Abstract
Transforming growth factor β (TGF-β) regulates many biological processes, and aberrant TGF-β signaling is implicated in tumor development. Smad3 is a central component of the TGF-β signaling pathway, and once activated, Smad3 forms complexes with Smad4 or other receptor-regulated Smads, which accumulate in the nucleus to transcriptionally regulate TGF-β target genes. Because Smad3 plays a significant role in mediating the activities of TGF-β, we examined its regulation during tumor development using a well characterized tumor model. We demonstrate that Smad3 levels are dramatically reduced in the tumorigenic cell line transformed with activated H-Ras compared with the normal parental epithelial cells. Interestingly, we also observe a cell cycle-dependent regulation of Smad3 in both cell types, with high Smad3 levels in quiescent cells and a significant drop in Smad3 protein levels in proliferating cells. Smad3 is regulated at the mRNA level and at the level of protein stability. In addition, functional analysis indicates that down-regulation of Smad3 levels is required for the tumor cells to proliferate in the presence of TGF-β, because ectopic expression of Smad3 in the tumorigenic cell line restores the growth inhibitory response to TGF-β. In contrast, expression of high levels of Smad3 did not interfere with the ability of these cells to undergo epithelial to mesenchymal transition upon TGF-β stimulation. Altogether, our results suggest that the level of Smad3 protein is an important determinant of the progression of tumorigenesis. High levels of Smad3 are required for the tumor suppressor activities of TGF-β, whereas lower levels are sufficient for the tumor promoting functions.
Keywords: Cancer Tumor Promoter, Protein Stability, Signal Transduction, SMAD Transcription Factor, Transcription Regulation, Transforming Growth Factor Beta (TGFbeta), Tumor Suppressor
Introduction
Members of the transforming growth factor β (TGF-β)3 superfamily regulate many diverse cellular behaviors including proliferation, differentiation, migration, and survival. TGF-β signals predominantly via a receptor complex comprising the type I receptor ALK5 (activin receptor-like kinase 5) and the type II receptor TβRII, both of which are serine/threonine kinases (1). Activation of ALK5 by the constitutively active TβRII leads to the phosphorylation of the receptor-regulated Smads Smad2 and Smad3 at their conserved C-terminal SSXS motif. A different subset of receptor-regulated Smads, Smad1 and Smad5, are also phosphorylated in response to TGF-β in many different cell lines, although the receptor complexes required for this are still under debate (2–4). Once activated, receptor-regulated Smads form complexes with each other and with the common Smad (co-Smad), Smad4. These Smad complexes accumulate in the nucleus, where they assemble with specific DNA-binding transcription factors, co-activators and co-repressors to regulate transcription (1).
TGF-β signaling is strongly implicated in cancer progression. At early stages of tumorigenesis, TGF-β signaling is thought to act as a tumor suppressor, as a result of its ability to arrest the growth of epithelial cells, and in some cases induce apoptosis (5, 6). However, during late stages of cancer progression, the activities of TGF-β switch from tumor suppressive to tumor promoting, enhancing tumor progression, invasion, and metastasis (7). This can occur through down-regulation, mutation, or deletion of core components of the TGF-β pathway such as the receptors or Smads or by alterations in downstream targets of the pathway that specifically eliminate the tumor suppressor activities of TGF-β (8). When the type II receptors are completely lost, as in some colorectal, ovarian, or head and neck cancers, the tumor cells themselves become completely nonresponsive to TGF-β, and the tumor promoting effects of TGF-β are elicited through effects on the tumor stroma (8). However, in other cases, such as when Smad4 is deleted or mutated, or when expression of other pathway components is down-regulated but not completely lost, the tumor cells remain responsive to TGF-β, which may promote their ability to migrate and invade (8, 9).
The role of Smad3 in cancer is particularly intriguing. It is a critical mediator of the cytostatic response to TGF-β. Evidence for this comes from the observation that Smad3-deficient mice show accelerated wound healing due to increased re-epithelialization, which can be attributed to impaired TGF-β growth inhibition of keratinocytes and increased recruitment of fibroblasts and macrophages (10). In addition, a variety of primary cells from Smad3-null mice are partially resistant to the growth inhibitory effects of TGF-β (11, 12). Smad3 has also been shown to suppress liver tumorigenesis by promoting apoptosis in tumor cells (13). Thus, Smad3 appears to have a tumor-suppressive function. However, Smad3 also plays a crucial role in the prometastatic activities of TGF-β. Reduction of Smad3 has been shown to strongly suppress metastasis to the lung of aggressive carcinoma cells (14), and Smad3 is essential for epithelial to mesenchymal transition (EMT), which is thought to be important for metastasis (15). This latter activity was demonstrated by the inability of TGF-β to induce EMT in primary tubular epithelial cells derived from kidneys of Smad3-null mice (16). Interestingly, down-regulation of Smad3 expression, though not a complete loss, was observed in Madin-Darby canine kidney cells undergoing EMT, which was concomitant with a resistance of these cells to TGF-β-induced growth inhibition (17). Thus, Smad3 is required for both the growth inhibitory and tumor-promoting activities of TGF-β. This dual role of Smad3 could explain why mutations in Smad3 in human tumors appear to be very rare and have only very recently come to light through genomic screening of human pancreatic, breast, and colorectal tumors (18, 19). A more common observation is the down-regulation of Smad3 expression in tumors, as has been observed in, for example, gastric cancer (20, 21).
Because Smad3 plays a significant role in mediating both the tumor suppressor and tumor-promoting activities of TGF-β, we decided to systematically study how its levels were regulated in a well studied tumor model, the EpH4/EpRas system (22). We observe a cell cycle-dependent regulation of Smad3 in both cell types, whereby high Smad3 levels are present in quiescent cells, and the levels drop significantly in proliferating cells. We also show that Smad3 levels are dramatically reduced in the H-Ras-transformed cell line (EpRas) compared with the normal parental epithelial cells (EpH4). We go on to show that Smad3 is regulated at the level of mRNA and also protein stability. Reduction in Smad3 levels in EpRas cells is essential for their ability to proliferate in the presence of TGF-β. In support of this, we demonstrate that ectopic expression of Smad3 in EpRas cells restores the growth inhibitory response to TGF-β but has no effect on the ability of these cells to undergo EMT.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatments, Transfections, and Generation of Cell Lines
All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, and in the case of EpRas cells, 500 μg/ml G418. Cells were induced at the indicated times with 2 ng/ml TGF-β1 (PreproTech) or 20 μg/ml cycloheximide (Sigma), as appropriate. For starvation assays, cells were maintained subconfluent in medium containing 0.1% serum for 72 h. Fresh medium containing 10% serum was then added with or without TGF-β. Samples for FACS and Western blotting were harvested after starvation and 20 h after release. For the establishment of FLAG-Smad3 EpRas cell lines, EpRas cells were transfected with EF-FLAG-Smad3 (23) together with a plasmid conferring puromycin resistance (pSUPER-retro-puro; OligoEngine) using LipofectamineTM 2000 Reagent (Invitrogen) according to the manufacturer's protocol. Stable transfectants were selected for resistance to puromycin (5 μg/ml) to generate clones expressing FLAG-Smad3. Clones were analyzed for their expression of FLAG-Smad3 by Western blotting.
Cell Lysis, Western Blotting, and Immunofluorescence
Whole cell extracts were prepared using radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 1 mm dithiothreitol, 25 mm NaF, 25 mm Na β-glycerophosphate, and protease inhibitors) or D0.4 extraction buffer (24). Western blotting was performed using standard procedures. The following antibodies were used: Smad3 (Zymed Laboratories Inc.); Smad2/3 and Grb2 (BD Biosciences); Smad4 (Clone B8), MCM7 (clone 141.2), JunB (clone C-11) and PAI-1 (clone C-9; Santa Cruz); phospho-Smad2 (Cell Signaling Technology); phospho-Smad3 (EP823Y, Epitomics); FLAG and FLAG-horseradish peroxidase (Sigma).
Immunofluorescence was carried out as described previously (25). Antigen detection was performed using anti-E-cadherin (BD Biosciences), anti-ZO1 (Zymed Laboratories Inc.) or Vimentin (clone LN-6; Sigma) followed by Alexa Fluor 488-conjugated goat anti-mouse IgG antibody or Texas Red conjugated goat anti-rabbit IgG antibody as appropriate. F-actin was detected with Texas Red-phalloidin (Alexa). Cells were mounted with Mowiol (Calbiochem), and fluorescence was observed with a Zeiss confocal LSM 510 microscope.
RNase Protection Assays
Cells were grown to confluency for 48 h (quiescent samples) or released from quiescence by trypsinization into fresh medium for 20 h. Cells were lysed, and extraction of RNA, probe preparation, and RNase protection assays were performed as described (26, 27). The Smad2 and -3 probes used for RNase protection were designed against the human sequences but cross-reacted with the mouse sequences and were as described (17). The γ-actin probe was as described previously (28).
Cell Cycle Analysis and EMT Studies
For EpH4 and EpRas cell G1/S progression studies, cells were seeded at high density, arrested in G0/G1 by confluency for 2 days, released by trypsinization, and replated at low density for up to 24 h before harvesting for FACS analysis and/or Western blotting. The growth inhibitory assays in EpH4, EpRas cells, and EpRas cell clones were performed essentially as described previously (29). Briefly, the cells were grown to confluency over 48 h, trypsin-released, and replated at a low density and then either left uninduced or induced with 2 ng/ml TGF-β. Samples for FACS analysis were collected 16, 18, or 20 h later.
To analyze cells for EMT progression, 5 × 104 EpH4, EpH4 cells depleted of Smad3, EpRas cells, or the FLAG-Smad3-expressing EpRas cell clones were plated out in 6-well plates and grown in the presence or absence of TGF-β1 (2 ng/ml). The medium was changed 1 day after seeding and then every other day. TGF-β was added to the cells upon medium change. Three days after plating, the cells were trypsinized and replated at equivalent density to day 1. Smad3 small interfering RNA transfection of EpH4 cells in the experiment shown in supplemental Fig. 6 was performed at days 1 and 4. Cells were grown for a total of 10 days, which required two trypsinizations, and then harvested 48 h after the final TGF-β addition. EMT was assayed by staining for E-cadherin, ZO-1, vimentin, and actin.
BrdUrd Incorporation Assay
EpH4, EpRas, and EpRas S3 C1 cells were plated at high density, arrested in G0/G1 by contact inhibition for 2 days, released by trypsinization, and replated at low density for up to 20 h, with or without 2 ng/ml TGF-β, in the presence of 10 mm bromodeoxyuridine (5-bromo-2-deoxyuridine, BrdUrd; Sigma). Cells were then fixed in 70% ethanol at 4 °C, treated with hydrochloric acid, incubated with an anti-BrdUrd antibody (Becton Dickinson) and secondary fluorescein isothiocyanate-conjugated antibody (DAKO) before FACS analysis.
RESULTS
Smad3 Expression Levels Are Regulated by Ras Activity and during the Cell Cycle
The regulation of Smad3 protein levels was investigated in a well characterized model system (22). The parental cells (EpH4 cells) are nontransformed mouse mammary gland epithelial cells, which are nontumorigenic and undergo growth inhibition and apoptosis in response to TGF-β. However, EpH4 cells that have been transformed by stable expression of oncogenic H-Ras (EpRas cells) do not arrest in response to TGF-β but instead undergo EMT, exhibit invasive growth in three-dimensional cultures, and form rapidly growing tumors in mice.
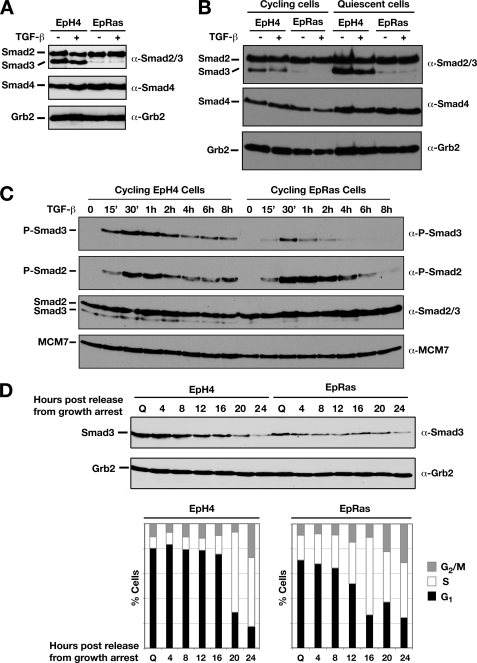
We first examined the levels of Smad3 as well as Smad2 and Smad4 in both cell lines. A striking difference was observed in the levels of Smad3 protein, whereby high levels of Smad3 were detected in EpH4 cells compared with low Smad3 levels observed in EpRas cells (Fig. 1A). In contrast, both Smad2 and Smad4 expression remained constant. Further analysis of Smad3 protein levels in these cells revealed that the expression pattern of Smad3 is also differentially regulated depending on the stage of the cell cycle. Quiescent cells, which have been arrested in G0/G1 by contact inhibition, and actively proliferating cells exhibited different Smad3 levels (Fig. 1B). This was most apparent in EpH4 cells, where high levels of Smad3 protein were detected in quiescent cells and dramatically decreased Smad3 levels were observed in the cycling cells (Fig. 1B, top panel). This trend was also observed in EpRas cells, although the levels of Smad3 in quiescent cells are still much lower than in EpH4 cells (Fig. 1B, top panel). Higher levels of Smad3 were also seen in quiescent EpH4 cells arrested by serum starvation compared with actively proliferating cells (supplemental Fig. 1).
FIGURE 1.
Smad3 expression levels are down-regulated in EpRas cells and are regulated during the cell cycle. A, confluent EpH4 and EpRas cells were treated ± 2 ng/ml TGF-β1 for 45 min. Whole cell extracts were prepared from confluent cells, and equal amounts of protein were analyzed by Western blotting using antibodies against the Smad2/3 and 4 and Grb2 as a loading control. B, Smad3 protein expression is regulated during the cell cycle. Synchronized, quiescent EpH4 and EpRas cells were prepared by contact inhibition for 72 h. Actively cycling, low confluency EpH4 and EpRas cells were prepared by synchronizing the cells by contact inhibition and then plating into fresh medium for 24 h. Cells were either unstimulated or treated with 2 ng/ml TGF-β1 for 1 h as indicated. Whole cell extracts were prepared, and equal amounts of protein were analyzed by Western blotting using antibodies against Smad2/3 and Smad4 and Grb2 as a loading control. C, cycling EpH4 and EpRas cells were treated with 2 ng/ml TGF-β for the times indicated. Whole cell extracts were blotted with antibodies against phosphorylated Smad2, phosphorylated Smad3, Smad2/3, and MCM7 as a loading control. D, EpH4 and EpRas cells were synchronized by contact inhibition and then plated into fresh medium for the number of hours indicated. Cells were harvested and analyzed by FACS, to determine the number of cells in G1 (black bar), S (white bar), or G2/M (gray bar) and by Western blotting, using antibodies against Smad3 and Grb2 as a loading control.
To investigate whether the low level of Smad3 protein in EpRas cells would potentially influence the functionality of the TGF-β signaling pathway, we assayed the phosphorylation of Smad2 and Smad3 over time in cycling cells. P-Smad3 levels were lower in EpRas cells compared with EpH4 cells, reflecting the lower levels of Smad3 protein in EpRas cells (Fig. 1C). The duration of Smad3 signaling was attenuated in the EpRas cells compared with the EpH4 cells. At 4 h post-TGF-β stimulation, the level of p-Smad3 in EpRas cells was barely detectable, whereas p-Smad3 was clearly observed in EpH4 cells (Fig. 1C). We also assayed the TGF-β induction of two Smad3-dependent target genes in these cell lines. Induction of both JunB and PAI-1 was substantially reduced in EpRas cells compared with EpH4 cells (supplemental Fig. 2).
We next examined the time course of down-regulation of Smad3 protein in cells transitioning from the quiescent to the proliferative state. Expression of Smad3 was examined in synchronous cultures of EpH4 and EpRas cells at different times after release from quiescence as they progressed toward S-phase. Western blot analysis revealed that Smad3 protein level was down-regulated over time at a steady rate (Fig. 1D). A high level of Smad3 expression was initially observed in EpH4 cells, but by 24 h after release, the Smad3 protein level was dramatically reduced (Fig. 1D). A similar profile was observed in EpRas cells, although the initial level of Smad3 was lower (Fig. 1D, top panels). FACS analysis of these cells at each time point revealed that the decrease in Smad3 expression does not precisely correlate with entry into S-phase, but rather, it appears to occur steadily throughout G1 (Fig. 1D, bottom panels).
These data indicate that Smad3 expression is regulated at two levels in this model system. First, Smad3 levels are down-regulated by a mechanism dependent on the continuous activation of Ras. In addition, Smad3 levels are modulated by the quiescent/cycling state of the cells.
Smad3 Is Regulated at the Level of mRNA in EpH4 and EpRas Cells
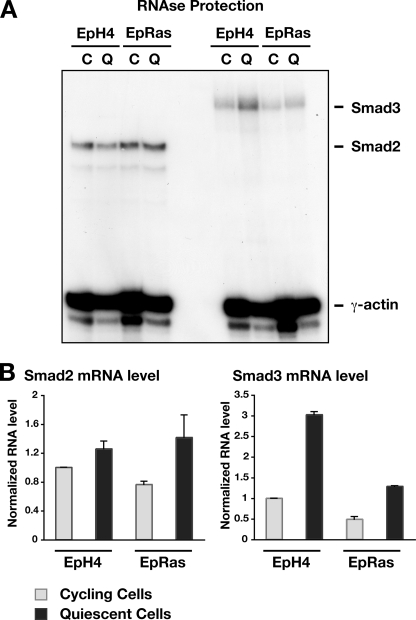
To gain insight into the mechanisms involved in Smad3 expression, we examined the levels of Smad3 mRNA in the different cell types, either quiescent or cycling. Smad3 mRNA levels in the quiescent and cycling EpH4 and EpRas cells were determined by quantitative RNase protection assays. Mouse Smad2 and Smad3 mRNA protected the radiolabeled RNA probe to generate a 230- and a 300-bp fragment, respectively. The γ-actin protection produced a smaller fragment of 65 bp (Fig. 2A). The quantitation of the RNase protection analysis revealed that Smad2 mRNA levels did not vary significantly from quiescent to cycling cells or between EpH4 and EpRas, which was expected from the Smad2 protein data (Fig. 2B, left). In contrast, Smad3 mRNA levels varied substantially, with a 3-fold decrease in Smad3 mRNA in cycling EpH4 cells compared with quiescent cells (Fig. 2B, right). The Smad3 mRNA profile was similar in EpRas cells, but there was an ∼2-fold decrease in Smad3 mRNA in EpRas cells compared with EpH4 cells. These results indicate that regulation of Smad3 mRNA levels contributes to the control of expression levels of Smad3 observed in quiescent versus cycling cells and also the decrease in Smad3 levels observed in EpRas cells compared with EpH4 cells.
FIGURE 2.
Smad3 is regulated at the level of transcription. A, mRNA levels of Smad2 and Smad3 were detected by RNase protection. Total RNA was isolated from both cycling (C) and quiescent (Q) EpH4 and EpRas cells. 20 μg of total RNA were analyzed by RNase protection with probes for Smad2 and Smad3. The protected fragments (Smad2 or Smad3) are indicated. γ-Actin was used as a loading control. Data are representative of two independent experiments. B, quantitation of RNase protection by phosphorimage analysis. Data are the means and S.D. of two separate experiments relative to appropriate controls.
Smad3 Is Additionally Regulated at the Level of Protein Stability
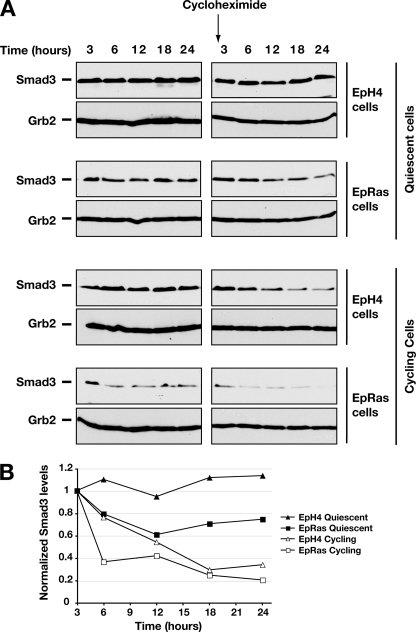
The differences in Smad3 mRNA levels were unlikely to account for the much more dramatic differences observed in Smad3 protein levels. Thus, to investigate whether other mechanisms, such as Smad3 stability, are also involved in the regulation of Smad3 protein levels, we determined the rate of degradation of Smad3 under different conditions. Cycling and quiescent EpH4 and EpRas cells were treated with the protein synthesis inhibitor cycloheximide to prevent translation of newly synthesized Smad3 mRNA. Cycloheximide prevented further progression of the cells through the cell cycle as determined by FACS analysis (data not shown), and, therefore, the stability of Smad3 could be assessed at these two stages in the cell cycle. In the control samples not treated with cycloheximide, Smad3 appears relatively stable in each condition, presumably because any degradation is balanced by resynthesis (Fig. 3, left panels). Smad3 protein levels were also stable in cycloheximide-treated samples of quiescent EpH4 cells over the 24-h period (Fig. 3A, right panels, and B). However, the Smad3 protein levels in the cycloheximide-treated quiescent EpRas decreased gradually over time (Fig. 3A, right panels, and B), indicating that Smad3 was undergoing degradation. This was even more striking in cycling EpH4 and EpRas cells (Fig. 3A, right panels, and B).
FIGURE 3.
Smad3 proteins are less stable in cycling cells than when cells are quiescent. A, synchronized, quiescent EpH4 and EpRas cells were prepared by contact inhibition for 72 h. Actively cycling, low confluency EpH4 and EpRas cells were prepared by synchronizing the cells by contact inhibition and subsequent release into fresh medium for 15 h. Cells were either untreated or treated with the protein synthesis inhibitor cyclohexamide (20 μg/ml) for 3–24 h as indicated. Whole cell extracts were prepared, and equal amounts of protein were analyzed by Western blotting, using antibodies against Smad3 and Grb2 as a loading control. B. The right panels of A were quantitated using ImageJ (NIH) software. The levels of Smad3 were corrected for the Grb2 loading control and normalized to the initial 3-h time point.
These data indicate that there is a higher turnover of Smad3 in EpRas cells, particularly in cycling cells, which is likely to account at least in part for the very low Smad3 levels seen in these cells. In contrast, Smad3 is most stable in quiescent EpH4 cells.
Re-expression of Smad3 Rescues the TGF-β-induced Growth Inhibitory Response in EpRas Cells
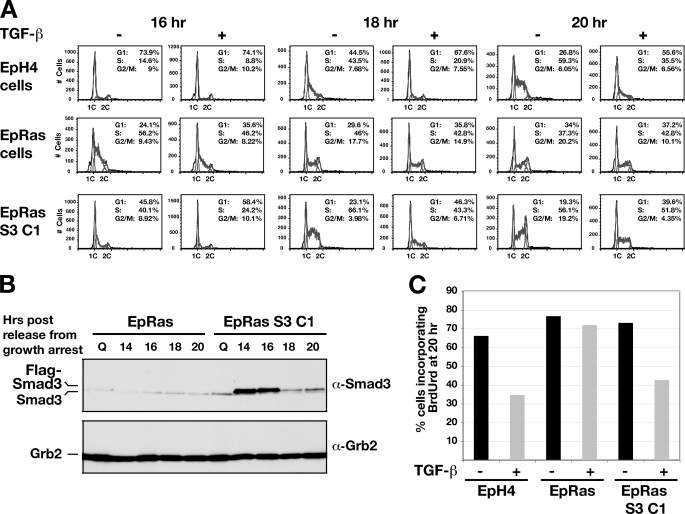
A major difference between EpH4 and EpRas cells is their functional response to TGF-β. In EpH4 cells, TGF-β has an antiproliferative effect, which is not observed in EpRas cells (Fig. 4A) (22). We demonstrated this by synchronizing cells by contact inhibition and then releasing them by trypsinization and replating in medium containing 10% serum in the absence and presence of TGF-β for 16, 18, or 20 h (29). At the 16-h time point, EpH4 cells were still predominantly in G1 regardless of the presence of TGF-β. At the 18- and 20-h time points, however, EpH4 cells entered S-phase in the absence of TGF-β, but the cells remained in G1 upon treatment with TGF-β (Fig. 4A, top row). In contrast, the EpRas cells, which exhibit a higher proliferation rate, had already entered S-phase 16 h after release, and progression through the cell cycle was not inhibited by the presence of TGF-β at any time point (Fig. 4A, second row).
FIGURE 4.
Stable expression of Smad3 restores the cell cycle arrest induced by TGF-β in EpRas cells. A, EpH4, EpRas, and EpRas S3 C1 cells were synchronized by contact inhibition and released by trypsinization into fresh medium in the presence or absence of TGF-β (2 ng/ml). Cells were collected at different time points and analyzed by FACS, to determine the number of cells in G1, S, and G2/M. Percentage of cells in each phase of the cell cycle are given. Cells were also collected for analysis by Western blotting (B). FLAG-Smad3 can be observed as a band running with a slightly lower mobility than endogenous Smad3 in extracts prepared from EpRas S3 C1 cells only. Grb2 serves as a loading control. C, cells were synchronized by contact inhibition then released by trypsinization into fresh medium with or without TGF-β in the presence of 10 mm BrdUrd. BrdUrd incorporation was measured by FACS analysis.
Having demonstrated that a key difference between EpH4 and EpRas cells is their level of Smad3, and given the importance of Smad3 for antiproliferative effects of TGF-β (supplemental Fig. 3), we investigated whether this response to TGF-β could be restored upon Smad3 re-expression in EpRas cells. To this end, we generated a clonal EpRas cell line (EpRas S3 C1) expressing FLAG-Smad3 and confirmed that the stably expressed FLAG-Smad3 was responsive to TGF-β (Fig. 4B and supplemental Fig. 4). The EpRas FLAG-Smad3 cells proliferate more slowly than EpRas cells, as seen by the fact that the percentage of cells in S-phase was reduced and the percentage of cells in G1 was increased, compared with the EpRas cells at 16 h post-release (Fig. 4A, third row). Most strikingly, the entry of the EpRas S3 C1 cells into S-phase was now inhibited by TGF-β. This was most evident at the 18-h time point, where a substantial reduction in the number of cells in S-phase and increase in the G1 population is observed in the presence of TGF-β (Fig. 4A, third row). Samples were taken in parallel to determine the level of Smad3 in EpRas and the EpRas Smad3 C1 cells. In highly quiescent cells, no FLAG-Smad3 is detected, as the cytomegalovirus promoter, which drives FLAG-Smad3 expression, is inactivated in these conditions.4 However, high levels of FLAG-Smad3 were detected 14 and 16 h after release from quiescence (Fig. 4B) and were slightly lowered thereafter but remain above the endogenous Smad3 levels of EpRas cells. A second independent FLAG-Smad3 expressing clone, exhibiting a lower level of Smad3 expression was also tested. In this case, the rescue of the growth inhibitory action of TGF-β was slightly less pronounced, correlating with the lower Smad3 expression level (supplemental Fig. 5A). The effect of TGF-β on cell cycle progression in these different cell types was also assayed by BrdUrd incorporation (Fig. 4C). The results clearly show that TGF-β inhibits cell cycle progression of EpH4 cells and EpRas FLAG-Smad3 cells but not EpRas cells. Taken together, these results suggest that the level of Smad3 dictates whether EpRas cells can respond to the antiproliferative effects of TGF-β.
High Smad3 Levels Do Not Interfere with TGF-β-induced EMT in EpRas Cells
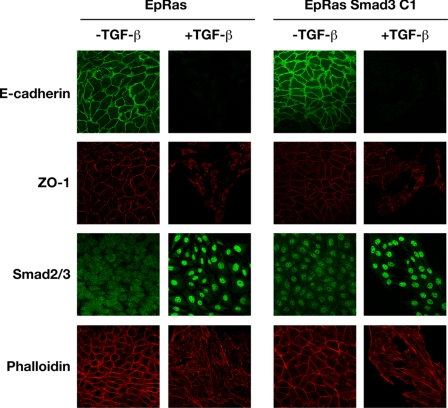
The predominant response of EpRas cells to TGF-β is to undergo EMT. This requires both activated Ras and TGF-β. It was previously shown in Madin-Darby canine kidney cells that Smad3 levels fall during the progression of EMT. However, low levels of Smad3 are not sufficient, as EpH4 cells depleted of Smad3 do not undergo EMT in response to TGF-β (supplemental Fig. 6). It was important to determine, however, whether increased Smad3 levels would interfere with the progression of EpRas cells through EMT. EpRas cells and the FLAG-Smad3-expressing EpRas cells were therefore assayed for their ability to undergo an EMT in response to TGF-β. Immunostaining of epithelial and mesenchymal markers in EpRas cells showed that in unstimulated cells, the adherens junction protein E-cadherin and the tight junction component ZO-1 are localized to intercellular junctions (Fig. 5, left panels). Staining of TGF-β-treated EpRas cells revealed a marked delocalization and down-regulation of ZO-1 and complete abolition of E-cadherin, revealing a loss in epithelial polarity indicative of EMT. In addition, phalloidin staining revealed a dramatic remodeling of actin to actin stress fibers in the presence of TGF-β, typical of migratory, mesenchymal cells (30). The FLAG-Smad3-expressing EpRas cell lines exhibited an identical phenotype, with loss of E-cadherin and down-regulation of ZO-1, suggesting that Smad3 expression in these cells does not interfere with the progression of EMT (Fig. 5, right panels and supplemental Fig. 5, B and C). These results establish that higher Smad3 levels do not interfere with progression of TGF-β-induced EMT in EpRas cells.
FIGURE 5.
Stable expression of Smad3 does not inhibit TGF-β-induced EMT in EpRas cells. EpRas and EpRas S3 C1 cells were plated out at low density and either grown in the presence or absence of TGF-β1 (2 ng/ml) for a total of 10 days as described under “Experimental Procedures.” Cells were then processed for immunofluorescence using an anti-E-cadherin antibody, to analyze adherens junctions or an anti-Zona Occludens 1 (ZO-1) antibody, to analyze tight junctions. Smad2/3 localization and actin reorganization was visualized with an anti-Smad2/3 antibody and Texas Red-conjugated phalloidin, respectively. The E-cadherin and ZO-1 staining was performed on one sample of cells, and the Smad2/3 and phalloidin staining on another. In the EpRas S3 C1 cells, FLAG-Smad3 can be seen in the nucleus in the absence of TGF-β, as has been described for EGFP-Smad3 (56).
DISCUSSION
Accumulating evidence has revealed that Smad3 plays an essential role in mediating the biological responses of TGF-β, and it has been implicated in both its antiproliferative and proinvasive effects. Here, we have shown that the expression of Smad3 protein is subject to regulation at the mRNA level as well as post-translationally, which results in a dramatic down-regulation of Smad3 in transformed EpRas cells compared with nontransformed EpH4 cells and also in a dramatic down-regulation of Smad3 in cycling versus quiescent cells. The low level of Smad3 accounts for the refractory response of the EpRas cells to the growth inhibitory activities of TGF-β, as this response is restored upon ectopic expression of Smad3.
Mechanism of Smad3 Regulation
We have shown that Smad3 is regulated at the mRNA level by signaling downstream of Ras and by the quiescent/cycling status of the cells. This is likely due to transcriptional regulation, but because we have only measured steady state mRNA levels, we cannot rule out effects on mRNA stability. To date however, there is very little known about regulatory elements controlling transcription of the Smad3 gene. Thus, an in depth analysis of the upstream sequences of the Smad3 gene is now important to identify elements required for the Ras-regulated transcriptional repression and the up-regulation in quiescent cells.
We have also shown that the stability of Smad3 protein is a major contributing factor in the observed differences in basal Smad3 levels between EpH4 and EpRas cells and also the differences in Smad3 levels between cycling and quiescent cells. We do not yet understand what dictates Smad3 stability in EpH4/EpRas cells. One possibility we investigated was that Smad3 degradation could be regulated by serine/threonine phosphorylation, as has been shown for Smad1, Myc, c-Jun, and cyclin E (31–33). Smad3 has been shown to be phosphorylated by ERK MAP kinase on Ser205, Ser208, and Thr179 in the linker region, and by CDK2/4 on Thr8 in the MH1 domain and Thr179 and Thr213 in the linker region (34, 35), and this has been shown to have a negative effect on Smad3 activity (34, 36, 37). Interestingly, the highest rate of Smad3 turnover we observed was in cycling EpRas cells, which would have the highest levels of CDK2/4 and ERK MAP kinases of the cell lines/conditions analyzed. However, mutation of any of the CDK or ERK MAP kinase sites in Smad3 independently or in various combinations had no stabilizing effect on Smad3 compared with wild type Smad3 in either EpH4 or EpRas cells (data not shown), ruling out a mechanism whereby CDK or ERK MAP kinase activity influences Smad3 stability through phosphorylation at these sites.
A recent study investigating basal Smad3 stability showed that Axin facilitates GSK3β-induced phosphorylation of Smad3 at Thr66 in the MH1 domain in HaCaT, mouse embryonic fibroblasts, and HepG2 cells (38). This triggers Smad3 ubiquitination and degradation, suggesting that GSK3β is a potential candidate for Smad3 regulation in EpRas cells. However, the activity of GSK3β is negatively regulated by Ras through phosphatidylinositol 3-kinase (39). Constitutive Ras activation would therefore be expected to lead to inhibition of GSK3β and thus reduce Smad3 turnover, contrary to what we observed. It therefore seems unlikely that GSK3β plays a role in Smad3 stability in EpRas cells. Consistent with this, treatment of EpRas cells with lithium chloride had no effect on Smad3 turnover (data not shown). Further work will be necessary to determine how Smad3 stability is regulated in EpH4/EpRas cells.
The Relevance of Smad3 Regulation in the EpH4/EpRas Tumor Model
The contribution of the signaling pathways downstream of H-Ras to induce the tumor-promoting activities of TGF-β has been clearly demonstrated in the EpH4/EpRas system (40). MAPK activity was shown to be important for TGF-β-induced EMT, tumorigenesis and metastasis, whereas Ras-induced phosphatidylinositol 3-kinase activity promoted cell scattering and rescued cells from TGF-β-induced cell-cycle arrest and apoptosis (40). However, the level at which these pathways cooperated and the molecular mechanisms governing these effects were largely unknown. Our current study reveals that prolonged Ras activity results in a reduction in Smad3 protein levels below the threshold required for growth inhibitory responses to TGF-β. In the EpRas cells, the decrease in Smad3 levels is sufficient to abrogate the responsiveness of these cells to TGF-β-induced growth inhibition, whereas the protumorigenic activities of TGF-β remain intact. Ectopic expression of Smad3 in these cells sensitizes them to this growth inhibitory response. Interestingly, endogenous Smad2 cannot compensate for the loss of Smad3, which is consistent with the distinct phenotypes of the Smad2- and Smad3-null mice (41), suggesting that these two proteins have nonredundant functions in the TGF-β pathway.
We further speculate that the decrease in Smad3 protein levels in EpRas cells may not only render the cells resistant to the antiproliferative effects of TGF-β but may also elicit Ras-induced transformation. In untransformed cells, oncogenes, like activated Ras and Raf, normally induce senescence (42), and in a mouse multistage skin carcinogenesis model, Ras-induced senescence has been shown to be mediated via Smad3 (43). However, upon acquisition of other oncogenic mutations, activated Ras and Raf induce transformation. Thus, it seems likely that transformation by Raf or Ras might be facilitated by down-regulation of Smad3, which may be induced by the expression of the activated Ras or Raf themselves, as in the Madin-Darby canine kidney and EpRas models (this work and Ref. 17), or through other distinct mechanisms such as promoter methylation.
We have also shown that Smad3 levels are dependent on the quiescent/cycling status of the EpH4 and EpRas cells. In EpH4 cells, this phenomenon restricts the antiproliferative effects of TGF-β to cells that are already in a quiescent state. TGF-β prevents quiescent cells re-entering the cell cycle, but it cannot induce arrest of EpH4 cells when added at other points in the cell cycle.4 We hypothesize that this might be important in vivo for cell cycle progression in the presence of low levels of autocrine TGF-β.
Regulation of Smad3 Levels in Cancer
Modulation of Smad3 expression has been observed in a wide spectrum of human cancers. Importantly, we find a correlation between colorectal tumor cells harboring activating Ras or Raf mutations and low levels of Smad3 relative to Smad2 (supplemental Fig. 7), suggesting that our observations in the EpH4/EpRas system are general. Interestingly, in tumor cells that also have an inactivating mutation in Smad4, Smad3 is readily detectable, suggesting that once Smad4 activity is lost there is no pressure to lose Smad3 (supplemental Fig. 7). It has also been previously reported that two of nine gastric cancer cell lines exhibit low Smad3 mRNA and protein levels, resulting in loss of some TGF-β responsiveness (20). Introduction of Smad3 into Smad3-deficient gastric cancer cell lines restores TGF-β-mediated p15 and p21 induction, as well as growth inhibition. Moreover, these Smad3-expressing cells showed dramatically reduced tumorigenicity in vivo, providing strong evidence that Smad3 has an important tumor suppressive function in the early stages of gastric carcinogenesis (20). In addition, Smad3 levels decrease during carcinogenesis in some tissues such as the breast, where the nuclear Smad3 level is reduced in high grade breast cancers (44). Furthermore, investigation of Smad3 protein levels in T-cell acute lymphoblastic leukemia patients revealed a complete loss of Smad3, which is not due to mutations in the Smad3 gene or alterations in the level of Smad3 mRNA expression (21).
Microarray analysis has also revealed a wealth of information regarding Smad3 expression in a range of cancers. In contrast to the Letterio study mentioned above (21), a detailed study on patients with leukemia demonstrated a reduction of Smad3 mRNA in T-cell acute lymphoblastic leukemia, and also found decreased Smad3 mRNA in B-cell acute lymphoblastic leukemia and acute myeloid leukemia tumors (45). Smad3 is also down-regulated in B-cell lymphomas (46–48) and in ovarian adenocarcinomas (serous and endometrioid) (49, 50). Three independent studies showed that Smad3 is down-regulated in prostate carcinomas (51–53), and moreover, the level of Smad3 is down-regulated to a greater extent in more aggressive, metastatic carcinomas (51, 53). From this evidence, it is tempting to speculate that Smad3 expression is further down-regulated in metastatic cells, to overcome the growth inhibitory and apoptotic activities of TGF-β at distant sites. It is possible that a minimal threshold level of Smad3 is required for TGF-β induced migration.
For many years, it was believed that Smad3 was not mutated in cancer, in contrast to Smad4, which is frequently mutated or deleted in pancreatic and colon cancer (54). However, Smad3 mutations have been very recently detected in colon, breast, and pancreatic cancers (18, 19, 55). These include a mutation in the MH2 domain (F260S) and extreme C-terminal serine (S422F) of Smad3, which are likely to interfere with the activity of Smad3 by perturbing the Smad3/Smad4 interaction (18).
In conclusion, Smad3 functions as both a negative and positive regulator of tumorigenesis. Here, we propose that the level of Smad3 protein is an important determinant of the TGF-β response and can dictate whether TGF-β acts as a tumor suppressor or tumor promoter. By regulation through a complex integration of signals, the level of Smad3, and, therefore, its activity, is altered. The high level of Smad3 is attributed to the antiproliferative effects of TGF-β, whereas a minimal level is sufficient for the migratory and invasive properties associated with TGF-β. Restoring high levels of Smad3, required for the growth inhibitory responses of TGF-β, could be critical in developing strategies to target metastatic tumors, which have lost responsiveness to the antiproliferative effects of TGF-β.
Supplementary Material
Acknowledgments
We thank the Cancer Research UK FACS lab for help with FACS analysis and the Cancer Research UK Cell Services for providing numerous cell lines. We thank Ian Tomlinson for the panel of colorectal cell lines. We thank Mike Howell, Laurence Levy, Bernhard Schmierer, Erik Sahai, and current members of the Hill lab for helpful discussions and advice and/or useful comments on the manuscript.
This work was supported by Cancer Research UK, a European Union Research Training Network Grant (MRTN-CT-2004-005428), and a Marie Curie Intra-European Fellowship PIEF-GA-2009-235995 (to P. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. 1–7, and additional references.
A. C. Daly, P. Vizan, and C. S. Hill, unpublished observations.
- TGF-β
- transforming growth factor β
- EMT
- epithelial to mesenchymal transition
- FACS
- fluorescence-activated cell sorting
- CDK
- cyclin-dependent kinase
- ERK
- extracellular-signal regulated kinase
- BrdUrd
- bromodeoxyuridine.
REFERENCES
- 1.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 2.Daly A. C., Randall R. A., Hill C. S. (2008) Mol. Cell Biol. 28, 6889–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu I. M., Schilling S. H., Knouse K. A., Choy L., Derynck R., Wang X. F. (2009) EMBO J. 28, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrighton K. H., Lin X., Yu P. B., Feng X. H. (2009) J. Biol. Chem. 284, 9755–9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhurst R. J., Derynck R. (2001) Trends Cell Biol. 11, S44–S51 [DOI] [PubMed] [Google Scholar]
- 6.Derynck R., Akhurst R. J., Balmain A. (2001) Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 7.Wakefield L. M., Roberts A. B. (2002) Curr. Opin. Genet. Dev. 12, 22–29 [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy L., Hill C. S. (2005) Mol. Cell Biol. 25, 8108–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashcroft G. S., Yang X., Glick A. B., Weinstein M., Letterio J. L., Mizel D. E., Anzano M., Greenwell-Wild T., Wahl S. M., Deng C., Roberts A. B. (1999) Nature Cell Biol. 1, 260–266 [DOI] [PubMed] [Google Scholar]
- 11.Datto M. B., Frederick J. P., Pan L., Borton A. J., Zhuang Y., Wang X. F. (1999) Mol. Cell Biol. 19, 2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., Deng C. (1999) EMBO J. 18, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y. A., Zhang G. M., Feigenbaum L., Zhang Y. E. (2006) Cancer Cell 9, 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian F., DaCosta Byfield S., Parks W. T., Yoo S., Felici A., Tang B., Piek E., Wakefield L. M., Roberts A. B. (2003) Cancer Res. 63, 8284–8292 [PubMed] [Google Scholar]
- 15.Thiery J. P. (2003) Curr. Opin. Cell Biol. 15, 740–746 [DOI] [PubMed] [Google Scholar]
- 16.Zavadil J., Böttinger E. P. (2005) Oncogene 24, 5764–5774 [DOI] [PubMed] [Google Scholar]
- 17.Nicolás F. J., Lehmann K., Warne P. H., Hill C. S., Downward J. (2003) J. Biol. Chem. 278, 3251–3256 [DOI] [PubMed] [Google Scholar]
- 18.Jones S., Zhang X., Parsons D. W., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S. M., Fu B., Lin M. T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) Science 321, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leary R. J., Lin J. C., Cummins J., Boca S., Wood L. D., Parsons D. W., Jones S., Sjöblom T., Park B. H., Parsons R., Willis J., Dawson D., Willson J. K., Nikolskaya T., Nikolsky Y., Kopelovich L., Papadopoulos N., Pennacchio L. A., Wang T. L., Markowitz S. D., Parmigiani G., Kinzler K. W., Vogelstein B., Velculescu V. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16224–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S. U., Kim H. T., Seong D. H., Kim Y. S., Park Y. S., Bang Y. J., Yang H. K., Kim S. J. (2004) Oncogene 23, 1333–1341 [DOI] [PubMed] [Google Scholar]
- 21.Wolfraim L. A., Fernandez T. M., Mamura M., Fuller W. L., Kumar R., Cole D. E., Byfield S., Felici A., Flanders K. C., Walz T. M., Roberts A. B., Aplan P. D., Balis F. M., Letterio J. J. (2004) N. Engl. J. Med. 351, 552–559 [DOI] [PubMed] [Google Scholar]
- 22.Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. (1996) Genes Dev. 10, 2462–2477 [DOI] [PubMed] [Google Scholar]
- 23.Inman G. J., Hill C. S. (2002) J. Biol. Chem. 277, 51008–51016 [DOI] [PubMed] [Google Scholar]
- 24.Marais R., Wynne J., Treisman R. (1993) Cell 73, 381–393 [DOI] [PubMed] [Google Scholar]
- 25.Nicolás F. J., Hill C. S. (2003) Oncogene 22, 3698–3711 [DOI] [PubMed] [Google Scholar]
- 26.Howell M., Hill C. S. (1997) EMBO J. 16, 7411–7421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierreux C. E., Nicolás F. J., Hill C. S. (2000) Mol. Cell Biol. 20, 9041–9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enoch T., Zinn K., Maniatis T. (1986) Mol. Cell Biol. 6, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petritsch C., Beug H., Balmain A., Oft M. (2000) Genes Dev. 14, 3093–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiery J. P., Sleeman J. P. (2006) Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 31.Sapkota G., Alarcón C., Spagnoli F. M., Brivanlou A. H., Massagué J. (2007) Mol. Cell 25, 441–454 [DOI] [PubMed] [Google Scholar]
- 32.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 33.Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr. (2005) Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 34.Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 35.Ross S., Hill C. S. (2008) Int. J. Biochem. Cell Biol. 40, 383–408 [DOI] [PubMed] [Google Scholar]
- 36.Kretzschmar M., Doody J., Timokhina I., Massagué J. (1999) Genes Dev. 13, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F. (2006) Cytokine Growth Factor Rev. 17, 9–17 [DOI] [PubMed] [Google Scholar]
- 38.Guo X., Ramirez A., Waddell D. S., Li Z., Liu X., Wang X. F. (2008) Genes Dev. 22, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massagué J. (2004) Nature 432, 298–306 [DOI] [PubMed] [Google Scholar]
- 40.Janda E., Lehmann K., Killisch I., Jechlinger M., Herzig M., Downward J., Beug H., Grünert S. (2002) J. Cell Biol. 156, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goumans M. J., Mummery C. (2000) Int. J. Dev. Biol. 44, 253–265 [PubMed] [Google Scholar]
- 42.Mooi W. J., Peeper D. S. (2006) N. Engl. J. Med. 355, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 43.Vijayachandra K., Lee J., Glick A. B. (2003) Cancer Res. 63, 3447–3452 [PubMed] [Google Scholar]
- 44.Jeruss J. S., Santiago J. Y., Woodruff T. K. (2003) Mol. Cell Endocrinol. 203, 185–196 [DOI] [PubMed] [Google Scholar]
- 45.Andersson A., Ritz C., Lindgren D., Edén P., Lassen C., Heldrup J., Olofsson T., Råde J., Fontes M., Porwit-Macdonald A., Behrendtz M., Höglund M., Johansson B., Fioretos T. (2007) Leukemia 21, 1198–1203 [DOI] [PubMed] [Google Scholar]
- 46.Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J., Jr., Lu L., Lewis D. B., Tibshirani R., Sherlock G., Chan W. C., Greiner T. C., Weisenburger D. D., Armitage J. O., Warnke R., Levy R., Wilson W., Grever M. R., Byrd J. C., Botstein D., Brown P. O., Staudt L. M. (2000) Nature 403, 503–511 [DOI] [PubMed] [Google Scholar]
- 47.Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. (2005) Nat. Genet. 37, 382–390 [DOI] [PubMed] [Google Scholar]
- 48.Haslinger C., Schweifer N., Stilgenbauer S., Döhner H., Lichter P., Kraut N., Stratowa C., Abseher R. (2004) J. Clin. Oncol. 22, 3937–3949 [DOI] [PubMed] [Google Scholar]
- 49.Hendrix N. D., Wu R., Kuick R., Schwartz D. R., Fearon E. R., Cho K. R. (2006) Cancer Res. 66, 1354–1362 [DOI] [PubMed] [Google Scholar]
- 50.Lancaster J. M., Dressman H. K., Whitaker R. S., Havrilesky L., Gray J., Marks J. R., Nevins J. R., Berchuck A. (2004) J. Soc. Gynecol. Investig. 11, 51–59 [DOI] [PubMed] [Google Scholar]
- 51.Lapointe J., Li C., Higgins J. P., van de Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Ekman P., DeMarzo A. M., Tibshirani R., Botstein D., Brown P. O., Brooks J. D., Pollack J. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsh J. B., Sapinoso L. M., Su A. I., Kern S. G., Wang-Rodriguez J., Moskaluk C. A., Frierson H. F., Jr., Hampton G. M. (2001) Cancer Res. 61, 5974–5978 [PubMed] [Google Scholar]
- 53.Yu Y. P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., Michalopoulos G., Becich M., Luo J. H. (2004) J. Clin. Oncol. 22, 2790–2799 [DOI] [PubMed] [Google Scholar]
- 54.Levy L., Hill C. S. (2006) Cytokine Growth Factor Rev. 17, 41–58 [DOI] [PubMed] [Google Scholar]
- 55.Ku J. L., Park S. H., Yoon K. A., Shin Y. K., Kim K. H., Choi J. S., Kang H. C., Kim I. J., Han I. O., Park J. G. (2007) Cancer Lett. 247, 283–292 [DOI] [PubMed] [Google Scholar]
- 56.Nicolás F. J., De Bosscher K., Schmierer B., Hill C. S. (2004) J. Cell Sci. 117, 4113–4125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.