Abstract
Insulin-like growth factor-1 (IGF-1) signaling system exerts a broad antiapoptotic function and plays a crucial role in resistance to anticancer therapies. Exposure of MCF-7 breast cancer cells to IGF-1 rapidly and transiently induced tyrosine phosphorylation and activation of phosphoinositide-dependent kinase-1 (PDK1). This was paralleled by Akt/protein kinase B and protein kinase C-ζ phosphorylation, at Thr308 and Thr410, respectively. IGF-1 treatment also enhanced PDK1 interaction with IGF-1 receptor (IGF-1R) in intact MCF-7 cells. Pulldown assays revealed that PDK1 bound IGF-1R in vitro and that the region encompassing amino acids 51–359 of PDK1 was necessary for the interaction. Synthetic peptides corresponding to IGF-1R C terminus amino acids 1295–1337 (C43) and to PDK1 amino acids 114–141 reduced in vitro IGF-1R/PDK1 interaction in a concentration-dependent manner. Loading of fluoresceinated-C43 (fluorescein isothiocyanate (FITC)-C43) into MCF-7 cells significantly reduced IGF-1R/PDK1 interaction and phosphorylation of PDK1 substrates. Moreover, FITC-C43 intracellular loading reverted the protective effect of IGF-1 on growth factor deprivation-induced cell death. Finally, the inhibition of IGF-1R/PDK1 interaction and signaling by FITC-C43 was accompanied by 2-fold enhanced killing capacity of cetuximab in human GEO colon adenocarcinoma cells and was sufficient to restore cell death in cetuximab-resistant cell clones. Thus, disruption of PDK1 interaction with IGF-1R reduces IGF-1 survival effects in cancer cells and may enhance cell death by anticancer agents.
Keywords: Cancer Therapy, Insulin-like Growth Factor (IGF), Peptides, Phosphatidylinositol-dependent Kinase-1 (PDK1), Phosphotyrosine Signaling, Signal Transduction
Introduction
The insulin-like growth factor (IGF)3 signaling system plays a key role in growth and development of many normal tissues and regulates overall growth of organisms (1). Type I insulin-like growth factor (IGF-1) has been identified as a regulator of cellular transformation and controls the acquisition of the tumorigenic phenotype by regulating multiple cellular functions that impact on the invasive/metastatic potential of cancer cells. These include cell survival, motility, invasion, growth potential in secondary organ sites, and the induction of angiogenesis (2). There are also several lines of evidence that dysregulation of the IGF-1 system is involved in resistance to certain anticancer therapies, including cytotoxic chemotherapy, hormonal agents, biological therapies, and radiation (3). The link between cancer and IGF signaling is also consistent with recent epidemiological studies showing an increased relative risk for the development of colon, prostate, breast, lung, and bladder cancers in individuals with circulating IGF-1 levels in the upper tertile of the normal range (4). These findings were confirmed in animal models, where reduced circulating IGF-1 levels result in significant reductions in cancer development, growth, and metastases, whereas increased circulating IGF-1 levels are associated with enhanced tumor growth (5).
Aberrant expression of the receptor for IGF-1 (IGF-1R) has been implicated in malignant transformation of cells (6, 7). IGF-1R is normally activated by binding of the secreted growth factor ligand to the extracellular domain. This, in turn, triggers a number of cellular signaling pathways, including the phosphatidylinositol 3-kinase (PI3K) pathway, the main mechanism by which IGF-1R protects cells from apoptosis (8, 9).
Phosphoinositide-dependent protein kinase 1 (PDK1) plays a crucial role in mediating signal transduction downstream of PI3K (10). PDK1 is a 64-kDa protein, of 556 amino acids, comprised of a serine/threonine kinase domain near the N terminus, a C-terminal pleckstrin homology (PH) domain, and an ATP-binding site located between the two domains (11). On the small lobe of the protein, a hydrophobic pocket, close to but distinct from the catalytic site, has been identified and characterized (12). Binding on this pocket of phosphorylated C-terminal motifs from other kinases allows allosteric regulation of PDK1 activity (13). PDK1 phosphorylates several protein kinases, including Akt/PKB and PKCs, at the activation loop phosphorylation sites (11, 14, 15). Phosphorylation of Ser241 at its own activation loop may be mediated by an intermolecular mechanism suggesting that dimerization and trans-phosphorylation may regulate PDK1 activity in cells (10, 11, 16). There is also evidence that PDK1 undergoes tyrosine phosphorylation in response to several growth factors. Studies with the tyrosine phosphatase inhibitor pervanadate and insulin indicate that full activation of PDK1 requires phosphorylation at Tyr373/376 (17–19). Consistent with its role in transformation, PDK1 is highly expressed in a large number of invasive human breast cancer cell lines and in ovarian cancers (20, 21).
Collectively, a large body of evidence identifies the IGF-1R/PDK1 system as a target for molecular therapy with potential benefits for a wide spectrum of human malignancies (3, 4, 22). Various strategies have been used to target components of this system in established cancer cell lines and animal models, and some of these strategies may be advancing to clinical use. Among them, down-regulation of IGF-1R by antisense oligonucleotides, antisense RNA, small interfering RNA, single chain antibody, full humanized anti-IGF-1R monoclonal antibodies, and specific kinase inhibitors have been attempted (8, 23). Some of these molecular strategies to target the IGF-1 system, however, have failed for the magnitude of toxicity, due, at least in part, to the cross-reactivity with the insulin system.
Here, we show that IGF-1R directly interacts with and activates PDK1. Synthetic peptides corresponding to the IGF-1R C terminus and to the PDK1 hydrophobic pocket selectively displace the interaction in vitro. Peptide loading into human cancer cell lines inhibits IGF-1 signaling via PDK1 and prevents IGF-1 prosurvival effects, thereby facilitating cell killing by anticancer agents.
EXPERIMENTAL PROCEDURES
Materials
Medium, serum, and antibiotics for cell culture were from Invitrogen (Invitrogen, Paisley, UK). Antibodies against PDK1, IGF-1 receptor α-subunit, IGF-1 receptor β-subunit, phospho-Thr308 Akt1/PKB, and Akt/PKBα were purchased from Upstate Biotech Millipore (Lake Placid, NY). PKCζ, phospho-Thr410 PKCζ, and extracellular signal-regulated kinase antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Phospho-Ser473 Akt, phospho-Ser241 PDK1, phospho-Tyr373/376 PDK1, and phospho-Thr202/Tyr204 ERK were obtained from Cell Signaling Technology (Danvers, MA). Recombinant human IGF-1 was from PeproTech (London, UK). The p-EBG-2T expression vector containing the cDNA of the human full-length PDK1 and the cDNA of PDK1 deletion mutants were a generous gift of Dr. D. R. Alessi (University of Dundee) and have been previously described (24). Protein A-Sepharose beads and enhanced chemiluminescence (ECL) reagents were from Pierce. Agarose-bound wheat germ agglutinin was from Vector Laboratories Inc. (Burlingame, CA), and SDS-PAGE reagents were from Bio-Rad. Nα-protected Fmoc-amino acid derivatives and coupling reagents for peptide synthesis were from Inbios (Pozzuoli, Italy). HPLC-grade solvents and trifluoroacetic acid were from LabScan (Stillorgan, Dublin, Ireland). 2-Deoxy-[14C]glucose was from PerkinElmer Life Sciences. All the other chemicals were from Sigma-Aldrich.
Cell Cultures
MCF-7 cells, L6 cells, and NIH-3T3 cells stably transfected with IGF-1 receptor (NIH-3T3IGF-1R) (25) were plated (6 × 103 cells/cm2) and grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 2 mm glutamine, 100 IU/ml penicillin, 100 IU/ml streptomycin. GEO and GEO-CR (26) colon cancer cells were cultured in McCoy's medium supplemented with 10% fetal bovine serum, 20 mm Hepes (pH 7.4), 2 mm glutamine. Cultures were maintained in humidified atmosphere of 95% air and 5% CO2 at 37 °C. L6 differentiation has been achieved as described previously (27).
Immunoblot and Immunoprecipitation Procedure
Cells were solubilized for 20 min at 4 °C with lysis buffer containing 50 mm HEPES, 150 mm NaCl, 10 mm EDTA, 10 mm Na4P2O7, 2 mm sodium orthovanadate, 50 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, pH 7.4, and 1% (v/v) Triton X-100. The lysates were clarified by centrifugation at 12,000 × g for 20 min at 4 °C. Proteins were separated by SDS-PAGE and blotted on Immobilon-P membranes (Millipore, Billerica, MA). Membranes were blocked for 1 h in TBS (10 mm Tris-HCl, pH 7.4, and 140 mm NaCl) containing 4% (w/v) bovine serum albumin and then incubated with the indicated antibodies. Detection of blotted proteins was performed by ECL according to the manufacturer's instructions. Immunoprecipitation experiments were performed as described previously (28). Densitometric analysis was performed using a Scion image analyzer. All the data were expressed as mean ± S.D.
IGF-1 Receptor Purification
IGF-1Rs were partially purified by wheat germ agglutinin affinity chromatography. Confluent monolayers of NIH-3T3IGF-1R cells (corresponding to 6–8 × 107 cells) were solubilized in lysis buffer. The insoluble material was separated by ultracentrifugation at 100,000 × g for 1 h at 4 °C. The supernatant was applied to a wheat germ agglutinin-Sepharose column pre-equilibrated with HNT buffer (50 mm HEPES, 150 mm NaCl, and 0.1% Triton) and the protease inhibitors described above. The column was extensively washed using the same buffer, and bound glycoproteins were eluted in the same buffer containing 0.3 m N-acetylglucosamine (29).
Pulldown Assay
PDK1-glutathione S-transferase (GST) fusion proteins were generated as described (24). Purified IGF-1R (50 μg of protein extract) was incubated in the presence of Sepharose-bound GST-PDK1 (2 μg) for 2 h at 4 °C. The beads were washed two times with HNT buffer and then resuspended in loading buffer, heated to 95 °C for 5 min, and centrifuged at 25,000 × g for 3 min. Supernatants were separated by SDS-PAGE followed by immunoblotting with appropriate antibodies.
In Vitro PDK1 Tyrosine Phosphorylation
Purified IGF-1R (50 μg of protein extract) was incubated in the presence of IGF-1 (100 ng/ml). Phosphorylation reactions were initiated by adding 2 mm CTP, 2 μm ATP, 10 mm HEPES, pH 7.4, 0.02% Triton X-100, 5 mm MnCl2, 7 mm MgCl2 (final concentrations) and prolonged for 30 min at 22 °C. Recombinant PDK1 was added for 15 min. The reaction was stopped on ice, and proteins were separated by SDS-PAGE and analyzed by Western blotting with phosphotyrosine antibodies.
Peptide Synthesis and Purification
Solid-phase peptide syntheses were carried out on a fully automated peptide synthesizer 433A (Applied Biosystems). Preparative reverse-phase-HPLC purifications were carried out on a Shimadzu LC-8A instrument equipped with a SPD-M10 AV detector and using a Phenomenex Luna-COMBI C18 column (50 × 22-mm inner diameter; 10 μm). LC-MS analyses were carried out on an LCQ DECA XP ion trap mass spectrometer (ThermoElectron, Milan, Italy) LC-MS columns were from ThermoElectron. Peptides were prepared by solid-phase synthesis following standard Fmoc/t-butyl protocols (30). To monitor cell delivery, N-terminal fluorescein labeling was achieved by on-resin treatment with fluorescein isothiocyanate (FITC). Peptide purity and identity were confirmed by LC-MS.
Peptide Loading into Cultured Cells
All the peptides (including FITC-conjugated peptides) were delivered onto semiconfluent MCF-7 cells by using the ProVectin protein delivery reagents method according to the manufacturer's instructions. Briefly, the dry film of ProVectin reagent found in a glass vial was dissolved with 250 μl of methanol and vortexed for 10-20 s. The manufacturer recommended 20 μl of ProVectin reagent for each 60-mm-diameter dish. The total amount of ProVectin needed was pipetted into an Eppendorf tube and left under a laminar flow hood to evaporate the solvent for at least 2 h at room temperature and then hydrated with PBS. Peptides, diluted in PBS, were added to the ProVectin reagent at different concentrations and let stand at room temperature for 5 min. The ProVectin/peptide complexes were added directly onto the cells cultured in serum-free DMEM for 24 h. Loading efficiency was about 65% for all peptides.
Cell Death Analysis
For detecting apoptosis, the cells were kept in the presence or in the absence of serum or incubated with 10 mm synthetic peptides, as indicated in the description of the individual experiments. Apoptosis was then assayed using the apoptosis ELISAPlus kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer's instructions or by cytometric analysis. For cytometry, cells were harvested and suspended in 0.3 ml of PBS. After adding 0.7 ml of cold absolute ethanol, cells were fixed and stored for at least 2 h at −20 °C. For analysis, cells were centrifuged, resuspended in PBS containing 0.1 mg/ml RNase A and 40 μg/ml propidium iodide, and incubated in dark for 30 min at room temperature. Samples were kept on ice, and the analysis was performed using FACS CyAn cytometer (Dako Cytomation). A minimum of 30,000 events was collected for each sample. Sub-G1 phase cells were identified using the Summit version 4.3 software (Dako).
Fluorescence Microscopy
Cells, loaded with FITC-peptides, were fixed with 3% formaldehyde for 15 min at 37 °C, washed three times with PBS, and incubated for 10 min at room temperature with 4′,6-diamidino-2-phenylindole diluted 1: 1000 in TBS. All slides were then washed three times with TBS 0.1% Tween and kept in dark until examined under a fluorescence microscope.
2-Deoxy-d-glucose Uptake
The measurement of 2-deoxy-d-[14C]glucose uptake was taken as a readout of glucose uptake by muscle cells, as described previously (31). Cells were incubated in serum-free DMEM supplemented with 0.2% (w/v) bovine serum albumin for 18 h in the presence or absence of C43 or scrambled peptide (SP) 10 μm. Cells were incubated in glucose-free 20 mm HEPES, pH 7.4, 140 mm NaCl, 2.5 mm MgSO4, 5 mm KCl, 1 mm CaCl2 (HEPES buffer) and exposed or not to 100 nm insulin for 30 min. Glucose uptake was measured by incubating cells with 0.15 mm 2-deoxy-d-[14C]glucose (0.5 μCi/assay) for 15 min in HEPES buffer. The reaction was terminated by the addition of 10 μm cytochalasin B, and the cells were washed three times with ice-cold isotonic saline solution prior to lysis in 1 m NaOH. Incorporated radioactivity was measured in a liquid scintillation counter.
Statistical Analysis
Data were analyzed with Statview software (Abacusconcepts) by one-factor analysis of variance. p values less than 0.05 were considered statistically significant.
RESULTS
IGF-1 Induces PDK1 Tyrosine Phosphorylation and Co-precipitation with IGF-1R in MCF-7 Cells
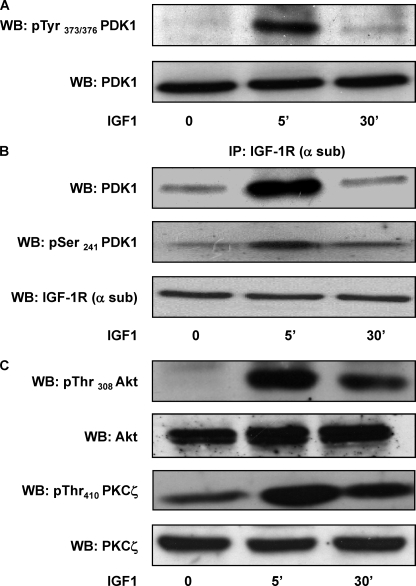
We analyzed tyrosine phosphorylation of PDK1 in response to IGF-1 in human breast cancer cells. MCF-7 cells were incubated with serum-free medium and treated with IGF-1 (100 ng/ml) for 5 and 30 min. Western blot analysis with antibodies specifically recognizing pTyr373/376-PDK1 revealed that IGF-1 rapidly induces tyrosine phosphorylation of endogenous PDK1 as compared with the control cells. Tyrosine phosphorylation returned close to the basal levels after 30 min, with no change in the total PDK1 levels (Fig. 1A). To gain further insights into the molecular mechanism of PDK1 phosphorylation, we tested the ability of IGF-1R to interact with PDK1. To this aim, MCF-7 cells were stimulated with IGF-1 (100 ng/ml), and cell lysates were precipitated with IGF-1R antibodies followed by blotting with PDK1 antibodies. As shown in Fig. 1B, an interaction between IGF-1R and PDK1 occurred after 5 min of IGF-1 exposure and was poorly detectable after 30 min, as in the case of tyrosine phosphorylation. Immunoblotting with specific antibodies also revealed that IGF-1R-associated PDK1 was phosphorylated at Ser241 (Fig. 1B). Moreover, stimulation of MCF-7 cells with IGF-1 increased phosphorylation of Akt/PKB and PKCζ at the PDK1 consensus sites, indicating the induction of PDK1 activity. Phosphorylation of both Akt/PKB and PKCζ remained well detectable up to 30 min, suggesting the existence of either a prolonged activation of PDK1 or a delayed activation of specific phosphatases in response to IGF-1. Control detection with Akt/PKB and PKCζ antibodies revealed no difference in the total levels (Fig. 1C).
FIGURE 1.
PDK1 tyrosine phosphorylation, IGF-1R co-precipitation, and substrate phosphorylation. A, MCF-7 cells were exposed to 100 ng/ml IGF-1 for 5 and 30 min, as indicated, and then solubilized as described under “Experimental Procedures.” Cell lysates (50 μg of protein/sample) were blotted with phospho-tyrosine 373/376-PDK1 antibodies (pTyr373/376-PDK1). To ensure the equal PDK1 transfer, membranes were blotted with PDK1 antibodies (PDK1). The filters were revealed by ECL and autoradiography. WB, Western blot. B, MCF-7 cell lysates (200 μg/sample) were immunoprecipitated (IP) with IGF-1R α-subunit antibodies followed by blotting with PDK1 antibodies and then reblotted with phospho-serine 241 PDK1 antibodies (pSer241-PDK1). To ensure equal immunoprecipitation, membranes were blotted with α-subunit-IGF-1R antibodies (α-sub-IGF-1R). Membranes were revealed by ECL and autoradiography. C, MCF-7 total cell lysates (50 μg of protein/sample) were blotted with phospho-threonine 308 Akt/PKB (pThr308-Akt/PKB) and phospho-threonine 410 PKCζ (pThr410-PKCζ) antibodies and then reblotted with Akt/PKB and PKCζ antibodies. Membranes were revealed by ECL and autoradiography. All the autoradiographs shown are representative of four independent experiments.
IGF-1R Directly Interacts with and Phosphorylates PDK1 in Vitro
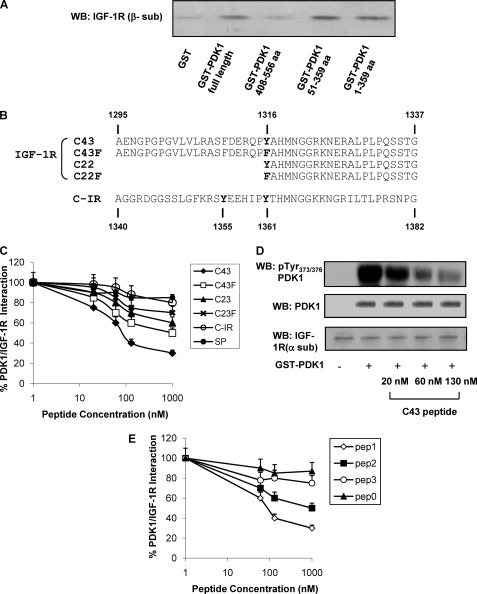
To determine the PDK1 portion involved in the interaction with IGF-1R, recombinant GST fused with full-length PDK1 protein (residues 1–556), two PDK1 mutants lacking the PH domain (corresponding to residues 1–359 and 51–359, respectively), and the PH domain alone (residues 408–556) were used in pulldown experiments with purified IGF-1R. IGF-1R was detectable in GST pulldown experiments with the full-length PDK1. The interaction was also well detectable for PDK1-(51–359) and PDK1-(1–359), whereas it was almost completely absent with PDK1-(408–556) (Fig. 2A).
FIGURE 2.
In vitro interaction of IGF-1R with PDK1. A, IGF-1R was partially purified from NIH3T3IGF-1R by wheat germ agglutinin affinity chromatography and subjected to pulldown using full-length GST-PDK1 and deletion mutants (amino acids (aa) 1–359, 51–359, and 408–556). Equal amounts of IGF-1R were incubated with each fusion protein and then eluted and analyzed by 7.5% SDS-PAGE; IGF-1R bound to fusion proteins was detected by Western blotting (WB) using anti-β-subunit IGF-1R antibodies. B, amino acid sequence alignment of synthetic peptides corresponding to IGF-1R C terminus fragment (C43; C22), to a mutated version, in which Tyr1316 has been substituted with Phe (C43F; C22F); and to C-IR. C, partially purified IGF-1R was incubated for 2 h at 4 °C with Sepharose-GST-PDK1 in the presence or in the absence of synthetic peptides (C43, C43F, C22, C22F, C-IR, and SP) at increasing concentrations as indicated. Pulled-down proteins were blotted with anti-IGF-1R (β-subunit) antibody, and the results were quantitated by laser densitometry. Error bars indicate mean ± S.D. D, equal amounts of purified IGF-1R were stimulated with 100 ng/ml IGF-1, as described under “Experimental Procedures,” and incubated or not with GST-PDK1 in the presence of increasing concentrations of C43, as indicated. Proteins were blotted with pTyr373/376-PDK1 antibodies. Membranes were reblotted with PDK1 and IGF-1R (α-subunit) antibodies, as shown on the bottom. Blots were revealed by ECL and autoradiography. E, partially purified IGF-1R was incubated for 2 h at 4 °C with Sepharose-GST-PDK1 in the presence or in the absence of synthetic peptides (pep1, pep2, pep3, and pep0) at increasing concentrations as indicated. Pulled-down proteins were blotted with anti-IGF-1R (β-subunit) antibody, and the results were quantitated by laser densitometry. All the autoradiographs shown are representative of at least four independent experiments.
It has been previously reported (19) that the region corresponding to the insulin receptor (IR) C terminus is critical for the interaction with PDK1. With this background, peptides corresponding to the C-terminal residues 1295–1337 of IGF-1R (C43), 1316–1337 (C22), and similar peptides featuring tyrosine replacement with phenylalanine (C43F and C22F) were synthesized (Fig. 2B). In pulldown assays (Fig. 2C), C43 peptide displayed the highest ability to reduce IGF-1R/PDK1 interaction (ED50 = 80 nm). At variance, C43F (ED50 = 1 μm), C22 (ED50 >1 μm), and C22F (ED50 >1 μm) were significantly less effective (Fig. 2C). Furthermore, a synthetic peptide corresponding to the IR C terminus (C-IR) and an SP did not significantly impair IGF-1R/PDK1 interaction (Fig. 2C). Similarly, co-incubation with C43 reduced tyrosine phosphorylation of PDK1 by activated IGF-1R in a concentration-dependent manner (Fig. 2D).
PDK1 shares with other kinases from the AGC (c-AMP-dependent, c-GMP-dependent, protein kinase C) family, a mechanism of regulation mediated by interaction with C-terminal hydrophobic motifs of substrate kinases. Binding on PDK1 of these motifs occurs on its hydrophobic pocket comprising α-helix 1B, α-helix 2C, and the β-strand 5, close to the catalytic site (13). We have therefore hypothesized that a similar mechanism of recognition takes place between the IGF-1R C-terminal tail and PDK1.
Based on PDK1 crystallographic structure (PDB code 3hrf), peptides corresponding to α-helix 1B (pep1, residues 114–123), to α-helix 2C (pep2, residues 123–141), and to β-strand 5 (pep3, residues 153–161) were produced and used in competition assays between PDK1 and IGF-1R (Fig. 2E). Interestingly, pep1 efficiently reduced GST-PDK1 interaction with IGF-1R (ED50 = 100 nm), suggesting that it could directly bind IGF-1R C terminus. Much lower efficiency was displayed by pep2 (ED50 = 1 μm) and pep3 (ED50 >1 μm). Also, an additional unrelated peptide containing four copies of the tetrapeptide Arg-Thr-Tyr (pep0), was used as a control in the binding competition experiment and displayed no significant variations (Fig. 2E).
IGF-1R C Terminus Peptides Reduce PDK1 Signaling and Prevent IGF-1 Protection from Apoptosis in MCF-7 Cells
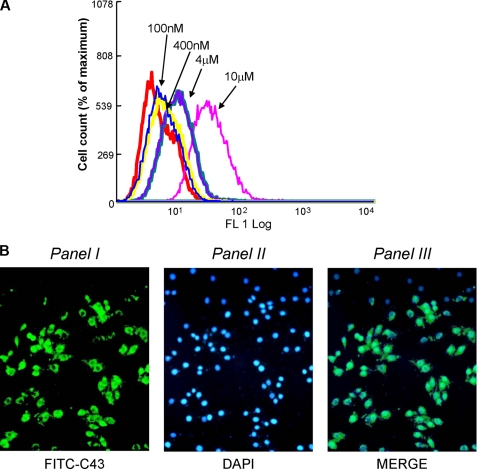
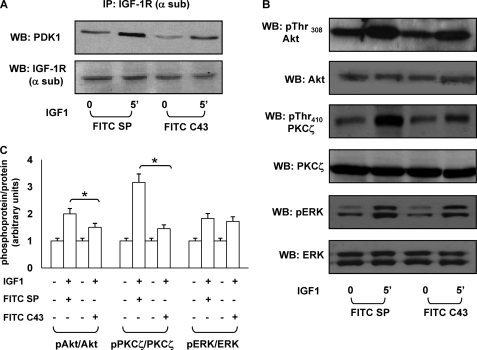
To evaluate the ability to reduce IGF-1R/PDK1 interaction in intact cells, C43 was conjugated to fluorescein isothiocyanate (FITC-C43) and loaded into MCF-7 cells at different concentrations (range: 100 nm–10 μm). Peptide loading was confirmed by FACS analysis (Fig. 3A) and fluorescence microscopy (Fig. 3B). Next, MCF-7 cells were loaded with 10 μm FITC-C43 and stimulated for 5 min with 100 ng/ml IGF-1. As shown in Fig. 4A, peptide loading decreased IGF-1R/PDK1 co-precipitation in response to IGF-1 as compared with a scrambled peptide (FITC-SP). Consistently, FITC-C43 loading was paralleled by reduced Akt/PKB and PKCζ phosphorylation in response to IGF-1 (Fig. 4, B and C). ERK1/2 phosphorylation was preserved instead.
FIGURE 3.
Transduction of FITC-C43 into MCF-7 cells. A, MCF-7 cells were incubated with increasing concentrations of the fluorescein-conjugated peptide (FITC-C43), as indicated. Peptide uptake in MCF7 cells was detected by FACS analysis of fluorescein-labeled cells. The experiment was repeated four times with comparable results. The graph shows one representative experiment. FL, FITC log. B, MCF-7 cells were incubated with FITC-C43 peptide (10 μm) and fixed in formaldehyde at 37 °C. Uptake was monitored by fluorescence with appropriate filters. Panel I shows the FITC-C43 peptide internalized in the cells with prevalent submembrane localization; panel II shows cell nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI) reagent; and panel III shows the overlay between the two images. These experiments were repeated four times with similar findings.
FIGURE 4.
Effect of C43 on IGF-1 signaling in intact MCF-7cells. A, MCF-7 cells, treated with FITC-SP or with FITC-C43, were starved for 16 h and stimulated for the indicated times with 100 ng/ml IGF-1. Cell lysates (200 μg/sample) were immunoprecipitated with anti-IGF-1R (α-sub) antibody, and then immunoprecipitated proteins (IP) were separated on SDS-PAGE, and Western blot (WB) analysis was performed with anti-PDK1 and anti-IGF-1R α-subunit antibodies. Blots were revealed by ECL and autoradiography. B, MCF-7 cells, loaded with FITC-SP or FITC-C43, were incubated for 5 min with 100 ng/ml IGF-1. Total cell lysates (50 μg of protein/sample) were blotted with pThr308-Akt/PKB, pThr410-PKCζ, or phospho-threonine 202/tyrosine 204 ERK1/2 (pERK) antibodies and then reblotted with Akt/PKB, PKCζ, or ERK1/2 antibodies. Membranes were revealed by ECL and autoradiography. The autoradiographs shown are representative of four independent experiments. C, filters obtained in B have been analyzed by laser densitometry as described under “Experimental Procedures.” Asterisks indicate statistically significant differences (*, p < 0.05). Error bars indicate mean ± S.D.
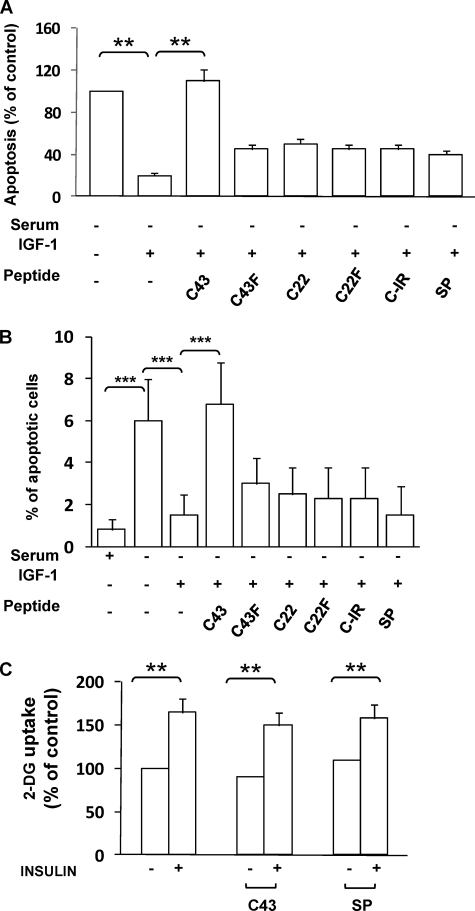
In the MCF-7 cells, exposure to IGF-1 for 16 h rescued cell death induced by growth factor deprivation (Fig. 5, A and B). Therefore, we wanted to prove the principle that IGF-1 protective effect on cell death could be reverted by disrupting the interaction between IGF-1R and PDK1. To this aim, 10 μm FITC-peptides (C43, C43F, C22, C22F, C-IR, and SP) were loaded into MCF-7 cells before the treatment with IGF-1. Interestingly, loading of C43 completely reverted IGF-1 protective action, whereas no significant effect was elicited by the other peptides (Fig. 5, A and B). Also, no significant change was induced by loading of any peptide alone (data not shown).
FIGURE 5.
Biological effect of reduced IGF-1R/PDK1 interaction in intact cells. A, MCF-7 cells, loaded with FITC-C43, -C43F, -C22, -C22F, -C-IR, or SP, were incubated in serum-free DMEM for 16 h in the presence or in the absence of IGF-1, and apoptosis was quantified by the ELISAPlus apoptosis detection kit as described under “Experimental Procedures.” Error bars represent mean ± S.D. of three different experiments in triplicate. Asterisks indicate statistically significant differences versus untreated cells (**, p < 0.01). B, MCF-7 cells, loaded with FITC-C43, -C43F, -C22, -C22F, -C-IR, or -SP, were incubated in serum-free DMEM for 16 h in the presence or in the absence of IGF-1. Cells were stained with propidium iodide, and the ratio of cells in sub-G1 phase was determined by cytometric analysis, as described under “Experimental Procedures.” Error bars represent mean ± S.D. of three different experiments in triplicate. Asterisks indicate statistically significant differences versus untreated cells (***, p < 0.001). C, L6 cells were incubated in serum-free medium for 16 h before exposure to 100 nm insulin for 30 min, as indicated. Alternatively, 10 μm C43 or SP were simultaneously added to serum-free medium in the presence or absence of insulin. Then the cells were assayed for 2-deoxy-glucose (2-DG) uptake as described under “Experimental Procedures.” Error bars represent mean ± S.D. of three different experiments in triplicate. Asterisks indicate statistically significant differences versus untreated cells (**, p < 0.01).
Effect of C43 on Glucose Uptake in L6 Cells
To investigate whether reduced IGF-1R/PDK1 interaction may affect glucose uptake in a skeletal muscle cell model, we have performed a 2-deoxy-glucose (2-DG) uptake assay in L6 myotubes in the absence or in the presence of 10 μm C43 or SP (Fig. 5C). No significant effect was achieved following treatment of L6 cells with either C43 or SP (Fig. 5C).
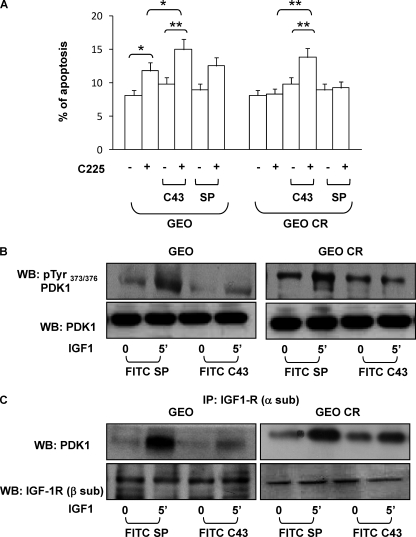
Synergic Effect of C43 and Cetuximab on GEO and GEO-CR Cells
It has been shown that switching to the IGF-1R pathway is a common mechanism to promote resistance to anti-EGFR treatment (32). The correlation between IGF-1R activation and acquired resistance to EGFR inhibitors has been demonstrated for several cancer cell lines (33). Therefore, we investigated whether C43 could affect the response to cetuximab (C225) in human colon adenocarcinoma cells (GEO cells) and in a clone of GEO cells resistant to cetuximab action (GEO-CR). FITC-C43 or FITC-SP (10 μm) was loaded into GEO and GEO-CR, and the entry was assessed by FACS and fluorescence microscopy (data not shown). In GEO cells, C225 increased cell death by 1.5-fold. FITC-C43 loading did not significantly change basal cell death as compared with the control but enhanced the effect of C225 up to 2-fold (Fig. 6A). As expected, C225 was ineffective in GEO-CR cells. However, when FITC-C43 was loaded into the cells, C225 effect was detectable at levels similar to those observed in GEO cells. No effect was elicited when FITC-SP was loaded in the cells (Fig. 6A).
FIGURE 6.
Effect of C43 on cetuximab-induced apoptosis and IGF-1 signaling in GEO and GEO-CR cells. A, 10 μm FITC-C43 or FITC-SP was loaded into GEO and GEO-CR cells. After a 24-h treatment with cetuximab (C225), the cells were harvested, fixed, and stained with propidium iodide. The ratio of cells in sub-G1 phase was determined by cytometric analysis, as described under “Experimental Procedures.” Error bars represent mean ± S.D. of three different experiments in triplicate. Asterisks indicate statistically significant differences versus untreated cells (*, p < 0.05; **, p < 0.01). B, GEO and GEO-CR cells, treated with FITC-SP or FITC-C43, were exposed to 100 ng/ml IGF-1, as indicated, and then solubilized. Cell lysates (50 μg of protein/sample) were blotted with pTyr373/376-PDK1 antibody. To ensure equal PDK1 transfer, membranes were further blotted with PDK1 antibodies. Blots were revealed by ECL and autoradiography. WB, Western blot; IP, immunoprecipitate. C, GEO and GEO-CR cells were loaded with FITC-SP or with FITC-C43 and then stimulated for the indicated times with 100 ng/ml IGF-1. Cell lysates (200 μg/sample) were immunoprecipitated with anti-IGF-1R (α-sub) antibody, and then immunoprecipitated proteins were separated on SDS-PAGE. Western blot analysis was performed with anti-PDK1 and anti-IGF-1R β-subunit antibodies. Blots were revealed by ECL and autoradiography. The autoradiographs shown in B and C are representative of four independent experiments.
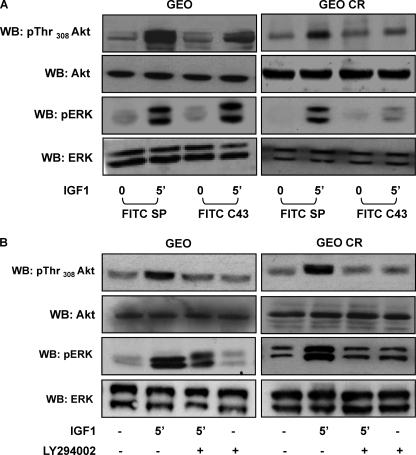
Next, we analyzed the effect of C43 on IGF-1 signaling in GEO and GEO-CR cells. IGF-1R total content and tyrosine phosphorylation were unchanged by the peptide (data not shown). In both cell lines, FITC-C43 loading decreased IGF-1-induced PDK1 tyrosine phosphorylation (Fig. 6B) and IGF-1R co-precipitation (Fig. 6C), whereas no effect was elicited by FITC-SP. We also analyzed Akt/PKB phosphorylation as a marker for PDK1 activity. As shown in Fig. 7A, FITC-C43, but not FITC-SP, reduced Akt/PKB phosphorylation in response to IGF-1 in both GEO and GEO-CR cells. Nevertheless, upon FITC-C43 loading, IGF-1-induced ERK1/2 phosphorylation was preserved in GEO cells but reduced in GEO-CR cells (Fig. 7A).
FIGURE 7.
Effect of C43 and LY294002 on Akt/PKB and ERK1/2 phosphorylation in GEO and GEO-CR cells. A, GEO and GEO-CR cells, loaded with FITC-SP or with FITC-C43, were stimulated with 100 ng/ml IGF-1 and then solubilized as described under “Experimental Procedures.” Total cell lysates (50 μg of protein/sample) were blotted with pThr308-Akt/PKB or pERK antibodies and then reblotted with Akt/PKB or ERK1/2 antibodies. Membranes were revealed by ECL and autoradiography. WB, Western blot. B, GEO and GEO-CR cells, treated or not with 25 μm LY294002 for 60 min, as indicated, were exposed to 100 ng/ml IGF-1 for 5 min and then solubilized. Cell lysates (50 μg of protein/sample) were blotted with pThr308-Akt/PKB or pERK antibodies and with Akt/PKB or ERK1/2 antibodies. Blots were revealed by ECL and autoradiography. The autoradiographs shown are representative of four independent experiments. All the autoradiographs shown are representative of four independent experiments.
To address whether IGF-1-induced PDK1 activity was needed for ERK1/2 activation, GEO-CR cells were pretreated with the PI3K inhibitor LY294002 and then stimulated with IGF-1 (Fig. 7B). In parallel with the expected reduction of Akt/PKB phosphorylation, IGF-1-stimulated ERK1/2 phosphorylation was also severely decreased by the presence of LY294002.
DISCUSSION
Targeting IGF-1R signaling pathways represents an attractive strategy for innovative therapeutic approaches (3, 34–36). Therefore, understanding the details of IGF-1R signaling may be beneficial to envision tailored strategies.
We have described that upon ligand binding, IGF-1R directly interacts with and phosphorylates PDK1 at Tyr373/376. The molecular interaction may account, at least in part, for the specificity of IGF-1 signals via PDK1. Indeed, recruitment of kinases in close proximity of specific ligand-receptor complexes may selectively direct their further action (14). The interaction between PDK1 and IGF-1R requires residues 51–359 of PDK1 and not the PH domain (amino acids 408–556), which is located at the C terminus of the molecule. In particular, the interaction is displaced by synthetic peptides corresponding to the α-helices, located within the hydrophobic pocket of PDK1, suggesting that this portion may directly bind to IGF-1R. It should be noted that binding of other C-terminal motifs to the PDK1 hydrophobic pocket has been identified as a mechanism of allosteric regulation of PDK1 activity (13). However, PDK1 substrate tyrosines are distal to the former portion. Thus, one may hypothesize that the PH domain of PDK1 binds to plasma membrane lipids (37, 38) and that the N-terminal portion interacts with the IGF-1R β-subunit, thereby locating the tyrosines in proximity of the IGF-1R catalytic domain. IGF-1R C-terminal residues are likely involved in PDK1 binding. Indeed, in vitro, a synthetic peptide corresponding to the IGF-1R C terminus (residues 1295–1337; C43) inhibits the interaction between the two molecules.
This hypothesis is further supported by the finding that intracellular loading of C43 peptide prevents co-precipitation of PDK1 with IGF-1R antibodies in both MCF-7 (breast cancer) and GEO (colon cancer) cells. Moreover, C43 loading in intact cells reduced IGF-1-stimulated phosphorylation of the PDK1 substrates Akt/PKB and PKCζ, suggesting that interaction with either IGF-1R or tyrosine phosphorylation is needed for eliciting downstream effects. Similarly, C43 loading is sufficient to impair IGF-1 antiapoptotic effect. This is consistent with previous findings indicating that PI3K activation is a relevant mechanism for IGF-1-mediated cell survival (39). This is also consistent with previous work by Surmacz et al. (40), demonstrating that the C terminus portion is required for the transforming potential of the IGF-1R. In addition, a peptide with the substitution of Tyr1316 with a phenylalanine (C43F) failed to be as effective as the wild-type C43. These observations suggest that Tyr1316 is necessary for the interaction with PDK1. However, neither a similar peptide, corresponding to the insulin receptor C terminus (containing a tyrosine in a similar position), nor shorter peptides containing or not Tyr1316 (residues 1316–1337; C22, C22F), were able to elicit comparable effects. It is noteworthy that the upper segment of C43 (residues 1295–1316) displayed weaker homology with the corresponding IR fragment as compared with the lower segment (Fig. 2B). Thus, one might speculate that the specificity of interaction resides in the amino acid stretch immediately upstream Tyr1316.
Reiss et al. (41) reported that a peptide encompassing residues 1282–1298 (peptide 2) induces apoptosis in mouse embryo fibroblasts with minor changes in IGF-1 signaling, suggesting that peptide 2 per se could elicit the activation of caspase cascade. Although we cannot exclude the possibility that C43 may directly impinge on the caspase pathway as well, our in vitro data indicate that the main mechanism could be related to its ability to interfere with IGF-1R/PDK1 interaction. Nevertheless, the other peptides at the concentration used (10 μm) slightly increased apoptosis in the absence of alterations in IGF-1 signaling, further exploiting the concept that the C terminus fragment of the IGF-1R may possess an intrinsic proapoptotic function (42, 43).
Previous findings indicated that IGF-1 alters sensitivity to several anticancer drugs by inhibiting apoptosis (44). More recent evidence also points to IGF-1 as a major candidate for the acquired resistance to tyrosine kinase inhibitors and monoclonal antibodies as cancer therapeutics (45, 46). Up-regulation of IGF-1 or IGF-1R (47), down-regulation of IGF-binding proteins (48), and heterodimerization of the IGF-1R with other tyrosine kinase receptors (32, 49) are putative mechanisms accounting for drug resistance. On the other hand, the use of monoclonal antibodies or inhibitors of IGF-1R tyrosine kinase, although effective in a number of human tumors (50), has encountered difficulties, at least in part, because of the cross-interference with insulin receptor signaling (51). One of the potential mechanisms involves the signaling through PDK1 by both pathways.
Indeed, PDK1, besides being a potential molecular target for cancer therapy (22), is largely involved in the control of glucose metabolism. In particular, it plays a critical role in insulin-stimulated glucose uptake (55), which partly attenuates the enthusiasm for PDK1 targeting. Indeed, pharmacologic or genetic blockade of PDK1 enhances apoptosis but severely affects glucose metabolism in animal models (52, 53). Moreover, 7-hydroxystaurosporine (UCN-01), a non-selective inhibitor of PDK1, has shown antiapoptotic activity in cancer cells (54) and has been tested in phase I trials (55, 56). Although 7-hydroxystaurosporine displays an acceptable toxicity profile, hyperglycemia is a relevant adverse event, possibly due to the major role exerted by PDK1 in regulation of insulin-stimulated glucose metabolism (55). We now show that pathway-selective strategies may help to discriminate between IGF-1R and IR signaling via PI3K/PDK1. In this regard, C43 loading into L6 skeletal muscle cells does not significantly inhibit insulin-stimulated glucose uptake (Fig. 5C).
Disrupting communication between IGF-1R and PDK1 by means of C43 is sufficient to prevent IGF-1 protection from apoptosis induced by growth factor deprivation. In addition, C43 exposure of GEO cells enhanced cell death following treatment with cetuximab and, importantly, restored cell killing in cetuximab-resistant clones. It has been recently suggested that combined inhibition of IGF-1 signals, which mainly occurs via PI3K, and EGF signals, which mainly occurs via ERK, results in a synergistic effect in colon cancer cells (57). We now observe that targeting the IGF-1R C terminus in a cetuximab-resistant clone of colon cancer cells, although per se not sufficient to induce cell death, blocked both PDK1 and ERK1/2 activation and restored sensitivity to cetuximab, supporting the hypothesis that both pathways are needed to escape cell death. The mechanism by which C43 may also interfere with ERK activation remains to be elucidated. Because no hyperexpression/hyperactivation of IGF-1 signaling components has been detected in GEO-CR cells, IGF-1R/EGFR hybrid formation could explain the differential impairment of the ERK pathway by C43. This is a less likely possibility, however, because C43 failed to inhibit EGF signaling in both GEO and GEO-CR cells (data not shown). Alternatively, as also described in other cell types (58), PDK1 may directly regulate the MEK/ERK pathway. Indeed, inhibition of PI3K activity simultaneously reduced Akt/PKB and ERK1/2 phosphorylation in GEO-CR cells, suggesting that PDK1 may act upstream of MEK in this cell line. How this occurs in GEO-CR and not in GEO cells, however, remains unclear. In addition, in drug-resistant cells, IGF-1R may have acquired the ability through its C terminus to recruit different signaling complexes, finally leading to ERK1/2 activation (59).
Thus, we have provided the proof of principle that targeted disruption of IGF-1R/PDK1 interaction prevents antiapoptotic action of IGF-1 and may facilitate cell killing by anticancer agents. Moreover, selectively interfering with IGF-1R signaling via PDK1 restores cell death in cetuximab-resistant colon cancer cells.
Acknowledgments
We thank Dr. Dario R. Alessi (University of Dundee) for generously providing PDK1 constructs and Prof. Massimo Santoro for critical reading of the manuscript.
This work was supported by grants from the European Community FP6 EUGENE2 (Grant LSHM-CT-2004-512013) and the FP7 PREPROBEDIA (Grant 201681), the European Federation for the Study of Diabetes, the Associazione Italiana per la Ricerca sul Cancro, and the Ministero dell'Università e della Ricerca Scientifica (PRIN and FIRB Grants RBRN07BMCT). The financial support of Telethon — Italy is also gratefully acknowledged.
- IGF
- insulin-like growth factor
- IGF-1R
- IGF receptor
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- IR
- insulin receptor
- C-IR
- IR C terminus
- SP
- scrambled peptide
- PDK1
- phosphoinositide-dependent kinase-1
- FITC
- fluorescein isothiocyanate
- PI3K
- phosphatidylinositol 3-kinase
- PH
- pleckstrin homology
- PKB
- protein kinase B
- PKC
- protein kinase C
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/ERK kinase
- DMEM
- Dulbecco's modified Eagle's medium
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorting
- HPLC
- high pressure liquid chromatography
- LC-MS
- liquid chromatography-mass spectrometry
- pep
- peptide.
REFERENCES
- 1.Samani A. A., Yakar S., LeRoith D., Brodt P. (2007) Endocr. Rev. 28, 20–47 [DOI] [PubMed] [Google Scholar]
- 2.Samani A. A., Brodt P. (2001) Surg. Oncol. Clin. N. Am. 10, 289–312 [PubMed] [Google Scholar]
- 3.Ryan P. D., Goss P. E. (2008) Oncologist 13, 16–24 [DOI] [PubMed] [Google Scholar]
- 4.LeRoith D., Helman L. (2004) Cancer Cell. 5, 201–202 [DOI] [PubMed] [Google Scholar]
- 5.Wu Y., Cui K., Miyoshi K., Hennighausen L., Green J. E., Setser J., LeRoith D., Yakar S. (2003) Cancer Res. 63, 4384–4388 [PubMed] [Google Scholar]
- 6.Zhang L., Zhou W., Velculescu V. E., Kern S. E., Hruban R. H., Hamilton S. R., Vogelstein B., Kinzler K. W. (1997) Science 276, 1268–1272 [DOI] [PubMed] [Google Scholar]
- 7.Ouban A., Muraca P., Yeatman T., Coppola D. (2003) Hum. Pathol. 34, 803–808 [DOI] [PubMed] [Google Scholar]
- 8.Pollak M. (2008) Nat. Rev. Cancer 8, 915–928; Correction (2009) Nat. Rev. Cancer.9, 224 [DOI] [PubMed] [Google Scholar]
- 9.Chitnis M. M., Yuen J. S., Protheroe A. S., Pollak M., Macaulay V. M. (2008) Clin. Cancer Res. 14, 6364–6370 [DOI] [PubMed] [Google Scholar]
- 10.Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell. Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 11.Biondi R. M., Komander D., Thomas C. C., Lizcano J. M., Deak M., Alessi D. R., van Aalten D. M. (2002) EMBO J. 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biondi R. M., Cheung P. C., Casamayor A., Deak M., Currie R. A., Alessi D. R. (2000) EMBO J. 19, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindie V., Stroba A., Zhang H., Lopez-Garcia L. A., Idrissova L., Zeuzem S., Hirschberg D., Schaeffer F., Jørgensen T. J., Engel M., Alzari P. M., Biondi R. M. (2009) Nat. Chem. Biol. 5, 758–764 [DOI] [PubMed] [Google Scholar]
- 14.Casamayor A., Morrice N. A., Alessi D. R. (1999) Biochem. J. 342, 287–292 [PMC free article] [PubMed] [Google Scholar]
- 15.Toker A., Newton A. C. (2000) J. Biol. Chem. 275, 8271–8274 [DOI] [PubMed] [Google Scholar]
- 16.Wick M. J., Ramos F. J., Chen H., Quon M. J., Dong L. Q., Liu F. (2003) J. Biol. Chem. 278, 42913–42919 [DOI] [PubMed] [Google Scholar]
- 17.Grillo S., Grémeaux T., Casamayor A., Alessi D. R., Le Marchand-Brustel Y., Tanti J. F. (2000) Eur. J. Biochem. 267, 6642–6649 [DOI] [PubMed] [Google Scholar]
- 18.Park J., Hill M. M., Hess D., Brazil D. P., Hofsteenge J., Hemmings B. A. (2001) J. Biol. Chem. 276, 37459–37471 [DOI] [PubMed] [Google Scholar]
- 19.Fiory F., Alberobello A. T., Miele C., Oriente F., Esposito I., Corbo V., Ruvo M., Tizzano B., Rasmussen T. E., Gammeltoft S., Formisano P., Beguinot F. (2005) Mol. Cell. Biol. 25, 10803–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Z., Zeng X., Waldman T., Glazer R. I. (2003) Cancer Res. 63, 5370–5375 [PubMed] [Google Scholar]
- 21.Ahmed N., Riley C., Quinn M. A. (2008) Br. J. Cancer 98, 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peifer C., Alessi D. R. (2009) Biochem. J. 417, e5–7 [DOI] [PubMed] [Google Scholar]
- 23.Riedemann J., Macaulay V. M. (2006) Endocr. Relat. Cancer 13, S33–43 [DOI] [PubMed] [Google Scholar]
- 24.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 25.Kato H., Faria T. N., Stannard B., Roberts C. T., Jr., LeRoith D. J. (1993) J. Biol. Chem. 268, 2655–2661 [PubMed] [Google Scholar]
- 26.Ciardiello F., Bianco R., Caputo R., Caputo R., Damiano V., Troiani T., Melisi D., De Vita F., De Placido S., Bianco A. R., Tortora G. (2004) Clin. Cancer Res. 10, 784–793 [DOI] [PubMed] [Google Scholar]
- 27.Caruso M., Miele C., Formisano P., Condorelli G., Bifulco G., Oliva A., Auricchio R., Riccardi G., Capaldo B., Beguinot F. (1997) J. Biol. Chem. 272, 7290–7297 [DOI] [PubMed] [Google Scholar]
- 28.Formisano P., Oriente F., Miele C., Caruso M., Auricchio R., Vigliotta G., Condorelli G., Beguinot F. (1998) J. Biol. Chem. 273, 13197–13202 [DOI] [PubMed] [Google Scholar]
- 29.Formisano P., Sohn K. J., Miele C., Di Finizio B., Petruzziello A., Riccardi G., Beguinot L., Beguinot F. (1993) J. Biol. Chem. 268, 5241–5248 [PubMed] [Google Scholar]
- 30.Fields G. B., Noble R. L. (1990) Int. J. Pept. Protein Res. 35, 161–214 [DOI] [PubMed] [Google Scholar]
- 31.Klip A., Logan W. J., Li G. (1982) Biochim. Biophys. Acta 687, 265–280 [DOI] [PubMed] [Google Scholar]
- 32.Jones H. E., Gee J. M., Hutcheson I. R., Knowlden J. M., Barrow D., Nicholson R. I. (2006) Endocr. Relat. Cancer 13, S45–51 [DOI] [PubMed] [Google Scholar]
- 33.Tortora G., Bianco R., Daniele G., Ciardiello F., McCubrey J. A., Ricciardi M. R., Ciuffreda L., Cognetti F., Tafuri A., Milella M. (2007) Drug Resist. Updat. 10, 81–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller B. S., Yee D. (2005) Cancer Res. 65, 10123–10127 [DOI] [PubMed] [Google Scholar]
- 35.Huang F., Greer A., Hurlburt W., Han X., Hafezi R., Wittenberg G. M., Reeves K., Chen J., Robinson D., Li A., Lee F. Y., Gottardis M. M., Clark E., Helman L., Attar R. M., Dongre A., Carboni J. M. (2009) Cancer Res. 69, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck E., Eyzaguirre A., Rosenfeld-Franklin M., Thomson S., Mulvihill M., Barr S., Brown E., O'Connor M., Yao Y., Pachter J., Miglarese M., Epstein D., Iwata K. K., Haley J. D., Gibson N. W., Ji Q. S. (2008) Cancer Res. 68, 8322–8332 [DOI] [PubMed] [Google Scholar]
- 37.Harris T. K. (2003) IUBMB Life 55, 117–126 [DOI] [PubMed] [Google Scholar]
- 38.Komander D., Fairservice A., Deak M., Kular G. S., Prescott A. R., Peter Downes C., Safrany S. T., Alessi D. R., van Aalten D. M. (2004) EMBO J. 23, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentinis B., Baserga R. (2001) Mol. Pathol. 54, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surmacz E., Sell C., Swantek J., Kato H., Roberts C. T., Jr., LeRoith D., Baserga R. (1995) Exp. Cell Res. 218, 370–380 [DOI] [PubMed] [Google Scholar]
- 41.Reiss K., Yumet G., Shan S., Huang Z., Alnemri E., Srinivasula S. M., Wang J. Y., Morrione A., Baserga R. (1999) J. Cell. Physiol. 181, 124–135 [DOI] [PubMed] [Google Scholar]
- 42.Hongo A., Yumet G., Resnicoff M., Romano G., O'Connor R., Baserga R. (1998) Cancer Res. 58, 2477–2484 [PubMed] [Google Scholar]
- 43.Liu Y., Lehar S., Corvi C., Payne G., O'Connor R. (1998) Cancer Res. 58, 570–576 [PubMed] [Google Scholar]
- 44.Dunn S. E., Hardman R. A., Kari F. W., Barrett J. C. (1997) Cancer Res. 57, 2687–2693 [PubMed] [Google Scholar]
- 45.Wang Q., Greene M. I. (2008) J. Clin. Invest. 118, 2389–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guix M., Faber A. C., Wang S. E., Olivares M. G., Song Y., Qu S., Rinehart C., Seidel B., Yee D., Arteaga C. L., Engelman J. A. (2008) J. Clin. Invest. 118, 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahari D., Yamada Y., Okita N. T., Honda T., Hirashima Y., Matsubara J., Takashima A., Kato K., Hamaguchi T., Shirao K., Shimada Y., Shimoda T. (2009) Oncology 76, 42–48 [DOI] [PubMed] [Google Scholar]
- 48.Gallego R., Codony-Servat J., García-Albéniz X., Carcereny E., Longarón R., Oliveras A., Tosca M., Augé J. M., Gascón P., Maurel J. (2009) Endocr. Relat. Cancer 16, 311–317 [DOI] [PubMed] [Google Scholar]
- 49.Nahta R., Yuan L. X., Zhang B., Kobayashi R., Esteva F. J. (2005) Cancer Res. 65, 11118–11128; Correction (2008) Cancer Res.68, 9566 [DOI] [PubMed] [Google Scholar]
- 50.Barnes C. J., Ohshiro K., Rayala S. K., El-Naggar A. K., Kumar R. (2007) Clin. Cancer Res. 13, 4291–4299 [DOI] [PubMed] [Google Scholar]
- 51.Sachdev D., Singh R., Fujita-Yamaguchi Y., Yee D. (2006) Cancer Res. 66, 2391–2402 [DOI] [PubMed] [Google Scholar]
- 52.Hashimoto N., Kido Y., Uchida T., Asahara S., Shigeyama Y., Matsuda T., Takeda A., Tsuchihashi D., Nishizawa A., Ogawa W., Fujimoto Y., Okamura H., Arden K. C., Herrera P. L., Noda T., Kasuga M. (2006) Nat. Genet. 38, 589–593 [DOI] [PubMed] [Google Scholar]
- 53.Duronio V. (2008) Biochem. J. 415, 333–344 [DOI] [PubMed] [Google Scholar]
- 54.Seynaeve C. M., Stetler-Stevenson M., Sebers S., Kaur G., Sausville E. A., Worland P. J. (1993) Cancer Res. 53, 2081–2086 [PubMed] [Google Scholar]
- 55.Kortmansky J., Shah M. A., Kaubisch A., Weyerbacher A., Yi S., Tong W., Sowers R., Gonen M., O'reilly E., Kemeny N., Ilson D. I., Saltz L. B., Maki R. G., Kelsen D. P., Schwartz G. K. (2005) J. Clin. Oncol. 23, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 56.Jimeno A., Rudek M. A., Purcell T., Laheru D. A., Messersmith W. A., Dancey J., Carducci M. A., Baker S. D., Hidalgo M., Donehower R. C. (2008) Cancer Chemother. Pharmacol. 61, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Y. P., Patil S. B., Panasiewicz M., Li W., Hauser J., Humphrey L. E., Brattain M. G. (2008) Cancer Res. 68, 8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato S., Fujita N., Tsuruo T. (2004) J. Biol. Chem. 279, 33759–33767 [DOI] [PubMed] [Google Scholar]
- 59.Michel J. J., Townley I. K., Dodge-Kafka K. L., Zhang F., Kapiloff M. S., Scott J. D. (2005) Mol. Cell. 20, 661–672 [DOI] [PubMed] [Google Scholar]