Abstract
The adaptor protein APPL1 (adaptor protein containing pleckstrin homology (PH), phosphotyrosine binding (PTB), and leucine zipper motifs) was first identified as a binding protein of AKT2 by yeast two-hybrid screening. APPL1 was subsequently found to bind to several membrane-bound receptors and was implicated in their signal transduction through AKT and/or MAPK pathways. To determine the unambiguous role of Appl1 in vivo, we generated Appl1 knock-out mice. Here we report that Appl1 knock-out mice are viable and fertile. Appl1-null mice were born at expected Mendelian ratios, without obvious phenotypic abnormalities. Moreover, Akt activity in various fetal tissues was unchanged compared with that observed in wild-type littermates. Studies of isolated Appl1−/− murine embryonic fibroblasts (MEFs) showed that Akt activation by epidermal growth factor, insulin, or fetal bovine serum was similar to that observed in wild-type MEFs, although Akt activation by HGF was diminished in Appl1−/− MEFs. To rule out a possible redundant role played by the related Appl2, we used small interfering RNA to knock down Appl2 expression in Appl1−/− MEFs. Unexpectedly, cell survival was unaffected under normal culture conditions, and activation of Akt was unaltered following epidermal growth factor stimulation, although Akt activity did decrease further after HGF stimulation. Furthermore, we found that Appl proteins are required for HGF-induced cell survival and migration via activation of Akt. Our studies suggest that Appl1 is dispensable for development and only participate in Akt signaling under certain conditions.
Keywords: Development Differentiation, Gene/Knockout, Oncogene, Organisms/Mouse, Phosphorylation/Phosphatases/Serine-Threonine, Protein/Protein-Protein Interactions, Signal Transduction/Protein Kinases/Serine/Threonine
Introduction
The adaptor protein containing pleckstrin homology (PH), phosphotyrosine binding (PTB), and leucine zipper motifs (APPL)5 was first identified as an AKT2-interacting protein by yeast two-hybrid screening (1). APPL1, also dubbed DIP13α (DCC-interacting protein 13α) (2), comprises three domains, i.e. Bar, PH, and PTB motifs. Whereas all of these domains can bind to lipids, each has unique binding preferences, which theoretically enables APPL to bind to various signaling proteins. The membrane binding-bending feature of the APPL protein permits it to be trafficked among many subcellular compartments (3). Moreover, the APPL Bar domain interacts with its own PH domain to form a unique Bar-PH structure that distinguishes APPL from other Bar domain-containing molecules (4). We previously showed that APPL1 can bind to AKT2 through its PTB domain (1), and subsequent co-immunoprecipitation studies showed that APPL1 also binds to AKT1, AKT3, and the p110 catalytic subunit of phosphatidylinositol 3-kinase. APPL1 does not bind to phosphorylated (active) AKT, and our initial report did not ascertain whether binding to APPL1 affects AKT activity (1). Subsequent work identified a second APPL protein, APPL2, which shares a high homology with APPL1 as well as some of the same binding partners (5).
The function of APPL proteins was first demonstrated in HeLa cells, where APPL1 and APPL2 were found to represent key signaling links from the endosome to the nucleus by switching and activating binding partners from the small GTPase Rab5 on endosomes to the nucleosome remodeling and histone deacetylase multiprotein complex NuRD-MeCP1. Furthermore, knock down of APPL protein inhibited DNA synthesis and resulted in cell cycle arrest (6). Structural analysis revealed that APPL binds to Rab5 mainly through its PH domain, but the Bar domain at the other side of the dimer also binds to Rab5 (7). A broader role for APPL in signal transduction was soon discovered when various groups found that APPL proteins are implicated in nerve growth factor and follicle stimulating hormone signaling, as well as in lipid and glucose metabolism. Two groups independently reported that APPL1 tethers GIPC1 to the nerve growth factor receptor TrkA upon nerve growth factor stimulation in PC12 cells, which is necessary for downstream activation of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) and AKT and subsequent neurite outgrowth (8, 9). In addition, APPL1 and APPL2 were shown to be associated with the follicle stimulating hormone receptor when overexpressed in HEK293 cells, suggesting that APPL may play a role in reproduction (10, 11). APPL suppresses androgen receptor function by regulating AKT activity (12). APPL has also been shown to interact with the adiponectin receptor and thereby participates in glucose and lipid metabolism as well as vasodilation. Mao et al. (13) showed that overexpression of Appl1 in C2C12 myoblasts increases adiponectin-induced p38 MAPK activation, whereas knock down of Appl1 inhibits p38 activation. Appl1 knockdown also caused a moderate reduction in insulin-induced Akt activation in these cells, although no effect on cell proliferation was reported (13). However, no evidence was found for a link between human diabetes and genetic variation in the APPL1 locus (14). In endothelial cells, adiponectin can activate AMP-activated protein kinase through APPL1 to provide a survival signal (15). Alternatively, adiponectin activates the ERK pathway through APPL-dependant Ras activation (16). Nevertheless, APPL mediates the adiponectin-induced phosphorylation of endothelial nitric-oxide synthase, and the subsequent production of nitric oxide that triggers endothelium-dependent vasodilation (17). In adipocytes, knock down of APPL1 suppresses AKT phosphorylation, 2-deoxyglucose uptake, and Glut4 translocation (18).
APPL has also been implicated in some human pathological conditions such as Lowe syndrome. Recently, the inositol 5-phosphatase OCRL (Oculocerebrorenal Syndrome of Lowe) was found to be recruited by APPL1 at early endosomes. Whether this binding is essential for the enzymatic activity of OCRL was not documented, although all known point mutations in the OCRL gene in Lowe syndrome patients do abolish its interaction with APPL1 (19, 20).
Information gathered to date indicates that APPL is involved in multiple steps in cell signaling networks from very upstream, such as conveying an activated receptor signal, to far downstream, such as its involvement with NuRD/MeCP1 in the nucleus (6). It seems that under certain conditions, perturbation of this adaptor protein interferes with cell proliferation or even cell survival. This was apparent in zebrafish, where knock down of Appl1 or Appl2 alone was sufficient to trigger massive apoptosis, which was thought to be due to blockage of Akt signaling specifically to Gsk3β, but not to Tsc2 (21). The complexity of the function of APPL became even more intertwined when APPL was found to be essential for a certain type of apoptotic signaling. Interaction between APPL and the tumor suppressor DCC (deleted in colorectal cancer) was necessary for the latter to exert its pro-apoptotic function in the colon cancer cell line DLD1(2). Thus, APPL proteins participate in a wide array of cell signaling pathways, and their role appears to be highly cell-type specific.
To clarify the physiologic role of APPL, we generated and characterized an Appl1 knock-out mouse model. Lest a possible fetal lethal phenotype be observed, and for studies of tissue specific roles of Appl1, we targeted the Appl1 allele by a loxP strategy. To understand how the ubiquitous loss of Appl1 impacts embryonic development, we crossed the Appl1 floxed mice with EIIA-Cre mice. Surprisingly, the homozygous Appl1 knock-out mice developed normally to adulthood, and loss of Appl1 had growth factor-selective effects on Akt signaling in Appl1-null murine embryonic fibroblasts (MEFs).
EXPERIMENTAL PROCEDURES
Generation of Conditional Appl1-deficient Mice
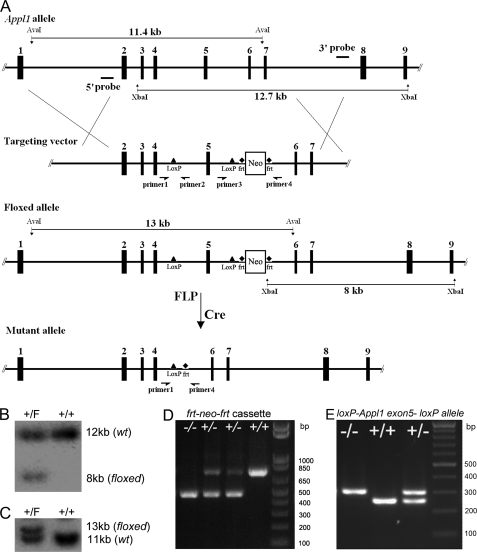
Exon 5 of the mouse Appl1 gene was chosen for deletion, because its absence causes a reading frameshift resulting in the predicted generation of multiple stop codons following exon 4 of the mRNA. A probe from exon 5 of the Appl1 gene was used to screen a 129Sv mouse genomic library (Stratagene, La Jolla, CA). A positive clone containing ∼15.5 kb of Appl1 genomic sequence was identified. The 9.3-kb sequence from this clone encompassing exons 2 to 7 of Appl1 was released by restriction digestion with PvuII, which was then inserted into pBluescript vector. An oligo with the loxP sequence and an EcoRI site was inserted into the BamHI site upstream of exon 5. Another loxP site with the Frt-flanked neomycin-resistance gene (neo) cassette sequence derived from plasmid pK-11/pM-30 was cloned into the vector downstream of exon 5, between EcoRV and KpnI sites. The targeting vector was linearized with XhoI and electroporated into RW4 ES cells. G418-resistant clones were then screened by Southern blot and PCR analysis. Genomic DNA from the positive ES clones was digested with XbaI (for the 3′ probe) and AvaI (for the 5′ probe). These two probes were used for Southern blotting to identify the correctly targeted ES clones. PCR with specific primers for the neo cassette sequence were used to confirm homologous recombination (Fig. 1A).
FIGURE 1.
Conditional targeting of the murine Appl1 gene. A, diagram of Appl1 locus and targeting vector showing the position of probes (5′ probe and 3′ probe) used for Southern blotting. B, Southern blot analysis of ES cell DNA used to identify correctly floxed clones by using the 3′ probe. F, floxed allele. C, Southern blot analysis using the 5′ probe. D, PCR genotyping (using primers 3 and 4) of tail DNA from mouse in which neo cassette has been removed. E, PCR genotyping (using primers 1, 2, and 4) of tail DNA from mouse in which Appl1 exon 5 has been excised. Primers used in D and E correspond to sites shown in A.
The positive ES clones were injected into C57BL/6 blastocysts, and chimeras were crossed with C57BL/6 mice. Germ line transmission was verified by PCR and sequence analysis. The heterozygous Appl1-floxed mice were crossed with FLP mice to remove the Frt-flanked neo cassette. Then, the Appl1-floxed, neo-negative mice were crossed with EIIA-Cre mice, and excision of the floxed allele was confirmed by a specific PCR. The homozygous Appl1 knock-out mice were subsequently generated from these mice. Experiments were performed in the 129Sv background.
Southern Blot Analysis
DNA extraction and electrophoresis, gel transfer, and probe labeling were performed according to standard protocols. For heterozygously floxed Appl1 clones, the 3′ probe distinguished a 12.7-kb band for the wild-type allele and an 8-kb band for the floxed allele with XbaI-digested genomic DNA (Fig. 1B). The 5′ probe recognized an 11.4-kb (wild-type allele) band and a 13-kb (floxed allele) band (Fig. 1C).
PCR Genotyping Analysis
Primers surrounding the neo cassette sequence were used to identify the excised neo allele (PCR product size 470 bp; forward sequence: CTG TTT CAG CTA GTC TGC ATT C; reverse sequence: AGT GCT GGA TGT GCT GAA GC), and the wild-type allele (PCR product size, 715 bp; Fig. 1D). After crossing the targeted mice with the EIIA-Cre mice, a primer from intron 5 (before loxP; forward: AGC AAT ACA CTA CCA GAA AAT CTA C), a primer from intron 4 (before loxP; forward: AAG TGA TAG GGG TCA GAT CC), and a primer from intron 4 (after loxP; reverse: TGT GAG TTT ATT CTC ATT ATA CAT ACC) were used for genotyping the knock-out allele (knock-out allele, 315 bp; wild-type allele, 249 bp; Fig. 1E).
Preparation of MEFs
Heterozygous Appl1 knock-out mice were crossed, and day E13.5 embryos were collected. MEFs were cultured as described before (22). MEFs at passages 1 to 5 were used for the assays. Akt1−/− MEFs were harvested from an Akt1 knock-out fetus at day E13.5 (23).
Cell Viability and Proliferation Assay
Cell viability was measured using a MTS-based CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega, Madison WI). MEFs at passage 1 were seeded at 3 × 103 cells/well in 96-well plates. Cells were maintained under normal conditions or serum starved and treated with HGF at the indicated concentrations. OD values at 490 nm were measured at 48 h after seeding, using a 96-well microplate reader (Bio-Rad). In addition, cell growth curves were ascertained to measure the cell proliferation rate. MEFs were seeded in 24-well plates at 3 × 104 cells/well, and then cells were collected every day for 7 days. The cell numbers were converted by measuring the total cell protein at A595 using Bradford reagent (Bio-Rad).
Gene Expression Knockdown by siRNA
Stealth siRNAs against mouse Appl2 and control siRNAs were purchased from Invitrogen. The siRNAs were transfected into primary MEFs (100 pg/2 × 106 cells) using Nucleofector Kit MEF2 (Amaxa Inc., Gaithersburg, MD) and program T-20. In addition, the same siRNAs were transfected into 3T3 cells by using Nucleofector Kit R and program U-30. Alternatively, 3T3 cells were transfected with siRNAs by using Lipofectamine RNAiMAX reagent (Invitrogen). Cells were used for experiments 72 h after transfection.
Growth Factor Stimulation
MEFs (3 × 105) were plated in 60-mm dishes and then incubated overnight. Cells were initially starved by incubating in DMEM supplemented with 0.05% FBS for 16 h, followed by stimulation with 10% FBS or the indicated growth factors (EGF, insulin, and HGF; R&D Systems, Minneapolis, MN). Alternatively, cells were starved for 30 min and treated with growth factors.
Western Blot Analysis
Total tissue or cellular protein was extracted by using 1× cell lysis buffer supplemented with 2 mm phenylmethylsulfonyl fluoride (Cell Signaling Technology, Danvers, MA). Cell debris was removed by centrifuging at 15,000 × g for 15 min at 4 °C. Protein concentration was determined using Bradford reagent. Then 50 μg/well of proteins were loaded into Tris glycine-buffered SDS-PAGE gels (Invitrogen). Separated protein was then transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). Antibodies against Appl1, p-Akt, Akt1, Akt2, Akt3, p-Gsk3, Gsk3, p-Tsc2, Tsc2, p-S6k, S6k, p-MET, Met, PARP, cleaved Caspase 3, Caspase 6, and Caspase 7 were purchased from Cell Signaling. Antibodies against Appl1, Appl2, glyceraldehyde-3-phosphate dehydrogenase, QM, cleaved Caspase 9, and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Density of protein signal was quantified by using AlphaEaseFC (Alpha Innotech Corporation, CA).
Co-immunoprecipitation
ExactaCruz IP kits were purchased from Santa Cruz Biotechnology. In brief, 1 mg of protein lysates were pre-cleared with beads and then incubated with IP matrix-antibody overnight in a cold room on a rotator. Beads were washed three times with lysis buffer and then resuspended in 2× loading buffer for electrophoresis. To analyze the percentage of Akt-Appl1 complex in a total cellular Akt or Appl1 pool, a semi-quantitative approach was adopted (24). To determine the percentage of the Appl1-associated Akt, 5% IP-Akt was loaded along with IP-Appl1 in a Western blot and probed with anti-Akt antibody. Similarly, the reciprocal IP was used to determine the amount of Akt-bound Appl1.
Plasmid Construction and Retroviral Infection of MEFs
HA-tagged human APPL1 cDNA was released from pcDNA3 (1) and blunt end-ligated into pMSCV (Clontech, Mountain View, CA) at the HpaI site. Tandem affinity purification (TAP) sequence was excised from pcDNA4/TO/TAP at KpnI and XhoI sites and semi-blunt-end ligated into pMSCV at BglII and XhoI sites (25). Mouse Appl1 and Appl2 full-length cDNAs were amplified using Pfx (Invitrogen) and cloned into pMSCV-Tap vector (Appl1 forward, CCT TCG CCA CGA TGC CGG; reverse, TAC GCT GTG CCA CAT CCC AGG G; Appl2 forward, CCG CCG TGG ACA AGC TCC TGC, reverse, AAT GAG TTA TGC TTC AGA TTC TGC GCC). Retroviruses were produced by co-transfecting 293T cells with pEcoPac. Supernatant was collected after 24 h, and MEFs were infected for 5 h at a multiplicity of infection of 1.
Transwell Migration Assay
Cell mobility was measured using a Boyden chamber assay. In short, cell migration was determined by transwell mobility through a 0.8-μm PET membrane (BD Bioscience). Alternatively, cell invasion was measured by transwell mobility through Matrigel (BD Bioscience). HGF or EGF was used as a chemoattractant in the lower chamber. After 22 h, cells that had migrated through the membrane were stained with Diff-Quik stain (Dade Behring, Inc., Newark, DE) and counted.
RESULTS
Appl1 Is Ubiquitously Expressed and Associated with Akt Family Members in Mouse Tissues
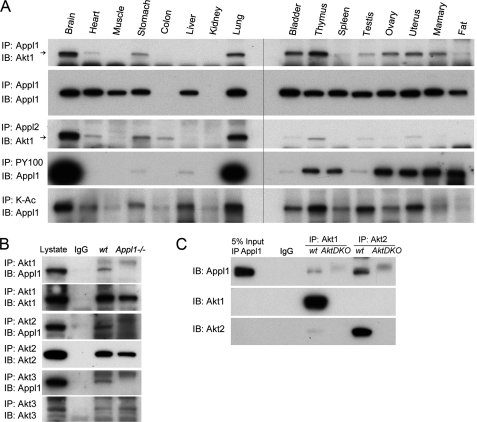
We examined the expression patterns of Appl1 protein in multiple tissues, with the goal of subsequently predicting where the effects of Appl1 loss might be most obvious in Appl1 knock-out mice (supplemental Fig. S1A). By using an antibody recognizing the COOH-terminal end of the Appl1 protein, we found that Appl1 expression is expressed ubiquitously and at similar levels in most tissues, except for the kidney, which had very low Appl1 protein expression. We also probed membranes containing proteins from various tissues with antibodies against Appl1-interacting proteins Akt1, -2, and -3, and Akt1 and Akt2 exhibited an expression pattern similar to that of Appl1 (supplemental Fig. S1B). Akt1 was found to interact with Appl1 in most tissues, as shown by co-immunoprecipitation (co-IP), suggesting that certain modifications may be required for tissue-specific interaction (Fig. 2A). Akt1 was also found to bind to Appl2 in a pattern similar to which it binds to Appl1. To address the nature of the modification of the Appl1 protein in different tissues, we used phosphorylation-specific and acetylation-specific antibodies for co-IP. As expected, Appl1 was phosphorylated and/or acetylated in a tissue-specific manner, suggesting different cellular roles (Fig. 2A).
FIGURE 2.
A, Akt1 binds to Appl1 and Appl2 in a tissue-specific manner. Akt1 interacts with Appl1 in multiple tissues as demonstrated by using co-immunoprecipitation/immunoblot analysis. Akt1 and Appl2 also interact in various tissues. Co-IP also identifies tissue-specific phosphorylation and acetylation of the Appl1 protein, using phosphotyrosine (PY100)- and lysine acetylation (K-Ac)-specific antibodies. B, co-IP in wild-type and Appl1 knock-out MEFs. Akt1/2/3 were immunoprecipitated, and Western blot analysis was performed to detect Appl1. C, co-IP in Akt1/2 double knock-out (DKO) MEFs. Akt1 and Akt2 were immunoprecipitated, and immunoblotting (IB) was performed to detect Appl1.
The specificity of Appl1 binding to Akt1/2/3 was unambiguously proven by using wild-type MEFs and Appl1−/− MEFs in an IP experiment (Fig. 2B). Similarly, the use of MEFs from wild-type and Akt1/2 double knock-out mice demonstrated the specificity of the interaction between Appl1 and Akt (Fig. 2C).
Generation of Conditionally Targeted Appl1 Mice
The loxP/Cre strategy was adopted to develop conditional Appl1 knock-out mice (Fig. 1A). In brief, exon 5 of the Appl1-targeted allele was flanked by a pair of loxP sites by homologous recombination in ES cells. The positive ES cells were screened by PCR, and the correctly targeted ES clones were further identified by Southern blot analysis (Fig. 1, B and C). Chimeric mice were then generated, and the heterozygous loxP-Appl1 exon 5-loxP-Frt-neo-Frt mice were first crossed to FLP mice to remove the neo sequence (Fig. 1D). Then, the floxed Appl1 mice were crossed to EIIA-Cre mice to delete exon 5 of the Appl1 gene (Fig. 1E). Although the mutant mRNA is stable as shown by semi-quantitative reverse transcription-PCR (supplemental Fig. S2A), the loss of exon 5 causes a reading frameshift that generates multiple stop codons after exon 4 of the messenger RNA and is predicted to produce a N′ Appl1 protein of 97 amino acids of ∼11 kDa.
Appl1 Is Dispensable for Embryonic and Postnatal Development as well as Akt Signaling in Tissues
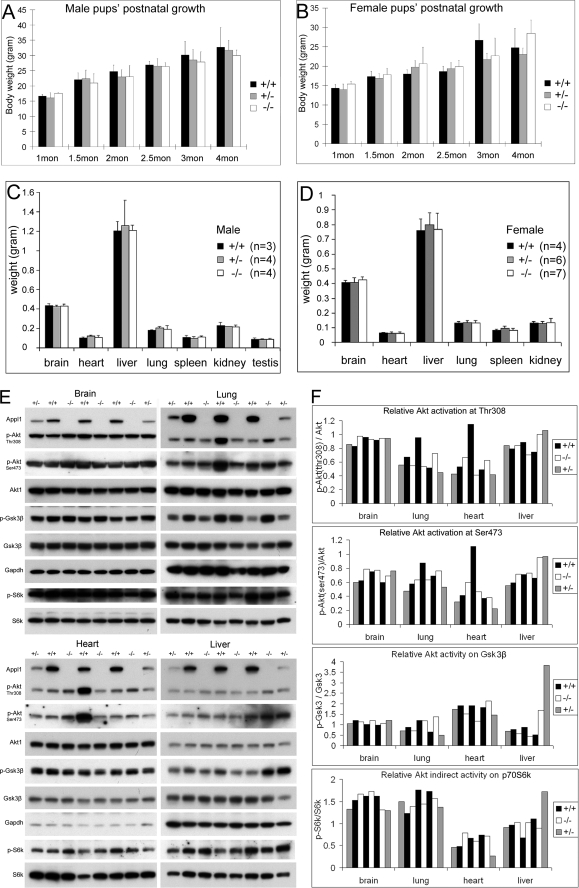
The heterozygous Appl1 knock-out mice were crossed to obtain homozygous knock-out pups. Unexpectedly, mice with complete loss of Appl1 were viable, with no visible abnormality observed. At birth, pups had a normal Mendelian distribution of +/+, +/−, and −/− genotypes (Table 1). Moreover, pups at different postnatal stages thrived and developed normally, with no difference in gross body weight compared with wild-type littermates (Fig. 3, A and B). Furthermore, 3-month-old mice with each of the three genotypes had similar organ weights (Fig. 3, C and D). Moreover, reproductive function also did not appear to be affected, as the litter sizes from the different mating sets were virtually identical (Table 2). To assess the effect of Appl1 loss on the hematopoietic system, peripheral blood was analyzed. No obvious defect was found in blood from Appl1-null mice (supplemental Table S1).
TABLE 1.
Genotypic analysis of offspring from 27 Appl1+/− x Appl1+/− crosses
| Gender | Wild-type | Heterozygous (+/−) | Homozygous (−/−) |
|---|---|---|---|
| Female | 30 (28%) | 56 (52%) | 22 (20%) |
| Male | 22 (22%) | 51 (50%) | 29 (28%) |
| Combined | 52 (25%) | 107 (51%) | 51 (24%) |
FIGURE 3.
Appl1-null mice show normal postnatal development as well as Akt signaling. A, weights of male wild-type, Appl1+/−, and Appl1−/− pups at 1, 1.5, 2, 2.5, 3, and 4 months of age. B, weights of female wild-type, Appl1+/−, and Appl1−/− pups at different postnatal times. For each bar, measurements were obtained from 3 to 19 mice. C and D, weights of brain, heart, liver, lung, spleen, kidney, and testis from male (C) and female (D) wild-type, Appl1+/− and Appl1−/− mice, each at 3 months of age. Error bars indicate S.D. E, fetal brain, lung, heart, and liver from one litter at E18.5 were homogenized and submitted to immunoblot analysis of Akt1/p-Akt and Gsk3β/p-Gsk3β. F, the signal density of phosphorylated Akt at Thr-308 and Ser-473 was quantified and normalized to that of the total Akt. Also, the relative Akt activity on Gsk3β and p70 S6k was quantified and normalized.
TABLE 2.
Litter sizes from matings of Appl knock-out mice and wild-type littermates
| Mating set | Litter size (mean ± S.D.) | Number of litters |
|---|---|---|
| +/+ x +/+ | 8.2 ± 1.67 | 18 |
| +/− x +/− | 7.37 ± 1.87 | 29 |
| +/− x −/− | 8 ± 2.54 | 10 |
| −/− x −/− | 7.25 ± 1.9 | 8 |
To evaluate the in vivo role of Appl1 on Akt stability and activation, various adult mouse tissues from wild-type, Appl1+/−, and Appl1−/− mice were subjected to immunoblotting. Akt1 protein level in various tissues was stable regardless of Appl1 loss, suggesting that binding of Appl1 to Akt does not affect its half-life (supplemental Fig. S2B). On the other hand, no compensating up-regulation of Appl2 was observed (supplemental Fig. S2B). To further validate that this model is an authentic Appl1 knock-out, we compared it with another Appl1 knock-out model in which Appl1 exon 1 is disrupted by a GeneTrap strategy and has no detectable Appl1 mRNA (data not shown). Antibodies against the amino terminus, midportion, and carboxyl terminus of Appl1 did not detect Appl1 protein of any length in mouse tissues from these two knock-out mouse models, indicating they are true Appl1-null models (supplemental Fig. S2C). These findings also suggest that the N′ Appl1 protein, if produced, is unstable, because it is not detectable. The levels of phosphorylated Akt and Erk varied in adult mice due possibly to individual biological rhythms (data not shown). This can be explained, because Akt signaling serves as a circadian output (26). Therefore, we isolated tissues from multiple embryos at E18.5 days from the same pregnant mouse. Surprisingly, the levels of phosphorylated Akt, as detected with phospho-specific antibodies against Akt Thr-308 and Ser-473, as well as phosphorylated Gsk3β and p70 S6k in the organs tested, including brain, heart, lung, and liver, did not any show consistent variation among fetuses with different Appl1 genotypes (Fig. 3, E and F, and supplemental Fig. 3S, A–C).
Appl1 Is Not Required for Cell Proliferation under Normal Culture Conditions
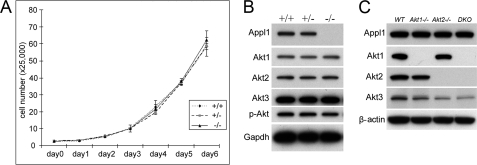
To investigate the effect of Appl1 loss on cellular function, wild-type, Appl1+/−, and Appl1−/− MEFs from E13.5-day embryos were isolated. MEFs were seeded in 6-well plates and collected every 24 h for 7 days. Proliferation of primary MEFs from Appl1+/− and Appl1−/− was similar to that of wild-type MEFs (Fig. 4A). Expression of Akt1, Akt2, and Akt3 proteins as well as phosphorylated Akt was unaltered in Appl1+/− and Appl1−/− MEFs compared with that in wild-type cells (Fig. 4B). In another experiment, we found that the half-life of Appl1 in MEFs from Akt1−/−, Akt2−/−, or Akt1/2 double knock-out mice did not differ from that of wild-type MEFs (Fig. 4C). Because Appl1 and Appl2 can bind to Akt in a similar manner (Fig. 2A), it is likely that the role of Appl1 is fully compensated by the presence of Appl2.
FIGURE 4.
Appl1 knock-out MEFs have normal cell proliferation rate and Akt signaling under normal culture conditions. A, cell proliferation in primary wild-type, Appl1+/−, and Appl1−/− MEFs cultured in DMEM with 10% FBS. Cells were collected daily and counted. B, immunoblot analysis of Akt1/2/3 and p-Akt in the same set of MEFs at passage 1. C, Appl1 protein levels in Akt1−/−, Akt2−/−, and Akt1/2 double knock-out (DKO) MEFs.
To further validate the redundant role played by Appl1 and Appl2, we performed a co-IP assay to identify additional Appl-binding proteins at the endogenous level. Because the available Appl2 antibody works better on human samples, we chose HEK293T cells as well as human ovarian cancer cell lines A2780 and IGR-OV1 for protein extraction. Although the APPL1/APPL2 protein ratio is different in cell lines from different tissues, APPL2, like APPL1, binds to AKT1, AKT2, and AKT3 (supplemental Fig. S4). Moreover, APPL1 and APPL2 bind weakly to GSK3β and TSC2, two major substrates of AKT. However, APPL1 and APPL2 did not associate with each other, nor did either bind to phosphorylated AKT. Also, no binding to mammalian target of rapamycin or ERK was detected.
Appl1 Loss Attenuates Akt Activation in a Growth Factor-specific Manner
To dissect the significance of the interaction between Appl1 and Akt, we performed growth factor stimulation studies. Wild-type and Appl1 knock-out MEFs were starved in DMEM with 0.05% FBS for 16 h and then incubated with fresh medium containing 10% FBS for 5–60 min. Western blot analysis was performed to detect p-Akt, Akt1, Akt2, p-Gsk3β, Gsk3β, and glyceraldehyde-3-phosphate dehydrogenase. Phospho-Akt (Ser-473 and Thr-308) levels at early time points were slightly lower in Appl1−/− MEFs. We also noted a slight difference in the phosphorylation level of the downstream target Gsk3β (supplemental Fig. S5A). We next tested for changes in Akt activation following growth factor stimulation. By incubating MEFs in medium under low serum conditions, Akt phosphorylation was suppressed within 90 min, but to our surprise returned to elevated levels after 3–16 h. Meanwhile, phosphorylation of p42/44 ERK remained suppressed (supplemental Fig. S5B). This unexpected reactivation of Akt may be due to a cell rescue response in MEFs, as complete serum deprivation triggers a stronger response that involves both Akt and Erk activation (supplemental Fig. S5C). Moreover, the Akt pathway inhibitor LY294002 and the ERK pathway inhibitor PD98059 accelerate serum starvation-induced apoptosis, as demonstrated by reduced cell viability, enhanced activation of caspase 9, 3, 6, and 7, as well as PARP cleavage (supplemental Fig. S5, D–F). No caspase 8 cleavage was detected, indicating that serum starvation causes cell death through an intrinsic apoptotic pathway.
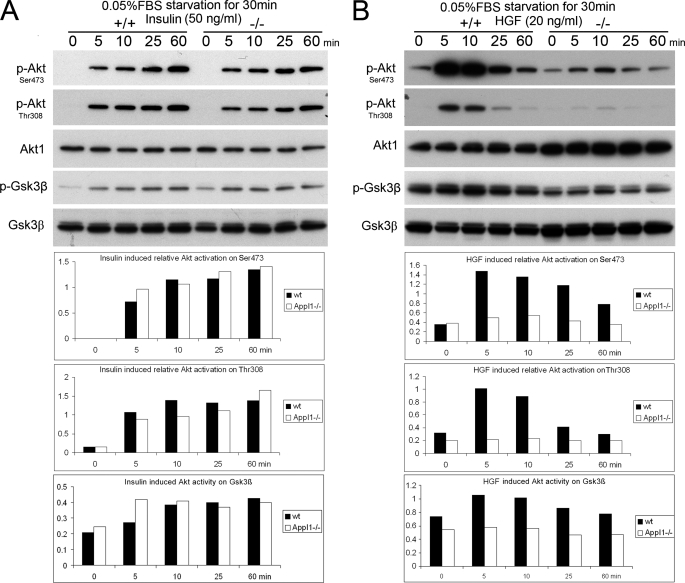
Therefore, we choose 30 min starvation in DMEM containing 0.05% FBS before growth factor addition. After starving the cells for 30 min, wild-type and Appl1−/− MEFs were treated with insulin (Fig. 5A), HGF (Fig. 5B), or EGF (supplemental Fig. S5G) at the indicated concentrations. Loss of Appl1 expression was found to cause a slight reduction in EGF-triggered Akt activation (phosphorylation of Thr-308 and Ser-473) and its activity on Gsk3β, but Appl1 loss did not appear to have an effect on insulin-induced Akt activation. Interestingly, Akt activation was apparently and consistently compromised in Appl1−/− cells upon stimulation with HGF, as was phosphorylation of downstream Gsk3β, Tsc2, and p70 S6K (Fig. 5B and data not shown).
FIGURE 5.
Loss of Appl1 has growth factor-dependent effects on Akt activation in MEFs. A, wild-type and Appl1-null MEFs were starved with DMEM containing 0.05% FBS for 30 min and then treated with 50 ng/ml of insulin, or B, 20 ng/ml of HGF for the indicated time. Expression of phosphorylated Akt (at Thr-308 and Ser-473) and total Akt as well as p-Gsk3β and total Gsk3β was determined by immunoblotting and quantified.
Appl1 Facilitates HGF-induced Akt Activation
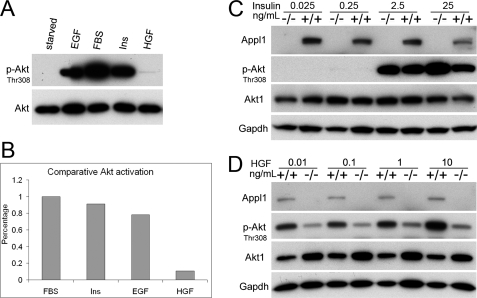
Because the p-Akt levels induced by 10% FBS and 20 ng/ml of EGF or 50 ng/ml of insulin was much higher than that induced by 20 ng/ml of HGF (Fig. 6, A and B), we questioned whether Appl1 plays a critical role in low level Akt activation. To address this possibility, we treated starved MEFs with EGF, insulin, and/or HGF at various concentrations. Medium to high concentrations of insulin (2.5 and 25 ng/ml) robustly triggered Akt activation, whereas low concentrations (25 and 250 pg/ml) of insulin did not (Fig. 6C). The same phenomenon was observed with EGF (supplemental Fig. S6), suggesting that insulin-like growth factor receptor/InsR and EGFR signaling require a higher threshold for activation and tend to trigger only strong activation of Akt. Intriguingly, HGF can induce weak Akt activation at a wide range of concentrations (10 pg/ml, 100 pg/ml, 1 ng/ml, and 10 ng/ml; Fig. 6D), suggesting that the HGF receptor, Met, has a low activating threshold in MEFs. Moreover, HGF-induced p-Akt levels were consistently weaker in Appl−/− MEFs than in wild-type MEFs. This effect was not caused by facilitating Met autophosphorylation; moreover, Appl1 does not bind to Met, nor does Appl1 deficiency cause reduced Met protein levels (data not shown). Our data implies that Appl1 facilitates activation of Akt induced by weak upstream signaling from Met. This effect may not be necessary or may be masked when strong upstream signals are transduced from receptors such as EGFR and insulin-like growth factor receptor/InsR, which trigger abundant activation of Akt.
FIGURE 6.
Appl1 facilitates low level activation of Akt upon HGF stimulation. A, MEFs were starved with medium containing 0.05% FBS for 30 min and then incubated for 10 min in medium containing 10% FBS, 50 ng/ml of insulin, 20 ng/ml of EGF, or 20 ng/ml of HGF to activate Akt. B, relative level of phosphorylated Akt was quantified. C and D, effect of various concentrations of different growth factors on the activation of Akt. Serum-starved MEFs were treated with insulin (D) (25 pg/ml, 250 pg/ml, 2.5 ng/ml, and 25 ng/ml, or HGF (E) (10 pg/ml, 100 pg/ml, 1 ng/ml, and 10 ng/ml).
Limited Amounts of Appl1 or Appl2 Proteins Are Bound to Akt
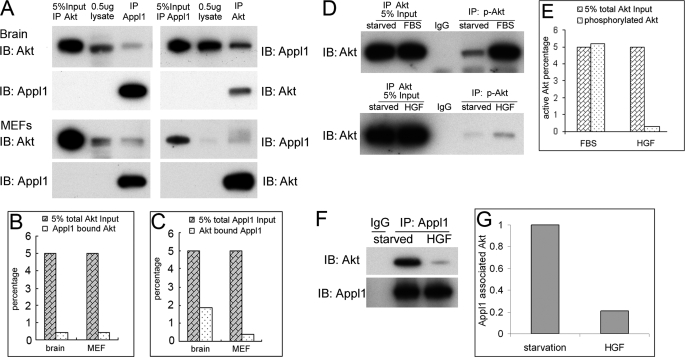
To shed further light on the growth factor-selective role of Appl1 in Akt activation, we next performed semi-quantitative co-IP between Appl1 and Akt in mouse brain and MEF cells (Fig. 7A). Not surprisingly, only trace amounts of Akt were associated with Appl1. In brain, only 0.4% of Akt was found to bind to Appl1, and 1.9% of Appl1 was bound to Akt (Fig. 7, B and C). Similarly, in MEFs, only 0.4% of Akt binds to Appl1, and only 0.4% of Appl1 binds to Akt. These findings appear to downplay the significance of the effect of Appl1 on Akt. In fact, upon stimulation with 10% FBS, ∼5% of total cellular Akt was activated, whereas only ∼0.3% of Akt was phosphorylated by 20 ng/ml of HGF after 10 min (Fig. 7, D and E). Interestingly, upon HGF stimulation, the Appl1-bound Akt amount was reduced, but not abolished. The binding-dissociating kinetics might enable Appl1 to bind to new native Akt (Fig. 7, F and G). Collectively, our data suggest that the limited amount of Appl1 and Akt that forms a complex may reside in certain cellular compartments that facilitate Akt activation only when the upstream signal is weak.
FIGURE 7.
Only low amounts of Akt bind to Appl1. A, co-IP was performed to determine the relative amount of Akt-Appl1 complex in wild-type brain and MEF cells, using a semiquantitative assessment. B, percentage of Appl1 bound to Akt in brain and MEF cells. C, percentage of Akt bound to Appl1. D, amount of Akt activation induced by 10% FBS or 20 ng/ml of HGF in MEFs 10 min after treatment, as determined by IP. E, percentage of Akt activation triggered by FBS or HGF in MEF cells. F and G, the amount of Appl1-bound Akt under serum starvation conditions or following stimulation with HGF for 10 min. IB, immunoblot.
Appl Proteins Are Not Required for MEF Survival under Normal Culture Conditions
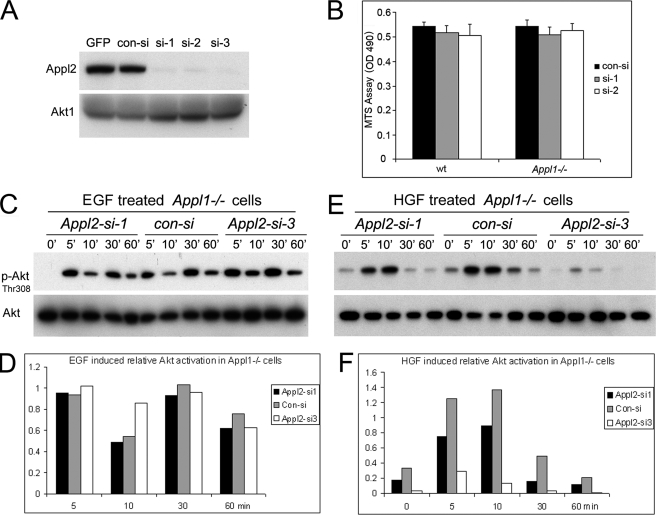
The possible redundancy between the two Appl proteins may account for the normal phenotype seen in Appl1−/− mice and the grossly normal signal transduction observed in Appl1-null cells. Because Appl2 knock-out mice are not yet available to test this notion in vivo, we used an RNA interference approach to knock down Appl2 in Appl1−/− MEFs. Three siRNA against mouse Appl2 were first tested on NIH 3T3 cells stably expressing TAP-tagged mouse Appl2, because the available Appl2 antibody does not recognize endogenous Appl2 in MEFs. All three oligos effectively depleted TAP-Appl2 protein as well as its mRNA (Fig. 8A and supplemental Fig. S7, A and B). These siRNAs were then nucleofected into primary wild-type and Appl1−/− MEFs and showed similar knockdown efficiency on endogenous Appl2 mRNA (supplemental Fig. S7C). Surprisingly, Appl1−/− MEFs in which Appl2 was knocked down proliferated normally, as shown by the MTS assay (Fig. 8B). Moreover, Appl2 knockdown did not affect cell cycle progression or bromodeoxyuridine incorporation of Appl1−/− cells (supplemental Fig. S7D and data not shown). The fact that Appl is dispensable for cell survival is understandable, however, given that the activation pattern of Akt is apparently unaltered in Appl1-null/Appl2 knockdown cells compared with that observed in control-transfected cells after EGF stimulation (Fig. 8, C and D). We next used HGF to stimulate these cells, questioning whether the HGF/Met signaling pathway would be more attenuated by Appl2 knockdown. As predicted, Appl1−/− MEFs in which Appl2 was knocked down with siRNA had weaker activation of Akt and downstream Tsc2 upon addition of HGF than did Appl1-null MEFs transfected with control siRNA (Fig. 8, E and F, and data not shown). Similarly, Appl2 knockdown also reduces HGF-induced Akt activation in wild-type MEFs (supplemental Fig. S7, E and F). Thus, although Appl1 and Appl2 are functionally redundant under certain circumstances, they are dispensable for cell survival under normal culture conditions.
FIGURE 8.
Appl2 knockdown fails to affect cell survival or diminish Akt activity in Appl1−/− MEFs under normal culture condition. A, stealth Appl2 siRNAs were transfected into NIH 3T3 cells stably expressing a TAP-tagged mouse Appl2 cDNA by Nucleofection. The knockdown effect 72 h after transfection was assessed by Western blotting using anti-Appl2 antibody. B, assessment of cell viability 72 h after transfection of wild-type and Appl1−/− MEFs with Appl2 siRNAs. Cell viability was evaluated by MTS assay. Error bars indicate S.D. C and D, Akt activity in EGF-stimulated Appl1−/− MEFs transfected with Appl2 siRNA. Cells were serum starved for 30 min and stimulated with 20 ng/ml of EGF, and then lysates were collected for immunoblotting. E and F, immunoblot analysis of Akt activity in HGF-stimulated Appl1−/− MEFs transfected with Appl2 siRNA.
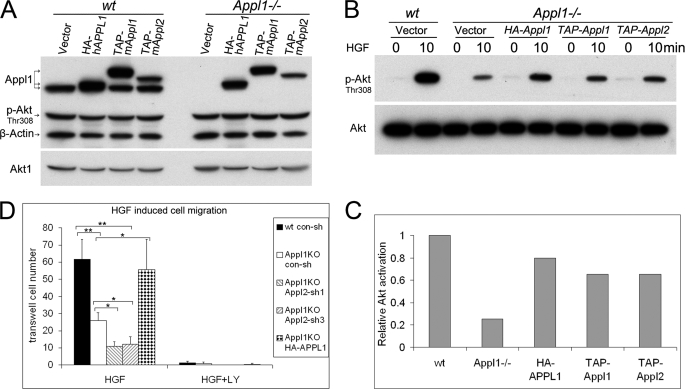
Appl1-null Cells Have a Defect in HGF-induced Cell Migration, Which Can Be Rescued by Human APPL1 cDNA
An HA-tagged human full-length APPL1 cDNA, a TAP-tagged mouse full-length Appl1 cDNA, and a TAP-tagged mouse full-length Appl2 cDNA were individually transduced into Appl1−/− MEFs through retroviral infection. The p-Akt level was not altered in wild-type cells overexpressing Appl1 or Appl2 under standard culture conditions (Fig. 9A). To determine whether re-expression of Appl1 can rescue the Appl1 phenotype upon HGF stimulation, wild-type, Appl1−/−, and the rescued Appl1−/− MEFs were serum starved for 30 min, followed by stimulation with 10 ng/ml of HGF. Activation of Akt was largely restored in Appl1−/− MEFs expressing either human APPL1 or mouse Appl1 (Fig. 9, B and C). Interestingly, overexpression of the TAP-tagged mouse Appl2 also restored phospho-Akt levels to some extent. These findings suggest that Appl1 and Appl2 have redundant roles under these conditions. To measure HGF (10 ng/ml)-induced MEF cell migration, a Boyden chamber assay was used. Appl1−/− cells showed reduced transwell migration, which was rescued by reintroduction of the human APPL1 cDNA. Moreover, Appl2 knockdown further decreased the mobility of Appl1−/− cells. Furthermore, HGF-triggered MEF migration was found to be Akt dependent in that LY294002 completely abolished transwell movement (Fig. 9D and supplemental Fig. S8). On the other hand, cell migration was only slightly affected in Appl1−/− cells stimulated with 10 ng/ml of EGF (supplemental Fig. S8).
FIGURE 9.
Appl1-null cells show defects in HGF-induced cell migration, which can be rescued by a human APPL1 cDNA expression plasmid. A, Appl1−/− MEFs were infected by a retrovirus harboring a HA-tagged human APPL1 cDNA, a TAP-tagged mouse Appl1 cDNA, or a TAP-tagged mouse Appl2 cDNA. Immunoblot analysis was conducted on cells grown under standard culture conditions, and a rabbit anti-Appl1 antibody was used for the detection of Appl1 as well as TAP. B, wild-type (wt), Appl1-null cells, and Appl1-null cells expressing exogenous Appl1/2 were starved for 30 min and then stimulated with 10 ng/ml of HGF, and then activation of Akt was evaluated by immunoblotting and quantified (C). D, HGF (10 ng/ml)-induced cell migration was measured by Boyden chamber assay 22 h after seeding (*, p < 0.01; **, p < 0.001). Error bars indicate S.D.
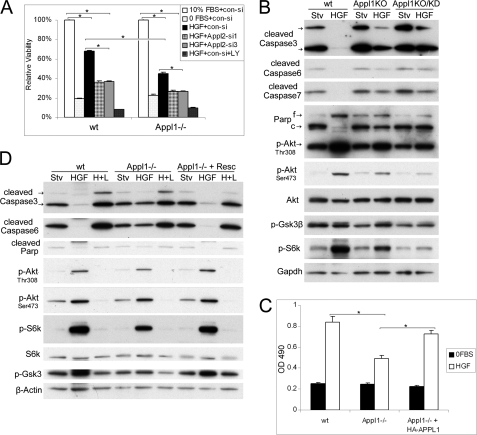
Appl Proteins Are Required for HGF-induced Cell Survival
Because HGF can activate Akt at low concentrations (Fig. 6D), we wanted to investigate the significance of the HGF-Appl-Akt link in cell survival under rigid conditions with sparse growth factors. Wild-type, Appl-null, and Appl1-null/Appl2 knockdown MEFs were incubated in serum-free medium or serum-free medium supplemented with 100 pg/ml of HGF. After incubating for 24 h, Appl1-null cells showed impaired survival. Moreover, Appl1-null/Appl2 knockdown cells showed a further decrease in their response to HGF (Fig. 10A).
FIGURE 10.
Appl proteins are required for HGF-induced cell survival. A, MTS assay was performed to measure viability of wild-type (wt), Appl1-null, and Appl1-null/Appl2 knockdown MEFs under serum starvation conditions, starvation followed by HGF (100 pg/ml) supplementation, or starvation with HGF and 5 μm LY294002 for 24 h (*, p < 0.01). B, apoptotic events triggered by serum starvation as well as Akt signaling were analyzed by immunoblotting. C, HGF-induced cell survival was measured in wild-type (wt), Appl1-null, and Appl1-null/HA-APPL1 cells 30 h after incubation. D, caspase activation and Akt signaling analyzed by Western blotting. Abbreviations: Stv., starvation; f and c, full-length and cleaved PARP; KD, knockdown of Appl2 with siRNA; H+L, HGF and LY294002.
Cell viability correlates with cellular apoptotic events. Three executioner caspases (3, 6, and 7) are actively cleaved during apoptosis, which is inversely correlated with the level of active Akt induced by HGF (Fig. 10B and supplemental Fig. S9A). On the other hand, the reintroduction of human APPL1 cDNA restored HGF-induced cell survival in Appl1−/− cells (Fig. 10C). These cells show increased Akt activity and attenuated apoptotic events such as caspase cleavage (Fig. 10D). The pro-survival effect of Appl is possibly through the Akt pathway, because inhibition of Akt by 5 μm LY294002 reversed the HGF-sustained cell survival in all cell types tested (Fig. 10, A and D).
To determine whether Appl1 itself participates in apoptotic pathways, wild-type and Appl1−/− cells were treated with UV to inflict DNA damage, tumor necrosis factor α to trigger an extrinsic apoptosis pathway, or tunicamycin to initiate ER stress-induced cell death. No difference was observed between the two cell types (supplemental Fig. S9B). Similarly, UV exposure induced similar levels of p53 activation, caspase 3 cleavage, and PARP cleavage in wild-type and Appl1-null cells (supplemental Fig. S9C).
DISCUSSION
The APPL adaptor protein family is comprised of two structurally homologous members, i.e. APPL1 and APPL2. Our data indicate that Appl1 expression is widely distributed in various mouse tissues, suggesting that this protein is involved in certain broadly relevant cellular events. Appl2 exhibits a similar tissue distribution to that of Appl1, suggesting that these related proteins may have at least partially redundant roles. We also found that Appl1 protein is subjected to multiple post-translational modifications, including phosphorylation and acetylation, in a tissue-specific manner, suggesting that fine tuning of the role of Appl1 exists in different types of cells.
APPL1 was first discovered as a binding partner of AKT2 (1). Such binding was speculated to expedite the membrane recruitment of AKT2 upon mitogenic stimulation. Subsequently, APPL was found to bind to Rab5 in the endosomal compartment and to bind to the nucleosome remodeling and histone deacetylase complex, NuRD-MeCP1. Such interactions are essential for cell proliferation, because knockdown of APPL1 or APPL2 by siRNA in HeLa cells caused DNA synthesis inhibition (6). The same research group also demonstrated a vital role for Appl in zebrafish. Knockdown of Appl1 or Appl2 by antisense morpholinos in zebrafish triggered massive apoptosis that resulted in embryonic lethality. This was thought to be due to blockage of Akt signaling to Gsk3 (21).
We had decided to use a conditional knock-out strategy to circumvent the possibility of embryonic lethality. However, we found that mice with ubiquitous loss of Appl1 expression were born at normal Mendelian ratios, and no obvious phenotypic abnormality in postnatal development was observed. It should be noted, however, that an effect caused by loss of a gene in cancer cells may be different from that observed in a mouse model. For example, AKT2 is amplified in pancreatic and ovarian cancer cells, and knock down of AKT2 inhibits cancer cell proliferation and tumor growth in vivo (27). However, Akt2 is dispensable for mouse organogenesis and postnatal growth in Akt2 knock-out mice, which instead show defects in glucose homeostasis (28). Interestingly, knock-out of Akt1 in the mouse results in small body size, and Akt3 knock-out mice have small brain size (23). This is because Akt1, Akt2, and Akt3 are partially redundant, and Akt1/2, Akt1/3, or Akt2/3 double knock-out mice have a more profound defect in embryonic development and postnatal survival (29). Likewise, because we have shown that APPL1 and APPL2 share the same binding partners, the in vivo role of Appl1 could be compensated for by the presence of Appl2. Moreover, whether APPL is a pro-survival or a pro-apoptotic factor in cancer cells is still controversial (2). In lower vertebrates such as zebrafish, Appl1 and Appl2 appear to have non-redundant roles, because knockdown of either of these genes results in apoptosis (21), which differs from our knock-out mouse model. Moreover, unlike in zebrafish, the level of phosphorylated Akt and Gsk3 was unaltered in multiple knock-out mouse tissues. These findings suggest that compensatory pathways may have evolved in higher mammals.
We have also confirmed the dispensable role of Appl1 for mouse development and reproduction in another Appl1 knock-out model we have recently generated, in which exon 1 of Appl1 is disrupted by a gene trap strategy.6 Although it is unclear why Appl1 deficiency does not affect phosphorylation of Akt in mouse tissues, Appl1 is required in a growth factor-selective manner in MEFs. Loss of Appl1 does not have an obvious effect on serum-, EGF-, and insulin-induced Akt activation in Appl1−/− MEFs. However, Appl1−/− cells do have compromised Akt activation upon HGF stimulation. Interestingly, Akt activation by HGF is ∼10-fold weaker than that triggered by serum, EGF, or insulin. On the other hand, Akt can be activated by a low concentration HGF, but not by low levels of EGF or insulin. Moreover, in MEFs, the amount of activated Akt (0.3%) induced by HGF approximates the amount of Appl1 bound to Akt (0.4%), suggesting that Appl1 may only facilitate Akt activation when the upstream signal is sparse. However, we recognize that MEF cells are not an ideal system to investigate the function of Appl1 in the Akt signaling pathway. Therefore, it will be necessary to revisit this signaling link in the future, once certain functionally differentiated cell types with high Appl1-Akt interaction are discovered.
The notion that Appl1 and Appl2 are functionally redundant is strengthened by the fact that these two proteins bind to the same partners, including all three Akt isoforms, phosphatidylinositol 3-kinase, Gsk3, and Tsc2. To date, more than 30 Akt substrates have been discovered that mediate the pleiotropic roles of Akt in cell survival, growth, proliferation, migration, angiogenesis, and metabolism (30). In future studies, it will be important to ascertain whether the three Akt isoforms can traverse to where different substrates reside through differentially post-translationally modified Appl. In addition, it will be interesting to determine whether Appl1 and Appl2 have different preferences with regard to different Akt substrates.
Moreover, we found that depletion of Appl2 could further attenuate the HGF-stimulated activation of Akt in Appl1 knock-out cells. Ectopic HGF signaling has been reported to transform NIH 3T3 cells and promote metastasis in cancer cells (31). Our data indicates that Appl1 can augment HGF-induced cell migration and survival. Intriguingly, a low concentration of HGF can prevent serum starvation-triggered cell death, suggesting that Appl1 is crucial in HGF-regulated cell survival under harsh conditions when major growth factors are not available. Notably, Appl1/2 loss/knockdown does not adversely affect cell survival and proliferation under normal culture conditions where growth factors are abundant. Nevertheless, at this time we cannot rule out the possibility of an effect on Akt or other signaling pathway activation that might occur in vivo in certain adverse environmental conditions, such as under stress or following stimulation. Because the interaction between Appl1 and Akt occurs in a tissue-specific manner, and Appl1 protein itself is subjected to tissue-specific modifications, in vivo investigations are needed to determine the role(s) of Appl1 in homeostasis maintenance.
Also, because of their redundant roles, we predict that Appl2 knock-out mice will have a normal phenotype. If so, it will be important to determine whether Appl1/2 double knock-out mice exhibit pronounced embryonic or postnatal defects.
Supplementary Material
Acknowledgments
We thank Dr. Antonio Di Cristofano (Albert Einstein College of Medicine, Bronx, NY) for advice regarding the targeting strategy; Dr. Gary Lyons (University of Wisconsin) for conducting in situ hybridization demonstrating ubiquitous expression of Appl1 mRNA in tissues from wild-type mice (not shown), and Dr. Emmanuelle Nicolas (Fox Chase Cancer Center) for performing real-time PCR. We also thank Erin Neumann-Domer (Fox Chase Cancer Center) for maintaining mouse colonies. The following Fox Chase Cancer Center shared facilities were used in the course of this work: Laboratory Animal, Transgenic Mouse, Cell Culture, and Genomic Facilities.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77429 (to J. R. T.) and CA06927 (to Fox Chase Cancer Center) from the NCI and an appropriation from the Commonwealth of Pennsylvania.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S9, and Table S1.
Y. Tan, H. You, and J. R. Testa, manuscript in preparation.
- APPL
- adaptor protein containing pleckstrin homology, phosphotyrosine binding, and leucine zipper motifs
- PH
- pleckstrin homology
- PTB
- phosphotyrosine binding
- MAPK
- mitogen-activated protein kinase
- OCRL
- Oculocerebrorenal Syndrome of Lowe
- MEF
- murine embryonic fibroblast
- ERK
- extracellular signal-regulated kinase
- Gsk3β
- glycogen synthase kinase 3β
- ES
- embryonic stem
- HGF
- hepatocyte growth factor
- MTS
- methanethiosulfonate
- siRNA
- small interfering RNA
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- EGF
- epidermal growth factor
- IP
- immunoprecipitation
- TAP
- tandem affinity purification
- HA
- hemagglutinin
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1.Mitsuuchi Y., Johnson S. W., Sonoda G., Tanno S., Golemis E. A., Testa J. R. (1999) Oncogene 18, 4891–4898 [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Yao F., Wu R., Morgan M., Thorburn A., Finley R. L., Jr., Chen Y. Q. (2002) J. Biol. Chem. 277, 26281–26285 [DOI] [PubMed] [Google Scholar]
- 3.Chial H. J., Wu R., Ustach C. V., McPhail L. C., Mobley W. C., Chen Y. Q. (2008) Traffic 9, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Mao X., Dong L. Q., Liu F., Tong L. (2007) Structure 15, 525–533 [DOI] [PubMed] [Google Scholar]
- 5.Habermann B. (2004) EMBO Rep. 5, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R. G., Zerial M. (2004) Cell 116, 445–456 [DOI] [PubMed] [Google Scholar]
- 7.Zhu G., Chen J., Liu J., Brunzelle J. S., Huang B., Wakeham N., Terzyan S., Li X., Rao Z., Li G., Zhang X. C. (2007) EMBO J. 26, 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varsano T., Dong M. Q., Niesman I., Gacula H., Lou X., Ma T., Testa J. R., Yates J. R., 3rd, Farquhar M. G. (2006) Mol. Cell. Biol. 26, 8942–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin D. C., Quevedo C., Brewer N. E., Bell A., Testa J. R., Grimes M. L., Miller F. D., Kaplan D. R. (2006) Mol. Cell. Biol. 26, 8928–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nechamen C. A., Thomas R. M., Dias J. A. (2007) Mol. Cell. Endocrinol. 260–262, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nechamen C. A., Thomas R. M., Cohen B. D., Acevedo G., Poulikakos P. I., Testa J. R., Dias J. A. (2004) Biol. Reprod. 71, 629–636 [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Lin H. K., Altuwaijri S., Xie S., Wang L., Chang C. (2003) J. Biol. Chem. 278, 16820–16827 [DOI] [PubMed] [Google Scholar]
- 13.Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ-Roberts C. Y., Hong J. Y., Kim R. Y., Liu F., Dong L. Q. (2006) Nat. Cell Biol. 8, 516–523 [DOI] [PubMed] [Google Scholar]
- 14.Staiger H., Machicao F., Machann J., Schick F., Kuulasmaa T., Laakso M., Fritsche A., Stefan N., Haring H. U. (2007) Diabetes Med. 24, 817–822 [DOI] [PubMed] [Google Scholar]
- 15.Chandrasekar B., Boylston W. H., Venkatachalam K., Webster N. J., Prabhu S. D., Valente A. J. (2008) J. Biol. Chem. 283, 24889–24898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M. H., Klein R. L., El-Shewy H. M., Luttrell D. K., Luttrell L. M. (2008) Biochemistry 47, 11682–11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng K. K., Lam K. S., Wang Y., Huang Y., Carling D., Wu D., Wong C., Xu A. (2007) Diabetes 56, 1387–1394 [DOI] [PubMed] [Google Scholar]
- 18.Saito T., Jones C. C., Huang S., Czech M. P., Pilch P. F. (2007) J. Biol. Chem. 282, 32280–32287 [DOI] [PubMed] [Google Scholar]
- 19.Erdmann K. S., Mao Y., McCrea H. J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., De Camilli P. (2007) Dev. Cell 13, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCrea H. J., Paradise S., Tomasini L., Addis M., Melis M. A., De Matteis M. A., De Camilli P. (2008) Biochem. Biophys. Res. Commun. 369, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schenck A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., Zerial M. (2008) Cell 133, 486–497 [DOI] [PubMed] [Google Scholar]
- 22.Tan Y., Wu C., De Veyra T., Greer P. A. (2006) J. Biol. Chem. 281, 17689–17698 [DOI] [PubMed] [Google Scholar]
- 23.Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., Hay N. (2001) Genes Dev. 15, 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dooher J. E., Lingappa J. R. (2004) J. Virol. 78, 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox D. M., Du M., Guo X., Siu K. W., McDermott J. C. (2002) BioTechniques 33, 267–268, 270 [DOI] [PubMed] [Google Scholar]
- 26.Ko M. L., Jian K., Shi L., Ko G. Y. (2009) J. Neurochem. 108, 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng J. Q., Ruggeri B., Klein W. M., Sonoda G., Altomare D. A., Watson D. K., Testa J. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3636–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B., 3rd, Kaestner K. H., Bartolomei M. S., Shulman G. I., Birnbaum M. J. (2001) Science 292, 1728–1731 [DOI] [PubMed] [Google Scholar]
- 29.Dummler B., Tschopp O., Hynx D., Yang Z. Z., Dirnhofer S., Hemmings B. A. (2006) Mol. Cell. Biol. 26, 8042–8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D., Dan H. C., Park S., Yang L., Liu Q., Kaneko S., Ning J., He L., Yang H., Sun M., Nicosia S. V., Cheng J. Q. (2005) Front. Biosci. 10, 975–987 [DOI] [PubMed] [Google Scholar]
- 31.Rong S., Segal S., Anver M., Resau J. H., Vande Woude G. F. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4731–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.