Abstract
Five nontruncating missense APOB mutations, namely A31P, G275S, L324M, G912D, and G945S, were identified in heterozygous carriers of familial hypobetalipoproteinemia (FHBL) in the Italian population. To test that the FHBL phenotype was a result of impaired hepatic secretion of mutant apoB proteins, we performed transfection studies using McA-RH7777 cells stably expressing wild type or mutant forms of human apolipoprotein B-48 (apoB-48). All mutant proteins displayed varied impairment in secretion, with G912D the least affected and A31P barely secreted. Although some A31P was degraded by proteasomes, a significant proportion of it (although inappropriately glycosylated) escaped endoplasmic reticulum (ER) quality control and presented in the Golgi compartment. Degradation of the post-ER A31P was achieved by autophagy. Expression of A31P also decreased secretion of endogenous apoB and triglycerides, yet the impaired lipoprotein secretion did not lead to lipid accumulation in the cells or ER stress. Rather, expression of genes involved in lipogenesis was down-regulated, including liver X receptor α, sterol regulator element-binding protein 1c, fatty acid synthase, acetyl-CoA carboxylase 1, stearoyl-CoA desaturase 1, and lipin-1. These results suggest that feedback inhibition of hepatic lipogenesis in conjunction with post-ER degradation of misfolded apoB proteins can contribute to reduce fat accumulation in the FHBL liver.
Keywords: Diseases/Atherosclerosis, Lipoprotein/Apolipoprotein Genes, Lipoprotein/Secretion, Metabolism/Lipogenesis, Protein/Binding/Lipid, Protein/Structure
Introduction
Familial hypobetalipoproteinemia (FHBL)5 (OMIM 107730) is an autosomal co-dominant disorder characterized by low plasma levels (<5th percentile for age and sex) of low density lipoprotein (LDL) cholesterol and apolipoprotein B (apoB) (1). The genetic basis for FHBL is heterogeneous and is often associated with mutations within the APOB gene (1). The APOB gene encodes both apoB-100 and apoB-48, the latter being a truncated form representing the N-terminal 48% of the full-length apoB-100 (2, 3). In rats and mice, both apoB-100 and apoB-48 are synthesized in the liver (4). The majority of APOB gene mutations in FHBL are nonsense, frameshift, or splicing variants that lead to various C-terminally truncated apoB species (5). However, nonsynonymous, nontruncating mutations (e.g. L343V and R463W) (6, 7) do occur in FHBL. Transfection studies showed that apoB-100 and apoB-48 bearing the L343V or R463W mutation were secreted inefficiently and exhibited increased endoplasmic reticulum (ER) retention (6, 7). Unlike truncation mutants (ranging from apoB-15 to apoB-94) that are often secreted normally (8, 9), the nontruncation mutants are impaired in their secretion.
The nontruncating mutations identified in FHBL to date are all located within the N-terminal region composed of multiple anti-parallel α-helices (residues 292–593) (10, 11). This region, together with the preceding β-barrel (the first 264 residues), is known as a modeled βα1 domain of apoB based on its homology to lipovitellin (10). This homology model predicts that the βα1 domain of apoB consists of β-barrel and α-helical domain plus two β-sheets termed C sheet (residues 611–782) and A sheet (residues 783–930), respectively. The βα1 domain is known to be important for effective secretion from transfected cells, because recombinant apoB segments lacking this domain were either secreted poorly or not secreted at all (12, 13). The βα1 domain can bind to microsomal triglyceride transfer protein (MTP), an apoB-specific molecular chaperone that facilitates lipid recruitment during VLDL assembly (14). There are at least three MTP binding sites within the βα1 domain (residues 2–154, 430–570, and 512–721) (10, 15, 16). Although some studies have shown that secretion of the βα1 domain is independent of MTP (see Ref. 17 and references cited within), there is no evidence showing that apoB-100 or apoB-48 can be secreted without the activity of MTP.
VLDL assembly and maturation involve multiple lipidation steps that occur during trafficking of VLDL precursors throughout the ER/Golgi compartments (18). The assembly process is highly dependent on the availability of lipids, mainly triacylglycerol (TAG). The activities of TAG synthesis enzymes, including DGAT1 (diacylglycerol acyltransferase 1) (19) and lipin-1 (20), have been shown to control VLDL assembly/secretion. Increased de novo lipogenesis also contributes significantly to VLDL assembly/secretion. Regulation of hepatic lipogenesis requires a network of nuclear receptors that coordinately govern expression of the lipogenesis enzymes acetyl-CoA carboxylase 1 (ACC1), fatty acid synthase (FAS), and SCD1 (stearoyl-CoA desaturase) (21). Transcription factors and co-factors regulating lipogenesis gene expression and VLDL production include liver X receptor α (LXRα) (22), SREBP-1 (sterol regulatory element-binding protein 1), and PGC-1 (peroxisome proliferator-activated receptor-γ coactivator) family members PGC-1α and PGC-1β (23), among others.
Because of the importance of the βα1 domain in VLDL assembly/secretion, we have searched for naturally occurring mutations within this domain that could affect apoB secretion. Recently, through molecular diagnosis of 110 Italian subjects with clinical diagnosis of FHBL, we discovered eight subjects carrying novel missense mutations, five of which occurred within the βα1 domain. The present studies showed that mutation A31P within the β-barrel severely impaired apoB secretion, and cells expressing the A31P mutant exhibited suppressed lipogenesis.
EXPERIMENTAL PROCEDURES
Materials
Cell culture reagents were purchased from Invitrogen. Reagents for recombinant DNA experiments were obtained from New England Biolabs (Pickering, Canada). [2-3H]Glycerol (9.6 Ci/mmol) and [35S]methionine/cysteine (1,000 Ci/mmol) were obtained from GE Healthcare. [3H]Acetic acid (0.1 Ci/mmol) was obtained from PerkinElmer Life Sciences. Rabbit anti-mouse apoE antibody (used for immunoprecipitation) was obtained from BioDesign (Saco, ME). Horseradish peroxidase-linked anti-goat antibody and 3-methyladenine were obtained from Sigma. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibodies were obtained from Amersham Biosciences. MG132 was obtained from Calbiochem. Chemiluminescent substrates were purchased from Roche Applied Science. Trypsin was obtained from Promega (Madison, WI). Antibodies against FAS and giantin (Abcam (Cambridge, MA)); ACC, phospho-ACC (at Ser79), calnexin, and actin (Millipore (Temecula, CA)); eIF2a and phospho-eIF2α (at Ser52) (Cell Signaling Technology (Danvers, MA)); C/EBP-homologous protein (CHOP or GADD153) (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)); and GRP78 (glucose-regulated protein 78) (Assay Designs (Ann Arbor, MI)) were purchased from the respective suppliers. Monoclonal antibody 1D1 against human apoB was a gift of Drs. Yves Marcel and Ross Milne (University of Ottawa Heart Institute). Anti-MTP antibody was a gift of Dr. Carol Shoulders (Medical Research Council Clinical Sciences Centre, Hammersmith Hospital, London, UK). Polyclonal antiserum against rat VLDL proteins (used for immunoprecipitation of rat apoB-100) was produced in our laboratory.
The FHBL Subjects
The FHBL subjects were identified as probable heterozygous carriers during a survey of 110 consecutive individuals referred to the hospital for the presence of fatty liver associated with low plasma levels of LDL cholesterol (LDL-C) and apoB (1). The diagnosis of definite FHBL was based on three criteria: (i) LDL-C < 70 mg/dl and apoB < 50 mg/dl; (ii) exclusion of secondary hypocholesterolemia; and (iii) vertical transmission of the lipid and apoB phenotype in family. The diagnosis of probable heterozygous FHBL was made when reliable family data were unavailable. Sequencing of the APOB gene revealed pathogenic mutations in 54 subjects (1). The majority of the mutations were located in the coding sequence of APOB. Fifty-one subjects were found to carry mutations causing the formation of truncated apoBs of various sizes (1). Three subjects were carriers of the previously described missense mutation R463W (6), known to be the cause of FHBL. Five subjects (four with definite and one with probable FHBL) were heterozygous for rare non-conserved APOB gene mutations, namely G945S, A31P, L324M, G912D, and G275S (1). The present study focused on these five mutations.
Analysis of APOB Gene
Genomic DNA was isolated from peripheral blood (NucleoSpin Blood L, Macherey-Nagel, Duren, Germany). The entire APOB gene coding region, including the 5′-flanking region and at least 50 base pairs of intronic sequence at each intron-exon boundary (GenBankTM accession number AY324608.1, GI:32187678) were amplified by PCR and sequenced as previously described (24). Sequences were detected on an ABI PRISM 3100 DNA sequencer, and results were analyzed with ABI PRISM SeqScape software (Applied Biosystems, Warrington, UK). Only a few members of the five kindred were available for DNA analysis. The APOB gene mutations found in the probands were screened in family members available for the study by sequencing the appropriate gene regions (25). Sequence variants were confirmed by using a second independent amplification of the affected DNA region and resequencing in both directions. One hundred control subjects randomly selected among normolipidemic individuals of both sexes were screened for the mutations of the APOB gene found in FHBL patients by direct sequencing of the appropriate gene regions or by restriction enzyme digestion of PCR products. APOB gene mutations are described according to mutation nomenclature (available on the Human Genome Variation Society Web site) (26, 27). Nucleotide numbers are derived from APOB cDNA sequence (GenBankTM accession number NM_000384). Amino acid sequence changes in apoB are described according to the National Centre for Biotechnology Information reference sequence (NP_000375.2, GI:105990532). The apoE genotype was performed as described (28). Informed consent was obtained from all probands (or in the case of children from their parents) and their family members as well as from control subjects. The study protocol was approved by the ethics committee of the University of Modena and Reggio Emilia and is in accordance with ethical standards as formulated by the Helsinki Declaration of 1975 (revised in 1983).
Analysis of Plasma Lipids and Lipoproteins
Blood samples were collected after an overnight fast. Plasma total cholesterol, TAG, high density lipoprotein cholesterol, LDL-C, apoA-I, and apoB levels were determined with enzymatic methods (Roche Applied Science) by using a Hitachi 912 autoanalyzer. Plasma lipoproteins were also separated by density gradient ultracentrifugation (24). Isolated lipoprotein fractions were precipitated in 10% trichloroacetic acid and extracted with ethanol diethyl ether (3:2, v/v), and apolipoproteins were separated by SDS-PAGE (3.5/5–10% gel for apoB and 5–20% gel for other proteins) (24).
Preparation of Expression Plasmids
The expression plasmids encoding human apoB-48 or apoB-17 prepared previously (7) were used as templates to prepare mutants using the QuikChange® II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic primers used to introduce the A31P, G275S, L324M, G912D, and G945S mutations were listed in supplemental Table 1. The expression plasmids were purified by the Qiagen plasmid purification kit (Qiagen, Milano, Italy), and the APOB coding regions were authenticated by sequencing.
Cell Culture, Transfection, and Detection of Recombinant ApoB
Stable expression of all of the recombinant apoB-48 proteins was achieved using McA-RH7777 cells as described previously (29). The cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 10% horse serum. Stable clones were selected with G418 (400 μg/ml), and those that showed secretion of apoB-48 mutants at similar levels were maintained in G418 (200 μg/ml) for experiments. Expression of apoB-48 and apoB-17 was determined by Western blotting of conditioned media using monoclonal antibody 1D1 that has an epitope at amino acids 401–582 of human apoB-100 (8).
Lipoprotein Fractionation
Lipoproteins presented in the microsomal lumen or secreted into the media were fractionated into VLDL1, VLDL2, and other lipoproteins as described previously (30).
Subcellular Fractionation of Microsomal Membranes
Subcellular fractionation of ER and Golgi microsome and analysis of endoglycosidase H (Endo H) sensitivity of apoB-48 proteins associated with the microsomes were performed as described previously (31).
Co-immunoprecipitation
Cells were cultured in DMEM containing 20% FBS and 0.4 mm oleate for 2 h and harvested in 1 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 20% sucrose, protease inhibitor mixture). The lysate samples were incubated for 1 h at 4 °C by gentle mixing, and the insoluble materials were removed by centrifugation (13,000 rpm, 4 °C, 15 min). An aliquot of the supernatant (750 μg of protein) was mixed with lysis buffer to a final volume of 1 ml. The samples were precleared by incubation with protein A-Sepharose CL-4B beads (2 h, 4 °C) prior to incubation with monoclonal antibody 1D1 that had been coupled to CNBr-activated Sepharose 4B beads as described previously (30). After overnight incubation at 4 °C, the proteins were eluted and subjected to SDS-PAGE and Western blot analysis to quantify apoB, MTP, and GRP78.
Pulse-chase Analysis of ApoB
Post-translational stability and secretion efficiency of apoB (recombinant apoB-48 and endogenous apoB-100) were determined by pulse-chase experiments with [35S]methionine/cysteine as described previously (7). In some experiments, the post-translational stability and secretion of apoB-48 mass were determined in the presence of cycloheximide (10 μm) (32). Cells (in 60-mm dishes) were cultured in DMEM containing 20% FBS and 0.4 mm oleate for 0, 1, 2, and 4 h. Cycloheximide was added to the media at time 0. The cell and medium samples were collected at the indicated times, and the recombinant apoB-48 proteins were recovered from cells and media, respectively, by immunoprecipitation. After SDS-PAGE and Western blotting, the intensity of apoB-48 bands was quantified by scanning densitometry.
Metabolic Labeling of Lipids
Synthesis and secretion of TAG and phosphatidylcholine (PC) were determined by metabolic labeling of cells with either [3H]glycerol for 1 and 2 h in DMEM containing 20% FBS and 0.4 mm oleate or [3H]acetic acid for up to 8 h in DMEM containing 20% FBS. Extraction and analysis of lipids by thin layer chromatography were performed as described previously (20).
Protein Sample Preparation for Mass Spectrometry
Total cell lysates were resolved by SDS-PAGE. Proteins were visualized by silver staining, and bands of interest were excised and subjected to in-gel digestion as described previously (33) with modifications. The gel pieces (∼1 mm3) were washed with 50 mm NH4HCO3 and subsequently dehydrated in 50% acetonitrile (v/v) and 25 mm NH4HCO3 (at room temperature, 15 min). After aspirating the supernatant, the gel pieces were dried under vacuum and treated with 10 mm dithiothreitol (in 50 mm NH4HCO3) at 56 °C for 15 min to reduce disulfide bonds. Free -SH groups were alkylated with 100 mm iodoacetamide (in 50 mm NH4HCO3) for 15 min in darkness at room temperature. The gel pieces were dehydrated again in 50% acetonitrile (v/v) as described above and subjected to trypsin digestion (20 ng of trypsin/μl in 50 mm NH4HCO3) at 37 °C overnight. Peptides were extracted from the gel pieces with 5% formic acid (v/v) and 50% acetonitrile (v/v) with sonication in ice-cold aqueous solution and dried under vacuum. The peptide samples were stored at −20 °C prior to mass spectrometric analysis.
Liquid Chromatography-MS/MS and Dada Analysis
The liquid chromatography-MS/MS was performed as described previously (34). The resulting peptides from the gel bands were resolved in 5% formic acid and loaded on a 200 μm × 50-mm fused silica precolumn packed in house with 5 cm of 5-μm YMC ODS-A C18 beads (Waters Co. (Milford, MA)) using an 1100 micro-HPLC system (Agilent Technologies, Santa Clara, CA). Following a desalting step, the flow was split, and peptides were eluted through a second 75 μm × 50-mm column packed with the same beads at ∼200 nl/min. The peptides were eluted using a 2-h gradient (5–80% acetonitrile with 0.1% formic acid) into an ESI LTQ linear ion trap mass spectrometer (Thermo Electron, Waltham, MA). MS/MS spectra were acquired in a data-dependent acquisition mode that automatically selected and fragmented the 10 most intense peaks from each mass spectrum generated. The acquired MS/MS spectra were searched against the mouse International Protein Index protein sequence data base (version 3.52, 39,906 protein sequences; European Bioinformatics Institute) augmented with the reversed sequence of each entry in the forward data base. Mascot 2.2.02 (Matrix Science) was used to search the protein sequence data base. The precursor and fragment mass tolerances for the LTQ data were set at 2.0 and 0.8 Da, respectively. Mascot cut-off scores were set to 30. Peptides ranked with a probability-based Mowse (expect) p < 0.05 were accepted. The false positive rate was controlled at <1%. The identified proteins with the expected molecular weight and at least two non-redundant peptides were accepted after rigorous filtration as described above.
Real-time RT-PCR
Isolation of RNA from cells, reverse transcription, and relative mRNA concentration determination by real-time RT-PCR were performed as described previously (20). Cyclophilin A was used as the real-time RT-PCR control. The primers used for RT-PCR were obtained from Sigma, and the sequences of the primers are listed in supplemental Table 2.
Molecular Modeling
The homology model of the N-terminal 930 amino acids of apoB-100 (apoB-20.5) was constructed using MODELLER 7.7 using standard parameters (35) and the coordinates of lamprey lipovitellin (36). The sequences of apoB20.5 and lipovitellin were aligned using the Blast-2-Sequence program (37). Graphic representation was achieved by MOLMOL (38).
RESULTS
Clinical Phenotyping and Genotyping of Five Heterozygous FHBL Subjects
Table 1 summarizes the clinical and biochemical features of members of the five FHBL kindreds examined. All probands and some of the respective family members had fatty liver documented by abdominal liver ultrasound examination (as described in Refs. 24 and 39). Detailed description of the kindreds is available as supplemental material. Sequencing of the APOB gene revealed that each proband was heterozygous for single nucleotide substitutions causing novel non-conservative amino acid substitutions in mature apoB. They are c.2914 G → A in exon 19 (G945S) in FHBL-36 (supplemental Fig. S1A), c.2816 G → A in exon 18 (G912D) in proband FHBL-39 (supplemental Fig. S1B), c.1051 C → A in exon 9 (L324M) in proband FHBL-44 (supplemental Fig. S1C), c.172 G → C in exon 3 (A31P) in proband FHBL-45 (supplemental Fig. S1D), and c.904 G → A in exon 8 (G275S) in proband FHBL-46 (not shown in supplemental Fig. S1). All of the mutations occurred within the βα1 domain (Fig. 1A) (10, 11). These mutations co-segregated with the FHBL lipid phenotype in the families and were not found in a large group of normolipidemic healthy control subjects. However, the small number of family members in the respective kindred precluded the feasibility of performing genetic linkage analysis. All of the amino acids involved in the mutations, with the exception of Gly912, are highly conserved in mammalian species.
TABLE 1.
Clinical and biochemical features of FHBL kindreds
BMI, body mass index; TC, total cholesterol; HDL-C, high density lipoprotein cholesterol; NR, normal range (aspartate aminotransferase (AST), 0–35 units/liter; alanine aminotransferase (ALT), 0–35 units/liter); Norm, normal; NT, not tested; +, presence of fatty liver detected by ultrasound examination.
| Subject ID | Age | BMI | TC | TAG | HDL-C | LDL-C | ApoB | ApoA-I | AST | ALT | Fatty liver | ApoB mutation | ApoE genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| years | kg/m2 | mg/dl | mg/dl | mg/dl | mg/dl | mg/dl | mg/dl | units/liter | units/liter | ||||
| Kindred FHBL-36 | |||||||||||||
| I.1 | 70 | NT | 152 | 39 | 61 | 83 | 71 | 141 | NR | NR | Norm | E3E3 | |
| I.2 | 70 | NT | 203 | 89 | 51 | 134 | 106 | 135 | NR | NR | G945S | E3E3 | |
| II.1 | 30 | NT | 121 | 86 | 40 | 64 | 55 | 118 | NR | NR | G945S | E3E3 | |
| II.2 | 36 | 19.1 | 72 | 30 | 50 | 16 | 18 | 110 | 45 | 89 | + | G945S | E3E3 |
| III.1 | 3 | NT | 129 | 47 | 44 | 73 | 79 | 123 | NR | NR | No. | E3E3 | |
| Kindred FHBL-39 | |||||||||||||
| I.1 | 76 | NT | 210 | 107 | 43 | 146 | 110 | 130 | NR | NR | Norm | NT | |
| I.2 | 69 | NT | 152 | 50 | 70 | 72 | 60 | 150 | G912D | NT | |||
| II.1 | 52 | NT | 208 | 188 | 26 | 144 | 111 | 100 | NR | NR | Norm | NT | |
| II.2 | 44 | NT | 197 | 46 | 61 | 126 | 98 | 140 | NR | NR | Norm | NT | |
| II.3 | 47 | NT | 78 | 57 | 33 | 36 | 29 | 102 | 39 | 47 | + | G912D | E3E3 |
| II.4 | 49 | NT | 207 | 93 | 45 | 143 | 113 | 141 | NR | NR | Norm | NT | |
| III.1 | 22 | NT | 145 | 81 | 29 | 99 | 90 | 108 | NR | NR | Norm | NT | |
| III.2 | 18 | 24.9 | 109 | 36 | 53 | 50 | 39 | 120 | 11 | 19 | + | G912D | E3E3 |
| Kindred FHBL-44 | |||||||||||||
| I.1 | 41 | NT | 126 | 52 | 56 | 60 | 43 | 135 | 77 | 47 | + | L324M | E3E3 |
| I.2 | 39 | NT | 186 | 121 | 41 | 121 | 77 | 127 | NR | NR | Norm | E2E3 | |
| II.1 | 9 | 14.3 | 85 | 59 | 30 | 43 | 30 | 80 | 47 | 37 | + | L324M | E2E3 |
| FHBL-45 | |||||||||||||
| II.1 | 65 | NT | 199 | 69 | 44 | 141 | 86 | 127 | NR | NR | Norm | NT | |
| II.2 | 71 | NT | 115 | 50 | 64 | 41 | 26 | 141 | 40 | 52 | + | A31P | E3E3 |
| II.3 | 73 | NT | 119 | 109 | 37 | 60 | 38 | 118 | 65 | 70 | + | A31P | NT |
| III.1 | 34 | 24.7 | 92 | 37 | 46 | 39 | 28 | 109 | 31 | 68 | + | A31P | E3E4 |
| FHBL-46 | 28 | 81 | 58 | 41 | 28 | 28 | 111 | 81 | 63 | + | G275S | E3E3 | |
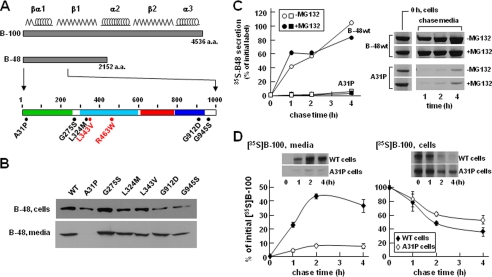
FIGURE 1.
Transfection analysis of apoB-48 mutants. A, schematic diagram of apoB-48 construct. The top bar shows apoB-100, with the predicted locations of βα1, β1, α2, β2, and α3 domains shown above. The middle bar depicts apoB-48 with the expanded N-terminal 1,000 amino acids shown at the bottom. The β barrel (residues 1–264), α-helical bundle (residues 292–593), C-sheet (residues 611–782), and A-sheet (residues 783–930) regions are highlighted in green, cyan, red, and blue, respectively. Positions of the five nonsynonymous mutations are indicated in black. Two previously reported mutations (L343V and R463W) (6, 7) are shown in red. B, transient assay of apoB-48 expression and secretion. McA-RH7777 cells were transfected with the indicated apoB-48-expressing plasmids (8 μg). Two days post-transfection, the cells were incubated with fresh DMEM containing 20% FBS and 0.4 mm oleate for 4 h. Cells and media were collected and subjected to SDS-PAGE and Western blot analysis for human apoB-48. The mutant L343V construct reported previously (6) was used as transfection control. WT, wild type apoB-48. C, pulse-chase analysis of apoB-48 secretion. Stable cell lines expressing apoB-48wt or apoB-48A31P mutant were pulse-labeled with [35S]methionine/cysteine in methionine/cysteine-free media with or without MG132 for 30 min and chased in normal DMEM with or without MG132 for up to 4 h. Both pulse and chase media contained 20% FBS and 0.4 mm oleate. The 35S-labeled apoB-48 was recovered from cells at the end of pulse labeling (0 h chase) and also recovered from media at 1, 2, and 4 h chase time. Secretion efficiency is expressed as percentage of the initial radiolabeled apoB-48. Representative fluorograms are shown on the right. D, pulse-chase analysis of endogenous apoB-100 secretion. The experiment was performed in essentially the same manner as in C, except endogenous [35S]apoB-100 in the media (left panel) and cells (right panel) during chase was analyzed. Data represent the average of two independent experiments. Representative fluorograms of apoB-100 are shown as an inset.
Transfection Studies of ApoB-48 Mutants Revealed Impaired Secretion from McA-RH7777 Cells
We tested the effect of the identified FHBL mutations on apoB-48 secretion by transfection studies using McA-RH7777 cells that retain the ability to assemble and secrete VLDL (30, 40). Transient assay with apoB-48 bearing the FHBL mutations gave correct expression in the transfected cells (Fig. 1B, top). Except for A31P, all of the apoB-48 proteins were detected in the media (Fig. 1B, bottom). The effect of the A31P mutation on apoB secretion was also tested in apoB-17 that had little lipid-binding ability (9). Transient assay with apoB-17A31P showed robust expression in the cells but no secretion (data not shown). Thus, the effect of A31P mutation on apoB secretion is independent of lipidation.
The lack of apoB-48A31P secretion was further confirmed using stable cell lines. Pulse-chase analysis showed that although apoB48wt was secreted efficiently, the A31P mutant was barely detectable in the media (Fig. 1C). Including the proteasome inhibitor MG132 had little effect on apoB-48 secretion efficiency, although cell-associated A31P protein was increased at the end of the 30-min pulse labeling (see bands labeled 0 h, cell in Fig. 1C). Expression of A31P severely impaired secretion of endogenous rat apoB-100 (Fig. 1D) but not apoE (data not shown).
Post-translational stability of the A31P mutant was determined in cells cultured with cycloheximide (to block protein synthesis). Although cellular apoB-48wt was decreased rapidly (t½ ∼ 1 h) as a consequence of efficient secretion, A31P mutant showed prolonged intracellular retention (t½ > 2 h), although it was not secreted (Fig. 2A). The addition of MG132 partially blocked decay of cellular A31P (Fig. 2B) but not apoB-48wt (data not shown). These results suggest that a significant proportion of A31P mutant was degraded by proteasomes.
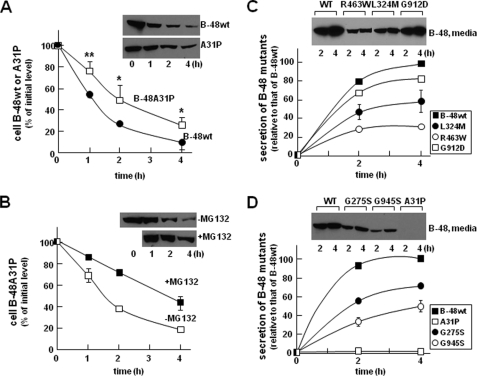
FIGURE 2.
Analysis of post-translational stability and secretion efficiency of apoB-48. A, stable apoB-48wt- or apoB-48A31P-expressing cells were incubated with DMEM containing 20% FBS and 0.4 mm oleate for 1, 2, and 4 h. Cycloheximide (10 μm) was included in the media at the beginning of incubation. At the indicated time points, the cells were lysed, and apoB-48 proteins were analyzed by SDS-PAGE/Western blotting. The intensity of apoB-48 bands was semiquantified by scanning densitometry. Data are presented as percentage of initial level (at time 0). Each data point is the average of four independent experiments. **, p < 0.01; *, p < 0.05 (Student's t test of apoB-48A31P versus apoB-48wt). Error bars, ±S.D. (n = 4). B, effect of MG132 on post-translational stability of apoB-48A31P. The experiment was performed in essentially the same manner as described in A, except that one group of apoB-48A31P cells were incubated in the same media supplemented with MG132. Each data point is the average of two independent experiments. C and D, secretion efficiency of other mutants. The experiments were performed in essentially the same manner as described in the legend to Fig. 1C, except stable cell lines expressing G275S (D), L324M (C), G912D (C), or G945S (D), in addition to A31P (D), were used. Data points for apoB-48wt, A31P, L324M, and G945S were the average of two independent experiments, and those for G275S and G912D were performed once. The previously reported R463W-expressing cells (6) were included as controls (C). WT, wild type.
Similar experiments were performed with stable cell lines expressing the other FHBL mutants. It was shown previously and confirmed here that the R463W mutant form of apoB-48 was secreted poorly as compared with apoB-48wt (Fig. 2C). Likewise, L324M (Fig. 2C), G275S (Fig. 2D), G912D (Fig. 2C), and G945S (Fig. 2D) mutants were also secreted at reduced levels. Among these, the G912D mutation had the least effect on apoB-48 secretion as compared with A31P. These results indicate that the hypobetalipoproteinemia phenotypes of the five kindreds, such as the reduced plasma cholesterol, TAG, and apoB concentrations (Table 1), are not caused solely by impaired secretion of the mutant apoB or apoB-containing lipoproteins. The t½ of cellular apoB for most mutants was similar to that of apoB-48wt (data not shown), except A31P showed markedly prolonged intracellular retention (Fig. 2A). These data suggested that, unlike mutations within the α-helical domain or β-sheets that only moderately affected apoB-48 secretion, the A31P mutation that occurred within the β-barrel almost entirely blocked apoB-48 secretion. The following experiments focused on A31P mutant.
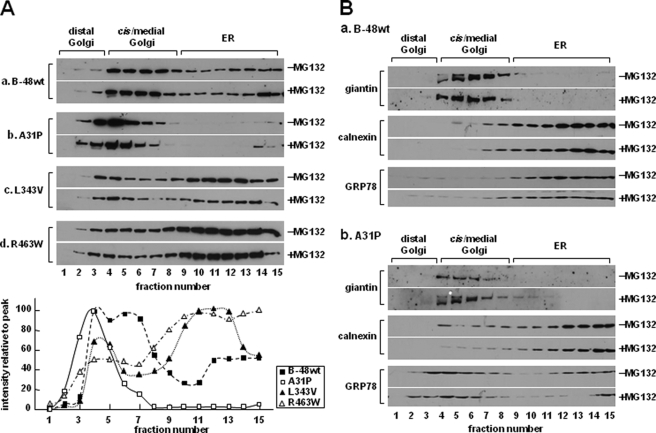
Distribution of ApoB-48A31P within ER/Golgi Compartment
The lack of A31P mutant secretion might be attributable to its inability to escape ER quality control. To test this, we determined ER/Golgi distribution of A31P mutant. At steady state, apoB-48wt was distributed across the entire ER/Golgi secretory pathway (Fig. 3A, panel a). In contrast, the A31P mutant was predominantly located in Golgi compartments (Fig. 3A, panel b), which was different form L343V or R463W mutants that showed ER retention (panels c and d). The addition of MG132 to the media did not alter distribution of any of the proteins (Fig. 3A, +MG132). Thus, the absence of A31P within the ER was unlikely to be attributable to rapid ER-associated degradation. Distribution of ER (calnexin) and Golgi (giantin) marker proteins was unchanged between apoB-48wt and A31P cells (Fig. 3B), indicting that physicochemical properties of the secretory machinery were intact. It was noted that distribution of GRP78 was skewed toward cis/medial Golgi fractions in A31P cells (Fig. 3B, compare GRP78 blots between panels a and b). The similar distribution between apoB-48A31P and GRP78 indicates that they may coexist in the same compartments.
FIGURE 3.
Distribution of apoB-48 proteins within the ER/Golgi secretory compartment. A, cells stably expressing apoB-48wt (a) or A31P (b), L343V (c), or R463W (d) mutants were cultured in DMEM (containing 20% FBS and 0.4 mm oleate) with or without MG132 for 1 h. The cells were homogenized, and the microsomes were separated into distal Golgi, cis/medial Golgi, and ER fractions by ultracentrifugation in a Nycodenz gradient. The apoB-48 proteins in each fraction were detected by SDS-PAGE/Western blotting. Bottom, semiquantification of apoB-48 (in −MG312 blots) by scanning densitometry. The value of each data point is plotted as percentage of peak value (set at 100%) across the Nycodenz gradient. B, distribution of Golgi marker (giantin), ER marker (calnexin), and GRP78 across the Nycodenz gradient. The experiments were performed exactly as described for A. Repetition of the experiments yielded identical results.
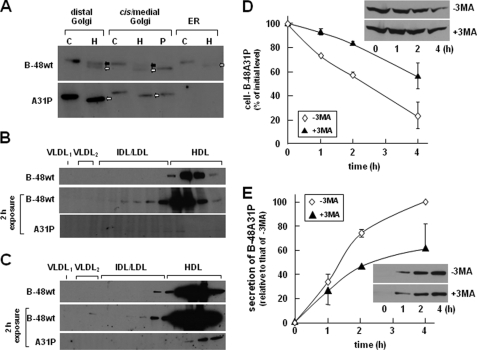
The nature of A31P accumulation within the Golgi compartment was further determined by its Endo H sensitivity and lipid association. The Endo H-resistant forms of apoB-48wt were presented within the distal and cis/medial Golgi (Fig. 4A, top panel, closed arrows), representing the lumenal lipoprotein-associated proteins. The Endo H-sensitive apoB-48wt species were present in the ER as well as in the distal and cis/medial Golgi (Fig. 4A, top, open arrows), which represented the membrane-associated proteins (31). In contrast, A31P mutant was Endo H-sensitive in both distal and cis/medial Golgi fractions (Fig. 4A, bottom, open arrows). Like what was shown in Fig. 3A, A31P mutant was barely detectable in the ER (Fig. 4A, bottom). These results suggest that although it was inappropriately glycosylated, the A31P mutant had escaped the ER quality control.
FIGURE 4.
Endo H digestion of apoB-48wt and A31P mutant. A, Endo H digestion of apoB-48 associated with microsomes. Cells were incubated in DMEM containing 20% FBS and 0.4 mm oleate for 2 h and homogenized to obtain microsomes. The microsomal membranes were fractionated by Nycodenz gradient ultracentrifugation, as described in the legend to Fig. 3A, and distal Golgi (fractions 1–3), cis/medial Golgi (fractions 4–8), and ER (fractions 9–15) were pooled. The apoB-48 proteins were immunoprecipitated from the pooled membranes and subjected to Endo H digestion (lanes marked H). An aliquot of apoB-48 recovered from the cis/medial Golgi was also subjected to peptide:N-glycosidase F digestion (lane marked P). Control samples (lanes marked C) were treated with no glycosidase. The digested samples were resolved by SDS-PAGE (5% gel), and apoB-48 proteins were detected by Western blot analysis. Endo H-resistant and Endo H-sensitive species are indicated with closed and open arrows, respectively. Density distribution of apoB-48 associated with lipoproteins within the microsomal lumen (B) and with lipoproteins secreted into the media (C) was determined by cumulative rate flotation ultracentrifugation. The apoB-48 proteins were detected by Western blot analysis (1 min exposure). The blots for apoB-48A31P were overexposed (2 h) in order to visualize the trace quantities of the mutant protein. Repetition of the experiments yielded identical results. IDL, intermediate density lipoproteins; HDL, high density lipoproteins. D, effect of inhibiting the autophagosome/lysosome system. The experiment was performed similarly to that in Fig. 2B, except that cells were pretreated with or without 10 mm 3-methyladenine (3MA) in DMEM containing 20% FBS for 25 min and then further incubated for 1, 2, and 4 h in the presence of cycloheximide (10 μm) in DMEM. E, secretion of apoB-48A31P from cells treated with or without 3-methyladenine. Data are the average of two independent experiments.
To ascertain that A31P protein was absent in the microsomal lumen, we analyzed lumenal lipoproteins. As expected, apoB-48wt was readily detectable in the lumen as high density lipoprotein (Fig. 4B, top). However, A31P mutant was barely detectable in the lumen unless the Western blot exposure time was extended (Fig. 4B, bottom). Notably, the trace amount of A31P proteins presented in the lumen was also associated with high density lipoprotein fractions. However, the trace amount of A31P proteins secreted from the cells was not associated with lipoproteins and rather was sedimented to d = 1.20 g/ml fractions (Fig. 4C). These results suggest that the aberrantly folded A31P protein, which escaped ER quality control and had been partially lipidated, was degraded prior to secretion by a post-ER mechanism.
Recently, autophagy has been described to play a role in post-ER degradation of apoB. We tested the effect of 3-methyladenine treatment on intracellular A31P (in the presence of cycloheximide) and found that blocking autophagosome formation indeed increased A31P protein stability (Fig. 4D). However, secretion of A31P protein was decreased from cells treated with 3-methyladenine (Fig. 4E). These results suggest that the A31P mutant, either in the membrane-bound or lipoprotein-associated form, might be degraded through the autophagosome pathway.
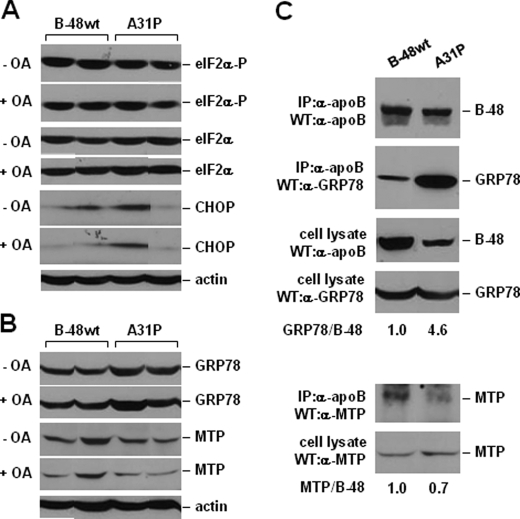
Consideration was given to the possibility that the impaired mutant apoB secretion was a consequence of ER stress (41, 42). However, there was no consistent difference in serine 52 phosphorylation of eIF2α or CHOP expression between multiple apoB-48wt and A31P clones (Fig. 5A). Thus, expression of the A31P mutant did not lead to ER stress (at least not the PERK/eIF2α branch) or apoptosis. The levels of GRP78 and MTP also did not show a consistent difference between apoB-48wt and A31P cells (Fig. 5B). However, binding of GRP78 to A31P mutant, as measured by co-immunoprecipitation, was increased by 4.6-fold as compared with apoB-48wt (Fig. 5C). This result, together with the co-existence of A31P and GRP78 in Golgi (Fig. 3), suggests that A31P might have escaped ER retention through tight association with GRP78. Binding of MTP to A31P mutant was decreased by 30% as compared with apoB-48wt (Fig. 5C, bottom panels), suggesting that MTP-assisted lipidation of the mutant protein is compromised.
FIGURE 5.
Analysis of ER stress and co-immunoprecipitation of MTP and GRP78 with apoB. A and B, Western blots of eIF2α, phospho-eIF2α, CHOP, GRP78, and MTP. Cells were cultured with DMEM containing 20% FBS with or without 0.4 mm oleate for 2 h. The cells were lysed, and equal amounts of proteins were resolved by SDS-PAGE. C, cells were cultured with DMEM containing 20% FBS plus 0.4 mm oleate for 2 h, and cell lysate were subjected to immunoprecipitation (IP) using monoclonal anti-human apoB antibody. After SDS-PAGE, apoB, MTP, and GRP78 were detected by Western blotting (WB). The amount of MTP and GRP78 associated with human apoB-48 was quantified by densitometry, and the ratios of MTP/B-48 and GRP78/B-48 are presented below the blots. The ratio obtained from samples derived from the B48wt-expressing cells was set at 1.0. WT, wild type.
Suppressed Lipogenesis Gene Expression in A31P Cells
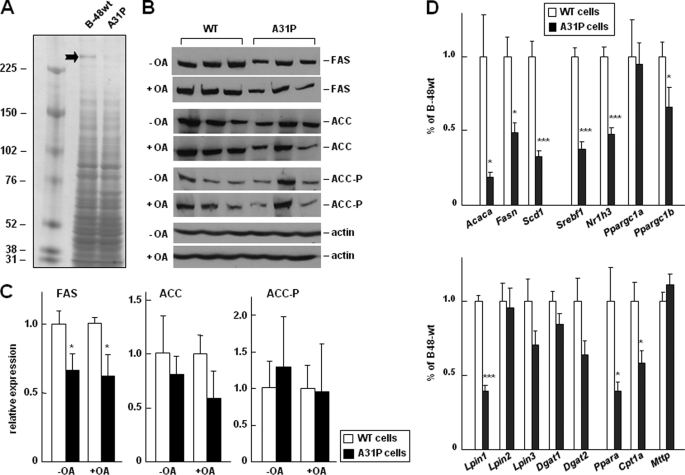
To gain insights into global changes in protein expression in A31P cells, we analyzed total proteins in cell lysate resolved by one-dimensional gel. One band of >250 kDa showed a marked decrease in A31P cells as compared with apoB-48wt (Fig. 6A). Mass spectrometric analysis of this band identified enzymes involved in lipogenesis, FAS, and ACC (Table 2). Western blot analysis confirmed that FAS expression was significantly decreased and ACC was modestly decreased in the A31P cells (Fig. 6, B and C), although the decrease in ACC was not consistently observed in all A31P clones; thus, the decrease was not statistically significant. Phosphorylation of ACC was comparable between apoB-48wt and A31P cells (Fig. 6, B and C). The levels of FAS and ACC expression in apoB-48wt cells were identical to that in nontransfected parental cells (data not shown). These results suggest that lipogenesis in A31P cells was suppressed.
FIGURE 6.
Changes in expression of genes involved in lipogenesis. A, identifying proteins differentially expressed between apoB-48wt and A31P cells. Cell lysates were resolved by SDS-PAGE (3–8% gel), and proteins were visualized by silver staining. The marked bank was excised, in-gel digested, and subjected to liquid chromatography-MS/MS analysis as described under “Experimental Procedures.” Proteins identified in the band, including FAS and ACC, are listed in Table 2. B, Western blot analysis of FAS, ACC, and phospho-ACC (ACC-P) in three stable clones for apoB-48wt or A31P mutant that had been treated with or without oleate (OA) for 2 h. Actin was used as loading control. C, semiquantification of the FAS, ACC, and ACC-P bands (shown in B) by scanning densitometry. The experiments were repeated, and identical results were obtained. D, real-time RT-PCR analysis of relative abundance of mRNAs of genes involved in lipogenesis (top), fatty acid oxidation, and VLDL synthesis (bottom). Total RNAs were isolated from three stable clones for apoB-48wt or A31P mutant. The average value of the three stable clones was used to calculate relative expression of each gene in A31P cells as compared with that in apoB-48wt cells (set as 1.0). ***, p < 0.001; **, p < 0.01; *, p < 0.05 (Student's t test of B-48A31P versus B-48wt). Acaca, ACC; Nr1h, LXRα; Srebf1, SREBP-1c. WT, wild type.
TABLE 2.
Proteins that showed reduction in A31P cells
| Protein name | Massa | IPI numberb | Function | Protein identification |
|||
|---|---|---|---|---|---|---|---|
| Scorec | Peptidesd | Unique peptidese | Coveragef | ||||
| kDa | % | ||||||
| Similar to GCN1 general control of amino acid synthesis 1-like 1 isoform 2 | 293.1 | IPI00767139 | Protein binding | 908 | 14 | 13 | 6.85 |
| Fatty acid synthase | 272.7 | IPI00200661 | Fatty acid biosynthesis | 3,907 | 51 | 45 | 24.47 |
| Isoform 1 of acetyl-CoA carboxylase 1 | 265.4 | IPI00194102 | Ligase | 414 | 8 | 7 | 4.56 |
| Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | 243.5 | IPI00365582 | Carbamoyl-phosphate synthetase; aspartate carbamoyltransferase activity; dihydroorotase activity | 1,157 | 17 | 16 | 9.35 |
a Predicted molecular mass based on an unprocessed precursor protein.
b IPI, international protein index protein sequence database (European Bioinformatics Institute) for rats.
c Protein score calculated by Mascot software.
d The total number of peptides assigned to the protein.
e Unique peptides (i.e. nonredundant) assigned to the protein.
f Sequence coverage, expressed as the number of amino acids spanned by the assigned peptides as a percentage of the sequence length.
We then determined expression of other lipogenesis genes by analyzing mRNAs using real-time RT-PCR. As shown in Fig. 6D (top), levels of the Fasn and Acaca mRNA were indeed decreased in A31P cells as compared with that in apoB-48wt cells. Likewise, the mRNA of Scd1, another important gene involved in lipogenesis, was also significantly decreased. Moreover, transcription factors involved in lipogenesis, including Nr1h3 (LXRα), Sresbf1 (SREBP-1c), and Ppargc1b (PGC-1β), but not Ppargc1a (PGC-1α), also decreased in A31P cells. Determination of expression of genes involved in TAG synthesis showed that only lpin1 was decreased significantly, whereas lpin2, lpin3, Dgat1, and Dgat2 were not (Fig. 6D, bottom). These results agree with a recent report showing that lipin-1 expression in hepatic cells is regulated by SREBP-1c (43). Western blot analysis confirmed that the lipin-1 protein level in A31P cells was markedly decreased as compared with apoB-48wt cells (data not shown).
We also determined the Pparα and Cpt1α genes involved in fatty acid oxidation and found that transcription of both genes was decreased in A31P cells as compared with apoB-48wt cells (Fig. 6D, bottom). The level of Mttp mRNA was comparable between apoB-48wt and A31P cells (Fig. 6D, bottom), in agreement with the unchanged MTP protein levels in cell lysates (Fig. 5C).
Decreased Lipogenesis and Lipid Secretion in A31P Cells
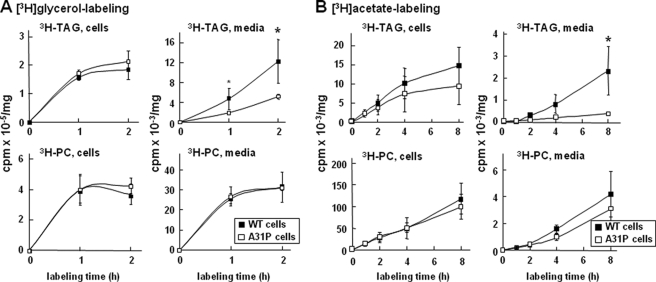
Finally, we determined the impact of A31P mutant expression on glycerolipid synthesis by [3H]glycerol labeling. The rate of [3H]glycerol into cell-associated glycerolipids (i.e. TAG and PC) was not altered in A31P cells (Fig. 7A), consistent with the unaltered Dgat1 and Dgat2 mRNA levels (Fig. 6D, bottom). As expected, secretion of [3H]glycerol-labeled TAG was decreased from A31P cells (Fig. 7A), probably attributable to inhibited secretion of both the A31P mutant and the endogenous apoB-100. The de novo synthesis of lipid was also determined by [3H]acetic acid labeling. The rate of [3H]acetic acid incorporation into cellular TAG was decreased (although because of cell variations the difference did not reach statistical significance) (Fig. 7B), probably related to decreased Fasn and Acaca expression. Incorporation of [3H]acetic acid into medium TAG was significantly decreased in A31P cells (Fig. 7B), consistent with decreased apoB secretion from these cells. The rate of [3H]acetic acid incorporation into PC was similar between A31P and apoB-48wt cells.
FIGURE 7.
Analysis of lipid synthesis and secretion. A, three stable clones for apoB-48wt or A31P mutant were plated at 2 × 106 cells/60-mm dish. The next day, the cells were labeled with [3H]glycerol (10 μCi in 2 ml) in DMEM containing 20% FBS and 0.4 mm oleate for 1 and 2 h. Lipids were extracted from cells and media, respectively, at the indicated times and resolved by thin layer chromatography. Radioactivity associated with 3H-labeled TAG and PC was quantified by scintillation counting. Each data point is the average of three stable clones for apoB-48wt or A31P. B, the experiments were performed in essentially the same manner as in A, except that [3H]acetic acid (20 μCi in 2 ml) in DMEM containing 20% FBS was used for labeling (up to 8 h). *, p < 0.05 (Student's t test of B-48A31P versus B-48wt).
DISCUSSION
Nonsynonymous, Nontruncating Mutations within APOB in Human FHBL
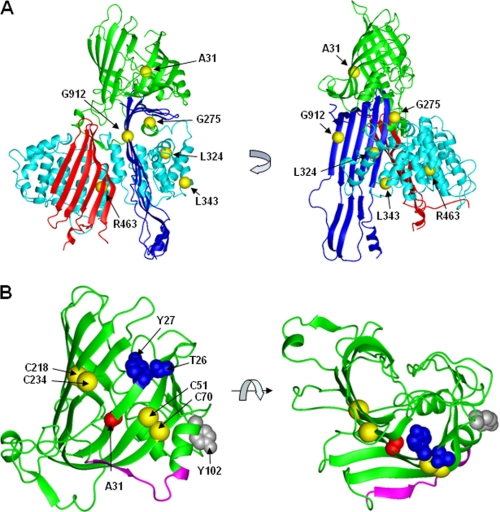
The present study examined the effect of five novel nonsynonymous, nontruncating mutations, A31P, G275S, L324M, G912D, and G945S. All of the mutations occur within the predicted βα1 domain and exhibited varied inhibition on apoB-48 secretion. Although mutations within the α-helical domain (e.g. G275S, L324M, L343V, and R463W) or β-sheets (e.g. G912D and G945S) only moderately impaired apoB secretion (the extent of inhibition varied from 30 to 80% of normal) (Fig. 2), the A31P mutation that occurred within the β-barrel entirely blocked apoB secretion. Unexpectedly, the secretion-incompetent A31P mutant was not retained within the ER like other mutants, such as L343V or R463W, and rather was unusually accumulated in the Golgi (Fig. 3A). These results suggest that the β-barrel may possess a unique function in governing apoB trafficking and secretion. The modeled β-barrel (the first 264 residues) is composed of 10 β-strands that are not completely closed (Fig. 8B). It is also predicted that there is an amphipathic α-helix within the partially closed barrel and an undefined loop (residues 268–300) connecting the β-barrel and downstream α-helical domain (44). The β-barrel contains one proteoglycan binding site (residues 84–94) in apoB-48 associated with chylomicrons or LDL (45), and its N-terminal part can also bind to MTP (15). The present work with naturally occurring FHBL A31P mutation reveals that the β-barrel may also be critical for apoB-48 secretion. The potent effect of A31P mutation on apoB secretion is reminiscent of previous mutagenesis studies showing that two disulfide bonds (Cys51–Cys70 and Cys218–Cys234) located within the β-barrel were required for apoB secretion (46, 47). Homologous modeling shows that Ala31 and these two disulfide bonds are aligned on the same surface of the β-barrel (Fig. 8B). Recently identified mutation in two hypocholesterolemia subjects, namely Thr26_27delinsAsn and Y102C, also occur within the β-barrel (Fig. 8B) (1). Our observation that A31P mutation abolishes secretion of apoB-17 that has no lipoprotein assembly capacity suggests strongly that proper folding of the β-barrel must precede lipid recruitment. The markedly increased association between GRP78 and A31P (Fig. 5C) indicates that the mutation caused gross misfolding, probably because the proline residues disrupted local folds. The local structural folds of this region essential for conferring apoB secretion competence need to be further determined with these other mutations within the β-barrel.
FIGURE 8.
Modeled structure of apoB-100. A, proposed homology model of the N-terminal 930 amino acids (apoB-20.5). The β barrel, α-helical bundle, C-sheet, and A-sheet are highlighted in green, cyan, red, and blue, respectively, as in Fig. 1A. Ala31, Gly275, Leu324, Leu343, Arg463, and Gly912 are shown as yellow spheres. B, the β-barrel region. The Cα backbone is shown as a ribbon. The side chain of the FHBL mutant residue Ala31 and positions of two mutant residues found in human hypocholesterolemia (Thr26-Tyr27 and Tyr102) (1) are highlighted in red, blue, and gray, respectively, in a space-filling representation. The Cα atoms of residues Cys51-Cys70 and Cys218-Cys234 that are involved in disulfide bonds are shown as yellow spheres (46). The backbone of the putative proteoglycan binding site (residues 84–94) (45) is shown in magenta. The homology model for apoB-20.5 was constructed using MODELLER 7.7 based on the coordinates of lamprey lipovitellin (Protein Data Bank code 1lpv). Graphics were prepared using MOLMOL.
Unusual Golgi Accumulation of the A31P Mutant
It is noteworthy that although some A31P mutant apoB-48 proteins was degraded by the proteasomes (presumably through an ER-associated mechanism), a significant amount of the mutant protein escaped ER retention and presented in the distal compartment of the secretory pathway. This is in sharp contrast to FHBL mutations occurring in the α-helical domain, such as L343V and R463W, that showed extended ER retention (Fig. 3A). Apparently, the A31P mutant proteins escaped the ER quality control mechanism (48). The observation that only a small amount of A31P that escaped from the ER was presented in the lumen (as high density lipoprotein) suggests that the misfolded proteins remained in association with the membranes. Currently, whether or not the membrane-associated A31P mutants had assembled some lipids is unknown; nor is it clear if they were retained within the Golgi by proteins other than GRP78. Examination of the minute amount of A31P proteins secreted into the media showed they were mostly nonlipoprotein entities (d > 1.20 g/ml) (Fig. 4C). These results suggest that those A31P mutants containing lipoprotein assembly intermediates were degraded by a post-ER degradation mechanism, probably autophagosome (49, 50), prior to secretion.
Suppressed Lipogenesis in FHBL A31P Mutant-expressing Cells
Another intriguing observation made in the present study was markedly suppressed lipogenesis in A31P cells; thus, the expressions of SREBP-1c, together with its target genes Fasn, Acaca, Scd1, and lpin1 were all down-regulated (Fig. 6). Suppressed hepatic lipogenesis has also been observed in two mouse FHBL models producing truncated apoB proteins equivalent to human apoB-38.9 or apoB-27.6 (51–53). Like what occurred in A31P cells, the livers expressing apoB-38.9 or apoB-27.6 also exhibited reduced SREBP-1c, FAS, and SCD1 (51). Thus, suppression of lipogenesis may occur in FHBL with either truncating or nontruncating apoB mutations. Suppressed lipogenesis in FHBL hepatocytes may explain the abnormally low secretion efficiency of endogenous rat apoB-100 from the transfected cells resulting from the limited lipid substrate availability (Fig. 1D).
How expression of FHBL mutant A31P could result in down-regulation of SREBP-1c (and LXRα as well) is unclear. One possibility may involve lipin-1. The dual function lipin-1 plays a role as PAP1 (phosphatidate phosphatase 1) (54) as well as a transcription coactivator for PGC-1α and PPARα (55). Not surprisingly, decreased Lpin1 in A31P cells was accompanied with reduced Pparα and Cpt1α (Fig. 6D). In McA-RH7777 cells, lipin-1 is located in both cytosol and membrane (ER and nuclear) fractions (20). We found that the lipin-1 mass in total cell lysate was markedly decreased in A31P cells as compared with apoB-48wt control (data not shown). Decreased lipin-1 (acting as PAP1) would thus be expected to result in low TAG synthesis. However, a [3H]glycerol labeling experiment suggested that TAG synthesis was normal in A31P cells (Fig. 7A). From a transcription coactivator point of view, decreased lipin-1 in A31P cells could have an impact on PGC-1α action, leading to lowered lipogenesis, although the expression of PGC-1α or -1β was little or only marginally affected (Fig. 6D). It is reported recently that lipin-1 expression is regulated by SREBP-1c (43). It is likely that expression of A31P mutant has perturbed the regulatory circuit among SREBP-1c, PPARα, LXRα and lipin-1, resulting in profound suppression of lipogenesis. The effect of A31P mutation on lipogenesis needs to be confirmed in knock-in mice harboring the Apob mutation.
Supplementary Material
Acknowledgments
We thank Dr. Gordon Jiang for helpful discussions on structure-function relationships within human apoB-100, Drs. Yves Marcel and Ross Milne for the monoclonal antibody 1D1, and Dr. Carol Shoulders for anti-MTP antibody.
This work was supported in part by National Institutes of Health Grant HL-26335 (to C. J. M.), Heart and Stroke Foundation of Ontario Grant T-4643 (to Z. Y.), Genome Canada Grant 2008-OGI-TD-01 (to D. F.), Canadian Institutes of Health Research (CIHR) Grant MOP-158235 (to L. W.), and Telethon Foundation Onlus, Italy Grant GGP05042 (to P. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Fig. S1.
- FHBL
- familial hypobetalipoproteinemia
- LDL
- low density lipoprotein(s)
- apo
- apolipoprotein
- apoB-48wt
- wild type apoB-48
- VLDL
- very low density lipoprotein(s)
- MTP
- microsomal triglyceride transfer protein
- TAG
- triacylglycerol
- LDL-C
- LDL cholesterol
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- ER
- endoplasmic reticulum
- Endo H
- endoglycosidase H
- PC
- phosphatidylcholine
- ACC
- acetyl-CoA carboxylase
- FAS
- fatty acid synthase
- LXRα
- liver X receptor α
- MS/MS
- tandem mass spectrometry
- HPLC
- high pressure liquid chromatography
- RT
- reverse transcription.
REFERENCES
- 1.Tarugi P., Averna M., Di Leo E., Cefalù A. B., Noto D., Magnolo L., Cattin L., Bertolini S., Calandra S. (2007) Atherosclerosis 195, e19–e27 [DOI] [PubMed] [Google Scholar]
- 2.Chan L. (1992) J. Biol. Chem. 267, 25621–25624 [PubMed] [Google Scholar]
- 3.Fisher E. A., Ginsberg H. N. (2002) J. Biol. Chem. 277, 17377–17380 [DOI] [PubMed] [Google Scholar]
- 4.Yao Z., McLeod R. S. (1994) Biochim. Biophys. Acta 1212, 152–166 [DOI] [PubMed] [Google Scholar]
- 5.Schonfeld G. (2003) J. Lipid Res. 44, 878–883 [DOI] [PubMed] [Google Scholar]
- 6.Burnett J. R., Shan J., Miskie B. A., Whitfield A. J., Yuan J., Tran K., McKnight C. J., Hegele R. A., Yao Z. (2003) J. Biol. Chem. 278, 13442–13452 [DOI] [PubMed] [Google Scholar]
- 7.Burnett J. R., Zhong S., Jiang Z. G., Hooper A. J., Fisher E. A., McLeod R. S., Zhao Y., Barrett P. H., Hegele R. A., van Bockxmeer F. M., Zhang H., Vance D. E., McKnight C. J., Yao Z. (2007) J. Biol. Chem. 282, 24270–24283 [DOI] [PubMed] [Google Scholar]
- 8.McLeod R. S., Zhao Y., Selby S. L., Westerlund J., Yao Z. (1994) J. Biol. Chem. 269, 2852–2862 [PubMed] [Google Scholar]
- 9.Yao Z. M., Blackhart B. D., Linton M. F., Taylor S. M., Young S. G., McCarthy B. J. (1991) J. Biol. Chem. 266, 3300–3308 [PubMed] [Google Scholar]
- 10.Mann C. J., Anderson T. A., Read J., Chester S. A., Harrison G. B., Köchl S., Ritchie P. J., Bradbury P., Hussain F. S., Amey J., Vanloo B., Rosseneu M., Infante R., Hancock J. M., Levitt D. G., Banaszak L. J., Scott J., Shoulders C. C. (1999) J. Mol. Biol. 285, 391–408 [DOI] [PubMed] [Google Scholar]
- 11.Jiang Z. G., Gantz D., Bullitt E., McKnight C. J. (2006) Biochemistry 45, 11799–11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gretch D. G., Sturley S. L., Wang L., Lipton B. A., Dunning A., Grunwald K. A., Wetterau J. R., Yao Z., Talmud P., Attie A. D. (1996) J. Biol. Chem. 271, 8682–8691 [DOI] [PubMed] [Google Scholar]
- 13.McLeod R. S., Wang Y., Wang S., Rusiñol A., Links P., Yao Z. (1996) J. Biol. Chem. 271, 18445–18455 [DOI] [PubMed] [Google Scholar]
- 14.Segrest J. P., Jones M. K., Dashti N. (1999) J. Lipid Res. 40, 1401–1416 [PubMed] [Google Scholar]
- 15.Bradbury P., Mann C. J., Köchl S., Anderson T. A., Chester S. A., Hancock J. M., Ritchie P. J., Amey J., Harrison G. B., Levitt D. G., Banaszak L. J., Scott J., Shoulders C. C. (1999) J. Biol. Chem. 274, 3159–3164 [DOI] [PubMed] [Google Scholar]
- 16.Hussain M. M., Bakillah A., Nayak N., Shelness G. S. (1998) J. Biol. Chem. 273, 25612–25615 [DOI] [PubMed] [Google Scholar]
- 17.Dashti N., Manchekar M., Liu Y., Sun Z., Segrest J. P. (2007) J. Biol. Chem. 282, 28597–28608 [DOI] [PubMed] [Google Scholar]
- 18.Olofsson S. O., Borèn J. (2005) J. Intern. Med. 258, 395–410 [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki T., Sasaki E., Kakinuma C., Yano T., Miura S., Ezaki O. (2005) J. Biol. Chem. 280, 21506–21514 [DOI] [PubMed] [Google Scholar]
- 20.Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D. N., Yao Z. (2009) J. Lipid Res. 50, 47–58 [DOI] [PubMed] [Google Scholar]
- 21.Musso G., Gambino R., Cassader M. (2009) Prog. Lipid Res. 48, 1–26 [DOI] [PubMed] [Google Scholar]
- 22.Grefhorst A., Elzinga B. M., Voshol P. J., Plösch T., Kok T., Bloks V. W., van der Sluijs F. H., Havekes L. M., Romijn J. A., Verkade H. J., Kuipers F. (2002) J. Biol. Chem. 277, 34182–34190 [DOI] [PubMed] [Google Scholar]
- 23.Lin J., Yang R., Tarr P. T., Wu P. H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., Newgard C. B., Spiegelman B. M. (2005) Cell 120, 261–273 [DOI] [PubMed] [Google Scholar]
- 24.Tarugi P., Lonardo A., Gabelli C., Sala F., Ballarini G., Cortella I., Previato L., Bertolini S., Cordera R., Calandra S. (2001) J. Lipid Res. 42, 1552–1561 [PubMed] [Google Scholar]
- 25.Di Leo E., Magnolo L., Lancellotti S., Crocè L., Visintin L., Tiribelli C., Bertolini S., Calandra S., Tarugi P. (2007) J. Med. Genet. 44, 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Dunnen J. T., Antonarakis S. E. (2000) Hum. Mutat. 15, 7–12 [DOI] [PubMed] [Google Scholar]
- 27.den Dunnen J. T., Paalman M. H. (2003) Hum. Mutat. 22, 181–182 [DOI] [PubMed] [Google Scholar]
- 28.Fasano T., Cefalù A. B., Di Leo E., Noto D., Pollaccia D., Bocchi L., Valenti V., Bonardi R., Guardamagna O., Averna M., Tarugi P. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 677–681 [DOI] [PubMed] [Google Scholar]
- 29.Blackhart B. D., Yao Z. M., McCarthy B. J. (1990) J. Biol. Chem. 265, 8358–8360 [PubMed] [Google Scholar]
- 30.Wang Y., McLeod R. S., Yao Z. (1997) J. Biol. Chem. 272, 12272–12278 [DOI] [PubMed] [Google Scholar]
- 31.Tran K., Thorne-Tjomsland G., DeLong C. J., Cui Z., Shan J., Burton L., Jamieson J. C., Yao Z. (2002) J. Biol. Chem. 277, 31187–31200 [DOI] [PubMed] [Google Scholar]
- 32.Borén J., Graham L., Wettesten M., Scott J., White A., Olofsson S. O. (1992) J. Biol. Chem. 267, 9858–9867 [PubMed] [Google Scholar]
- 33.Abu-Farha M., Lambert J. P., Al-Madhoun A. S., Elisma F., Skerjanc I. S., Figeys D. (2008) Mol. Cell Proteomics 7, 560–572 [DOI] [PubMed] [Google Scholar]
- 34.Zhou H., Hou W., Denis N. J., Zhou H., Vasilescu J., Zou H., Figeys D. (2009) J. Proteome Res. 8, 556–566 [DOI] [PubMed] [Google Scholar]
- 35.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 36.Thompson J. R., Banaszak L. J. (2002) Biochemistry 41, 9398–9409 [DOI] [PubMed] [Google Scholar]
- 37.Tatusova T. A., Madden T. L. (1999) FEMS Microbiol. Lett. 174, 247–250 [DOI] [PubMed] [Google Scholar]
- 38.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 39.Lancellotti S., Zaffanello M., Di Leo E., Costa L., Lonardo A., Tarugi P. (2005) J. Hepatol. 43, 188–191 [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Tran K., Yao Z. (1999) J. Biol. Chem. 274, 27793–27800 [DOI] [PubMed] [Google Scholar]
- 41.Su Q., Tsai J., Xu E., Qiu W., Bereczki E., Santha M., Adeli K. (2009) Hepatology 50, 77–84 [DOI] [PubMed] [Google Scholar]
- 42.Ota T., Gayet C., Ginsberg H. N. (2008) J. Clin. Invest. 118, 316–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishimoto K., Nakamura H., Tachibana K., Yamasaki D., Ota A., Hirano K., Tanaka T., Hamakubo T., Sakai J., Kodama T., Doi T. (2009) J. Biol. Chem. 284, 22195–22205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson P. E., Manchekar M., Dashti N., Jones M. K., Beigneux A., Young S. G., Harvey S. C., Segrest J. P. (2005) Biophys. J. 88, 2789–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flood C., Gustafsson M., Richardson P. E., Harvey S. C., Segrest J. P., Borén J. (2002) J. Biol. Chem. 277, 32228–32233 [DOI] [PubMed] [Google Scholar]
- 46.Huang X. F., Shelness G. S. (1997) J. Biol. Chem. 272, 31872–31876 [DOI] [PubMed] [Google Scholar]
- 47.Tran K., Borén J., Macri J., Wang Y., McLeod R., Avramoglu R. K., Adeli K., Yao Z. (1998) J. Biol. Chem. 273, 7244–7251 [DOI] [PubMed] [Google Scholar]
- 48.Dejgaard S., Nicolay J., Taheri M., Thomas D. Y., Bergeron J. J. (2004) Curr. Issues Mol. Biol. 6, 29–42 [PubMed] [Google Scholar]
- 49.Pan M., Maitin V., Parathath S., Andreo U., Lin S. X., St Germain C., Yao Z., Maxfield F. R., Williams K. J., Fisher E. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5862–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. (2006) Mol. Biol. Cell 17, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin X., Schonfeld G., Yue P., Chen Z. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 476–482 [DOI] [PubMed] [Google Scholar]
- 52.Lin X., Chen Z., Yue P., Averna M. R., Ostlund R. E., Jr., Watson M. A., Schonfeld G. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 290, G1170–G1176 [DOI] [PubMed] [Google Scholar]
- 53.Chen Z., Fitzgerald R. L., Schonfeld G. (2002) J. Biol. Chem. 277, 14135–14145 [DOI] [PubMed] [Google Scholar]
- 54.Carman G. M., Han G. S. (2006) Trends Biochem. Sci. 31, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Cell Metab. 4, 199–210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.