Abstract
Satellite cells/myoblasts account for the majority of muscle regenerative potential in response to injury and muscular adaptation to exercise. Although the ability to influence this process would provide valuable benefits for treating a variety of patients suffering from muscle loss, the regulatory mechanisms of myogenesis are not completely understood. We have tested the hypothesis that transforming growth factor-β-activated kinase 1 (TAK1) is an important regulator of skeletal muscle formation. TAK1 is expressed in proliferating C2C12 myoblasts, and its levels are reduced upon differentiation of myoblasts into myotubes. In vivo, TAK1 is predominantly expressed in developing skeletal muscle of young mice. However, the expression of TAK1 was significantly up-regulated in regenerating skeletal muscle of adult mice. Overexpression of a dominant negative mutant of TAK1 or knockdown of TAK1 inhibited the proliferation and differentiation of C2C12 myoblasts. TAK1 was required for the expression of myogenic regulatory factors in differentiating myoblasts. Genetic ablation of TAK1 also inhibited the MyoD-driven transformation of mouse embryonic fibroblasts into myotubes. Inhibition of TAK1 suppressed the differentiation-associated activation of p38 mitogen-activated protein kinase (MAPK) and Akt kinase. Overexpression of a constitutively active mutant of MAPK kinase 6 (MKK6, an upstream activator of p38 MAPK) but not constitutive active Akt restored the myogenic differentiation in TAK1-deficient mouse embryonic fibroblasts. Insulin growth factor 1-induced myogenic differentiation was also found to involve TAK1. Collectively, our results suggest that TAK1 is an important upstream regulator of skeletal muscle cell differentiation.
Keywords: Cell/Differentiation, Development Differentiation/Muscle, Signal Transduction/Protein Kinases/MAP, Tissue/Organ Systems/Muscle/Skeletal, Insulin-like Growth Factor (IGF), Akt, Myogenesis, TAK1, p38 MAPK
Introduction
Skeletal myogenesis is a complex cascade of events that involves the specification and differentiation of muscle precursor cells or myoblasts, their fusion to form primary and secondary myotubes, and their subsequent maturation into myofibers (1). This process is required not only for the development of skeletal muscle but also for postnatal growth and the regeneration of myofibers after injury (1, 2). Myogenesis is regulated by the sequential expression of myogenic regulatory factors (MRFs),2 a group of basic helix-loop-helix transcription factors that includes Myf-5, MyoD, myogenin, and MRF4 (3, 4). The commitment of myoblasts to differentiate into myotubes is influenced by a number of autocrine or paracrine factors. Insulin-like growth factor (IGF) and low serum conditions promote myogenic differentiation (5). Conversely, the presence of fetal bovine serum, fibroblast growth factors, transforming growth factor-β (TGF-β), myostatin, and proinflammatory cytokines block the process of differentiation of myoblasts into myotubes (6–9).
Myogenic differentiation requires the coordinated actions of multiple signaling pathways that regulate cell cycle withdrawal and specify myogenesis (i.e. activates MRFs). Several growth factors and hormones bind to their respective cell surface receptors and, thus, activate various intracellular signaling pathways leading to either proliferation or differentiation of myoblasts. Activation of Ras-Raf-MEK-extracellular signal-regulated kinase (ERK) and nuclear factor κB (NF-κB) pathways generally stimulate cell proliferation (10–13) and coordinately inhibit differentiation, in part by inactivation of MyoD (14, 15). In contrast, the activation of p38 mitogen-activated protein kinase (MAPK) pathway stimulates myogenesis (16). The activity of p38 MAPK is increased during myogenesis, and its inhibition blocks the expression of select muscle-specific genes and formation of multinucleated myotubes (17–23). During myogenesis, the activation of p38 MAPK promotes cell cycle exit by inducing the expression of a cyclin-dependent kinase inhibitor, p21, which facilitates terminal differentiation of muscle precursor cells (16, 20, 24). The p38 MAPK-mediated phosphorylation also plays a critical role in chromatin remodeling and the activation of key myogenic transcription factors during myogenesis (16). Several proteins including the BAF60 subunit of the SWI/SNF chromatin remodeling complex, RNA decay-promoting factor KH-type splicing regulatory protein, and different isoforms of myocyte enhancer factor 2 are the known phosphorylation targets of p38 MAPK during myogenesis (25–27).
In addition to p38 MAPK, the activation of phosphoinositide 3-kinase/Akt kinase is also increased during myogenesis. Inhibition or activation of phosphoinositide 3-kinase/Akt demonstrated that whereas it is not needed for early stages of myogenesis (elongation and alignment), its activation is required for myoblast fusion and terminal differentiation (28–30). The phosphoinositide 3-kinase/Akt is also essential for the myogenic actions of IGFs, which acts both upstream and downstream of myogenin (10, 31, 32). Akt kinase is a downstream target of phosphoinositide 3-kinase, the activation of which increases during myogenesis (33). The importance of functional Akt in myogenesis has been demonstrated using a dominant negative Akt mutant that inhibited myotube formation (28). However, the upstream signaling mechanisms leading to the activation of p38 MAPK and/or Akt during myogenesis remain unknown.
TGF-β-activated kinase 1 (TAK1), a member of the MEK kinase (MAP3K) family, was originally identified as a key regulator of TGF-β-induced activation of MAPK (34). However, several recent studies have shown that TAK1 is also an important component of several cell signaling pathways leading to the activation of NF-κB and activator protein-1 in response to diverse cytokines, microbial products, and cellular stress (35–39). Activated TAK1 complex, which also contains TAB1 and TAB2 proteins, has been shown to phosphorylate MAPK kinase 6 (MKK6), the upstream activator of p38 MAPK (40–42). TAK1 knock-out mice were embryonically lethal, suggesting that TAK1 plays a critical role during development (35). Indeed, several recent studies using tissue-specific knock-out mice have demonstrated that TAK1 has important functions in innate and adaptive immune responses (43–45), vasculature development (46), differentiation and prevention of apoptosis in keratinocytes (38, 47), survival of hematopoietic cells in bone marrow and hepatocytes in liver (48), and morphogenesis, growth, and maintenance of cartilage (49). However, the role of TAK1 in skeletal muscle progenitor cell survival, proliferation, and their differentiation into myofibers has not been yet determined.
In this study we have investigated the role and the mechanisms by which TAK1 regulates myogenic differentiation. Our study demonstrates that TAK1 is essential for the proliferation and differentiation but not survival of myoblasts. Furthermore, our results suggest that TAK1 is an upstream activator of p38 MAPK during myogenic differentiation, and it is also required for myogenic actions of IGF-I.
EXPERIMENTAL PROCEDURES
Materials
Dulbecco's modified Eagle's medium, fetal bovine serum, and Lipofectamine 2000 transfection reagent were obtained from Invitrogen. Protease inhibitor mixture and horse serum were from Sigma. Effectene transfection reagent was obtained from Qiagen (Valencia, CA). Antibody against phospho-p38 (Thr180/Tyr182) was purchased from Millipore. Antibodies against phospho-IκBα (Ser32), phospho-Akt (Ser473), phospho-TAK1 (Ser412), phospho-AMPK (Thr172), and total TAK1, Akt, p38, AMP-activated protein kinase (AMPK), TAB1, and TAB2 were obtained from Cell Signaling Technology (Beverly, MA). MyoD and myogenin antibodies were purchased from BD Pharmingen. MF-20 antibody was obtained from the Developmental Studies Hybridoma Bank, University of Iowa. Cardiotoxin was purchased from Calbiochem. A Creatine kinase (CK) activity assay kit was obtained from Stanbio Laboratory (Boerne, TX). An Annexin V-EGFP apoptosis detection kit was purchased from BioVision (Mountain View, CA).
Animals
Wild-type (strain: C57BL10ScSn and C57BL6) and mdx (strain: C57BL10ScSn DMDmdx) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were housed in the animal facility of the University of Louisville School of Medicine under conventional conditions with constant temperature and humidity and fed a standard diet. For muscle regeneration studies, animals were anesthetized, and a single dose of cardiotoxin (100 μl of a 10 μm stock in 0.9% saline solution) was injected intramuscularly into the right tibial anterior (TA) muscle of 3-month-old C57BL6 mice. As a control, the contralateral TA muscle was injected with sterile 0.9% saline in the same animal. After injection, animals were returned to their cages and allowed food and water ad libitum. Five days after cardiotoxin injection, the TA muscles were isolated and examined for the expression of TAK1. All experiments with animals were approved by the Institutional Animal Care and Use Committee of the University of Louisville.
Cell Culture
C2C12 (a mouse myoblastic cell line) were obtained from the American Type Culture Collection (Rockville, MD). 293T, a human embryonic kidney cell line, was purchased from Stratagene (La Jolla, CA). TAK1+/+ and TAK1−/− mouse embryonic fibroblasts (MEFs) were kindly provided by Prof. S. Akira (Osaka University, Osaka, Japan). The cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Differentiation in C2C12 cultures was induced by replacing the growth medium with differentiation medium (2% horse serum in Dulbecco's modified Eagle's medium) as described (50, 51).
Plasmid Constructs
Plasmid constructs encoding dominant negative (dn) mutant (HA-TAK1 K63W) and TAB1 cDNA were kind gifts from Dr. J. Ninomiya-Tsuji (North Carolina State University, Raleigh, NC). Construction of pcDNA3-MyoD plasmid has been described previously (51). Plasmid encoding constitutively active MKK6 (pcDNA3-caMKK6), muscle creatine kinase (MCK)-luciferase (pMCK-Luc), and pBABE-puro were purchased from Addgene (Cambridge, MA). pUSEamp plasmid encoding Myc-tagged constitutively active Akt cDNA was purchased from Upstate Biotechnology (Lake Placid, NY). Skeletal α-actin-luciferase (pSK-Luc) plasmid has been previously described (50, 51).
Short Hairpin RNA (shRNA)
Validated plasmids encoding shRNA for murine TAK1 (accession no. NM_172688) and negative control were purchased from SABiosciences (Frederick, MD). Two target sequences of the TAK1 used were TGA GAG GAA GGC TTT CAT TGT and CTT GGA TGG CGC CTG AAG TAT. Control shRNA plasmid contained the sequence GGA ATC TCA TTC GAT GCA TAC. All shRNA plasmids were amplified by transformation in Escherichia coli and purified using the endotoxin-free maxi plasmid kit (Qiagen).
Stable Transfection
The plasmids were introduced in C2C12 myoblasts using Effectene or Lipofectamine 2000 transfection reagents. Because shRNA plasmids contained a puromycin-resistant gene, the transfected cells were selected in the presence of 1.6 μg/ml puromycin (Sigma) for at least 72 h. The knockdown of TAK1 was confirmed by performing a Western blot using TAK1 antibody. For achieving stable transfection of dnTAK1 plasmid, C2C12 myoblasts were also transfected with pBabe-puro plasmid in a 1:10 ratio followed by their selection in growth medium containing puromycin (1.6 μg/ml).
Evaluation of Apoptosis
Annexin V specifically binds to phosphatidylserine, a plasma membrane lipid that rapidly relocalizes from the inner leaflet to the outer leaflet in cells that are undergoing apoptosis. C2C12 myoblasts stably transfected with control shRNA or TAK1 shRNA in six-well tissue culture plates were harvested using cold phosphate-buffered saline (PBS) and washed twice in PBS. Double staining with annexin V-EGFP and propidium iodide was carried out using the annexin V-EGFP apoptosis kit according to the manufacturer's recommendations (Biovision Inc., Mountain View, CA) and then analyzed by fluorescence-activated cell sorting. The percentage cells positive for EGFP-annexin V in each culture was measured.
Transient Transfection and Reporter Gene Activity
Cells plated in 24-well tissue culture plates were transfected with different plasmids using Effectene transfection reagent according to the protocol suggested by the manufacturer (Qiagen). Transfection efficiency was controlled by cotransfection of myoblasts with Renilla luciferase encoding plasmid pRL-TK (Promega). Specimens were processed for luciferase expression using a Dual luciferase assay system with reporter lysis buffer per the manufacturer's instructions (Promega). Luciferase measurements were made using a luminometer (Berthold Detection Systems).
Construction and Use of Dominant Negative TAK1 Adenovirus
Adenoviral vector encoding hemagglutinin-tagged dn mutant TAK1 (HA-TAK1 K63W) cDNA was constructed using a method as described (52). Briefly, the TAK1K63W cDNA was isolated from pCMV-HA-TAK1K63W and cloned into pAdTrack-CMV vector. The positive clones were linearized by the restriction endonuclease PmeI and cotransformed with the supercoiled adenoviral vector AdEasy-1 into E. coli strain BJ5183 (Stratagene). Recombinant adenoviral constructs were selected and digested with restriction PacI and finally transfected into packaging cell line 293T. Production of adenovirus in 293T cells was observed after 6–7 days and was monitored by expression of green fluorescence protein in viral plaques. The cells were collected 7–8 days after transfection; the adenoviruses were released by two freeze-thaw cycles and amplified by infecting 293T cells in one 100-mm tissue culture plate. After 3 days the adenoviruses were harvested and further amplified by infecting 293T cells. The amplified adenoviruses were harvested 3 days later, purified by centrifugation in CsCl, and stored at −80 °C in storage buffer (5 mm Tris-Cl (pH 8.0), 50 mm NaCl, 0.05% bovine serum albumin, and 25% glycerol). The titer of the virus was determined by infecting 293T cells with serial dilutions of adenoviruses and monitoring the viral plaques for expression of green fluorescence protein. Construction of adenoviral vector encoding MyoD has been previously described (51). All adenoviruses were used at a multiplicity of infection 50 particles/cell.
Western Blot
A Western blot was performed following a standard method as described (50, 51). All antibodies were used at a dilution of 1:1000. The band intensities on immunoblots were quantified using ImageQuant TL software (GE HealthCare).
RNA Isolation and Real-time PCR
Isolation of total RNA from skeletal muscle tissues or C2C12 cells and real-time PCR were done following a protocol as previously described (50, 51, 53). The primers were designed using Vector NTi XI software (Invitrogen) and purchased from Integrated DNA Technologies (San Diego, CA). The sequences of the primers were as follows: TAK1, 5′-GTC ATC CAG CCC TAG TGT CAG AAT-3′ (forward) and 5′-TTC TTT GGA GTT TGG GCA CG-3′ (reverse); myocyte enhancer factor 2D, 5′-ACA AAG TCA TCC CTG CCA AGT CTC-3′ (forward) and 5′-CGC TGG GCA TTG TTC AAA TG-3′ (reverse); MyoD, 5′-TGG GAT ATG GAG CTT CTA TCG C-3′ (forward) and 5′-GGT GAG TCG AAA CAC GGA TCA T-3′ (reverse); myogenin, 5′-CAT CCA GTA CAT TGA GCG CCT A-3′ (forward) and 5′-GAG CAA ATG ATC TCC TGG GTT G-3′ (reverse); glyceraldehyde-3-phosphate dehydrogenase, 5′-ATG ACA ATG AAT ACG GCT ACA GCA A-3′ (forward) and 5′-GCA GCG AAC TTT ATT GAT GGT ATT-3′ (reverse).
CK Assay
CK activity in cell extracts was measured using a CK activity assay kit as described (50, 51).
Immunofluorescence
Immunofluorescence was performed as previously described (50, 51). In brief, cells grown in 24-well plates were fixed with 3.7% paraformaldehyde followed by permeabilization with 0.1% Triton X-100. After three washings with PBS, the cells were blocked with 1% bovine serum albumin in PBS for 1 h and then incubated with MF-20 antibody (specific for myosin heavy chain fast (MyHCf) type protein) at a 1:100 dilution in PBS for 2 h. The cells were washed again with PBS, incubated with goat anti-mouse IgG-Alexa 546 (2 μg/ml) for 1 h, and counterstained with 4′,6-diamidino-2-phenylindole for 2 min. Stained cells were analyzed under a fluorescence microscope (Nikon). Pictures were captured using a digital camera and software. The images stained with MyHCf and 4′,6-diamidino-2-phenylindole were finally merged using Nikon Element software.
Proliferation Assay
C2C12 myoblasts were plated in 96-well plate and transfected with either TAK1 shRNA or dominant negative TAK1 plasmid vector. After 48 h of transfection, the proliferation of myoblasts was evaluated using BrdUrd dye uptake method following a protocol suggested by manufacturer (Roche Applied Science). Briefly, 20 μl of BrdUrd labeling solution was added to each well for 3 h. The microtiter plates were then centrifuged, cells were dried with a hair dryer and fixed, and the DNA was denatured to make the incorporated BrdUrd more accessible for detection by the antibody. The monoclonal anti-BrdUrd peroxidase-conjugated antibody was then added to the cultures and incubated for 2 h at room temperature. After three washing steps, the bound peroxidase was detected by addition of its substrate and quantified by measuring absorbance at a wavelength of 450 nm.
Statistical Analysis
Results are expressed as the mean ± S.D. Student's t test was used to compare quantitative data. A value of p < 0.05 was considered statistically significant unless otherwise specified.
RESULTS
Using C2C12 myoblasts and mouse embryonic fibroblasts (MEF) deficient in functional TAK1 protein, here we have investigated the role of TAK1 in myogenesis.
TAK1 Is Expressed in Proliferating Myoblasts and Skeletal Muscle of Young Mice
TAK1 is a cytoplasmic protein that forms a complex with TAB1 and TAB2 proteins for its activation (35). Before examining the role of TAK1 in skeletal muscle proliferation and differentiation, we first monitored the expression levels of TAK1, phospho-TAK1 (pTAK1), TAB1, and TAB2 protein in C2C12 myoblasts after incubation in differentiation medium (DM). C2C12 myoblasts expressed high levels of TAK1, pTAK1, TAB1, and TAB2 proteins. However, the levels of TAK1 and pTAK1 in C2C12 cultures were reduced to 50.6 ± 3.5 and 43.2 ± 4.5%, respectively, after 96 h of incubation in DM (Fig. 1A). Similarly, the protein levels of TAB1 and TAB2 were reduced to 41.6 ± 4.6 and 33.2 ± 5.6%, respectively, after 96 h of induction of differentiation in C2C12 cultures. The reduction in the protein levels of TAK1, TAB1, or TAB2 coincided with the appearance of MyHCf, a biochemical marker for muscle differentiation (50), in C2C12 cultures (Fig. 1A).
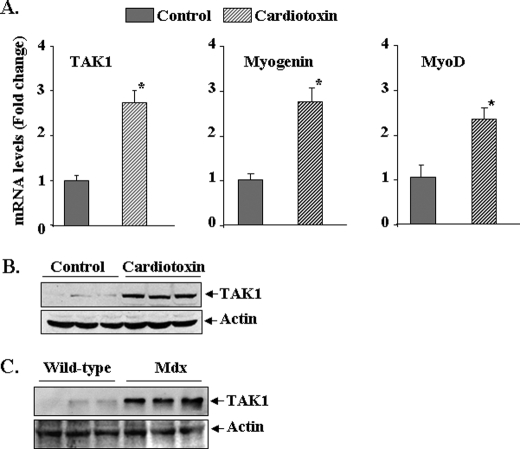
FIGURE 1.
Expression of TAK1 in cultured myoblasts and the skeletal muscle of mice. A, C2C12 myoblasts were incubated in DM (2% horse serum in Dulbecco's modified Eagle's medium) for the indicated time periods, and the levels of TAK1, phospho-TAK1, TAB1, TAB2, and MyHCf were measured by Western blot. Representative immunoblots and quantification from five independent experiments presented here show that TAK1, phosphorylated TAK1, TAB1, and TAB2 are highly expressed in C2C12 myoblasts, and their levels are reduced upon differentiation of myoblasts into myotubes. B, shown are protein levels of TAK1, phospho-TAK1, TAB1, and TAB2 in gastrocnemius muscle of mice of different ages. n = 4 for each age group.
We also measured the protein levels of TAK1, TAB1, and TAB2 in the skeletal muscle at different stages of mouse development. Interestingly, 1- or 2-week-old mice expressed high levels of total TAK1, pTAK1, TAB1, and TAB2 in gastrocnemius muscle. However, the expression of these proteins was diminished in adult mice (i.e. 5 and 8 weeks). The protein levels of TAK1, pTAK1, TAB1, TAB2 in 8-week-old mice were 35.6 ± 4.1, 22.3 ± 2.6, 38.7 ± 5.6, and 59.7 ± 5.3%, respectively, compared with those of 1-week-old mice. These results suggest that TAK1 complex may be important for the development of skeletal muscle in young animals (Fig. 1B).
TAK1 Expression Is Increased in Regenerating Skeletal Muscle of Adult Mice
To further understand the role of TAK1 in skeletal muscle formation, we monitored its expression in regenerating skeletal muscle. The tibial anterior (TA) muscle of 3-month-old C57BL6 mice was given intramuscular injections of saline alone or cardiotoxin, a well established model of skeletal muscle injury and regeneration (17). After 5 days, the TA muscles were isolated, and the mRNA levels of TAK1 and muscle regeneration markers myogenin and MyoD were determined by quantitative real-time-PCR. Interestingly, mRNA levels of TAK1 were significantly increased in regenerating TA muscle compared with contralateral control muscle (Fig. 2A). Similarly, protein levels of TAK1 were also found to be significantly increased in regenerating muscle fibers after injury (Fig. 2B).
FIGURE 2.
Expression of TAK1 in regenerating TA muscle in vivo. A, TA muscle of 3-month-old C57BL6 mice was injected with saline alone or cardiotoxin as described under “Experimental Procedures.” After 5 days the TA muscle was isolated and processed for RNA isolation and measurement of mRNA levels for TAK1, myogenin, and MyoD by real-time-PCR. Data presented here show that the mRNA levels of TAK1 as well as myogenin and MyoD are significantly increased in cardiotoxin-injected regenerating TA muscle compared with contralateral saline-injected TA muscle (n = 3). *, p < 0.01, value significantly different from controls. B, representative immunoblots from two independent experiments presented here show that the protein levels of TAK1 are significantly increased in TA muscle 5 days after cardiotoxin injection. C, shown are protein levels of TAK1 in gastrocnemius muscle of 8-week-old wild-type and mdx mice measured by Western blot. The levels of TAK1 are noticeably higher in mdx mice compared with wild-type mice. There was no difference in the levels of unrelated protein actin.
Skeletal muscle degeneration and regeneration is a common feature in mdx (a mouse model of Duchenne muscular dystrophy) mice (54). By performing Western blots, we measured the protein levels of TAK1 in the skeletal muscle of 8-week-old control and mdx mice. Interestingly, the level of TAK1 protein was again found to be increased in gastrocnemius muscle of mdx mice compared with age-matched control mice (Fig. 2C). These data further support the inference that TAK1 may play an important role in the proliferation and/or differentiation of myogenic cells.
TAK1 Is Required for the Proliferation but Not Survival of Myoblasts
Because the expression of TAK1 was high in proliferating C2C12 myoblasts and in regenerating muscle of mice, we next investigated whether TAK1 is important for C2C12 myoblast proliferation. C2C12 myoblasts were stably transfected with plasmid vector expressing either TAK1 shRNA or dominant negative TAK1 (dnTAK1), and proliferation was measured using the BrdUrd dye uptake method. As shown in Fig. 3A, stable transfection of C2C12 myoblasts with TAK1 shRNA reduced the levels of TAK1 protein and significantly inhibited their proliferation. Similarly, stable transfection with dnTAK1 plasmid also significantly reduced the proliferation of C2C12 myoblasts (Fig. 3B).
FIGURE 3.
Role of TAK1 in proliferation of C2C12 myoblasts. A, C2C12 myoblasts (10,000 cells/well) were plated in 24-well plate and transfected with vector alone or TAK1 shRNA plasmid. Cells were selected in the presence of puromycin (1.5 μg/ml), and the proliferation of myoblasts was measured using the BrdUrd uptake method as described under “Experimental Procedures.” Data presented here show that transfection of C2C12 myoblasts with TAK1 shRNA significantly reduced its protein levels and inhibited proliferation. *, p < 0.01, values significantly different from control shRNA transfected cells. B, stable transfection of C2C12 myoblasts with a dominant negative TAK1 (dnTAK1) plasmid also inhibited the proliferation of C2C12 myoblasts. #, p < 0.01, values significantly different from control shRNA transfected myoblasts. C, knockdown of TAK1 using RNAi technique did not affect the percentage of apoptotic cells in C2C12 myoblasts assayed using the annexin V-EGFP apoptosis detection kit and fluorescence-activated cell sorting method.
Published reports suggest that TAK1 is important for the survival of a number of cell types including keratinocytes, hepatocytes, and hematopoietic cells (38, 47, 48). We next sought to determine whether TAK1 is required for the survival of C2C12 myoblasts. We did not find any significant difference in the number of apoptotic cells between control and TAK1 shRNA-transfected myoblasts assessed using the annexin V-EGFP apoptosis kit and fluorescence-activated cell sorting analysis (Fig. 3C). Furthermore, there was no significant difference in cellular viability between control and TAK1 shRNA-transfected C2C12 myoblasts 24 h after incubation in differentiation medium (data not shown), indicating that TAK1 may be important for the proliferation of myoblasts but not for their survival.
TAK1 Is Required for the Differentiation of C2C12 Myoblasts into Myotubes
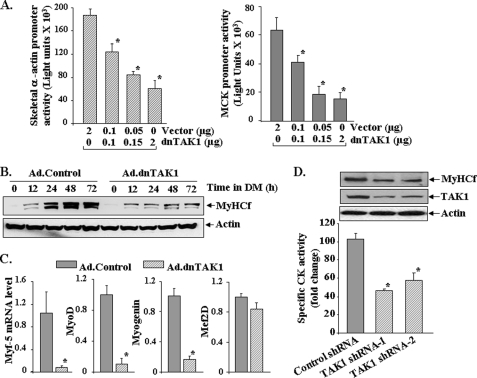
We next investigated whether TAK1 has a role in myogenic differentiation. C2C12 myoblasts were transfected with increasing amounts of a dominant negative TAK1 (dnTAK1) plasmid along with skeletal α-actin (pSK-Luc) or muscle creatine kinase (pMCK-Luc) luciferase reporter plasmid in a 1:10 ratio. The medium of the cells was replaced by differentiation medium, and the cells were incubated for an additional 72 h followed by measurement of luciferase activity in cell extracts. As shown in Fig. 4A, transfection of C2C12 myoblasts with dnTAK1 plasmid inhibited the activation of both skeletal α-actin and muscle creatine kinase promoters in a dose-dependent manner. We also studied the effect of ectopic expression of dnTAK1 using adenoviral vector in the differentiation of C2C12 myoblasts. Transduction with Ad.dnTAK1 significantly inhibited the expression of MyHCf in C2C12 myoblasts upon incubation in differentiation medium (Fig. 4B). Because myogenic differentiation is regulated by sequential expression of MRFs, we also investigated the effects of overexpression of dominant negative TAK1 on the expression levels of various MRFs. As shown in Fig. 4C, overexpression of dnTAK1 drastically reduced the mRNA levels of Myf-5, MyoD, and myogenin but not myocyte enhancer factor 2D in C2C12 cultures measured 72 h after the induction of differentiation. Furthermore, knockdown of TAK1 using an RNAi technique also reduced the expression of muscle differentiation markers CK and MyHCf in C2C12 cultures (Fig. 4D). Collectively, these results suggest that functional TAK1 is required for the differentiation of C2C12 myoblasts.
FIGURE 4.
Involvement of TAK1 in differentiation of C2C12 myoblasts. A, C2C12 myoblasts were transiently transfected with increasing amounts of dominant negative TAK1 (dnTAK1) plasmid along with either pSK-Luc or pMCK-Luc plasmid in a 1:10 ratio. After 24 h the cells were incubated in DM, and the luciferase activity in cell extracts was measured. Representative data from two independent experiments (each done in triplicate) presented here show that dnTAK1 inhibits the activation of both skeletal α actin and muscle creatine kinase promoters in a dose-dependent manner. *, p < 0.05, values significantly different from corresponding C2C12 cultures transfected with vector only. B, C2C12 myoblasts were transduced (multiplicity of infection 1:50) with control (Ad.Control) or dominant negative TAK1 (Ad.TAK1) adenoviral vectors for 24 h. The cells were then incubated in DM for indicated time intervals, and the expression of MyHCf was measured by Western blot. Representative immunoblots presented here show that dnTAK1 inhibits the expression of MyHCf without affecting the levels of an unrelated protein actin in C2C12 cultures. C, -fold difference is shown in the mRNA levels of Myf-5, MyoD, myogenin, and myocyte enhancer factor 2D (Mef2D) in Ad.control and Ad.dnTAK1-transduced C2C12 myoblasts 72 h after incubation in DM measured by real-time PCR technique. *, p < 0.01, values significantly different from C2C12 myoblasts transduced with Ad.control vector. D, C2C12 myoblasts were transfected with control or either of the two TAK1 shRNA plasmids, each containing a different target sequence for TAK1 knockdown. The cells were selected in the presence of puromycin (1.6 μg/ml) for 72–96 h followed by incubation in differentiation medium for 72h. CK activity in cell extracts was measured using the CK activity assay kit. The levels of MyHCf and TAK1 were measured by Western blot. Data presented here show that knockdown of TAK1 inhibits the expression of CK and MyHCf in C2C12 cultures. *, p < 0.01, values significantly different from control shRNA transfected C2C12 myoblasts.
Inactivation of TAK1 Inhibits the MyoD-induced Differentiation of MEF into Myotubes
MyoD is an important muscle-specific transcription factor that is considered as a “master switch” gene for skeletal muscle formation (1). Expression of MyoD can convert several other cell types into skeletal muscle (55, 56). To further investigate the role of TAK1 in myogenesis, we employed TAK1+/+ and TAK1−/− MEF. As shown in Fig. 5A, transduction with Ad.MyoD adenoviral vector led to the formation of multinucleated myotubes in TAK1+/+ MEF cultures. However, the myotube formation was significantly impaired in TAK1−/− MEFs. We also measured MyoD-driven myogenic differentiation in TAK1+/+ and TAK1−/− MEF by biochemical methods. The expression of MyHCf was significantly reduced in TAK1−/− MEF compared with TAK1+/+ MEF (Fig. 5B). Similarly, the levels of CK were also found to be significantly reduced in TAK1−/− MEF compared with TAK1+/+ MEF (Fig. 5C). Furthermore, we also evaluated myogenic differentiation in MyoD-expressing MEFs by studying the transcriptional activation of skeletal α-actin promoter, which controls the expression of several structural genes in skeletal muscle (1). As shown in Fig. 5D, the activation of skeletal α-actin promoter was significantly reduced in TAK1−/− MEF compared with TAK1+/+ MEF.
FIGURE 5.
Deletion of TAK1 inhibits MyoD-induced differentiation in fibroblasts. TAK1+/+ and TAK1−/− MEF were transduced with Ad.MyoD for 24 h at multiplicity of infection 50. The cells were then incubated in DM for different time intervals. A, myotube formation was measured after 48 h of incubation in DM by performing immunofluorescence using MF20 antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole. The top photomicrographs (green fluorescence protein) show equal transduction of TAK1+/+ and TAK1−/− MEF by Ad.MyoD vector. The bottom photomicrographs show that myotube formation (red color) is significantly reduced in TAK1−/− MEF cultures compared with TAK1+/+. B, Western blot showed that the levels of MyHCf were reduced in TAK1−/− MEF compared with TAK1+/+ after incubation in DM. Immunoblots also show equal levels of MyoD protein in both TAK1+/+ and TAK1−/− MEF and the presence of truncated TAK1 protein in TAK1−/− MEF. Wt, wild type. C, levels of CK were also found to be significantly reduced in TAK1−/− MEF compared with TAK1+/+ MEF after 24 and 48 h of incubation in DM. *, p < 0.01, values significantly different from TAK1+/+ MEF at corresponding time point. D, TAK1+/+ and TAK1−/− MEFs were transfected with pcDNA3-MyoD plasmid along with pSK-Luc plasmid in a 1:10 ratio for 24 h. Cells were then incubated in DM for 48 h, and the activation of skeletal α-actin promoter was monitored by measuring luciferase activity. Data presented here show a significant reduction in the activation of skeletal α actin promoter in TAK1−/− MEF compared with TAK1+/+ MEF. #, p < 0.01, value significantly different from TAK1−/− MEF.
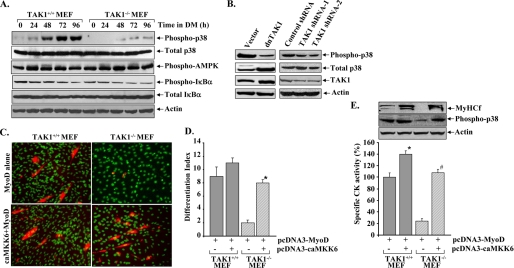
TAK1 Is Required for the Activation of p38 MAPK during Myogenic Differentiation
After establishing that TAK1 is essential for myogenic differentiation, we next investigated the mechanisms of action of TAK1 during muscle formation. Published reports suggest that depending on the type of stimuli, TAK1 can cause the activation of several downstream signaling pathways including p38 MAPK, AMPK, and NF-κB (35–39). We studied the activation of p38 MAPK, AMPK, and NF-κB in MyoD-expressing TAK1+/+ and TAK1−/− MEFs at different time points after induction of differentiation. As shown in Fig. 6A, the phosphorylation of p38 MAPK was significantly increased in MyoD-expressing TAK1+/+ MEFs after incubation in DM. In contrast, the phosphorylation of p38 MAPK was significantly blocked in TAK1−/− MEF (Fig. 6A). There was no significant difference in the levels of total p38MAPK between Ad.MyoD-transduced TAK1+/+ and TAK1−/− MEFs. Furthermore, there was also no difference in the levels of phosphorylated AMPK or total or phosphorylated IκBα protein between TAK1+/+ MEF and TAK1−/− MEF. These data suggest that TAK1 functions through the activation of p38 MAPK but not AMPK or NF-κB during myogenesis.
FIGURE 6.
TAK1 regulates myogenic differentiation through the activation of p38 MAPK. A, TAK1+/+ and TAK1−/− MEF were transduced with Ad.MyoD for 24h followed by incubation in DM for the indicated time intervals. Analysis of cell extracts by Western blot showed that the phosphorylation of p38 MAPK protein was completely blocked in TAK1−/− MEF compared with TAK1+/+. There was no difference in total p38, phosphorylated AMPK, and total or phosphorylated IκBα levels between TAK1+/+ and TAK1−/− MEFs. B, C2C12 myoblasts were stably transfected with vector alone or plasmid expressing either dominant negative TAK1 (dnTAK1) or TAK1 shRNAs and incubated in differentiation medium for 72 h. The levels of total and phospho-p38 MAPK and TAK1 were measured by Western blot. Representative immunoblots presented here show that overexpression of either dnTAK1 protein or TAK1 shRNAs inhibited the levels of phosphorylated p38 in C2C12 cultures. C, TAK1+/+ and TAK1−/− MEF were transiently transfected with either pcDNA3-MyoD alone or with pcDNA3-caMKK6 plasmid for 24 h followed by incubation in differentiation medium for additional 72 h. Myotube formation was monitored by immunocytochemistry using MF-20 antibody and 4′,6-diamidino-2-phenylindole. Representative photomicrographs presented here show that transfection with caMKK6 restored the myotube formation in TAK1−/− MEF cultures. D, shown is quantification of the differentiation index in TAK1+/+ and TAK1−/−MEF cultures transfected with MyoD along with caMKK6 or without caMKK6. *, p < 0.01, values significantly different from TAK1−/− MEF cultures without caMKK6. E, levels of CK measured using a CK activity assay kit and MyHCf and phospho-p38 protein (Western blot) were also found to be significantly increased in TAK1−/− MEF transfected with caMKK6. *, p < 0.05, values significantly different from TAK1+/+ MEFs without caMKK6. #, p < 0.05, values significantly different from TAK1−/− MEFs transfected with no caMKK6.
We also investigated the role of TAK1 in the phosphorylation of p38 MAPK in C2C12 myoblasts. As shown in Fig. 6B, overexpression of dnTAK1 or knockdown of TAK1 using RNAi considerably reduced the phosphorylation of p38 MAPK in C2C12 myoblasts upon their incubation in DM.
To further validate whether TAK1 functions through the activation of the p38 MAPK pathway during myogenic differentiation, we next investigated whether forced activation of p38 MAPK can restore myogenic differentiation in TAK1−/− MEF. TAK1+/+ and TAK1−/− MEF were cotransfected with MyoD plasmid along with a plasmid expressing a constitutively active mutant of MKK6 (caMKK6, i.e. MKK6S207E, T211E also known as MKK6EE), a kinase that directly phosphorylates and activates p38 MAPK (57). Overexpression of caMKK6 protein restored the MyoD-induced myotube formation in TAK1−/− MEF (Fig. 6C), which was also confirmed by measuring the differentiation index (% of nuclei in MyHCf-stained cells) in these cultures (Fig. 6D). Similarly, the levels of CK, MyHCf, and phosphorylated p38 MAPK were also found to be significantly increased in caMKK6-transfected TAK1−/− MEFs (Fig. 6E). These data suggest that TAK1 is an essential component of a signaling pathway that leads to the activation of p38 MAPK and the formation of multinucleated myotubes.
Functional TAK1 Is Needed for the Activation of Akt during Myogenic Differentiation
Next, we sought to investigate whether TAK1 is involved in the activation of Akt kinase. We observed that the phosphorylation of Akt was also reduced in MyoD-expressing TAK1−/− MEFs compared with TAK1+/+ MEFs after incubation in differentiation medium (Fig. 7A). The phosphorylation of Akt was also lower in cultured C2C12 myoblasts transfected with dnTAK1 plasmid (Fig. 7B), suggesting that TAK1 may also be involved in the activation of Akt during myogenesis.
FIGURE 7.
Role of TAK1 in activation of Akt during myogenic differentiation. A, TAK1+/+ and TAK1−/− MEF were transduced with Ad.MyoD for 24 h followed by incubation in DM for different time intervals. Levels of phosphorylated and total Akt were measured by Western blotting. Data presented here show that the phosphorylation of Akt is blocked in TAK1−/− MEF compared with TAK1+/+ MEF upon incubation in DM. B, C2C12 myoblasts were transfected with vector alone or dnTAK1 and incubated in differentiation medium for 72 h. Representative immunoblots show that the overexpression of dnTAK1 inhibits the levels of phosphorylated Akt in C2C12 cultures. C, TAK1+/+ and TAK1−/− MEF were transiently transfected with either pcDNA3-MyoD alone or with pUSEamp-caAkt plasmid for 24 h followed by incubation in DM for 72 h. Data presented here show that the expression of caAkt did not affect with the levels of CK or MyHCf in TAK1−/− MEF. D, TAK1+/+ and TAK1−/− MEF were transiently transfected with either pcDNA3-MyoD alone or with pcDNA3-caMKK6 plasmid followed by incubation in differentiation medium for 72 h. A representative immunoblot presented here shows that transfection with caMKK6 increased the levels of phosphorylated Akt in TAK1−/− MEF.
We next sought to determine whether ectopic expression of constitutively active Akt (caAkt) can rescue myogenic differentiation in TAK1−/− MEFs expressing MyoD. The expression of caAkt did not affect the differentiation of TAK1−/−MEF upon expression of MyoD. There was no change in the levels of either CK or MyHCf after transfection of TAK1−/− MEF with MyoD and caAkt (Fig. 7C).
A recent study has demonstrated that p38 MAPK induces the phosphorylation of Akt kinase in differentiating myoblasts (58). To understand whether the reduced phosphorylation of Akt in TAK1−/− MEF was a result of inhibition of p38 MAPK or TAK1 regulates the activation of p38 MAPK and Akt independently during myogenic differentiation, we investigated the effects of overexpression of a caMKK6 on the phosphorylation of Akt kinase. Interestingly, caMKK6 considerably increased the phosphorylation of Akt kinase not only in TAK1+/+ but also in TAK1−/− MEF (Fig. 7D).
IGF-I-induced Myogenic Differentiation Is Mediated by TAK1
IGF-I is a potent stimulator of skeletal muscle cell proliferation and differentiation (1). We also investigated whether IGF-I induces myogenic differentiation through the activation of TAK1. TAK1+/+ and TAK1−/− MEF were transduced with Ad.MyoD followed by their incubation in DM with or without IGF-I. Differentiation of MEF into myotubes was monitored by measuring the levels of CK in cell extracts. As shown in Fig. 8A, treatment with IGF-I significantly increased CK levels in TAK1+/+ MEF. However, there was no significant difference in the levels of CK between untreated and IGF-I-treated TAK1−/− MEF (Fig. 8A). Similarly, treatment with IGF-I did not induce the expression of muscle protein MyHCf and myogenin in TAK1−/− MEFs, further confirming that TAK1 is required for the stimulatory effects of IGF-I in myogenesis (Fig. 8B).
FIGURE 8.
IGF-I augments myogenesis through the activation of TAK1. TAK1+/+ and TAK1−/− MEF were transduced with Ad.MyoD for 24 h followed by incubation in DM with or without IGF-I (100 ng/ml). A, data presented here show that IGF-I significantly increased the levels of CK in TAK1+/+ but not TAK1−/− MEF. *, p < 0.01, values significantly different compared with TAK1+/+ MEF incubated without IGF-I. B, Western blot analysis showed that there was no increase in the levels of either MyHCf or myogenin between IGF-1-treated or untreated TAK1−/− MEF. C, TAK1+/+ and TAK1−/− MEF were treated with IGF-I (100 ng/ml) for the indicated time periods, and the levels of phosphorylated or total Akt and p38 MAPK were measured by Western blot. Representative immunoblots presented here show that IGF-I-induced phosphorylation of p38 MAPK (but not Akt) was blocked in TAK1−/− MEF compared with TAK1+/+ MEF.
Although TAK1 is implicated in the activation of multiple signal transduction pathways, the direct role of TAK1 in the activation of p38 MAPK or Akt in IGF-activated signaling pathways has not been previously identified. We employed TAK1+/+ and TAK1−/− MEF (without expression of MyoD) to investigate the involvement of TAK1 in IGF-induced activation of Akt and p38 MAPK. Interestingly, there was no difference in the levels of phosphorylation of Akt between TAK1+/+ and TAK1−/− MEF upon treatment with IGF-I. However, IGF-induced phosphorylation of p38 MAPK was significantly impaired in TAK1−/− MEF (Fig. 8C).
DISCUSSION
TAK1 is an important component of diverse cellular responses including innate and adaptive immune responses, survival of hematopoietic cells and hepatocytes, and growth and differentiation of epidermis (38, 43–45, 47, 48). TAK1 was originally identified as a key regulator of MAPK activation in TGF-β and bone morphogenetic protein signaling pathways (34). However, accumulating evidence suggests that TAK1 is activated in response to a number of stimuli such as proinflammatory cytokines, biomedical stress, bacterial products, and several members of TGF-β family proteins (35–39). Interestingly, many of these known activators of TAK1 also inhibit myogenic differentiation (50, 53, 59, 60), suggesting that TAK1 could be a negative regulator of myogenic differentiation. However, contrary to this assumption, we found that TAK1 is essential for both proliferation and differentiation of myogenic cells.
The important role of TAK1 in proliferation of myogenic cells is supported by our findings that TAK1 and its associated proteins TAB1 and TAB2 are highly expressed in proliferating C2C12 myoblasts (Fig. 1A), and the inhibition of TAK1 activity by RNAi or through overexpression of a dominant negative mutant of TAK1 significantly reduced proliferation of myoblasts (Fig. 3, A and B). Similar to cultured myoblasts, the expression of TAK1 was also significantly high in the skeletal muscle of neonatal mice, and the levels of TAK1 were reduced in adult mice (Fig. 1B). It is also noteworthy that the levels of TAK1 are increased in the models of muscle regeneration, further supporting the inference that TAK1 plays an important role in skeletal muscle homeostasis (Fig. 2). A significant number of differentiation-incompetent myoblasts undergo apoptosis after incubation in differentiation medium (1, 2). Although there are reports indicating that TAK1 is required for the survival of certain cell types (38, 47, 49), our results suggest that it does not have any role in the survival of proliferating myoblasts or after induction of differentiation (Fig. 3C).
Myogenesis involves not only the proliferation of myoblasts but also their terminal differentiation into myotubes (1, 2). Although the proliferation and differentiation are two antagonistic processes, the results of the present study suggest that in addition to stimulating proliferation, TAK1 also acts as an important molecular switch for the induction of differentiation in myoblasts. This is evident from our results that the ectopic expression of dominant negative TAK1 protein (Fig. 4, A and B) or knockdown of endogenous TAK1 using RNAi (Fig. 4D) inhibited the differentiation of C2C12 myoblasts. Differentiation of cells in skeletal muscle lineage is governed by the basic helix-loop-helix transcription factors of MyoD family (MyoD, Myf-5, myogenin, and MRF4) (1, 2). These proteins work in a coordinated fashion with additional transcription factors such as myocyte enhancer factor 2 to drive the muscle-specific gene expression and promote fusion of myoblasts into multinucleated myotubes (1). Previous studies using knock-out animals have suggested that MyoD and Myf-5 act redundantly at the early steps in myoblast specification (61), whereas MyoD is essential for muscle regeneration in adults (62). Myogenin acts downstream of MyoD and Myf-5, whereas MRF4 plays a more limited role in muscle formation in vivo (63–65). MyoD enhances the expression of a large number of muscle genes including myogenin, whereas its own expression is regulated through the activation of various cell signaling pathways (1, 16). Although the exact mechanisms by which TAK1 regulates muscle formation remain unknown, our data suggest that TAK1 is critical for the expression of MyoD family transcription factors (Fig. 4C). It is also of interest to note that whereas the inhibition of TAK1 drastically reduced the expression of Myf-5, MyoD, and myogenin in differentiating myoblasts, it did not affect the expression of MFF2 (Fig. 4C), indicating that TAK1 specifically regulates the expression of MyoD family transcription factors. Because overexpression of MyoD in TAK1−/− MEF failed to induce myogenesis (Fig. 5), it is possible that signaling through TAK1 is also essential for the transactivation of MyoD and/or other accessory factors.
Several recent studies using pharmacological agents and genetic mouse models have revealed that p38 MAPK and Akt signaling pathways play a prominent role in skeletal muscle formation and regeneration (17–23). Although the forced activation of either the p38 MAPK or Akt pathways is sufficient to accelerate myogenesis, it is effective only when the reciprocal pathway is functional (66). Because TAK1 can activate multiple cell signaling pathways in response to various extracellular stimuli, we determined the role of TAK1 in the activation of p38 MAPK, Akt, and also AMPK and NF-κB. Our results suggest that TAK1 specifically activates p38 MAPK during myogenic differentiation, and inhibition of p38 MAPK is responsible for the reduced muscle formation in TAK1-deficient cells (Fig. 6). This conclusion is supported by our data which demonstrate that the overexpression of a constitutive active MKK6 protein increases the myotube formation, expression of muscle-specific proteins, and the phosphorylation of p38MAPK in TAK1−/−MEF (Fig. 6, C–E). These results are also consistent with the published reports indicating that TAK1 can directly phosphorylate and activate MKK6 in response to different extracellular stimuli (40–42). It has been reported that Cdo, a cell surface protein with a long intracellular domain, through its interaction with scaffold protein JLP, promotes the activation of p38 MAPK during myogenesis both in vivo and in vitro (22, 23, 67, 68). Whether TAK1 directly interacts with Cdo-JLP complex and mediates the downstream activation of p38 MAPK remains to be determined.
Intriguingly, we also found that the inhibition of TAK1 reduces the activation of Akt kinase during myogenesis (Fig. 7). Although the exact mechanisms remain unknown, the inhibition of Akt in the absence of TAK1 could be a result of reduced activation of the p38 MAPK signaling pathway, which is also supported by our findings that the overexpression of constitutively active MKK6 protein increased the phosphorylation of Akt in TAK1-deficinet cells (Fig. 7D). Our results are also consistent with general premise that until the p38 MAPK pathway is active, overexpression of active Akt is not sufficient to augment differentiation of myoblasts (66). There is also evidence suggesting that p38 MAPK increases the activation of Akt during myogenic differentiation (58). Therefore, it is possible that during myogenesis, TAK1 is first activated, which stimulates the MKK6-p38-Akt cascade, and both p38 and Akt promote myogenesis by modulating the activity of independent downstream target proteins. In addition to TAK1/p38MAPK-dependent mechanisms, the increased phosphorylation of Akt could also be a result of increased production of growth factors, especially IGFs, in muscle cultures upon expression of caMKK6 in TAK1−/− MEFs.
IGF is the only group of growth factors that induces both proliferation and differentiation of myogenic cells besides inducing hypertrophy in myofibers (1, 5). Interestingly, IGF-I is a potent inducer of Akt and p38 MAPK signaling pathways in skeletal muscle cells (1, 16, 69). We postulated that IGF-I may use the TAK1-dependent signaling pathway to stimulate myogenic differentiation. Consistent with our hypothesis, we found that whereas IGF-I-stimulated myogenic differentiation in TAK1+/+ cells, it failed to rescue myogenic differentiation in TAK1-deficient MEF, suggesting that TAK1 is required for the myogenic actions of IGF-I (Fig. 8, A and B). The role of TAK1 in the IGF-I-induced activation of p38 MAPK and Akt has not been previously investigated. Our experiments studying the effects of IGF-I on the phosphorylation of Akt and p38 MAPK proteins have provided important information regarding the essential role that TAK1 plays in the IGF-I-induced activation of p38 but not in Akt (Fig. 8C). These data are consistent with general observations that the functional activation of both the p38 MAPK and Akt pathways is important to stimulate myogenic differentiation. These data also help explain why IGF-I could not augment myogenic differentiation in TAK1−/− MEF.
In summary, our study provides initial evidence that TAK1 is an essential regulator of myogenic cell proliferation and differentiation. More investigations are required, especially using skeletal muscle-specific knock-out mice to further validate whether TAK1 is important for the development and/or regeneration of myofibers and whether specific modulation of TAK1 can improve regeneration in various muscular disorders. TAK1 has also been found to be critical for the activation of various cell signaling pathways in response to muscle catabolic cytokines tumor necrosis factor-α and TWEAK (tumor necrosis factor-like weak inducer of apoptosis) (39, 53). Whether these cytokines function through the activation of TAK1-dependent pathways to induce atrophy in diverse muscle-wasting conditions is also not yet known. This is an area of interest for future investigations.
Acknowledgments
We thank Prof. Shizuo Akira and Dr. Osamu Takeuchi of Osaka University, Osaka, Japan for providing wild-type and TAK1-deficient mouse embryonic fibroblasts. We also thank Dr. Jun Ninomiya-Tsuji for providing dominant negative TAK1 and TAB1 plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AG129623 (to A. K.).
- MRF
- myogenic regulatory factor
- AMPK
- adenosine monophosphate-activated kinase
- ca
- constitutively active
- CK
- creatine kinase
- dn
- dominant negative
- IGF
- insulin-like growth factor
- MAPK
- mitogen-activated protein kinase
- MEF
- mouse embryonic fibroblast
- MKK6
- MAPK kinase 6
- MyHCf
- myosin heavy chain fast type
- PBS
- phosphate-buffered saline
- RNAi
- RNA interference
- shRNA
- short hairpin RNA
- TA
- tibial anterior
- TGF
- transforming growth factor
- TAK1
- TGF β-activated kinase1
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- DM
- differentiation medium
- MCK
- muscle creatine kinase
- EGFP
- enhanced green fluorescent protein
- p-
- phosphorylated
- BrdUrd
- bromodeoxyuridine.
REFERENCES
- 1.Chargé S. B., Rudnicki M. A. (2004) Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 2.Perry R. L., Rudnick M. A. (2000) Front. Biosci. 5, D750–D767 [DOI] [PubMed] [Google Scholar]
- 3.Sartorelli V., Caretti G. (2005) Curr. Opin. Genet. Dev. 15, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun K., Wold B. (1996) Curr. Opin. Cell Biol. 8, 877–889 [DOI] [PubMed] [Google Scholar]
- 5.Florini J. R., Ewton D. Z., Coolican S. A. (1996) Endocr. Rev. 17, 481–517 [DOI] [PubMed] [Google Scholar]
- 6.Husmann I., Soulet L., Gautron J., Martelly I., Barritault D. (1996) Cytokine Growth Factor Rev. 7, 249–258 [DOI] [PubMed] [Google Scholar]
- 7.Thomas M., Langley B., Berry C., Sharma M., Kirk S., Bass J., Kambadur R. (2000) J. Biol. Chem. 275, 40235–40243 [DOI] [PubMed] [Google Scholar]
- 8.Späte U., Schulze P. C. (2004) Curr. Opin. Clin. Nutr. Metab. Care 7, 265–269 [DOI] [PubMed] [Google Scholar]
- 9.Kollias H. D., McDermott J. C. (2008) J. Appl. Physiol. 104, 579–587 [DOI] [PubMed] [Google Scholar]
- 10.Coolican S. A., Samuel D. S., Ewton D. Z., McWade F. J., Florini J. R. (1997) J. Biol. Chem. 272, 6653–6662 [DOI] [PubMed] [Google Scholar]
- 11.Bennett A. M., Tonks N. K. (1997) Science 278, 1288–1291 [DOI] [PubMed] [Google Scholar]
- 12.Rommel C., Clarke B. A., Zimmermann S., Nuñez L., Rossman R., Reid K., Moelling K., Yancopoulos G. D., Glass D. J. (1999) Science 286, 1738–1741 [DOI] [PubMed] [Google Scholar]
- 13.Guttridge D. C., Albanese C., Reuther J. Y., Pestell R. G., Baldwin A. S., Jr. (1999) Mol. Cell. Biol. 19, 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry R. L., Parker M. H., Rudnicki M. A. (2001) Mol. Cell 8, 291–301 [DOI] [PubMed] [Google Scholar]
- 15.Guttridge D. C., Mayo M. W., Madrid L. V., Wang C. Y., Baldwin A. S., Jr. (2000) Science 289, 2363–2366 [DOI] [PubMed] [Google Scholar]
- 16.Lluís F., Perdiguero E., Nebreda A. R., Muñoz-Cánoves P. (2006) Trends Cell Biol. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 17.Chen S. E., Jin B., Li Y. P. (2007) Am. J. Physiol. Cell Physiol 292, C1660–C1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuenda A., Cohen P. (1999) J. Biol. Chem. 274, 4341–4346 [DOI] [PubMed] [Google Scholar]
- 19.de Angelis L., Zhao J., Andreucci J. J., Olson E. N., Cossu G., McDermott J. C. (2005) Dev. Biol. 283, 171–179 [DOI] [PubMed] [Google Scholar]
- 20.Wu Z., Woodring P. J., Bhakta K. S., Tamura K., Wen F., Feramisco J. R., Karin M., Wang J. Y., Puri P. L. (2000) Mol. Cell. Biol. 20, 3951–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zetser A., Gredinger E., Bengal E. (1999) J. Biol. Chem. 274, 5193–5200 [DOI] [PubMed] [Google Scholar]
- 22.Kang J. S., Bae G. U., Yi M. J., Yang Y. J., Oh J. E., Takaesu G., Zhou Y. T., Low B. C., Krauss R. S. (2008) J. Cell Biol. 182, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaesu G., Kang J. S., Bae G. U., Yi M. J., Lee C. M., Reddy E. P., Krauss R. S. (2006) J. Cell Biol. 175, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puri P. L., Wu Z., Zhang P., Wood L. D., Bhakta K. S., Han J., Feramisco J. R., Karin M., Wang J. Y. (2000) Genes Dev. 14, 574–584 [PMC free article] [PubMed] [Google Scholar]
- 25.Briata P., Forcales S. V., Ponassi M., Corte G., Chen C. Y., Karin M., Puri P. L., Gherzi R. (2005) Mol. Cell 20, 891–903 [DOI] [PubMed] [Google Scholar]
- 26.Lluís F., Ballestar E., Suelves M., Esteller M., Muñoz-Cánoves P. (2005) EMBO J. 24, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simone C., Forcales S. V., Hill D. A., Imbalzano A. N., Latella L., Puri P. L. (2004) Nat. Genet. 36, 738–743 [DOI] [PubMed] [Google Scholar]
- 28.Jiang B. H., Aoki M., Zheng J. Z., Li J., Vogt P. K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2077–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang B. H., Zheng J. Z., Vogt P. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14179–14183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaliman P., Viñals F., Testar X., Palacín M., Zorzano A. (1996) J. Biol. Chem. 271, 19146–19151 [DOI] [PubMed] [Google Scholar]
- 31.Kaliman P., Canicio J., Shepherd P. R., Beeton C. A., Testar X., Palacín M., Zorzano A. (1998) Mol. Endocrinol. 12, 66–77 [DOI] [PubMed] [Google Scholar]
- 32.Kaliman P., Canicio J., Testar X., Palacín M., Zorzano A. (1999) J. Biol. Chem. 274, 17437–17444 [DOI] [PubMed] [Google Scholar]
- 33.Fujio Y., Guo K., Mano T., Mitsuuchi Y., Testa J. R., Walsh K. (1999) Mol. Cell. Biol. 19, 5073–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. (1995) Science 270, 2008–2011 [DOI] [PubMed] [Google Scholar]
- 35.Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., Ghosh S. (2005) Genes Dev. 19, 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wald D., Commane M., Stark G. R., Li X. (2001) Eur. J. Immunol. 31, 3747–3754 [DOI] [PubMed] [Google Scholar]
- 37.Huangfu W. C., Omori E., Akira S., Matsumoto K., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 28802–28810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R. C., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 19610–19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar M., Makonchuk D. Y., Li H., Mittal A., Kumar A. (2009) J. Immunol. 182, 2439–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanafusa H., Ninomiya-Tsuji J., Masuyama N., Nishita M., Fujisawa J., Shibuya H., Matsumoto K., Nishida E. (1999) J. Biol. Chem. 274, 27161–27167 [DOI] [PubMed] [Google Scholar]
- 41.Moriguchi T., Kuroyanagi N., Yamaguchi K., Gotoh Y., Irie K., Kano T., Shirakabe K., Muro Y., Shibuya H., Matsumoto K., Nishida E., Hagiwara M. (1996) J. Biol. Chem. 271, 13675–13679 [DOI] [PubMed] [Google Scholar]
- 42.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 43.Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 44.Sato S., Sanjo H., Tsujimura T., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Takeuchi O., Akira S. (2006) Int. Immunol. 18, 1405–1411 [DOI] [PubMed] [Google Scholar]
- 45.Liu H. H., Xie M., Schneider M. D., Chen Z. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11677–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jadrich J. L., O'Connor M. B., Coucouvanis E. (2006) Development 133, 1529–1541 [DOI] [PubMed] [Google Scholar]
- 47.Sayama K., Hanakawa Y., Nagai H., Shirakata Y., Dai X., Hirakawa S., Tokumaru S., Tohyama M., Yang L., Sato S., Shizuo A., Hashimoto K. (2006) J. Biol. Chem. 281, 22013–22020 [DOI] [PubMed] [Google Scholar]
- 48.Tang M., Wei X., Guo Y., Breslin P., Zhang S., Zhang S., Wei W., Xia Z., Diaz M., Akira S., Zhang J. (2008) J. Exp. Med. 205, 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim J. H., Greenblatt M. B., Xie M., Schneider M. D., Zou W., Zhai B., Gygi S., Glimcher L. H. (2009) EMBO J. 28, 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dogra C., Changotra H., Mohan S., Kumar A. (2006) J. Biol. Chem. 281, 10327–10336 [DOI] [PubMed] [Google Scholar]
- 51.Dogra C., Hall S. L., Wedhas N., Linkhart T. A., Kumar A. (2007) J. Biol. Chem. 282, 15000–15010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava A. K., Qin X., Wedhas N., Arnush M., Linkhart T. A., Chadwick R. B., Kumar A. (2007) J. Biol. Chem. 282, 35113–35124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allamand V., Campbell K. P. (2000) Hum. Mol. Genet. 9, 2459–2467 [DOI] [PubMed] [Google Scholar]
- 55.Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. (1988) Science 242, 405–411 [DOI] [PubMed] [Google Scholar]
- 56.Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) Mol. Cell. Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabane C., Coldefy A. S., Yeow K., Dérijard B. (2004) Cell. Signal. 16, 1405–1415 [DOI] [PubMed] [Google Scholar]
- 59.Li H., Malhotra S., Kumar A. (2008) J. Mol. Med. 86, 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trendelenburg A. U., Meyer A., Rohner D., Boyle J., Hatakeyama S., Glass D. J. (2009) Am. J. Physiol. Cell Physiol. 296, C1258–C1270 [DOI] [PubMed] [Google Scholar]
- 61.Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 62.Sabourin L. A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M. A. (1999) J. Cell Biol. 144, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., Klein W. H. (1993) Nature 364, 501–506 [DOI] [PubMed] [Google Scholar]
- 64.Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993) Nature 364, 532–535 [DOI] [PubMed] [Google Scholar]
- 65.Olson E. N., Arnold H. H., Rigby P. W., Wold B. J. (1996) Cell 85, 1–4 [DOI] [PubMed] [Google Scholar]
- 66.Li Y., Jiang B., Ensign W. Y., Vogt P. K., Han J. (2000) Cell. Signal. 12, 751–757 [DOI] [PubMed] [Google Scholar]
- 67.Bae G. U., Kim B. G., Lee H. J., Oh J. E., Lee S. J., Zhang W., Krauss R. S., Kang J. S. (2009) Mol. Cell. Biol. 29, 4130–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cole F., Zhang W., Geyra A., Kang J. S., Krauss R. S. (2004) Dev. Cell 7, 843–854 [DOI] [PubMed] [Google Scholar]
- 69.Conejo R., Lorenzo M. (2001) J. Cell. Physiol. 187, 96–108 [DOI] [PubMed] [Google Scholar]