Abstract
Myosin II association with actin, which triggers contraction, is regulated by orchestrated waves of phosphorylation/dephosphorylation of the myosin regulatory light chain. Blocking myosin regulatory light chain phosphorylation with small molecule inhibitors alters the shape, adhesion, and migration of many types of smooth muscle and cancer cells. Dephosphorylation is mediated by myosin phosphatase (MP), a complex that consists of a catalytic subunit (protein phosphatase 1c, PP1c), a large subunit (myosin phosphatase targeting subunit, MYPT), and a small subunit of unknown function. MYPT functions by targeting PP1c onto its substrate, phosphorylated myosin II. Using RNA interference, we show here that stability of PP1c β and MYPT1 is interdependent; knocking down one of the subunits decreases the expression level of the other. Associated changes in cell shape also occur, characterized by flattening and spreading accompanied by increased cortical actin, and cell numbers decrease secondary to apoptosis. Of the three highly conserved isoforms of PP1c, we show that MYPT1 binding is restricted to PP1c β, and, using chimeric analysis and site-directed mutations, that the central region of PP1c β confers the isoform-specific binding. This finding was unexpected because the MP crystal structure has been solved and was reported to identify the variable, C-terminal domain of PP1c β as being the region key for isoform-specific interaction with MYPT1. These findings suggest a potential screening strategy for cardiovascular and cancer therapeutic agents based on destabilizing MP complex formation and function.
Keywords: Cancer, Cell/Migration, Cell/Motility, Cytoskeleton, Cytoskeleton/Actin, Phosphorylation/Cytoskeletal Proteins, Phosphorylation/Enzymes, Phosphorylation/Myosin
Introduction
Phosphorylation of smooth muscle and non-muscle myosin II is implicated in many physiological phenomena, including smooth muscle contraction, cell motility, and cytokinesis. Myosin phosphatase (MP)3 is responsible for dephosphorylation of the phosphorylated myosin light chain (1). Increases in the levels of cytosolic [Ca2+]i initiate smooth muscle contraction by triggering binding of calmodulin to myosin light chain kinase, which then phosphorylates myosin light chain, increasing cross-bridge cycling and the rate of tension development (2). Conversely, smooth muscle relaxation occurs via dephosphorylation of myosin light chain by MP.
MP directs cell migration by increasing contractile forces through regulating both myosin phosphorylation and actin assembly (3). Knockdown of myosin phosphatase targeting subunit 1 (MYPT1), one of the components of the MP complex, increases F-actin stress fibers and the number of focal adhesions (3). Contractile force exerted from the focal adhesion sites (cell bottom) is regulated by several kinases including Rho kinase. Less is known about how the cortical contractile force is regulated, although we suggested recently that it may in part be through the action of phospholipase D2, which inhibits MP-regulated myosin II-driven changes in cell shape during spreading (4).
MP undertakes other roles, including inhibiting apoptosis by dephosphorylating the histone deacetylase protein HDAC7, facilitating its nuclear localization and repression of Nur77, a proapoptotic gene (5). MP also blocks oncogenic signaling cascades through the dephosphorylation of other target proteins, such as merlin, which regulates the Ras-extracellular signal-regulated kinase (ERK) pathway (6, 7), and an MP inhibitor, CPI-17, has been shown to drive tumorigenic transformation (6). Consistent with these reports, MP is up-regulated in some cancer cell lines, and its knockdown using RNA interference decreases the cancer cell viability (8), suggesting it as a potential target for cancer therapeutics.
MP is composed of a complex of three proteins: a catalytic subunit (protein phosphatase 1c, PP1c), a large subunit (MYPT), and a 20-kDa small subunit (M20) of unknown function (Fig. 1A). By altering the phosphatase active site, the interaction with MYTP confers myosin specificity to PP1c. The main isoform in smooth muscle, MYPT1, complexes with only one of the three isoforms of PP1c, specifically PP1c β (1, 9), despite the extensive similarity it has to the α and γ isoforms throughout most of the protein. A crystal structure of the complex between PP1c β and residues 1–299 of MYPT1 has been solved (10), from which it was predicted that MYPT1 makes contact with the central region of PP1c β via the MYPT1 N-terminal arm (amino acids 1–34) and an RVXF motif (amino acids 35–38), and with the C terminus of PP1c β (amino acids Tyr305 and Tyr307) via the second group of ankyrin repeats in MYPT1. Because the PP1c isoforms differ most dramatically in the N-terminal 45 amino acid residues and in the C terminus (amino acids 302–328, with the rest of the protein sequences being more than 95% identical), the report also predicted that the PP1c β interaction with MYPT1 through the PP1c β C-terminal domain (e.g. residues Tyr305 and Tyr307) would constitute the basis for the selective complex formation of the β-isoform with MYPT1.
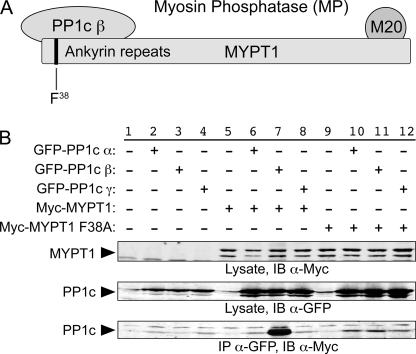
FIGURE 1.
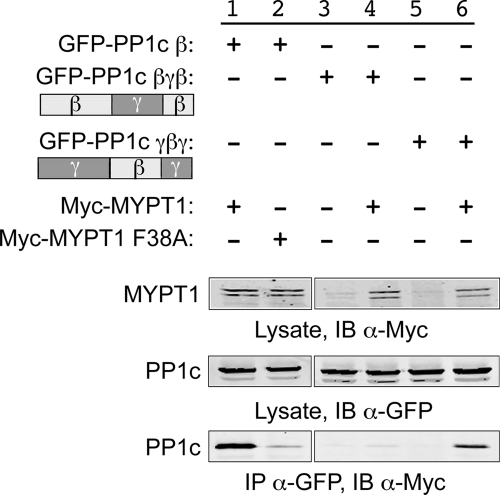
MP complex: specific interaction of MYPT1 with the β isoform of PP1c. A, organization of the individual components of MP is shown (see the Introduction for details). B, GFP-tagged expression plasmids for the three isoforms of PPIc and Myc-tagged expression plasmids for wild-type and F38A-mutated MYPT1 were co-expressed in HeLa cells for 1 day, lysed, and immunoprecipitated using anti-GFP antibodies. The immunoprecipitant was then analyzed using SDS-PAGE and Western blotting to assess the extent to which Myc-tagged MYPT1 was co-immunoprecipitated with the GFP-tagged PP1c proteins (bottom panel). A portion of the lysate was retained for each sample and electrophoresed and blotted (IB) separately to demonstrate the total levels of expression of the Myc-tagged MYPT1 and GFP-tagged PP1c proteins (top two panels). Experiments present in this and subsequent figures were repeated three or more times unless otherwise noted. Representative Western blots are shown.
In this study, we sought to explore further the basis for the isoform specificity of PP1c β complex formation with MYPT1. Unexpectedly, chimeric and mutational studies revealed that the central region of PP1c β mediates the isoform-selective interaction. Moreover, the mutational series generated a mutant that exhibits “weakened” complex formation, setting the stage for a rationalized screening to identify potential chemotherapeutic agents that could target this component of the cytoskeletal infrastructure through destabilizing the interaction between PP1c β and MYPT1.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
PP1 chimeric expression plasmids were created using a PCR-based strategy with pQCXIP-GFP. The PP1c β (T197Q, S232A, N236H/R237K, G280A) and PP1c γ (Q198T, A233S, H237N/K238R, A281G) site-directed mutants were generated by substituting corresponding amino acids in pQCXIP-GFP-PP1c γ or -PP1c β using a PCR-based strategy. All mutation and fusion constructs were confirmed by sequencing.
Cell Culture
CHO cells were cultured in F-12 nutrient mixture (Ham's) supplemented with 10% fetal bovine serum. MDA-MB-231, GP2-293, and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum.
Retroviral Infection
GP2-293 retroviral packaging cells were passaged into six 10-cm plates at a 1:7 ratio and transfected 24 h later with small hairpin RNA (shRNA) constructs plus vesicular stomatitis virus glycoprotein using Lipofectamine Plus reagent (Invitrogen). On the following day, the cells to be infected were passaged into a 6-well plate, and the next day, harvested virus was added (along with 2 μl of Polybrene (stock 6 mg/ml)). One day later, the cells were selected with puromycin.
Western Blotting
Cells cultured to near confluence in 60-mm tissue culture plates were rinsed with phosphate-buffered saline and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 150 mm NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 1 mm EGTA) containing protease inhibitor mixture (Roche Applied Science) as well as 1 mm Na3VO4 and 1 mm NaF. The lysates were subjected to 10% SDS-PAGE, transferred to nitrocellulose membrane, and probed with primary antibodies recognizing c-Myc epitope (monoclonal, 1:1000, 9B11; Cell Signaling, Beverly, MA), green fluorescent protein (GFP) (polyclonal, 1:1000; Abcam, Cambridge, MA), MYPT1 (polyclonal, 1:10,000; Covance, Princeton, NJ), PP1α (monoclonal, 1:500; Sigma), PP1γ (polyclonal; Stratagene), PP1δ (β) (polyclonal, 1:500; Upstate, Lake Placid, NY), or α-tubulin (B-5-1-2, 1:1000; Sigma), followed by secondary antibodies conjugated with Alexa Fluor 680 anti-rabbit (1:5000; Invitrogen) or IRDye 800 anti-mouse (1:5000; Rockland Immunochemicals, Gilbertsville, PA). Fluorescent signals were detected with an Odyssey infrared imaging system (LICOR Biosciences, Lincoln, NE) and quantitated using the instrumentation software.
PP1 Immunoprecipitation
PP1 was immunoprecipitated from cell lysates prepared as above using anti-c-Myc monoclonal antibody (M5546; Sigma) as described previously (11). In brief, the lysates were passed through a 255/8-gauge needle five times and spun at 4 °C at 14,000 × g for 10 min. Anti-c-Myc was then added to the supernatants in fresh Eppendorf tubes and incubated at 4 °C with gentle shaking for 1 h. A 50:50 slurry of protein A-Sepharose (Sigma) was then added to the supernatant, followed by another hour of incubation. The samples were then spun down at 14,000 × g for 1 min, and the supernatant was removed. The protein A-Sepharose was then washed three times with radioimmune precipitation assay buffer lacking protease inhibitors, resuspended in SDS-PAGE sample buffer, and boiled at 95 °C for 5 min. Western blotting was then performed as described above.
Live Cell Analysis
CHO cells were imaged using a Nikon Eclipse TS100 light microscope and photographed using a Nikon Digital Sight camera. Images were analyzed using NIS-Elements F v2.30 imaging software.
Immunofluorescence Microscopy
MDA-MB-231 breast cancer cells plated on coated coverslips were fixed in 3.7% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min. F-actin was visualized using rhodamine-conjugated phalloidin (Invitrogen) and imaged using a Leica TCS SP2 confocal microscope. Images were processed using Adobe Photoshop.
Protein Sequence Alignments
Protein sequence alignments were performed using the Network Protein Sequence Analysis: Multalin Alignment algorithm (12).
Statistics
At least three independent experiments were performed, unless indicated otherwise in the figure legends. Statistical analyses were performed using two-tailed equal variance Student's test. p values < 0.05 were considered to be statistically significant.
RESULTS
Specificity and Significance of the MYPT1 Interaction with PP1c β
We confirmed that MYPT1 interacts specifically with PP1c β using a co-immunoprecipitation (co-IP) approach (Fig. 1B). HeLa cells were co-transfected with enhanced GFP-tagged PP1c α, β, or γ, and c-Myc epitope-tagged MYPT1 or MYPT1 F38A. Mutation of the Phe38 residue in MYPT1 to alanine has been reported to suffice to disrupt the interaction of MYPT1 with PP1c β (9). Lanes 2–4 show samples in which only the GFP-tagged PP1c isoforms were expressed and subjected to co-IP using anti-c-Myc for the co-IP step and anti-GFP for the immunoblotting step; under these circumstances, the GFP-PP1c proteins were not nonspecifically pulled down. In contrast (lanes 6–8), co-expression of MYPT1 resulted in efficient co-IP of PP1c β, but not α or δ. Specificity of the co-IP was confirmed in lane 11, where it is shown that MYPT1 F38A was unable to co-immunoprecipitate PP1c β.
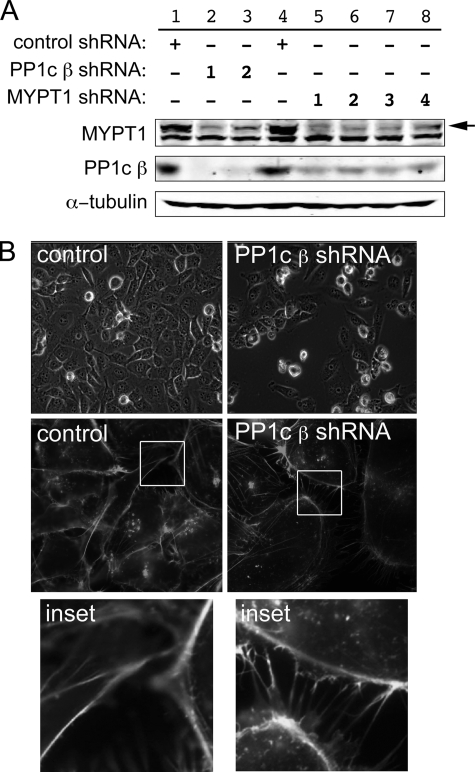
Specificity and significance of the interaction were examined in CHO cells for the endogenous proteins using a shRNA approach (Fig. 2). shRNA-mediated knockdown of PP1c β using two different targeting sequences (Fig. 2A, lanes 2 and 3) resulted in decreased expression of MYPT1, implying that MYPT1 stability requires a successful complex formation that cannot be performed by other potential partners extant in the cells (for example, other PP1c isoforms). Similarly, shRNA-mediated knockdown of MYPT1 using any of four different targeting sequences resulted in decreased levels of PP1c β expression, implying that PP1c β stability analogously requires complex formation specifically with MYPT1.
FIGURE 2.
Interdependence of PP1c β and MYPT1. A, CHO cells were infected with shRNAs directed against either PP1c β or MYPT1. 24 h following transfection, the cells were selected with blasticidin for an additional 2 days, rinsed with phosphate-buffered saline, lysed, and Western blotting was performed against the corresponding proteins using the indicated antibodies (see “Experimental Procedures”). Two independent shRNA sequences were used to knock down PP1c β and four for MYPT1, as indicated by the numbers in the legend. An immunoblot representative of two experiments with similar outcomes is shown. The top band seen in the MYPT1 Western blot (arrow) is MYPT1; the lower band is an unknown protein unspecifically immunostained by the primary antibody. B, morphological changes following PP1c β knockdown. MDA-MB-231 breast cancer cells were analyzed for changes in cellular morphology in the PP1c β knockdown cells using live cell imaging (top row) or using confocal microscopy with fixed cells stained with rhodamine-phalloidin to visualize F-actin (middle and bottom rows). 10 fields of cells were counted for each field for quantitation purposes. Figure is representative of two independent experiments.
Similar to the previously reported consequences of knockdown of MYPT1 (3), we found that PP1c β knockdown had readily apparent effects on cellular morphology and potentially on viability. At time points at which MDA-MB-231 cells expressing control shRNA had become confluent, dishes with cells expressing either of the two PP1c β shRNAs were subconfluent and were remarkable for many floating, presumably dead cells (Fig. 2B, top panels). Quantitatively, adherent PP1c β shRNA-expressing cells were 35% as numerous as control shRNA-expressing cells (45 cells/field versus 120 cells/field, 10 fields counted for each condition, n = 3, p < 0.01). The PP1c β shRNA-expressing cells that remained adherent were larger and more flattened (middle panels; 10.5 flattened cells/field for the PP1c β shRNA-expressing cells versus 5.2 flattened cells/field for the control shRNA-expressing cells, p < 0.01) and were characterized by large numbers of thin, F-actin-rich membrane extensions (bottom panels). Quantitatively, control cells exhibited three membrane extensions/cell versus 16/cell for the PP1c β shRNA-expressing cells (p < 0.01).
Identification of the Central Region of PP1c β as Being Key in Mediating the Isoform-specific Interaction with MYPT1
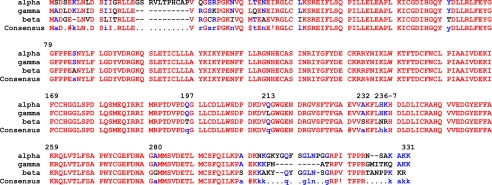
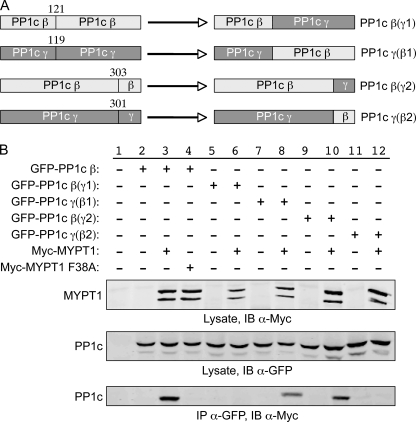
The α, β, and γ isoforms of PP1c exhibit variability in the N- and C-portions of the proteins but are highly similar over the central 85% of the protein (Fig. 3). Based on crystallographic studies, Terrak et al. (10) reported that the isoform-specific interaction between MYTP1 and PP1c β is mediated by the C-terminal region of PP1c β. To confirm and extend this report using a direct approach, we thus generated chimeric PP1c proteins between PP1c β and PP1c γ as shown in Fig. 4 and then repeated the co-IP pulldown assay performed in Fig. 1. As before, co-IP of PP1c β and MYPT1 was readily detectable (Fig. 4, lane 3), in contrast to the attempted co-IP of PP1c β and MYPT1 F38A (lane 4), where no interaction was observed. Consistent with the N terminus being unimportant in this context, the β(γ)1 allele, which contained the first 121 amino acids of PP1c β and the middle and remainder of PP1c γ, did not co-IP with MYPT1 (lane 6), whereas the reciprocal mutant, γ(β)1, did (lane 8). Unexpectedly though, the β(γ)2 construct, which derived its C-terminal region from PP1c γ, did co-IP with MYPT1 (lane 10), whereas the reciprocal construct γ(β)2, did not (lane 12). These findings suggested that the isoform specificity of the interaction arose from the highly conserved central region of the protein (amino acids 121–303) rather than via the variable C terminus. This possibility was then examined by generating chimeric isoforms in which the central regions of the β and γ isoforms were swapped (Fig. 5). Confirming the earlier results, the βγβ isoform (in which the central region was derived from PP1c γ) did not co-IP with MYPT1 (lane 4), whereas the reciprocal γβγ chimeric protein did (lane 6), ruling out a significant role for amino acids Tyr305 and Tyr307 in the PP1c β C terminus.
FIGURE 3.
Alignment of human PP1 isoforms. A sequence alignment of the three isoforms of human PP1c was generated using the Network Protein Sequence Analysis: Multalin Alignment (12). Sequence similarity is indicated in red, and semiconserved or nonconserved residues in blue or black, respectively.
FIGURE 4.
The C terminus of PP1c β does not mediate the isoform specificity of interaction with MYPT1. A, structure of PP1 chimera constructs. Based on the alignment of the three human PP1c isoforms, two sets of chimera constructs were generated as shown for PP1c β and PP1c γ. In the first set, PP1c was divided in half, and the C-terminal halves of PP1c β and PP1c γ were exchanged. These constructs were denoted PP1c β(γ1) and PP1c γ(β1). For the second set of constructs, the extreme C termini (last 30 amino acids) of PP1c β and PP1c γ were exchanged. These constructs were denoted PP1c β(γ2) and PP1c γ(β2). B, HeLa cells were transiently co-transfected with the GFP-tagged PP1c wild-type or chimeric plasmids and Myc-tagged wild-type or mutant MYPT1 as indicated. 48 h later, the cells were lysed, and Western blotting was performed against the corresponding proteins using the appropriate antibodies. Immunoprecipitation assays were performed as described in Fig. 1.
FIGURE 5.
The central region of PP1c β mediates isoform specificity of interaction with MYPT1. Chimeric constructs were generated and co-immunoprecipitated as shown and described in Fig. 1.
Isoform-specific Amino Acid Residues in PP1c β That Determine the Interaction with MYPT1
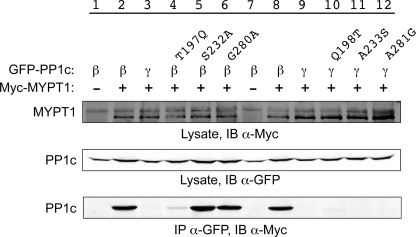
There are only six positions at which the amino acid sequences vary between PP1c β and PP1c γ in the central region (Fig. 3). We next used site-directed mutagenesis to examine their importance in the interaction. Mutation of Ser232 and Gly280 (and Gln213, not shown) to the corresponding amino acid in PP1c γ did not alter the efficiency of PP1c β and MYPT1 co-IP (Fig. 6, lanes 5 and 6). However, mutation of Thr197 to Gln almost completely eliminated the PP1c β-MYPT1 interaction (lane 4; residual interaction ∼4% of wild-type protein, n = 3 independent experiments). Conversely, substitution of the individual PP1c β amino acids into PP1c γ, including for Q198T, did not result in gain-of-interaction mutant alleles (lanes 10–12), ruling out a single determinative residue as being responsible for the isoform-specific interaction.
FIGURE 6.
Amino acid Thr197 is critical for interaction of PP1c β with MYPT1. Site-directed mutants of PP1c β and PP1c γ were generated and co-immunoprecipitated as shown and described in Fig. 1. For quantitation purposes, Western blot band density was measured using Odyssey Infrared imaging system software. The band density measurement for the PP1c β-MYPT1 co-IP, normalized to the total amount of MYPT1, was set at 100%, and the band density of the PP1c γ-MYPT1 co-IP was set at 0. The percentage of PP1c β T197Q interacting with MYPT1 was calculated in comparison. n = 3.
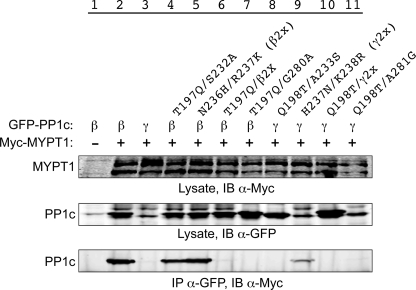
We then generated multimutants for analysis (Fig. 7), which yield interesting but more complex outcomes. Adding the S232A mutation, which in itself had no apparent effect on the interaction, to T197Q, rescued much of the loss of interaction observed for T197Q (Fig. 7, lane 4; 25% of control co-IP, n = 3). The double mutant N236H/R237K (β2x) interacted well with MYPT1 (lane 5, 65% of control interaction, n = 3), but adding this change to the T179Q mutation did not result in rescue of the T179Q loss of interaction (lane 6), and the same lack of rescue was observed for addition of the G280A mutation to T179Q (lane 7). Given these results, it was thus unexpected that mutation of two amino acids in PP1c γ (H237N and K238R (γ2x)) to the corresponding residues in PP1c β resulted in a partial gain-of-interaction outcome (lane 9, 3% of control interaction, n = 3), and further conversion of the sequence toward PP1c β (for example, Q198T/γ2x) again resulted in loss of the interaction (lane 10). Taken together, these results again suggest the lack of a single residue controlling the interaction and suggest instead that the residues unique to PP1c β position residues potentially in common between the different isoforms to interact effectively with MYPT1.
FIGURE 7.
MYPT1 binding capability of double and triple PP1c β and γ mutants. The experiment was performed as described above.
Placement of the Key Isoform-specific Residues on the PP1c β-MYPT1 Crystal Structure
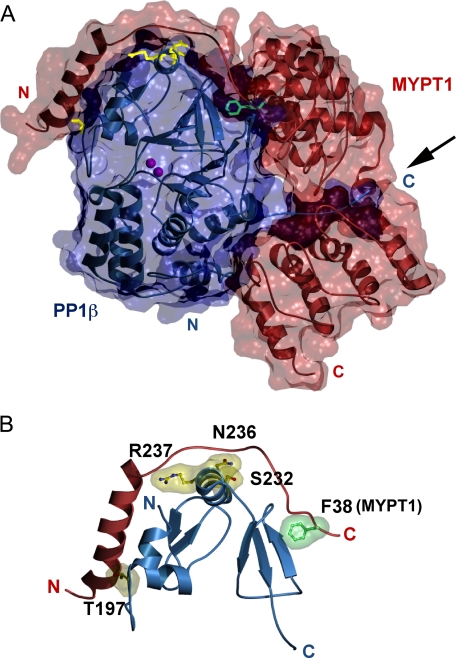
As described earlier, Terrak et al. (10) reported that the isoform-specific interaction between MYTP1 and PP1c β is likely mediated by the C-terminal region of PP1c β, which is indicated by the arrow in Fig. 8A. In contrast, we show here that this region is nondeterminative for the specificity of the interaction and that the key residues instead lie far away in the structure, at or near Thr197 through Ser232. Intriguingly, this stretch of residues constitutes the interaction surface of PP1c with the N-terminal arm of MYPT1, a region in MYPT1 thought to be key in mediating the interaction with myosin. Furthermore, this region neighbors the hydrophobic pocket in PP1c β (amino acids 242–293) that interacts with the MYPT1 RVXF motif. Phe38 in this motif (Fig. 8B) is essential for MP complex formation because the interaction is disrupted when it is mutated to alanine (9, 10). Interestingly, Gly280 is adjacent to the Phe38 binding pocket and, although not part of the interaction surface, substitution of this residue with alanine would necessarily imply a structural perturbation of the pocket.
FIGURE 8.
PP1c β residues that allow for interaction with MYPT1. A, space-filling model of the MYPT1 and PP1c β interaction. Residues 1–299 of MYPT1 are illustrated in red as previously published (Protein Data Bank code 1S70 (10), and full-length PP1c β is shown in blue. The secondary structure is displayed using a ribbon representation, and the molecular surface is shown as transparent. Manganese ions are illustrated in magenta. PP1c β residues 197, 232, 236, and 237 are illustrated in yellow. MYPT1 residue Phe38 is shown in green. Arrow, PPIc C terminus. B, interaction of the isoform specificity-determining region of PP1c β with the N-terminal arm of MYPT1. Residues 197, 232, 236, and 237 of PP1c β (yellow) and residue 38 of MYPT1 (green) are illustrated using a ball-and-stick representation, and their van der Waals surfaces are shown as transparent. The secondary structure is shown in red (MYPT1) or blue (PP1c β). Figures were made using MSMS (14) and povscript (15).
Three of the five remaining residues that are not conserved between PP1c isoforms (Ser232, Asn236, and Thr197) appear to mediate the PP1c β-MYTP1 interaction directly. Ser232 and Thr197 make hydrophobic contacts with MYTP1 residues, whereas Asn236 hydrogen-bonds with MYTP1 side chains and backbone. The N236H and T197Q substitutions would result in obvious steric clashes with MYTP1 in the conformation observed in the crystal structure, and, consistently, the single T197Q mutation was able to disrupt the interaction severely. None of the residues, however, appears to be individually responsible for the specificity of the interaction, and this is consistent with their observed positions in the crystal structure. The results with the multiple mutants confirm that subtle alterations to the interaction surface, rather than the presence or absence of specific contacts, are responsible for modulating binding specificity. Thus, the effects of different individual mutations can be partially compensatory, and it is the global structural changes that result from them that appear to determine the specificity of the interaction. For instance, whereas the PP1c β N236 and R237 side chains are not essential to maintain the interaction, the reciprocal mutation (of PP1c γ His237 and Lys238 to Asn and Arg, respectively) generated a gain-of interaction outcome.
DISCUSSION
In this study, we report the surprising finding that the isoform-specific interaction of PP1c β with MYPT1 is driven by interactions with just a few nonconserved residues in a central region of PP1c β that is otherwise 96% identical to PP1c γ, rather than by interactions with the nonconserved N and C termini. This finding is in contrast to the interpretation of a prior report on the co-crystal structure of PP1c β with MYPT1 (10). However, our work also supports and extends other aspects of the published report by demonstrating that the region in PP1c β that confers the isoform specificity is one known already to interact with MYPT1, at its N terminus. The RVXF has become recognized as a key motif in the more than 100 proteins that interact with the PP1 superfamily (13). However, because the hydrophobic region in PP1c that binds the RVXF motif is identical in the three PP1c isoforms, isoform-specific interaction cannot be mediated by this component of the complex formation. The four key residues identified, Thr197, Ser232, Asn236, and Arg237, are similarly unique in PP1c β compared with PP1c α, which also does not interact with MYPT1.
As described in the Introduction, MP undertakes diverse roles in transformation and growth and migration of cancer cells, and we illustrate the significance of this complex by showing changes in cell viability/adherence and morphology when PP1c β or MYPT1 is knocked down using shRNA. Small molecule compounds that target other components of the cytoskeleton as therapeutics have been developed, such as Taxol, which stabilizes the microtubule network, and having the ability to target MP could add depth in the clinical setting because Taxol is generally only effective initially, i.e. cancer cells eventually develop resistance to it. One approach would be to develop phosphatase inhibitors; however, it would be difficult to create a PP1c β-specific inhibitor, given its high degree of similarity to other PP1c isoforms, much less to the larger PP1 superfamily of phosphatases.
Another approach would be to identify small molecule compounds capable of disrupting the specific interaction between PP1c β and MYPT1 using high throughput screens. Finding such compounds could be challenging using approaches based on the wild-type, full-strength PP1c β-MYPT1 interaction. It is possible, however, that lead compounds could readily be identified, based on the series of mutants we describe in this report, which exhibit weakened interactions. Screens and counterscreens could be designed to disrupt the interaction between MYPT1 and residues on PP1c β such as Thr197, Thr197/Ser232, and Asn236/Arg237, which exhibit 4%, 25%, and 75% of the MYPT1 interaction compared with wild-type PP1c β to identify lead compounds that could then be improved through combinatorial chemistry optimization.
Acknowledgments
We thank Dr. Masumi Eto at Thomas Jefferson University for the Myc-MYPT1 constructs and Dr. Yusuf A. Hannun at Medical University of South Carolina for the GFP-PP1α, β, and γ. We also thank members of the Frohman and Du laboratories for technical assistance.
This work was supported by National Institutes of Health Grants GM071520 and GM084251 (to M. A. F.) and GM071475 (to G. D.).
- MP
- myosin phosphatase
- MYPT1
- myosin phosphatase targeting subunit 1
- PP1
- protein phosphatase 1
- CHO
- Chinese hamster ovary
- shRNA
- small hairpin RNA
- GFP
- green fluorescent protein
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1.Ito M., Nakano T., Erdodi F., Hartshorne D. J. (2004) Mol. Cell. Biochem. 259, 197–209 [DOI] [PubMed] [Google Scholar]
- 2.Dimopoulos G. J., Semba S., Kitazawa K., Eto M., Kitazawa T. (2007) Circ. Res. 100, 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia D., Stull J. T., Kamm K. E. (2005) Exp. Cell Res. 304, 506–517 [DOI] [PubMed] [Google Scholar]
- 4.Du G., Frohman M. A. (2009) Mol. Biol. Cell 20, 200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra M., Mahmoudi T., Verdin E. (2007) Genes Dev. 21, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin H., Sperka T., Herrlich P., Morrison H. (2006) Nature 442, 576–579 [DOI] [PubMed] [Google Scholar]
- 7.Jung J. R., Kim H., Jeun S. S., Lee J. Y., Koh E. J., Ji C. (2005) Mol. Cells 20, 196–200 [PubMed] [Google Scholar]
- 8.Schlabach M. R., Luo J., Solimini N. L., Hu G., Xu Q., Li M. Z., Zhao Z., Smogorzewska A., Sowa M. E., Ang X. L., Westbrook T. F., Liang A. C., Chang K., Hackett J. A., Harper J. W., Hannon G. J., Elledge S. J. (2008) Science 319, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eto M., Kirkbride J. A., Brautigan D. L. (2005) Cell Motil. Cytoskeleton 62, 100–109 [DOI] [PubMed] [Google Scholar]
- 10.Terrak M., Kerff F., Langsetmo K., Tao T., Dominguez R. (2004) Nature 429, 780–784 [DOI] [PubMed] [Google Scholar]
- 11.Corbalan-Garcia S., Margarit S. M., Galron D., Yang S. S., Bar-Sagi D. (1998) Mol. Cell. Biol. 18, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet F. (1988) Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. (2009) Chem. Biol. 16, 365–371 [DOI] [PubMed] [Google Scholar]
- 14.Sanner M. F., Olson A. J., Spehner J. C. (1996) Biopolymers 38, 305–320 [DOI] [PubMed] [Google Scholar]
- 15.Fenn T., Ringe D., Petsko G. (2003) J. Appl. Crystallogr. 36, 944–947 [Google Scholar]