Abstract
The p53 tumor suppressor pathway is activated by defective ribosome synthesis. Ribosomal proteins are released from the nucleolus and block human double minute-2 (Hdm2) that targets p53 for degradation. However, it remained elusive how abrogation of individual rRNA processing pathways contributes to p53 stabilization. Here, we show that selective inhibition of 18 S rRNA processing provokes accumulation of p53 as efficiently as abrogated 28 S rRNA maturation. We describe hUTP18 as a novel mammalian rRNA processing factor that is specifically involved in 18 S rRNA production. hUTP18 was essential for the cleavage of the 5′-external transcribed spacer leader sequence from the primary polymerase I transcript, but was dispensable for rRNA transcription. Because maturation of the 28 S rRNA was unaffected in hUTP18-depleted cells, our results suggest that the integrity of both the 18 S and 28 S rRNA synthesis pathways can be monitored independently by the p53 pathway. Interestingly, accumulation of p53 after hUTP18 knock down required the ribosomal protein L11. Therefore, cells survey the maturation of the small and large ribosomal subunits by separate molecular routes, which may merge in an L11-dependent signaling pathway for p53 stabilization.
Keywords: Cell/Cycle, RNA/Ribosomal Processing, RNA/Ribosomal RNA, RNA/Ribosomes, Tumor/Suppressor/p53, Tumor/Suppressor/Retinoblastoma
Introduction
Mammalian RNA polymerase I (pol I)3 transcribes a single 47 S precursor ribosomal RNA (rRNA). Endonucleolytic, exonucleolytic, and other RNA modification steps generate the mature 5.8 S, 18 S, and 28 S rRNAs (1). Processing of rRNA is exerted within preribosomal particles and therefore tightly connected with 40 S and 60 S subunit assembly. Ribosome biogenesis and thus successful doubling of the translational machinery are prerequisites for ongoing cell proliferation. Interestingly, the integrity of mammalian ribosome biogenesis is monitored by the p53 tumor suppressor pathway. A truncated version of Bop1 blocked 28 S rRNA maturation and elicited a p53-dependent cell cycle arrest (2). Bop1 is the core component of the PeBoW complex, consisting of Pes1, Bop1, and WDR12, which plays a crucial role in 28 S rRNA maturation (3). Moreover, selective abrogation of pol I transcription by low doses of actinomycin D (5–10 nm) provokes stabilization of p53. The large ribosomal subunit protein L11 was identified as an endogenous inhibitor of Hdm2/Mdm2 (4, 5). Hdm2 is an E3 ubiquitin ligase that targets p53 for degradation and is a key negative regulator of p53 (6). L11 was no longer incorporated into nascent ribosomes in actinomycin D-treated cells because of defective pol I transcription. L11 binds Hdm2 and inhibits its function resulting in accumulation of p53. Subsequently, the ribosomal proteins L5 and L23 were described as additional inhibitors of Hdm2 (7, 8). Similar results were obtained in 5-fluorouracil (5-FU)-treated cells (9, 10). The chemotherapeutic agent 5-FU is incorporated efficiently in newly synthesized RNA and potently blocks rRNA processing. 5-FU treatment caused an increase of ribosome-free L5, L11, and L23 mediating Mdm2 inhibition. L5, L11, and L23 are all components of the large 60 S ribosomal subunit. Recently, a ribosomal protein of the small 40 S subunit was isolated in a yeast two-hybrid screen as a novel Mdm2-binding protein. Ribosomal protein S7 overexpression caused Mdm2 inhibition and p53 accumulation (11). Apparently, ribosomal proteins of the small 40 S and large 60 S subunit are utilized in the Hdm2-p53 feedback circuit.
However, it remained elusive whether specific blockage of 40 S subunit maturation can cause accumulation of p53 as it has been demonstrated for selective impairment of 60 S subunit synthesis. Therefore, we targeted the mammalian small subunit (SSU) processome to inhibit the 18 S rRNA processing pathway selectively. In yeast, the SSU processome was identified as the macromolecular correlate of the terminal knob structure of nascent pol I transcripts that can be detected by electron microscopy (12). SSU processome factors are designated as UTPs (U3 small nucleolar RNA-associated proteins). Deletion of classical UTPs abrogates 18 S rRNA maturation, whereas transfer-UTPs are also required for pol I transcription (12). We describe hUTP18 as the putative mammalian homolog of yeast UTP18, a member of the classical UTPs. We investigated its function in ribosome biogenesis and particularly focused on the effect of hUTP18 depletion on p53 accumulation.
EXPERIMENTAL PROCEDURES
Tissue Culture
HeLa, H1299, 2fTGH, and RKO cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 8% CO2. 2fTGH cells were cultured at 5% CO2.
siRNA Transfection
6 × 104 H1299 or 2fTGH cells were seeded in 6-well plates. 7.5 μl of 20 μm control, Pes1-specific, or hUTP18-specific siRNAs (CureVac, Tübingen, Germany) or Dharmacon smart pools were used for transfection/well using Oligofectamine and Opti-MEM (Invitrogen). Cells were incubated for 5–6 h and transfected on two consecutive days. The following siRNA sequences (sense) were used: control (no cellular target), 5′-UUCUCCGAACGUGUCACGUdTdT; Pes1ORF, 5′-AGGUCUUCCUGUCCAUCAAdTdT; Pes1UTR, 5′-CCAGAGGACCUAAGUGUGAdTdT; hUTP18ORF, 5′- GCAAGGUUCUUUAUGUCUAdTdT; and hUTP18UTR, 5′-AGCCAGUAAUGUCUUAAUAdTdT. Dharmacon smart pools were: RPS7-1, GAGAUGAACUCGGACCUCAdTdT; RPS7-2, GGGCAAGGAUGUUAAUUUUdTdT; RPS7-3, CUAAGGAAAUUGAAGUUGGdTdT; RPS7-4, GCCGUACUCUGACAGCUGUdTdT; RPL11-1, GAACUUCGCAUCCGCAAACdTdT; RPL11-2, UAAAGGUGCGGGAGUAUGAdTdT; RPL11-3, AAAGCUAGAUACACUGUCAdTdT; and RPL11-4, AAAGAUUGCUGUCCACUGCdTdT.
Quantitative Reverse Transcriptase PCR
Total RNA was harvested and 1 μg of RNA was used for the reverse transcriptase reaction using random primers (Invitrogen). Equal amounts of cDNA were combined with the respective primer pair and the SYBR fast reaction mix (Roche Applied Science). A 7500 Fast system (Applied Biosystems) was used for the real-time PCR. The following primers were used in this study: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′- AAGGTGAAGGTCGGAGTCAA-3′ and 5′-AATGAAGGGGTCATTGATGG-3′; L11, 5′-CTTTTCATTTCTCCGGATGC-3′ and 5′-ATCTGTGTTGGGGAGAGTGG-3′; S7, 5′-CCATCTAGTTTGACGCGGAT-3′ and 5′-AATAAGCAAAAGCGTCCCAG-3′.
Metabolic Labeling of Nascent rRNA
H1299 or 2fTGH cells were incubated in phosphate-free Dulbecco's modified Eagle's medium/10% fetal bovine serum for at least 30 min and then incubated for 1 h in the presence 15 μCi/ml 32Pi. The metabolic labeling medium was then removed, and cells were further cultivated for 4–5 h in Dulbecco's modified Eagle's medium/10% fetal bovine serum. Total RNA was isolated using the RNeasy Mini kit (Qiagen). 1.5 μg of metabolically labeled total RNA was separated on a 1% agarose-formaldehyde gel. The gel was then dried on a Whatman paper using a regular gel drier (Bio-Rad) connected to a vacuum pump for 2–3 h at 80 °C. Metabolically labeled RNA was visualized by autoradiography.
Northern Blot Analysis of rRNA Intermediates
Total RNA of H1299 cells was isolated using Trifast (PeqLab, Erlangen, Germany). 5 μg of total RNA was separated on a 1% agarose-formaldehyde gel and blotted on Hybond N+ membranes (Amersham Biosciences). The following DNA oligonucleotides were 5′ end-labeled using T4 polynucleotide kinase in the presence of [γ-32P]ATP: ITS-1, 5′-CCTCCGCGCCGGAACGCGCTAGGTACCTGGACGGCGGGGGGGCGGACG;ITS-2, 5′-GCGGCGGCAAGAGGAGGGCGGACGCCGCCGGGTCTGCGCTTAGGGGGA; and 5′-external transcribed spacer leader sequence, 5′-CGGAGGCCCAACCTCTCCGACGACAGGTCGCCAGAGGACAGCGTGTCAGC. Membranes were preincubated with Church buffer at 65 °C for 3 h, and the radioactively labeled probes were then added overnight. Membranes were shortly washed in 1×SSC and 0.2×SSC.
Immunoblotting and Immunofluorescence
Adherent cells were washed once with phosphate-buffered saline and then directly lysed in 2× SDS loading buffer (100 mm Tris/HCl, 200 mm dithioerythritol, 4% SDS, 10 mm EDTA, 0.2% bromphenol blue, 20% glycerol). Whole cell lysates were separated by SDS-PAGE and blotted on nitrocellulose membranes (Amersham Biosciences). Immunodetection was performed with monoclonal antibodies directed against hUTP18 (3C4), Pes1 (8H11), and α-tubulin (Sigma-Aldrich), p21 (C-19; Santa Cruz Biotechnology), and DDX6 (NB200-191; Novus Biologicals). The monoclonal antibody against human Pes1 was described previously (3). Generation of the monoclonal antibody against hUTP18 is separately described within this section.
For indirect immunofluorescence, cells were grown on coverslides, fixed with ice-cold methanol, and air-dried. Unspecific binding was blocked with phosphate-buffered saline and 10% fetal bovine serum. Primary antibodies were incubated overnight at 4 °C in a humidified chamber. Cy3-labeled secondary antibodies (Dianova) were incubated for 1 h at room temperature. Nuclei were counterstained with DAPI (Sigma-Aldrich). Digital images were acquired using the Openlab acquisition software (Improvision) and a Zeiss Axiovert 200 M microscope (Carl Zeiss MicroImaging) with a 63 (1.15) plan oil objective connected to a CCD camera (Hamamatsu, ORCA-479).
Generation of Monoclonal Antibody against hUTP18
An internal peptide of hUTP18 (26RPDWKAGAGPGGPPQK41) was synthesized and coupled to keyhole limpet hemocyanin or ovalbumin (PSL; Heidelberg). Lou/c rats were immunized intraperitoneally and subcutaneously with 50 μg of peptide-keyhole limpet hemocyanin using CPG 2006 and Incomplete Freund's adjuvant. After 8 weeks, a boost of antigen was given intraperitoneally and subcutaneously. Three days later, fusion of P3X63-Ag8.653 myeloma cells with the rat spleen cells was performed according to standard procedures. Supernatants were tested in an enzyme-linked immunosorbent assay with the peptide coupled to ovalbumin. Peptide-specific antibodies were further analyzed by Western blotting using extracts from HeLa cells. The rat monoclonal hUTP18 antibody (3C4) is an IgG1 subtype.
RESULTS
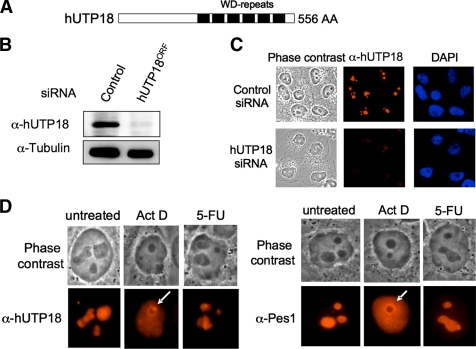
hUTP18 contains C-terminal WD repeats (Fig. 1A) that mediate protein-protein interactions and are frequently found in the nucleolar proteome (13, 14). We raised a monoclonal antibody against hUTP18 and confirmed its specificity by analyzing total cell lysates of control and hUTP18 siRNA-treated HeLa cells (Fig. 1B). A band at the expected molecular size of hUTP18 was clearly diminished by siRNA knock down. Further, we analyzed the subcellular localization of hUTP18 by indirect immunofluorescence (Fig. 1C). hUTP18 predominantly localized to the nucleolus and only faintly to the nucleoplasm. The specificity of the staining was verified by the analysis of hUTP18-depleted cells that exhibited a profound reduction in the signal intensity. Several chemotherapeutic agents disrupt the nucleolar structure (15). We compared the localization of Pes1, involved in the maturation of the 28 S rRNA, and hUTP18 upon treatment of actinomycin D and 5-FU (Fig. 1D). Low doses of the DNA intercalating agent actinomycin D (10 nm) specifically block pol I transcription due to the explicitly high GC content of the rDNA gene clusters. Actinomycin D treatment caused a rapid nucleoplasmic redistribution of hUTP18 and Pes1. In addition, hUTP18 and Pes1 accumulated in a rim around the nucleolus, and hUTP18 also formed cap-like structures adjacent to the rim (Fig. 1D). It is noteworthy that the structural integrity of the nucleolus remained intact in low dose actinomycin D-treated cells as can be concluded from the phase-contrast images. This suggests that nucleolar localization of hUTP18 and Pes1 is dependent on active pol I transcription. The antimetabolite 5-FU interferes with rRNA processing by incorporation into nascent rRNA transcripts. Even high doses of 5-FU (250 mm) had no obvious impact on the structural integrity of the nucleolus. We also observed no significant redistribution of Pes1 or hUTP18.
FIGURE 1.
Subcellular localization of hUTP18 in unstressed and stressed cells. A, schematic view of hUTP18 protein. WD repeats are indicated by black bars. B, verification of the specificity of the monoclonal α-hUTP18 antibody (3C4) by Western blot analysis of control and hUTP18ORF siRNA-treated HeLa cells. A tubulin blot is shown as control. C, verification of the specificity of the monoclonal α-hUTP18 antibody (3C4) by indirect immunofluorescence. Control and hUTP18ORF siRNA-treated HeLa cells were stained with α-hUTP18, and nuclei were counterstained with DAPI. Images were taken with identical exposure times. D, HeLa cells were treated with actinomycin D (Act D; 10 nm) or 5-FU (250 mm) for 6 h. Cells were stained with α-hUTP18 or α-Pes1 antibodies. White arrows in the images of actinomycin D-treated cells highlight cap-like (hUTP18) and rim-like structures (Pes1).
hUTP18 Is Required for 18 S rRNA Maturation and the Cleavage of the 5′-ETS Leader Sequence
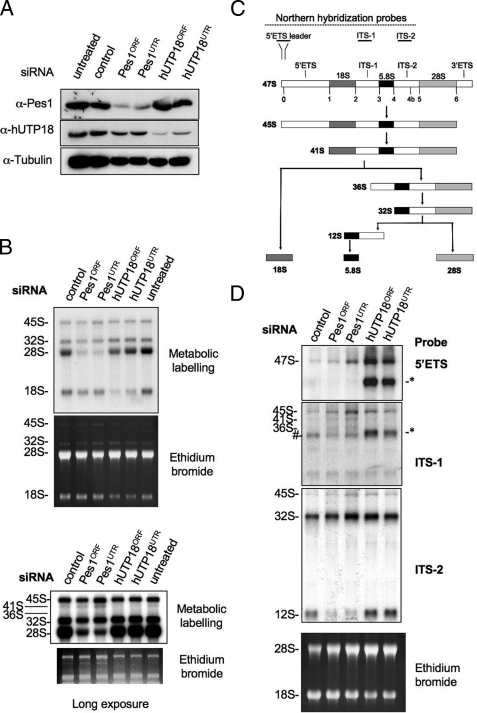
Next, we investigated the role of hUTP18 in ribosome biogenesis by RNA interference. Two hUTP18-specific siRNAs were used to confirm the specificity of the observed depletion phenotypes. Both hUTP18 siRNAs depleted the endogenous protein in H1299 cells; however, the 3′-UTR-specific siRNA was less efficient than the ORF targeting siRNA (Fig. 2A). Knock down of Pes1 by two individual siRNAs did not affect hUTP18 levels, thus allowing the selective inhibition of the SSU processome and the PeBoW complex. To study rRNA processing in hUTP18- or Pes1-depleted cells, we performed metabolic labeling of nascent rRNA. Pes1-depleted cells failed to produce mature 28 S rRNA (Fig. 2B), but production of the 18 S rRNA was unaffected (16). In contrast, knock down of hUTP18 selectively abrogated accumulation of 18 S rRNA. Noteworthy, the defect in hUTP18ORF-treated cells was more pronounced than in hUTP18UTR-treated cells, consistent with protein knockdown efficiencies (Fig. 2A) and thus arguing for a specific dose-dependent depletion phenotype. Longer exposures of the autoradiography and the ethidium bromide gel revealed an additional reduction of the less abundant 41 S and 36 S pre-RNA precursors, indicating an upstream defect of pre-rRNA processing in hUTP18 knockdown cells (Fig. 2C). In Pes1-depleted cells, these precursor rRNAs slightly accumulated consistent with a downstream rRNA processing defect. Our metabolic labeling assay is convenient to assess gross defects in 18 S and 28 S rRNA maturation, but it is not very sensitive for the detection of aberrant intermediates that are rapidly degraded by the nuclear exosome (17).
FIGURE 2.
Role of hUTP18 in rRNA processing. A, Western blot analysis of untreated, control, Pes1, or hUTP18 siRNA-treated H1299 cells using antibodies directed against Pes1, hUTP18, or tubulin. B, metabolic labeling of nascent rRNA in hUTP18-depleted cells visualized by autodiography. The ethidium bromide-stained gel is shown as loading control. A longer exposure is shown at the bottom. C, scheme of mammalian rRNA-processing pathways. Binding sites of Northern blot hybridization probes are depicted. D, Northern blot analysis of rRNA species in hUTP18-or Pes1-depleted cells using three different probes that bind to the 5′-ETS, ITS-1, and ITS-2 region of the rRNA. The ethidium bromide-stained gel is shown at the bottom as loading control.
In yeast, SSU processome function is crucial for the very early steps of pre-rRNA processing (12). To investigate the formation of aberrant rRNA species, we conducted Northern blot analysis using a 5′-ETS-specific hybridization probe (Fig. 2D). A scheme of mammalian rRNA processing pathways and the approximate location of the hybridization probes are shown in Fig. 2C. hUTP18-depleted cells accumulated the 47 S pre-rRNA and a novel 5′-ETS leader sequence containing pre-rRNA of ∼33 S–34 S. This aberrant form also hybridized with an ITS-1-specific but not with an ITS-2-specific probe. Therefore, this aberrant form represents a pre-rRNA containing the complete 5′-ETS, 18 S and ITS-1 region, but lacking the ITS-2, 28 S, and 3′-ETS sequences. As hUTP18 repression had no impact on 12 S rRNA levels, the aberrant rRNA precursor in hUTP18-depleted cells also lacks the 5.8 S rRNA region. Otherwise, it would be detected by the ITS-2-specific probe that also hybridizes to the 12 S rRNA. These findings are consistent with the metabolic labeling experiments that revealed abrogated formation of 18 S rRNA but intact maturation of the 28 S rRNA and 5.8S rRNA in hUTP18-depleted cells (Fig. 2B).
Conclusively, we found that hUTP18 is required for the cleavage of the 5′-ETS rRNA leader sequence and subsequent maturation of the 18 S rRNA. Pol I transcription was not affected in hUTP18 knockdown cells. Therefore, the role of hUTP18 in rRNA processing is compatible with a classical UTP factor of the SSU processome.
Selective Inhibition of 18 S rRNA Maturation Is Sufficient for a Functional p53 Response
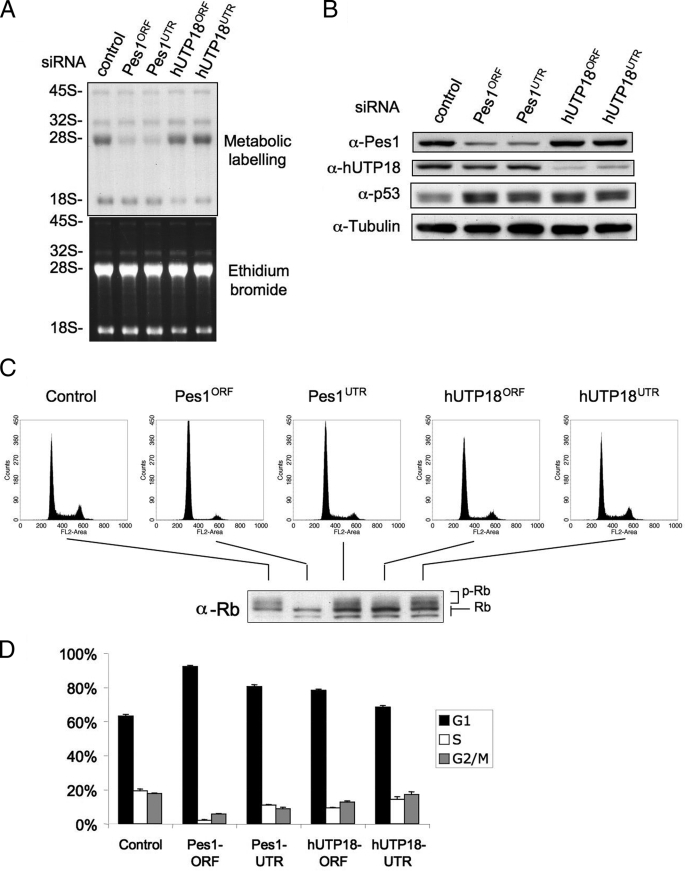
So far, we have shown that depletion of hUTP18 abrogates production of the 18 S rRNA, whereas maturation and export of the large ribosomal subunit are unaffected. Next, we tested whether specific inhibition of the 18 S rRNA synthesis pathway also triggers accumulation of endogenous p53. Several studies have demonstrated that interference with the ribosomal synthesis pathway induces a p53 response, most likely mediated by the inhibitory effect of nonincorporated ribosomal proteins on the ubiquitin ligase Hdm2 that targets p53 for degradation in unstressed cells (4, 15). Previous studies addressed either the blockage of pol I transcription that abrogates the synthesis of both ribosomal subunits or the impaired rRNA processing of the large subunit (2, 3, 18). We used p53 wild-type 2fTGH cells that are derivatives of the HT1080 human fibrosarcoma cell line. Cells were treated with control or Pes1-specific or hUTP18-specific siRNAs on 2 consecutive days and harvested for metabolic labeling of nascent rRNA or Western blotting 48 h later. Depletion of Pes1 and hUTP18 impaired 28 S and 18 S rRNA production, respectively (Fig. 3A). Protein levels of Pes1 and hUTP18 were also efficiently decreased (Fig. 3B). Importantly, p53 increased in Pes1 and hUTP18 knockdown cells. We conclude that specific inhibition of 18 S rRNA maturation is sufficient to mediate accumulation of p53. To substantiate our results further, we also investigated concomitant cell cycle changes.
FIGURE 3.
Depletion of hUTP18 induces accumulation of p53. A, 2fTGH cells were treated with control, Pes1, or hUTP18 siRNAs, and a metabolic labeling of nascent rRNA was subsequently performed. B, cells were treated as described in A, and total cell lysates were harvested for Western blot analysis. C, cells were treated as described in A, and DNA content was determined by propidium iodide staining using flow cytometry (FL2-Area). Experiments were performed in triplicate, and representative fluorescence-activated cell sorting blots are shown. The corresponding phosphorylation status was determined by Western blot analysis for total Rb. Hypo- and hyperphosphorylated forms of Rb can be discriminated by size as indicated. D, quantification of cell cycle distributions was derived from triplicate experiments shown in C. Error bar, S.D.
We assessed the cell cycle distribution by FACS analysis of the DNA content (Fig. 3C) and quantified the results of triplicate experiments (Fig. 3D). Pes1- or hUTP18-depleted cells had an increased G1 phase fraction at the expense of the S and G2/M phase population (Fig. 3D). Loss of retinoblastoma (Rb) phosphorylation is causally linked to a p53-mediated cell cycle arrest. Consistently, Pes1 and hUTP18 knockdown cells contained less phosphorylated Rb as determined by Western blot analysis of the samples used for cell cycle analysis (Fig. 3C).
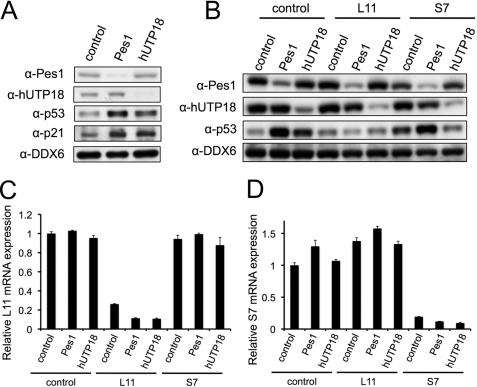
To confirm that knockdown of hUTP18 causes accumulation of p53 in other cell lines with wild-type p53, we transfected RKO colon cancer cells with control, Pes1, or hUTP18 siRNAs. Indeed, we also observed an increase of endogenous p53 upon loss of hUTP18 (Fig. 4A). Moreover, RKO cells exhibited a functional p53 response, as the established target p21 was likewise induced.
FIGURE 4.
Knockdown of hUTP18, Pes1, L7, and L11 in colon cancer (RKO) cells. A, RKO cells were transfected with Pes1, hUTP18, or control siRNAs. Protein levels were determined by Western blotting, and ubiquitously expressed DDX6 served as a loading control. B, RKO cells were transfected with the indicated combinations of siRNAs. Protein levels were determined by Western blotting. C and D, RKO cells were transfected with combinations of siRNAs as in B. Knockdown of L11 and S7 was confirmed by quantitative reverse transcriptase PCR. Relative expression levels of L11 and S7 are normalized to glyceraldehyde-3-phosphate dehydrogenase expression.
L11-dependent Stabilization of p53 after Knock Down of hUTP18
Recently, it was shown that the small subunit ribosomal protein S7 functions as an inhibitor of Hdm2 and therefore might be involved in the accumulation of p53 in hUTP18-depleted cells (11, 19). On the other hand, it was also reported that the large subunit ribosomal protein L11 mediates the accumulation of p53 in cells with defective small subunit biogenesis caused by ablation of the S6 ribosomal protein (20). This was due to a specific recruitment of ribosomal protein mRNAs to the polysomes in cells with abrogated small subunit production. An increase of L11 translation accounted for inhibition of Hdm2 and subsequent stabilization of p53. Therefore, we examined the role L11 and S7 in the response to nucleolar stress caused by loss of Pes1 and hUTP18. RKO cells were transfected with combinations of siRNAs targeting Pes1 or hUTP18 and L11 or S7 (Fig. 4, B–D). We found that loss of L11 prevented the accumulation of p53 in Pes1- and hUTP18-depleted cells. This result is consistent with the studies in S6-deficient cells. Apparently, L11 integrates the nucleolar stress response of defective large and small subunit biogenesis. Loss of S7 by itself appeared to induce p53 levels slightly, most likely by a mechanism similar to that of loss of S6 protein. Accumulation of p53 by knockdown of Pes1 was not affected by concomitant suppression of S7. Interestingly, concomitant suppression of S7 and hUTP18 failed to induce p53 synergistically. It rather compromised accumulation of p53. This result remains elusive as one would expect an intact L11-dependent mechanism of p53 stabilization. Definitely, this observation and the role of S7, which has been described as regulator and substrate of Mdm2 (20), requires further investigation.
DISCUSSION
In this study, we focused on the dissection of the two major mammalian rRNA processing pathways. We found that selective inhibition of either 18 S or 28 S rRNA maturation efficiently provoked accumulation of p53. Apparently, mammalian cells have acquired mechanisms that independently survey small and large ribosomal subunit production. It is conceivable that the integral measurement of the total ribosome number is an inappropriate readout to detect acute disturbances of the ribosome synthesis pathway as the large amounts of preexisting ribosomes by far outranges the number of newly synthesized ribosomes within a limited time frame. Experimental evidence has substantially supported this hypothesis. Ribosomal proteins of the large ribosomal subunits L5, L11, and L23 were found to function as inhibitors of Hdm2 when ribosome synthesis is blocked, e.g. by low dose actinomycin D or 5-FU treatment. Impaired ribosome maturation decreases the demand for ribosomal proteins and favors their interaction with Hdm2. Inhibition of Hdm2 subsequently results in reduced degradation and thus accumulation of p53. Thereby, cells are capable of responding instantly to stresses affecting the ribosome synthesis pathway.
Our results would suggest that ribosomal proteins or synthesis factors of the 40 S subunit also contribute to the nucleolar stress-induced p53 response because depletion of hUTP18 and selective inhibition of 18 S rRNA maturation caused accumulation of p53. Indeed, the ribosomal protein S7, a component of 40 S subunit, was recently found to bind and inhibit Hdm2/Mdm2 (11, 20). S7 is therefore a likely candidate to mediate a feedback signal to the Hdm2-p53 circuit in cells with impaired 18 S rRNA processing.
Knock down of S7 or L11 compromised the stabilization of p53 achieved by knock down of hUTP18, whereas knock down of S7 did not affect stabilization of p53 after knock down of Pes1. This result supports a previous report that inhibition of 18 S as well as 28 S rRNA maturation may merge in a common L11-dependent signaling pathway for p53 stabilization (20).
It will be interesting to unravel whether other 40 S ribosomal proteins or synthesis factors are also implicated in Hdm2 inhibition. Most of the studies that aimed to identify Hdm2-interacting factors were conducted in unstressed Hdm2-overexpressing cells or by yeast two-hybrid approaches (4, 5). In the view of our results, applying selective rRNA-processing defects would increase the likelihood of isolating ribosomal proteins or potentially also rRNA-processing factors that block Hdm2 function.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB684, SFB-Transregio5.
- pol I
- polymerase I
- Hdm
- human double minute
- Mdm
- mouse double minute
- 5-FU
- 5-fluorouracil
- SSU
- small subunit
- siRNA
- small interfering RNA
- ORF
- open reading frame
- UTR
- untranslated region
- Rb
- retinoblastoma.
REFERENCES
- 1.Mayer C., Grummt I. (2006) Oncogene 25, 6384–6391 [DOI] [PubMed] [Google Scholar]
- 2.Pestov D. G., Strezoska Z., Lau L. F. (2001) Mol. Cell. Biol. 21, 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hölzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G. W., Eick D. (2005) J. Cell Biol. 170, 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris S. L., Levine A. J. (2005) Oncogene 24, 2899–2908 [DOI] [PubMed] [Google Scholar]
- 7.Dai M. S., Lu H. (2004) J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 8.Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Mol. Cell. Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X. X., Dai M. S., Lu H. (2007) J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 11.Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 12.Gallagher J. E., Dunbar D. A., Granneman S., Mitchell B. M., Osheim Y., Beyer A. L., Baserga S. J. (2004) Genes Dev. 18, 2506–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith T. F., Gaitatzes C., Saxena K., Neer E. J. (1999) Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 14.Staub E., Fiziev P., Rosenthal A., Hinzmann B. (2004) Bioessays 26, 567–581 [DOI] [PubMed] [Google Scholar]
- 15.Rubbi C. P., Milner J. (2003) EMBO J. 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm T., Hölzel M., Rohrmoser M., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Eick D. (2006) Nucleic Acids Res. 34, 3030–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houseley J., LaCava J., Tollervey D. (2006) Nat. Rev. 7, 529–539 [DOI] [PubMed] [Google Scholar]
- 18.Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Gröne H. J., Schütz G., Grummt I. (2005) Mol. Cell 19, 77–87 [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Mol. Cell 14, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G. F., Bernardi R., Pandolfi P. P., Thomas G. (2009) Nat. Cell Biol. 11, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]