A, FLAG-tagged RRP1B was produced by in vitro transcription/translation in the presence of [35S]methionine and added to buffer containing either GST-E2F1 bound to glutathione-agarose or GST alone, nutated, washed, separated by SDS-PAGE, fixed, enhanced, and exposed to film. Equal loading of GST proteins is indicated by a parallel Coomassie stain. B, nuclei from U2OS and HCT116 cells were extracted, sonicated, lysed, and immunoprecipitated with E2F1 antibodies. Beads were washed, blotted, and probed with the indicated antibodies. C, FLAG-tagged RRP1B or RRP1B domain mutants were coexpressed with E2F1 in HEK293T cells. Cells were lysed, immunoprecipitated with anti-FLAG-agarose, washed, blotted, and probed with the indicated antibodies. D, FLAG-tagged RRP1B was expressed in HEK293T cells, lysed, and incubated with a control irrelevant protein GST-PDZ2, GST-E2F1, or GST-E2F1 mutants bound to glutathione-agarose overnight, washed, blotted, and probed with the indicated antibodies. Equal loading of GST proteins indicated by a parallel Coomassie stain. IB, immunoblot; IP, immunoprecipitation.
Next, we examined whether RRP1B could interact with E2F1 in vivo. We detected an endogenous interaction between E2F1 and RRP1B in nuclear extracts from both U2OS and HCT116 cells (Fig. 6B). DNA damage increased the interaction between RRP1B and E2F1, but this was due to induction of both E2F1 and RRP1B (data not shown).
We further investigated the ability of RRP1B to interact with E2F1 by dissecting the domains of interaction between RRP1B and E2F1. E2F1 was coexpressed with FLAG-tagged RRP1B or RRP1B N-terminal domain (aa 1–473), middle domain (aa 474–589), or C-terminal domain (aa 590–758); when cells were lysed and immunoprecipitated with FLAG, E2F1 was pulled down with full-length RRP1B, RRP1B-(1–473), and RRP1B-(590–758), indicating two separate domains of interaction (Fig. 6C). We also dissected the domains of interaction between RRP1B and E2F1. Purified GST-tagged full-length E2F1 or GST-tagged E2F1 domain mutants corresponding to the N terminus (aa 1–109), DNA binding domain (aa 110–284), marked box domain (aa 285–358), or Rb/dimerization domain (aa 359–437) were incubated with lysates from HEK293T cells in which FLAG-tagged RRP1B was overexpressed. Only GST-E2F1 and GST-E2F1-(110–284) were able to pull down FLAG-tagged RRP1B (Fig. 6D). Unlike TopBP1 (25) and 14-3-3τ (39), interaction between RRP1B and E2F1 was not perturbed by mutation of E2F1 serine 31 (data not shown), as expected because RRP1B does not interact with the N terminus of E2F1.
RRP1B and E2F1 Interact in the Nucleolus and Punctate Nucleoplasmic Foci
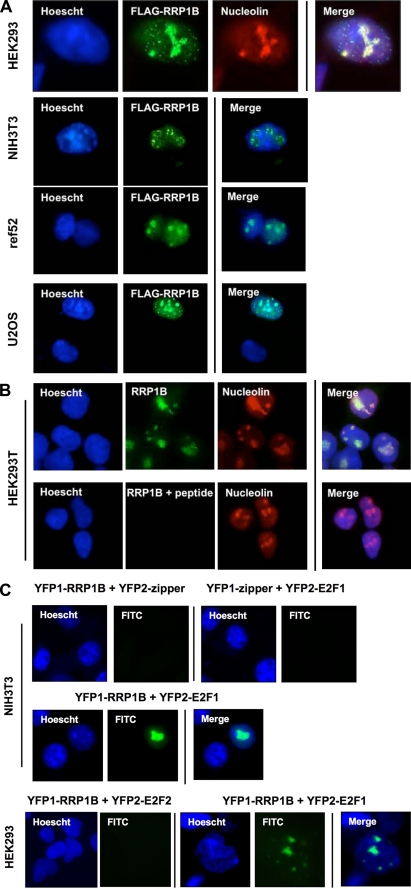
To further investigate the role of RRP1B in E2F1 regulation, we assayed the localization of RRP1B and E2F1. We overexpressed FLAG-tagged RRP1B in HEK293 cells and probed for intracellular localization using antibodies against FLAG. RRP1B was localized to areas within the nucleus corresponding to nucleolin staining, a marker for the nucleolus. In addition, punctate nucleoplasmic foci were also observed, which did not correspond to nucleolin staining (Fig. 7A). We observed similar patterning in other cell lines (Fig. 7A). We then assayed the endogenous localization of RRP1B; although our antibody could not detect endogenous RRP1B in several cell lines, in HEK293T cells, RRP1B was localized to the nucleolus, consistent with Fig. 7A (Fig. 7B).
FIGURE 7.
RRP1B localizes and interacts with E2F1 in nucleoli and punctate nucleoplasmic foci. A, the indicated cells were transfected with FLAG-RRP1B, fixed, probed with the indicated antibodies, nucleus-stained with Hoescht 33258, and mounted for microscopy. B, HEK293T cells were fixed and probed with the indicated antibodies or antibodies with neutralizing peptide, stained, and mounted as above. C, RRP1B and E2F1 were each cloned into vectors expressing one part each of YFP in a single continuous cDNA and transfected into the indicated cells. YFP subunits expressing a nonspecific leucine zipper or E2F2 were used as a negative control. Green fluorescence indicates colocalization of YFP subunits and the subcellular location of interaction. Cells were fixed in paraformaldehyde, nucleus-stained with Hoescht 33258, and mounted for immunofluorescence. FITC, fluorescein isothiocyanate.
We next investigated the localization of interaction between RRP1B and E2F1. We assayed the localization of interaction using a bifluorescence complementation assay (36). No fluorescence was seen when either RRP1B or E2F1 was coexpressed with a nonspecific leucine zipper control, but when both YFP-tagged RRP1B and E2F1 were coexpressed, fluorescence was seen within intracellular locations similar to those seen in Fig. 7A (Fig. 7C). Furthermore, no fluorescence was seen with coexpression of YFP-tagged RRP1B and E2F2. These results suggest that the RRP1B and E2F1 interaction is located within nucleoli and punctate nucleoplasmic foci.
DISCUSSION
With a role for E2F1 in apoptosis during either DNA damage response or thymocyte development, the molecular details that dictate the proapoptotic activity of E2F1 have drawn much attention. For example, association of Jab1 (42) and MCPH1/BRIT1 (37) has been identified to contribute to this activity (42), although how these interactions specifically lead to activation of E2F1-dependent apoptosis remains unclear. In this report, we identify the nucleolar protein RRP1B as an E2F1-specific target (Figs. 1 and 2), which in turn selectively up-regulates certain E2F1 target genes, such as caspase 3 and 7 and Apaf-1 (Figs. 4 and 5), and is required for E2F1-induced apoptosis (Fig. 3C). These data unravel a novel function for RRP1B and identify it as one of the factors that activate the proapoptotic activity of E2F1.
The nucleolar localization of RRP1B is also worth noting (Fig. 7, A and B). Although the role of the nucleolus in ribosome production is well known, a role for the nucleolus in cancer, including in regulation of cellular proliferation and apoptosis, has only recently been established (43, 44). We now show RRP1B as an example of a multifunctional nucleolar protein that regulates apoptosis through E2F1-medicated transcription. A role for nucleolar and ribosomal proteins in transcriptional regulation has also only been recently recognized (45–47). Two nucleolar proteins have been extensively investigated in transcriptional regulation. Nucleophosmin was the first histone chaperone identified (48) and has been shown to bind to histone acetyltransferases (49, 50) and regulate transcriptional activity through GCN5 (51), AP2α (52), c-Myc (40), and the androgen receptor (53). Nucleolin is a histone chaperone with FACT-like activity (54) and regulates transcriptional activity of pRb (55), KLF2 (56), AP-1 (57), c-Myc (41), and IRF-2 (58). Other nucleolar and ribosomal proteins involved in transcriptional regulation through binding of chromatin include RPS3 in NFκB-dependent transcription (59), L11 in c-Myc-dependent transcription (60), Nopp140 (61), ApLLP (62), and Drosophila ribosomal proteins (63). To these examples, we now add RRP1B as a specific regulator of transcription by a nucleolar protein in a manner similar to that seen in nucleolin- or nucleophosmin-regulated transcription.
Another nucleolar protein that is induced by E2F1 but also regulates E2F1 is ARF. ARF binds to MDM2 to activate the growth-suppressive functions of p53 but can also exert its tumor suppressor activity independently of p53; for example, ARF has been shown to inhibit the transcriptional activity of E2F1 through regulation on both E2F and DP1 (64, 65). More recently, ARF has been shown to inhibit ribosomal RNA processing and to specifically interact with the rRNA promoter (66) and inhibit rRNA transcription by blocking upstream binding factor phosphorylation (67). These inhibitory functions toward E2F1 by ARF are in contrast to the promoting function by RRP1B, at least in the aspect of certain E2F1 target gene expression and the rRNA promoter activity.
RRP1B binds together with E2F1 on the chromatin of specific E2F1 target genes (Fig. 4, D and E); however, the mechanism by which E2F1 transcriptional activity is controlled by RRP1B remains unclear. RRP1B does not contain any known DNA binding or transcriptional regulatory motifs; therefore, its role may be in binding to chromatin or in recruitment of chromatin modifiers. Nucleophosmin and nucleolin have been shown to directly bind histones and act as histone chaperones to regulate transcription (48, 54). Consistent with these examples, a recent study has shown RRP1B to bind generally to chromatin, including to general chromatin components, such as histone H1X (68). However, because our data suggest selective and promoter-specific regulation of E2F1 target genes, it may be more likely that general binding of RRP1B to histones is uninvolved in regulation of E2F1 target genes. Alternatively, RRP1B may recruit histone modifiers, such as histone acetyltransferases, to up-regulate E2F1 target genes. This is similar to the mode of action seen for both nucleophosmin and nucleolin, which recruit GCN5 and P/CAF respectively, to specific promoters for transcriptional regulation (51, 58). Consistent with this hypothesis, RRP1B has been shown to bind acetylated lysine 5 of histone 4 and other nonubiquitous chromatin-binding proteins (68). Further investigation of the ability of RRP1B to recruit chromatin modifiers is warranted. On the other hand, binding of RRP1B to the DNA binding domain of E2F1 suggests other alternative mechanisms of E2F1 regulation. For example, the ETS-related transcription factor GABPγ1 has been shown to bind to the E2F1 DNA binding domain and negatively regulate the ability of E2F1 to transduce caspase-3 and caspase-7 (69). pRb also appears to have an independent binding E2F1 ability; although pRb does bind other E2Fs, a separate domain within pRb is capable of binding E2F1 at a site outside of the C-terminal Rb/dimerization domain. This site includes the E2F1 DNA binding domain (70) to which RRP1B also binds. Therefore, the possibility exists for RRP1B to participate in regulation of E2F1 apoptosis by displacing negative cofactors. Investigation into a potential role of RRP1B, GABPγ1, or pRb competitive interactions may be of interest.
We also show that RRP1B is localized to the nucleolus and punctate nucleoplasmic foci in multiple cell lines (Fig. 7, A and B). These data are consistent with other studies showing localization of the RRP1, a RRP1B homolog, to the nucleolus (21, 22), and also with proteomic studies suggesting nucleolar localization (71–73). However, our results are inconsistent with a recent study suggesting localization of RRP1B to the nucleoplasm and nuclear lamina to the exclusion of the nucleolus (68); this disparity may result from differences in cell lines used.
One possible reason for the selective ability of RRP1B to regulate particular E2F1 target genes is the localization of gene promoters during interphase in proximity to the nucleolus. The rRNA promoter, an E2F1- and RRP1B-regulated promoter (Fig. 4D), is situated within nucleolar organizing regions inside the nucleolus (74). Whether the promoters of caspase-3, caspase-7, or RRP1B are located within or near the nucleolus remains to be determined. RRP1B was also observed to be localized with E2F1 in punctate nucleoplasmic foci. Although the type and nature of these foci are unknown, regulation of E2F1 target genes unrelated to ribosome biogenesis, such as caspase-3 or caspase-7, may be localized to these foci. Finally, because the nucleolus is not membrane-bound, proteins may freely enter and exit the nucleolus into the nucleoplasm; regulation of E2F1 target gene promoters may be situated within the nucleoplasm as a consequence.
Identification of RRP1B as a promoter of apoptosis may also suggest an explanation for the observation of higher survival in breast cancers with an expression profile driven by high RRP1B expression (23, 68). RRP1B may be an important factor in apoptotic response to genotoxic agents and aberrant proliferation (Fig. 3, A–C); therefore, it is possible that increased survival seen in breast cancers with high RRP1B expression may be due to increased responsiveness to genotoxic therapy. It would be interesting to see whether expression profiles seen in RRP1B overexpression also show increases in E2F1-dependent target genes involved in apoptosis.
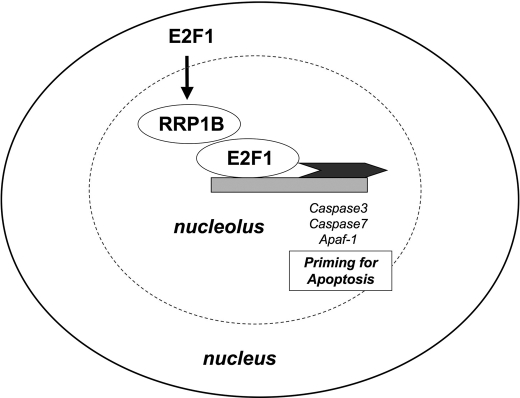
In summary, we have identified RRP1B as a novel specific target of E2F1 involved in the regulation of apoptosis. Loss of RRP1B expression inhibits the cellular apoptotic response to genotoxic agents as well as E2F1 overexpression. RRP1B selectively regulates E2F1 target gene expression through binding with E2F1 on target gene promoters. These data suggest that RRP1B is a new specificity factor for E2F1-mediated apoptosis (Fig. 8). Furthermore, we have identified a novel nucleolar protein in regulation of apoptosis through binding of chromatin.
FIGURE 8.
Proposed model of regulation of E2F1 and RRP1B. RRP1B is specifically stimulated by E2F1 expression. RRP1B then binds E2F1 at specific E2F1 promoters, acting as a cofactor for expression of those specific E2F1 targets to up-regulate E2F1-mediated apoptosis.
Acknowledgments
We greatly appreciate the gift of rRNA promoter plasmids from Dr. Paule Séité (University of Poitiers), caspase-7 promoter plasmid from Dr. Zaher Nahle (Washington University), and GST-NHERF-PDZ2 from Fannie Lin (University of Alabama at Birmingham (UAB)). We also thank Marion Spell at the UAB CFAR (Center for AIDS Research) Flow Cytometry Core Facility and Enid Keyser at the UAB Arthritis and Musculoskeletal Center Flow Cytometry Core Facility for assistance with flow cytometry analysis. We also thank Dr. Susan Nozell (UAB) for technical assistance with ChIP assays.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM008361 (to J. C. P.) and CA100857 (to W.-C. L.). This work was also supported by Department of Defense Breast Cancer Research Program Grants W81XWH-09-1-0338 (to W.-C. L.) and W81XWH-06-1-0708 (to J. C. P.). This work is submitted in partial fulfillment of the requirements for the UAB Cell Biology Graduate Program (J. C. P.).
- FBS
- fetal bovine serum
- YFP
- yellow fluorescent protein
- siRNA
- small interfering RNA
- 7-AAD
- 7-amino-actinomycin
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ChIP
- chromatin immunoprecipitation
- GST
- glutathione S-transferase
- rRNA
- ribosomal ribonucleic acid
- aa
- amino acids
- PBS
- phosphate-buffered saline
- TK
- thymidine kinase
- CMV
- cytomegalovirus
- SiScr
- nonspecific siScramble.
REFERENCES
- 1.Dimova D. K., Dyson N. J. (2005) Oncogene 24, 2810–2826 [DOI] [PubMed] [Google Scholar]
- 2.Iaquinta P. J., Lees J. A. (2007) Curr. Opin. Cell Biol. 19, 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh J. K., Fredersdorf S., Kouzarides T., Martin K., Lu X. (1997) Genes Dev. 11, 1840–1852 [DOI] [PubMed] [Google Scholar]
- 4.Hunt K. K., Deng J., Liu T. J., Wilson-Heiner M., Swisher S. G., Clayman G., Hung M. C. (1997) Cancer Res. 57, 4722–4726 [PubMed] [Google Scholar]
- 5.Phillips A. C., Bates S., Ryan K. M., Helin K., Vousden K. H. (1997) Genes Dev. 11, 1853–1863 [DOI] [PubMed] [Google Scholar]
- 6.Field S. J., Tsai F. Y., Kuo F., Zubiaga A. M., Kaelin W. G., Jr., Livingston D. M., Orkin S. H., Greenberg M. E. (1996) Cell 85, 549–561 [DOI] [PubMed] [Google Scholar]
- 7.Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N. J. (1996) Cell 85, 537–548 [DOI] [PubMed] [Google Scholar]
- 8.Meng R. D., Phillips P., El-Deiry W. S. (1999) Int. J. Oncol. 14, 5–14 [PubMed] [Google Scholar]
- 9.Pruschy M., Wirbelauer C., Glanzmann C., Bodis S., Krek W. (1999) Cell Growth Differ. 10, 141–146 [PubMed] [Google Scholar]
- 10.Lin W. C., Lin F. T., Nevins J. R. (2001) Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens C., Smith L., La Thangue N. B. (2003) Nat. Cell Biol. 5, 401–409 [DOI] [PubMed] [Google Scholar]
- 12.Ianari A., Gallo R., Palma M., Alesse E., Gulino A. (2004) J. Biol. Chem. 279, 30830–30835 [DOI] [PubMed] [Google Scholar]
- 13.Pediconi N., Ianari A., Costanzo A., Belloni L., Gallo R., Cimino L., Porcellini A., Screpanti I., Balsano C., Alesse E., Gulino A., Levrero M. (2003) Nat. Cell Biol. 5, 552–558 [DOI] [PubMed] [Google Scholar]
- 14.Lissy N. A., Davis P. K., Irwin M., Kaelin W. G., Dowdy S. F. (2000) Nature 407, 642–645 [DOI] [PubMed] [Google Scholar]
- 15.Irwin M., Marin M. C., Phillips A. C., Seelan R. S., Smith D. I., Liu W., Flores E. R., Tsai K. Y., Jacks T., Vousden K. H., Kaelin W. G., Jr. (2000) Nature 407, 645–648 [DOI] [PubMed] [Google Scholar]
- 16.Moroni M. C., Hickman E. S., Lazzerini Denchi E., Caprara G., Colli E., Cecconi F., Müller H., Helin K. (2001) Nat. Cell Biol. 3, 552–558 [DOI] [PubMed] [Google Scholar]
- 17.Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., Vousden K. H. (1998) Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]
- 18.Müller H., Bracken A. P., Vernell R., Moroni M. C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J. D., Helin K. (2001) Genes Dev. 15, 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabian G. R., Hopper A. K. (1987) J. Bacteriol. 169, 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horsey E. W., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr. (2004) RNA 10, 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savino T. M., Bastos R., Jansen E., Hernandez-Verdun D. (1999) J. Cell Sci. 112, 1889–1900 [DOI] [PubMed] [Google Scholar]
- 22.Savino T. M., Gébrane-Younès J., De Mey J., Sibarita J. B., Hernandez-Verdun D. (2001) J. Cell Biol. 153, 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford N. P., Qian X., Ziogas A., Papageorge A. G., Boersma B. J., Walker R. C., Lukes L., Rowe W. L., Zhang J., Ambs S., Lowy D. R., Anton-Culver H., Hunter K. W. (2007) PLoS Genet. 3, e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke E., Barik S. (2003) Methods Mol. Biol. 226, 525–532 [DOI] [PubMed] [Google Scholar]
- 25.Liu K., Lin F. T., Ruppert J. M., Lin W. C. (2003) Mol. Cell. Biol. 23, 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remy I., Montmarquette A., Michnick S. W. (2004) Nat. Cell Biol. 6, 358–365 [DOI] [PubMed] [Google Scholar]
- 27.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 29.Ayrault O., Andrique L., Séité P. (2006) Exp. Cell Res. 312, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 30.Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 31.Johnson D. G., Ohtani K., Nevins J. R. (1994) Genes Dev. 8, 1514–1525 [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Slansky J. E., Myers D. J., Drinkwater N. R., Kaelin W. G., Farnham P. J. (1994) Mol. Cell. Biol. 14, 1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeGregori J., Leone G., Miron A., Jakoi L., Nevins J. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K., Luo Y., Lin F. T., Lin W. C. (2004) Genes Dev. 18, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.E S., Lai Y. J., Tsukahara R., Chen C. S., Fujiwara Y., Yue J., Yu J. H., Guo H., Kihara A., Tigyi G., Lin F. T. (2009) J. Biol. Chem. 284, 14558–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerppola T. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S. Z., Lin F. T., Lin W. C. (2008) EMBO. Rep. 9, 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kel-Margoulis O. V., Romashchenko A. G., Kolchanov N. A., Wingender E., Kel A. E. (2000) Nucleic Acids Res. 28, 311–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B., Liu K., Lin F. T., Lin W. C. (2004) J. Biol. Chem. 279, 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z., Boone D., Hann S. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González V., Guo K., Hurley L., Sun D. (2009) J. Biol. Chem. 284, 23622–23635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallstrom T. C., Nevins J. R. (2006) Genes Dev. 20, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggero D., Pandolfi P. P. (2003) Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 44.Maggi L. B., Jr., Weber J. D. (2005) Cancer Invest. 23, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindström M. S. (2009) Biochem. Biophys. Res. Commun. 379, 167–170 [DOI] [PubMed] [Google Scholar]
- 46.Warner J. R., McIntosh K. B. (2009) Mol. Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 48.Laskey R. A., Honda B. M., Mills A. D., Finch J. T. (1978) Nature 275, 416–420 [DOI] [PubMed] [Google Scholar]
- 49.Swaminathan V., Kishore A. H., Febitha K. K., Kundu T. K. (2005) Mol. Cell. Biol. 25, 7534–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shandilya J., Swaminathan V., Gadad S. S., Choudhari R., Kodaganur G. S., Kundu T. K. (2009) Mol. Cell. Biol. 29, 5115–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou Y., Wu J., Giannone R. J., Boucher L., Du H., Huang Y., Johnson D. K., Liu Y., Wang Y. (2008) J. Biol. Chem. 283, 5728–5737 [DOI] [PubMed] [Google Scholar]
- 52.Liu H., Tan B. C., Tseng K. H., Chuang C. P., Yeh C. W., Chen K. D., Lee S. C., Yung B. Y. (2007) EMBO Rep. 8, 394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Léotoing L., Meunier L., Manin M., Mauduit C., Decaussin M., Verrijdt G., Claessens F., Benahmed M., Veyssière G., Morel L., Beaudoin C. (2008) Oncogene 27, 2858–2867 [DOI] [PubMed] [Google Scholar]
- 54.Angelov D., Bondarenko V. A., Almagro S., Menoni H., Mongélard F., Hans F., Mietton F., Studitsky V. M., Hamiche A., Dimitrov S., Bouvet P. (2006) EMBO J. 25, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grinstein E., Shan Y., Karawajew L., Snijders P. J., Meijer C. J., Royer H. D., Wernet P. (2006) J. Biol. Chem. 281, 22223–22235 [DOI] [PubMed] [Google Scholar]
- 56.Huddleson J. P., Ahmad N., Lingrel J. B. (2006) J. Biol. Chem. 281, 15121–15128 [DOI] [PubMed] [Google Scholar]
- 57.Samuel S., Twizere J. C., Beifuss K. K., Bernstein L. R. (2008) Mol. Carcinog. 47, 34–46 [DOI] [PubMed] [Google Scholar]
- 58.Masumi A., Fukazawa H., Shimazu T., Yoshida M., Ozato K., Komuro K., Yamaguchi K. (2006) Oncogene 25, 5113–5124 [DOI] [PubMed] [Google Scholar]
- 59.Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 60.Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) EMBO. J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiu C. M., Tsay Y. G., Chang C. J., Lee S. C. (2002) J. Biol. Chem. 277, 39102–39111 [DOI] [PubMed] [Google Scholar]
- 62.Kim H., Lee S. H., Han J. H., Lee J. A., Cheang Y. H., Chang D. J., Lee Y. S., Kaang B. K. (2006) Neuron 49, 707–718 [DOI] [PubMed] [Google Scholar]
- 63.Ni J. Q., Liu L. P., Hess D., Rietdorf J., Sun F. L. (2006) Genes Dev. 20, 1959–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Datta A., Sen J., Hagen J., Korgaonkar C. K., Caffrey M., Quelle D. E., Hughes D. E., Ackerson T. J., Costa R. H., Raychaudhuri P. (2005) Mol. Cell. Biol. 25, 8024–8036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datta A., Nag A., Raychaudhuri P. (2002) Mol. Cell. Biol. 22, 8398–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayrault O., Andrique L., Larsen C. J., Seite P. (2004) Oncogene 23, 8097–8104 [DOI] [PubMed] [Google Scholar]
- 67.Ayrault O., Andrique L., Fauvin D., Eymin B., Gazzeri S., Séité P. (2006) Oncogene 25, 7577–7586 [DOI] [PubMed] [Google Scholar]
- 68.Crawford N. P., Yang H., Mattaini K. R., Hunter K. W. (2009) J. Biol. Chem. 284, 28660–28673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hauck L., Kaba R. G., Lipp M., Dietz R., von Harsdorf R. (2002) Mol. Cell. Biol. 22, 2147–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dick F. A., Dyson N. (2003) Mol. Cell 12, 639–649 [DOI] [PubMed] [Google Scholar]
- 71.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 72.Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., Diaz J. J. (2002) Mol. Biol. Cell 13, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 74.Raska I., Shaw P. J., Cmarko D. (2006) Int. Rev. Cytol. 255, 177–235 [DOI] [PubMed] [Google Scholar]