Abstract
Ficolins are oligomeric innate immune recognition proteins consisting of a collagen-like region and a fibrinogen-like recognition domain that bind to pathogen- and apoptotic cell-associated molecular patterns. To investigate their carbohydrate binding specificities, serum-derived L-ficolin and recombinant H- and M-ficolins were fluorescently labeled, and their carbohydrate binding ability was analyzed by glycan array screening. L-ficolin preferentially recognized disulfated N-acetyllactosamine and tri- and tetrasaccharides containing terminal galactose or N-acetylglucosamine. Binding was sensitive to the position and orientation of the bond between N-acetyllactosamine and the adjacent carbohydrate. No significant binding of H-ficolin to any of the 377 glycans probed could be detected, providing further evidence for its poor lectin activity. M-ficolin bound preferentially to 9-O-acetylated 2-6-linked sialic acid derivatives and to various glycans containing sialic acid engaged in a 2-3 linkage. To further investigate the structural basis of sialic acid recognition by M-ficolin, point mutants were produced in which three residues of the fibrinogen domain were replaced by their counterparts in L-ficolin. Mutations G221F and A256V inhibited binding to the 9-O-acetylated sialic acid derivatives, whereas Y271F abolished interaction with all sialic acid-containing glycans. The crystal structure of the Y271F mutant fibrinogen domain was solved, showing that the mutation does not alter the structure of the ligand binding pocket. These analyses reveal novel ficolin ligands such as sulfated N-acetyllactosamine (L-ficolin) and gangliosides (M-ficolin) and provide precise insights into the sialic acid binding specificity of M-ficolin, emphasizing the essential role of Tyr271 in this respect.
Keywords: Carbohydrate/Binding Protein, Immunology/Innate Immunity, Methods/Site-directed Mutagenesis, Methods/Surface Plasmon Resonance, Protein/Ligand Binding, Ficolins, Sialic Acid Binding
Introduction
Innate immunity relies on the capacity of constitutive recognition molecules to sense characteristic pathogen-associated molecular patterns at the surface of microbes, which are often carbohydrates contributed by invariant components of microorganisms. Recognition of these microbial patterns elicits effector mechanisms aimed at containing early infection while instructing an appropriate adaptive immune response (1, 2). Among these innate immune molecules are the defense collagens, including the soluble oligomeric proteins C1q, mannan-binding lectin, and ficolins, which recognize both pathogens and potentially noxious elements from self, such as apoptotic cells (3). Ficolins constitute a family of evolutionarily conserved proteins present in species ranging from invertebrates to mammals, assembled from homotrimeric subunits comprising collagen-like triple helices and globular fibrinogen-like recognition domains (4). Subunit oligomerization involves disulfide linkages between cysteines located at the N-terminal end of the polypeptide chains (5). Three ficolins have been identified in humans: M-ficolin (ficolin-1), L-ficolin (ficolin-2), and H-ficolin (ficolin-3) (6–9). Rodents and pigs have only two ficolins that are orthologous to L- and M-ficolins (10). L- and M-ficolins are serum proteins with average concentrations of 3–5 and 18 μg/ml, respectively (11–14), whereas M-ficolin has been localized at the surface of blood monocytes and in secretory granules of neutrophils, monocytes, and lung epithelial cells (15–17). However, two recent studies have reported its detection in serum, with mean concentrations ranging from 0.06 to 1 μg/ml (18, 19). The three human ficolins share with mannan-binding lectin the ability to associate with mannan-binding lectin-associated serine protease-2. The resulting complexes trigger activation of the lectin complement pathway, a major effector of innate host defense, upon binding to suitable targets.
L-ficolin has been shown to recognize several strains of opportunistic capsulated bacteria (13) and apoptotic HL60, U937, and Jurkat cells (20, 21). It specifically binds to various ligands including acetylated compounds (22, 23), 1,3-β-d-glucan (24), lipoteichoic acid (25), the capsular polysaccharide of group B streptococci (26, 27), and DNA (21). H-ficolin is known to specifically recognize Aerococcus viridans (13, 28) and apoptotic Jurkat cells (20, 29) and to bind a few ligands such as d-fucose, galactose (30), and acetylated albumin (31, 32). Recombinant M-ficolin displays a binding preference for various acetylated compounds including sialic acid (16, 17) and has been shown to bind to Staphylococcus aureus and the smooth type of Salmonella typhimurium (17).
The crystal structures of the trimeric recognition domains of the three human ficolins alone and in complex with various ligands have been determined recently (30, 33, 34), revealing striking differences in their ligand recognition properties. A single ligand binding site was observed in M-ficolin, located close to the Ca2+-binding site in the outer part of the trimer and homologous to the GlcNAc2 binding pocket of the invertebrate tachylectin TL5A (35). The homologous sites in H- and L-ficolin were not found to bind acetylated ligands but accommodated d-fucose and galactose in the case of H-ficolin. Three additional binding sites were identified in L-ficolin, defining an extended charged binding surface able to recognize elongated polysaccharides such as 1,3-β-d-glucan through binding subsites for small acetylated compounds and neutral carbohydrates (30).
To further decipher and compare the carbohydrate binding specificities of the three human ficolins, these were used to probe the glycan array developed by the Consortium for Functional Glycomics, comprising several hundreds diverse natural and synthetic glycans (36). Our results confirm that the three ficolins have different specificities and provide new insights about the recognition properties of L- and M-ficolins. Site-directed mutagenesis was also used to identify the amino acid residues of the M-ficolin fibrinogen (FBG) domain involved in sialic acid binding.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
BSA glycoconjugates derivatized with GlcNAc, Gal, GalNAc, LacNAc, Neu5Acα2-3LacNAc, and Man were purchased from Dextra Laboratories (Reading, UK). Acetylated BSA, streptavidin, and 6-kDa heparin were from Sigma, and biotin-LC-hydrazine was from Pierce. Human serum L-ficolin and recombinant H-ficolin expressed in Chinese hamster ovary cells were purified as described previously, and their molar concentrations were determined using absorption coefficients (A1%, 1 cm at 280 nm) of 17.6 and 19.4 and Mr values of 406,000 and 396,000 (i.e. 12 polypeptide chains of Mr 33,600 and 33,000), respectively (37). The recombinant FBG domain of L-ficolin was expressed in a baculovirus/insect cells system and purified as described by Garlatti et al. (30). Its molar concentration was determined using an absorption coefficient of 2.2 and a Mr value of 78,000 (3 polypeptide chains of Mr 26,000).
Site-directed Mutagenesis
The expression plasmids coding for the mutants of full-length M-ficolin and of its FBG domain were generated using the QuikChangeTM XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. A pMT/Bip/V5-His A Drosophila expression system plasmid coding for full-length wild-type M-ficolin without the His tag (17) (kindly provided by Dr. T. Fujita) and a pNT-Bac baculovirus transfer vector coding for the FBG domain of M-ficolin (33) were used as templates. Mutagenic oligonucleotides were purchased from Eurogentec (Seraing, Belgium). The sequences of all mutants were confirmed by double-stranded DNA sequencing (Cogenics, Meylan, France).
Production and Purification of the Ficolins
The recombinant full-length M-ficolin variants were produced in Schneider S2 Drosophila cells after stable transfection with the pMT/Bip/V5-His A vector containing the cDNA of M-ficolin, as described by Liu et al. (17). Stable transfectants were expanded in Schneider's Drosophila medium supplemented with 10% (v/v) heat-inactivated fetal calf serum. Production of the M-ficolin variants was induced by adding 500 μm CuSO4 to the culture medium containing 50 μg/ml ascorbic acid (Sigma) without added serum, and the culture supernatants were harvested after 72 h incubation.
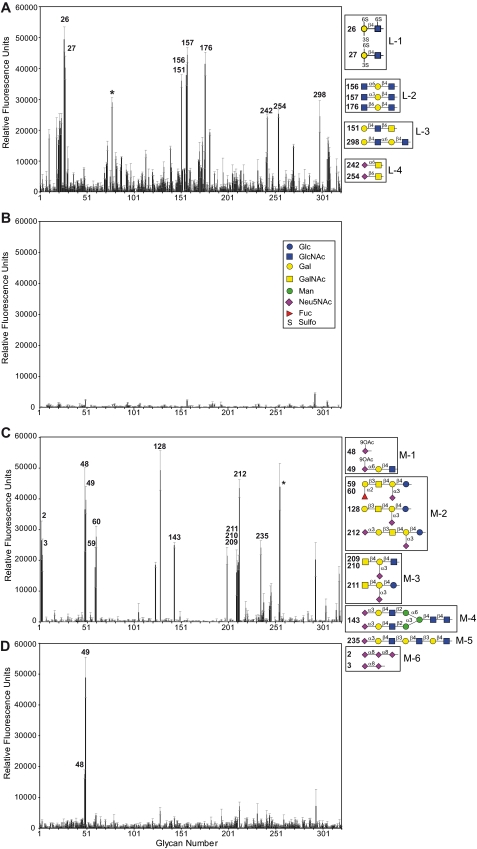
The M-ficolin variants were purified from the culture supernatants by affinity chromatography on a GlcNAc-agarose column (Sigma) equilibrated in 145 mm NaCl, 5 mm CaCl2, 20 mm Tris-HCl, pH 7.4. Elution of bound M-ficolin was performed by applying the same buffer containing 0.3 m GlcNAc. The purified proteins were dialyzed against 145 mm NaCl, 2 mm CaCl2, 50 mm triethanolamine-HCl, pH 7.4, and concentrated to 0.5–1.0 mg/ml. The concentration of purified wild-type M-ficolin was estimated using an absorption coefficient (A1%, 1 cm at 280 nm) of 17.6 and a protomer Mr value of 32,342 as determined by mass spectrometry. The same absorption coefficient and Mr values of 32,432, 32,360, 32,450, and 32,326 were used for the G221F, A256V, Y271F/A256V, and Y271F mutants, respectively. The molar concentrations were expressed relative to the protomers as the oligomeric state of the M-ficolin variants has not been determined precisely.
The recombinant baculovirus for expression of the Y271F mutant of the FBG domain of M-ficolin was generated using the Bac-to-BacTM system (Invitrogen) as described for the wild-type domain (33). The recombinant protein was produced in baculovirus-infected High Five insect cells and purified from the culture supernatant by ion-exchange chromatography as described previously (33).
SDS-PAGE Analysis of the Oligomerization State of the M-ficolin Variants
Ficolin samples were analyzed by SDS-PAGE under non-reducing conditions using 7.5% polyacrylamide gels as described previously (32).
SPR Analyses on Immobilized BSA Glycoconjugates
Analyses were performed using a BIAcore 3000 instrument (GE Healthcare). Acetylated BSA and BSA glycoconjugates were diluted to 25 μg/ml in 10 mm sodium formate, pH 3.0, and 10 mm sodium acetate, pH 4.0, respectively, and immobilized on the surface of a CM5 sensor chip (GE Healthcare) using the amine coupling chemistry in 10 mm Hepes, 150 mm NaCl, EDTA 5 mm, pH 7.4. Binding of L- and M-ficolins and of the M-ficolin variants to the immobilized BSA conjugates (3800–4200 RU) and to acetylated BSA (900 RU) was measured at a flow rate of 20 μl/min in 150 mm NaCl, 1 mm CaCl2, 20 mm Hepes, pH 7.4, containing 0.005% surfactant P20 (GE Healthcare). Regeneration of the surface was achieved by injection of 10 μl of 1 m sodium acetate, pH 7.2. Equivalent volumes of each protein sample were injected over a reference surface with immobilized BSA (4000 RU) to serve as blank sensorgrams for subtraction of the bulk refractive index background.
SPR Analyses on Immobilized Heparin
Streptavidin (200 μg/ml in 10 mm sodium acetate, pH 4.2) was immobilized on a CM4 sensor chip (GE Healthcare) using the amine-coupling chemistry until a coupling level of 2500 RU was reached. 6-kDa heparin was biotinylated as described previously (38) and captured on the streptavidin surface in 10 mm Hepes, 300 mm NaCl, 0.005% surfactant P20, pH 7.4, to reach a final level of 50 RU. L-, H-, and M-ficolins and the FBG domain of L-ficolin were injected over the heparin-bound surface at 20 μl/min in 50 mm triethanolamine-HCl, 145 mm NaCl, 0.005% surfactant P20, pH 7.4. Surfaces were regenerated with 10 μl of 1 m Na2SO4, with additional injections of 2 m NaCl when needed. A flow cell with 2500 RU streptavidin was used as a control surface.
SPR Data Evaluation
Data were analyzed by global fitting to a 1:1 Langmuir binding model of both the association and dissociation phases for at least five concentrations simultaneously (0.5–8 nm L-ficolin, 15–240 nm L-ficolin FBG domain, and 0.75–12 nm M-ficolin protomer) using the BIAevaluation 3.2 software (GE Healthcare). The apparent equilibrium dissociation constants (KD) were calculated from the ratio of the dissociation and association rate constants (koff/kon), and the maximal binding capacities (Rmax) were determined using the same model. Although interaction of oligomeric lectins with multivalent glycoconjugates is inherently more complex than a simple 1:1 binding model, data fitting using this model yielded satisfactory Chi2 values (below 4 in all cases) and was, thus, used for comparison purposes.
Glycan Array Screening
Purified ficolins were labeled with Alexa Fluor® 488 using the monoclonal antibody labeling kit according to the manufacturer's instructions (Molecular Probes). The incorporation of the fluorescent dye varied from 0.54 to 1.1 mol of dye/mol of ficolin monomer. Alexa Fluor® 488-labeled ficolins were used to probe the glycan arrays (Versions 3.0, 3.1, and 3.2) developed by the Consortium for Functional Glycomics. Ficolins were diluted to the desired concentration in standard binding buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2 containing 1% (w/v) BSA and 0.05% (v/v) Tween 20). Seventy microliters of protein were applied to the printed surface of the array and incubated at room temperature in a dark humidified chamber for 1 h before washing and reading in a PerkinElmer Life Sciences Microscanarray XL 4000 scanner. The average relative fluorescence units for binding to each glycan, printed in replicates of six, was calculated by averaging four values after removing the highest and lowest values from the six data points. The error bars in histograms represent the S.E., and % CV is the coefficient of variation (S.D./mean) calculated as %.
Assay of the Ability of the M-ficolin Variants to Trigger the Lectin Complement Pathway
Ficolin-deficient serum was obtained by incubating 3 ml of normal human serum from a healthy donor with 1 ml of acetylated BSA-Sepharose for 3 h at 4 °C as described by Lacroix et al. (32). Microtiter plates of 96 wells (Maxisorp Nunc) were coated with 50 μg/ml acetylated BSA in 10 mm NaHCO3, pH 9.6, for 2 h at room temperature. Wells were washed with phosphate-buffered saline (PBS; (136 mm NaCl, 2.7 mm KCl, 1.45 mm KH2PO4, 8 mm Na2HPO4, pH 7.2) and incubated for 1 h at room temperature with PBS containing 0.1% Tween 20 (w/v). The recombinant M-ficolin variants were added to the wells at different concentrations and incubated on ice for 1 h with ficolin-deficient serum diluted 1:25 in 5 mm sodium veronal, 145 mm NaCl, 5 mm CaCl2, 1.5 mm MgCl2, pH 7.5. The wells were washed with 5 mm sodium veronal, 145 mm NaCl, 5 mm EDTA, pH 7.5, and a rabbit anti-C4 polyclonal antibody (1:500 dilution) (Siemens Healthcare Diagnostics) was added to each well and incubated for 1 h at room temperature. After washing with 50 mm Tris-HCl, 150 mm NaCl, 0.05% (w/v) Tween 20, pH 8.0, a peroxidase-conjugated goat anti-rabbit polyclonal antibody (1:20,000 dilution) (Jackson ImmunoResearch) was added and incubated for 1 h at room temperature. After washing as mentioned, the plates were developed in the dark with tetramethylbenzidine (Sigma), the reaction was stopped with 1 n H2SO4, and absorbance was read at 450 nm.
Evaluation of the amounts of M-ficolin variants bound to immobilized acetylated BSA was performed as described above, except that a rabbit polyclonal antibody against the L-ficolin FBG domain, which cross-reacts with M-ficolin (32), was used for detection instead of the anti-C4 antibody. The amounts of M-ficolin variants added had to be reduced 50 times compared with the C4b deposition assay to avoid saturation of the signal.
Crystallization, Structure Determination, and Refinement
Crystals of the Y271F FBG domain of M-ficolin were obtained as described for the native domain (33). Briefly, the protein was concentrated in 145 mm NaCl, 50 mm Tris-HCl, pH 7.6, and if necessary supplemented with 50 mm GlcNAc. Crystals were obtained by the hanging-drop vapor diffusion method, with the well solution containing 11% polyethylene glycol 4000, 5% isopropanol, 0.1 m Hepes, pH 7.0. Diffraction data were collected on European Synchrotron Radiation Facility beam lines ID29 and BM30. Reflection data were processed using the XDS package (39). Crystals grew in the H3 space group (a = b = 73.550 Å, c = 124.760 Å, α = β = 90.00°, γ = 120.00°) or in the presence of GlcNAc in the monoclinic P1 space group (a = 59.526 Å, b = 59.696 Å, c = 59.700 Å, α = 76.99°, β = 76.96°, γ = 76.69°). The structure was solved by automated molecular replacement using Phaser (40) with the native FBG domain (PDB code 2JHM) as a search model. Refinement was done using Refmac5 (41) alternately with inspection of electron density maps and model corrections using Coot (42). Five percent of the reflections were not included in the refinement to monitor Rfree. The crystallographic collection data and final refinement statistics are listed in supplemental Table S1.
RESULTS
With a view to gain insight into their carbohydrate binding specificity, L-ficolin was purified from serum, and H- and M-ficolins were produced in recombinant form, as described under “Experimental Procedures.” SDS-PAGE analysis of the purified proteins yielded a single band at 33–34 kDa under reducing conditions and ladder-like patterns under non-reducing conditions (supplemental Fig. S1), as expected from the multimeric state of the proteins and consistent with previous analyses (17, 32).
SPR Spectroscopy Analysis of the Carbohydrate Binding Properties of Human Ficolins
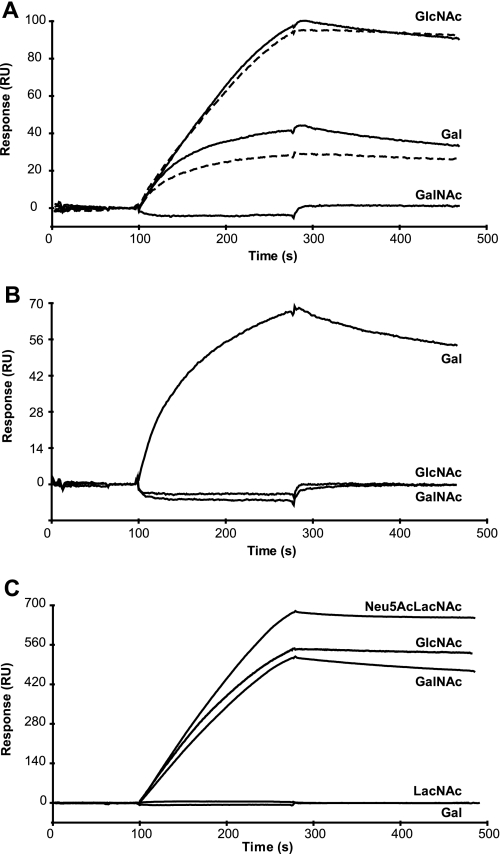
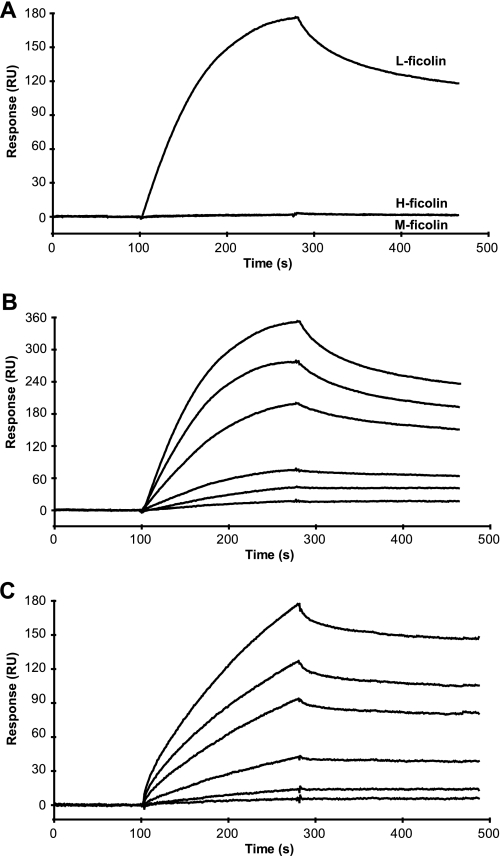
The ability of the ficolins to bind carbohydrate ligands was first investigated by SPR using immobilized BSA glycoconjugates selected according to the previously described ligand specificity of these proteins (17, 22, 30). In the presence of 1 mm CaCl2, L-ficolin bound to GlcNAc-BSA and to a lesser extent to Gal-BSA but not to GalNAc-BSA (Fig. 1A). Under the same conditions, H-ficolin bound significantly to Gal-BSA but not to GlcNAc- and GalNAc-BSA (Fig. 1B). For both ficolins, no significant interaction was observed with LacNAc- and Neu5Acα2-3LacNAc-BSA (not shown). As illustrated in Fig. 1C, M-ficolin strongly bound to and slowly dissociated from GlcNAc-, Neu5Acα2-3LacNAc-, and GalNAc-BSA, whereas no interaction was observed with the latter two carbohydrates lacking Neu5Ac and NAc, respectively. None of the ficolins interacted with Man-BSA (not shown), whereas all three strongly bound to acetylated BSA (Ref. 32 and Table 1). Replacement of Ca2+ by EDTA in the binding buffer abolished binding of H- and M-ficolins to their ligands (not shown) but had no effect on L-ficolin binding to GlcNAc-BSA and only slightly inhibited interaction with Gal-BSA (Fig. 1A). This indicates that binding to these carbohydrates likely involves ligand binding sites of L-ficolin that are distant from the Ca2+-binding site, in agreement with our previous structural data (30).
FIGURE 1.
Analysis by SPR spectroscopy of the interaction of the three human ficolins with various immobilized BSA glycoconjugates. L-ficolin (A) and H-ficolin (B) at a concentration of 4.2 μg/ml were injected over immobilized GlcNAc-, Gal-, and GalNAc-BSA (3800–4200 RU) in 150 mm NaCl, 20 mm Hepes, pH 7.4, containing 0.005% surfactant P20 at a flow rate of 20 μl/min in the presence of 1 mm CaCl2 (solid lines) or 1 mm EDTA (dashed lines). M-ficolin (C) at a concentration of 1.1 μg/ml was injected over immobilized GlcNAc-, Gal-, GalNAc-, LacNAc-, and Neu5Acα2-3LacNAc-BSA (3800–4200 RU) under the same conditions in the presence of 1 mm CaCl2. The specific signal shown was obtained by subtracting the background signal recorded over a surface with immobilized BSA.
TABLE 1.
Kinetic and dissociation constants for binding of the M-ficolin variants to immobilized BSA derivatives
a NA, data not analyzable due to the weakness of binding.
b Value not measurable due to the lack of binding.
Glycan Array Screening of the Carbohydrate Binding Specificity of Human Ficolins
Because only a limited range of carbohydrate ligands and BSA glycoconjugates is commercially available, the carbohydrate binding specificity of ficolins was further investigated using the glycan array screening facility provided by the Consortium for Functional Glycomics. The three ficolins were fluorescently labeled as described under “Experimental Procedures” and probed at concentrations of 1–200 μg/ml using the v3.0 and v3.1 versions of the glycan array which display 320 and 377 different glycan species, respectively.
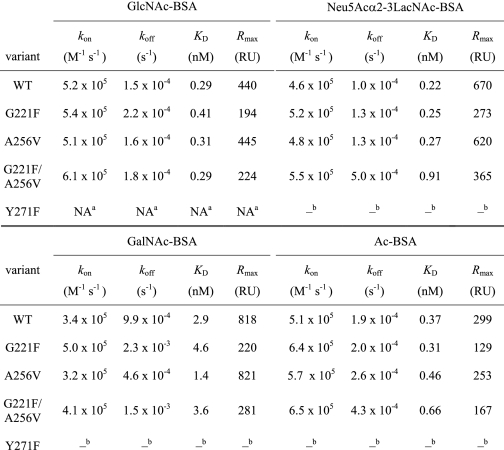
As illustrated in Fig. 2A using the v3.0 array version, L-ficolin (200 μg/ml) specifically bound to four groups of glycans. Group L-1 encompasses disaccharides with terminal disulfated Gal β-linked to non-sulfated or mono-sulfated GlcNAc (glycans 26 and 27). Group L-2 consists of GlcNAc-Gal-GlcNAc trisaccharides, with different α or β GlcNAc-Gal linkages (glycans 156, 157, and 175). The L-3 group comprises two glycans with terminal Gal-GlcNAc (LacNAc) β1–6-linked to GalNAc (151) or α1–6-linked to a second LacNAc unit (298). A fourth group (L-4) is composed of Neu5Acα2-6- or β2-6-linked to GalNAc (glycans 242 and 254). Binding to these same glycans was also observed when a different preparation of L-ficolin was probed using the v3.1 version of the glycan array (supplemental Fig. S2A). However, it should be mentioned that the binding profiles exhibited some variability, and therefore, the ligands considered to be reliable were those yielding reproducible binding values in three distinct experiments (two separate analyses using the same labeled ficolin sample plus one using a different sample). This excludes glycan 77 (Fucα1-4GlcNAc, indicated by a star in Fig. 2A) and glycans 48 and 49 (9-O-AcNeu5Ac and 9-O-AcNeu5Acα2-6Galβ1-4GlcNAc), which, respectively, yielded 100 and 73% of the maximal binding signal in one experiment (supplemental Fig. S2B) but less than 10% in the other two (Fig. 2A and supplemental Fig. S2A). Glycan 49 was recently reported to be the strongest ligand of L-ficolin in a glycan array screening based on a single analysis (43). Decreasing L-ficolin concentration to 100 μg/ml led to an overall decrease in binding to glycan 298 (21,400 RFU) and to glycans of groups L-1 (19,900–21,700 RFU) and L-2 (11,500–17,900 RFU), whereas no residual binding to group L-4 glycans was observed (supplemental Fig. S3). Taking into account the SPR data (Fig. 1A), this latter observation likely reflects a weak and probably not significant affinity of L-ficolin for sialylated ligands. Probing H-ficolin at a concentration of 200 μg/ml yielded no significant binding to any glycan of the v3.0 and v3.1 versions of the glycan array (Fig. 2B and supplemental Fig. S4).
FIGURE 2.
Glycan array screening of human ficolins. Version 3.0 of the printed array of the Consortium for Functional Glycomics was probed with L-ficolin (200 μg/ml) (A), H-ficolin (200 μg/ml) (B), M-ficolin (100 μg/ml) (C), and M-ficolin (1 μg/ml) (D). Error bars represent the means ± S.D. of four measurements. Glycans yielding significant binding are shown with their numbers and structures, and the glycan groups defined in the text are indicated. One representative experiment of three is shown. The asterisk indicates non-reproducible binding from one experiment to the other.
M-ficolin yielded similar binding patterns when probed at concentrations of 200 and 100 μg/ml (supplemental Fig. S5A and Fig. 2C, Version 3.0), with a lower background than L-ficolin. As illustrated in Fig. 2C, the carbohydrate ligands of M-ficolin could be divided into six groups, their common hallmark being the presence of sialic acid. Group M-1 encompasses glycans containing 9-O-acetylated sialic acid (9-O-AcNeu5Ac) either alone (48) or α2-6-linked to LacNAc (49). Group M-2 comprises gangliosides such as GM1 (128), fucosyl-GM1 with different spacer arms (59/60), and GD1a (212), exhibiting a Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glc core. Group M-3 comprises ganglioside GM2 (211) and the related glycans 209/210 and 389 (v3.2 array version, see Fig. 4), in which the GM2 glucose unit is substituted with GlcNAc and GlcNAcβ1-3GalNAc, respectively. The fourth group (M-4) corresponds to an α2-3 sialylated biantennary N-linked type glycan (143), whereas groups M-5 and M-6 encompass α2-3 sialylated polyLacNAc (235) and α2-8-linked di- and trisialic acids (2, 3), respectively. Interestingly, no binding was observed to glycans 129, 52, and 146, the non-sialylated counterparts of glycans 128, 143, and 235, respectively. Comparable results were obtained using the v3.1 version of the array (supplemental Fig. S5B). It should be mentioned that the length of the spacer between the glycan and the surface had an impact on M-ficolin binding. Thus, glycan 59 with a (CH2)2-NH2 linker yielded consistently lower signals than glycan 60 with the longer (CH2)5-NH2 linker, possibly due to a lower flexibility and/or decreased accessibility of the saccharide chain. Decreasing M-ficolin concentration to 10 μg/ml yielded decreased, but still significant binding to glycans of groups M-2 to M-4 and M-6 along with disappearance of binding to glycan 235 (supplemental Fig. S6). Binding to group M-1 glycans containing 9-O-AcNeu5Ac was maintained at a high level under these conditions and even when M-ficolin concentration was decreased to 1 μg/ml, reflecting high affinity binding (Fig. 2D).
FIGURE 4.
A, shown is SDS-PAGE analysis of the M-ficolin variants; Coomassie Blue staining of unreduced wild-type M-ficolin (lane 1) and mutants G221F (lane 2), A256V (lane 3), G221F/A256V (lane 4), and Y271F (lane 5). B, shown is analysis by SPR spectroscopy of the interaction of the M-ficolin variants with immobilized Neu5Acα2-3LacNAc-BSA. Immobilization of the BSA glycoconjugate and injection of 12 nm M-ficolin (protomer concentration) were performed as described under “Experimental Procedures.” WT, wild type. C, analysis of the ability of the M-ficolin variants to activate the lectin complement pathway. Increasing concentrations of each M-ficolin variant were added to ficolin-depleted human serum in microtiter wells coated with 50 μg/ml acetylated BSA, and the resulting mannan-binding lectin-associated serine protease-2 C4-cleaving activity was measured by a C4b deposition assay as described under “Experimental Procedures.” Results are expressed as the means ± S.D. of three independent experiments.
Binding of L-ficolin to Heparin
Our finding that L-ficolin bound sulfated galactose raised the possibility that this protein could bind to sulfated glycans such as heparan sulfates or heparin. To test this hypothesis, the ability of L-ficolin to interact with heparin was investigated using SPR spectroscopy. As shown in Fig. 3A, L-ficolin readily bound to immobilized heparin, whereas no interaction was observed in the case of H- and M-ficolins. Complete elution of the bound L-ficolin could be achieved by pulse injections of 1 m sodium sulfate. The kinetic parameters of the interaction were determined as described under “Experimental Procedures,” yielding association (kon) and dissociation (koff) rate constants of 1.9 × 107 m−1 s−1 and 3.4 × 10−3 s−1, respectively, with a resulting apparent KD of 0.17 nm. This value is in the same range as that determined previously for binding of L-ficolin to acetylated BSA (0.13 nm) (32). Additional experiments were carried out using the isolated FBG domain of L-ficolin, which was also found to interact in a dose-dependent manner with immobilized heparin (Fig. 3C). Analysis of the binding data yielded kon and koff values of 2.3 × 104 m−1 s−1 and 3.4 × 10−4 s−1, respectively, yielding a KD of 14.8 nm. The decrease in affinity arose mainly from a reduced kon value, which is consistent with the fact that the recognition domain lacks the binding avidity inherent in the intact oligomeric L-ficolin molecule. Interestingly, the complex between heparin and the FBG domain was more stable than that obtained for the full-length L-ficolin molecule. This is in contrast with previous results obtained for binding of the FBG domain to immobilized acetylated BSA (44).
FIGURE 3.
SPR analysis of the interaction of ficolins with immobilized heparin. A, L-, H- and M-ficolins were injected at a concentration of 1.7 μg/ml over immobilized 6-kDa biotinylated heparin (50 RU) as described under “Experimental Procedures.” B, L-ficolin was injected at 6 concentrations ranging from 0.25 to 4 nm (from bottom to top) over immobilized heparin. C, the FBG domain of L-ficolin was injected at 6 concentrations ranging from 15 to 240 nm (from bottom to top) over immobilized heparin. Results are representative of two independent experiments.
Production of M-ficolin Point Mutants
In light of the key role of the Tyr271 hydroxyl group of M-ficolin in the binding of acetyl groups, we have previously proposed that replacement of this residue by Phe in L-ficolin may explain its inability to bind N-acetylated ligands through its S1 site (30). We also hypothesized that accommodation of the relatively large sialic acid molecule in site S1 of M-ficolin may be conditioned by the presence in the vicinity of this site of two small residues, Gly221 and Ala256, which are replaced by bulkier residues in other ficolins with no sialic acid binding ability (33). To test these hypotheses, single mutants of full-length M-ficolin were generated in which Tyr271 was replaced by Phe, and Gly221 and Ala256 were replaced by the corresponding residues Phe and Val of L-ficolin. The double mutant G221F/A256V was also produced, and all mutants were purified by affinity chromatography on a GlcNAc-agarose column, as described for wild-type M-ficolin. SDS-PAGE analysis of the four mutants under non-reducing conditions yielded ladder-like patterns similar to those obtained for the wild-type protein, indicating that the mutations had no significant impact on their oligomerization state (Fig. 4A).
SPR Analysis of the Ligand Binding Properties of the M-ficolin Variants
The ligand binding capacity of the M-ficolin variants was first analyzed by SPR spectroscopy using GlcNAc-, Neu5Acα2-3LacNAc-, GalNAc-, and Ac-BSA as immobilized ligands and the M-ficolin variants as soluble analytes. Binding of M-ficolin to each of the BSA derivatives was not affected by mutation A256V as illustrated for Neu5Acα2-3LacNAc-BSA in Fig. 4B. Mutations G221F and G221F/A256V each resulted in comparable lower binding levels at the end of injection (about 50%). Mutation Y271F abolished interaction with Neu5Acα2-3LacNAc-, GalNAc-, and Ac-BSA and strongly inhibited binding to GlcNAc-BSA, thus precluding kinetic analysis (Fig. 4B and Table 1). The kinetic parameters of the interactions were determined as described under “Experimental Procedures.” As listed in Table 1, wild-type M-ficolin bound GlcNAc-, Neu5Acα2-3LacNAc-, and Ac-BSA with comparable high apparent affinities (KD values in the subnanomolar range), mainly arising from low koff values (1.0–1.9 × 10−4 s−1), likely reflecting binding avidity. A 10-fold higher KD value was obtained for GalNAc-BSA due to a faster dissociation of the complex (Fig. 1C and Table 1). The values for M-ficolin binding to acetylated BSA were in the same range as those reported previously for L-ficolin (32). As expected from the binding curves shown in Fig. 4B, the A256V mutant yielded kinetic parameters and Rmax values comparable with those obtained for the wild-type protein (Table 1). The G221F and G221F/A256V mutants consistently exhibited similar decreased Rmax values for all BSA derivatives, whereas both the kon and koff values were in the same range as those of the wild-type protein (Table 1). Although both variants containing the G221F mutation exhibited electrophoretic patterns comparable with those of the other variants (Fig. 4A), gel filtration analysis of these mutants revealed the presence of an additional peak corresponding to species eluting later than the high order oligomers contained in the major peak common to all variants (data not shown). If one assumes that molecules of different oligomeric states bind to the same targets on the chip, the decreased mean molecular mass of the G221F oligomers could account for the observed decrease of the Rmax values, which are expressed in RU. As judged from the SPR data, the G221F and A256V mutations had no significant impact on the apparent binding affinity of M-ficolin for acetylated molecules, including Neu5Acα2-3LacNAc. In contrast, mutation of Tyr271 to Phe clearly prevented binding to acetylated ligands, thus confirming its crucial role in the ligand binding activity of M-ficolin.
Ability of the M-ficolin Variants to Trigger Activation of the Lectin Complement Pathway
The ability of wild-type M-ficolin to trigger activation of the lectin pathway was measured as described previously for L- and H-ficolins by assaying its capacity to induce C4b deposition after incubation with ficolin-deficient serum in microplate wells coated with acetylated BSA (32). The addition of increasing amounts of M-ficolin yielded a signal that reached a maximum value at a concentration of 30 μg/ml (Fig. 4C). The A256V and G221F mutants were less efficient, with C4b deposition values at 30 μg/ml corresponding to 71 and 47%, respectively, of that measured for wild-type M-ficolin. In contrast, no detectable C4b deposition was observed using the Y271F mutant (Fig. 4C). These results confirm that the A256V and G221F mutants retain the ability to trigger complement activation after binding to acetylated BSA, whereas the Y271F mutant is not functional. Evaluation of the amount of M-ficolin bound to acetylated BSA confirmed that the complement activating capacity of the variants correlated with their binding ability. Although this variant did not induce C4b deposition, a weak binding signal could be observed with the Y271F mutant (supplemental Fig. S7). The decreased activating ability of the G221F mutant may arise from its different oligomeric composition, in accordance with its reduced binding level observed by SPR (Fig. 4B) and an enzyme-linked immunosorbent assay (supplemental Fig. S7).
Glycan Array Profiling of the M-ficolin Variants
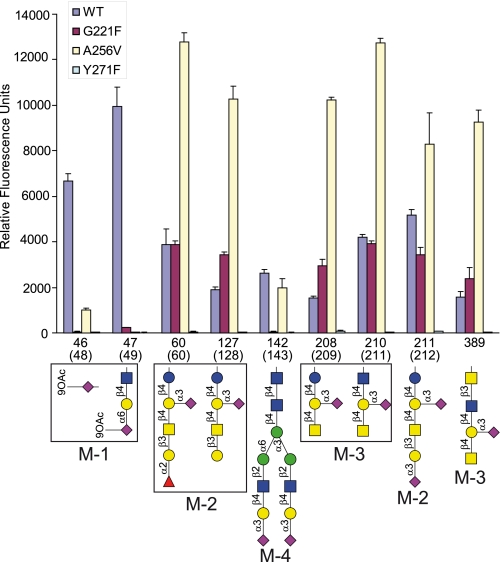
The effect of the three single mutations on the glycan binding specificity of M-ficolin was assessed using Version 3.2 of the glycan array. A first trial performed at the highest concentrations available for G221F and A256V (66.5 and 42 μg/ml, respectively) and at 50 μg/ml for Y271F revealed a strong decrease in binding of the former two mutants to group M-1 glycans (46 and 47), whereas the latter virtually lost binding ability to all glycans (supplemental Fig. S8, A–D). All variants were then assayed at a concentration of 5 μg/ml, and the data obtained for the nine major ligands (groups M-1 to M-4) are shown in Fig. 5 (see supplemental Fig. S9, A–D for the complete binding profiles). In line with the above results, no significant binding of the Y271F mutant to any glycan was observed at this concentration. Maximal binding of wild-type M-ficolin was obtained for the 9-O-acetylated sialic acid derivatives 46 and 47 (48 and 49 in the array Version 3.0), and binding of G221F and A256V to both glycans was strongly decreased or abolished. Otherwise, except for the sialylated biantennary N-linked type glycan 142 to which no significant binding was observed, the G221F mutant exhibited binding levels close to those of wild-type M-ficolin. Unexpectedly, whereas binding of A256V to glycan 142 was comparable with that of wild-type M-ficolin, interaction of this mutant with the other six ligands was consistently much higher. Thus, these results reveal a change in the specificity of the G221F and A256V mutants, resulting in a common loss of binding to 9-O-AcNeu5Ac derivatives and a decreased binding to sialylated biantennary N-linked type glycans.
FIGURE 5.
Effect of M-ficolin mutations on its ability to bind sialylated glycans. Binding of wild-type (WT) M-ficolin and its G221F, A256V, and Y271F mutants at a concentration of 5 μg/ml is shown for selected sialylated glycans of the array version 3.2. Error bars represent the means ± S.D. of four measurements. The glycan numbers of array version 3.0 are indicated in parentheses.
Structure of the Y271F Mutant FBG Domain
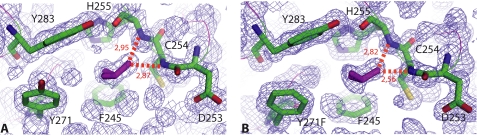
Because the Y271F mutation virtually abolished the glycan binding ability of M-ficolin, the question arose of whether this could be due to a modification of the overall folding of its N-acetyl binding pocket. To answer this question, the mutated FBG domain was produced in a baculovirus/insect cells system, and its x-ray structure was solved and refined to 1.21 Å resolution (supplemental Table S1). As expected, the protein is homotrimeric, and the overall fold of the protomer, including the Ca2+ binding site, is fully conserved (root mean square deviation = 0.08 Å, based on 217 Cα positions). Likewise, in the external S1 binding site, except for the replacement of Tyr271 by Phe, the architecture of the hydrophobic pocket, lined by Phe245, His255, Phe271, Ala272, and Tyr283, is conserved (Fig. 6, A and B). The major functional feature of the S1 binding site, i.e. the cis conformation of the Asp253-Cys254, peptide bond is also retained. As observed for the wild-type domain, an isopropanol molecule, present in the crystallization medium, binds to this side of the pocket, further indicating that its conformation is unchanged (Fig. 6, A and B). No ligand was observed in this pocket when crystallization was performed in the presence of GlcNAc. It may be concluded, therefore, that the lack of binding of N-acetylated compounds is a direct consequence of the absence of the tyrosine OH group, which provides H-bonding stabilization of the N-acetylated group in wild-type M-ficolin (33). Further analysis of the electron density in the co-crystallization experiments only showed poorly defined extra-density in a region corresponding to the internal site of L-ficolin. Thus, the slight residual GlcNAc binding ability observed for the Y271F M-ficolin mutant (Table 1) possibly arises from unspecific binding to this site.
FIGURE 6.
Comparative views are shown of the external S1 ligand binding site in wild-type M-ficolin (A) and in the Y271F mutant (B). A bound isopropanol molecule is shown in purple sticks. The red dashed lines represent hydrogen bonds. The electron density map (2mFo-DFc) is shown in blue.
DISCUSSION
These studies confirm that the three human ficolins exhibit striking differences in their carbohydrate binding specificities. Although H-ficolin was shown to bind to galactosylated BSA using SPR spectroscopy, no significant interaction with any of the glycans contained in the arrays probed in the present study could be detected. However, it has been shown recently that H-ficolin strongly binds to acetylated albumin (31, 32), which could indicate a specificity for yet unidentified non-carbohydrate acetylated ligands from pathogens or altered self. It should also be pointed out that currently available glycan arrays such as those used here are mostly dedicated to mammalian carbohydrates and, therefore, do not provide information regarding binding specificity toward polysaccharides of microbial origin. The glycan array profiles obtained for L- and M-ficolins show no evidence for common ligands; L-ficolin binds preferentially to saccharides with terminal Gal or GlcNAc residues, whereas M-ficolin specifically recognizes sialic acid and its 9-O-acetylated derivative. It should also be stressed that significant glycan binding could only be observed at high L-ficolin concentrations (>100 μg/ml), with binding of M-ficolin to its best ligands obtained at a 100-fold lower concentration, which is in the range of its recently reported serum concentration (19). This suggests that high affinity binding of L-ficolin to physiological ligands likely requires additional factors such as optimal spacing of repeated motifs.
The recognition specificity of L-ficolin for Gal and GlcNAc is in agreement with our SPR analyses performed on BSA glycoconjugates. However, glycan array screening reveals that L-ficolin interacts only weakly with these monosaccharides but binds preferentially to tri- and tetrasaccharides containing a terminal LacNAc or GlcNAc unit, provided that the linkage with the following carbohydrate is not of the β1–3 type (Table 2), in agreement with a recent report by Krarup et al. (43). Intriguingly, strong binding to a single LacNAc unit was observed only when the terminal Gal residue was substituted with two sulfate groups at positions 3 and 6 (glycan 27) (Table 2). In line with this observation, SPR analyses allowed us to confirm that L-ficolin is indeed able to bind with high affinity to heparin, a highly sulfated glycosaminoglycan, through its fibrinogen domain.
TABLE 2.
Binding of L-ficolin to selected carbohydrate structures; effects of sulfation of group L-1 glycans and of the GlcNAc-Gal/GalNAc linkage of groups L-2 and L-3 glycans
The values listed are those obtained in the glycan array analysis shown in Fig. 2A. CV, coefficient of variation. The O-sulfo groups of L-1 glycans and the GlcNAc-Gal and GlcNAc-GalNAc linkages of L-2 and L-3 glycans are indicated in bold characters.
| Glycan no. | Structure | Averagea | CVb |
|---|---|---|---|
| RFU | % | ||
| Group L-1 | |||
| 26 | [3OSO3][6OSO3]Galβ1-4[6OSO3]GlcNAcβ-Sp0c | 49,405 | 16 |
| 27 | [3OSO3][6OSO3]Galβ1-4GlcNAcβ-Sp0 | 40,226 | 19 |
| 287 | [3OSO3][4OSO3]Galβ1-4GlcNAcβ-Sp0 | 5,431 | 22 |
| 39 | [4OSO3][6OSO3]Galβ1-4GlcNAcβ-Sp0 | 982 | 60 |
| 286 | [3OSO3]Galβ1-4[6OSO3]GlcNAcβ-Sp0 | 1,990 | 36 |
| 36 | [3OSO3]Galβ1-4GlcNAcβ-Sp8 | 5,873 | 10 |
| 152 | Galβ1-4GlcNAcβ-Sp0 | 7,035 | 7 |
| Group L-2 | |||
| 156 | GlcNAcα1-3Galβ1-4GlcNAcβ-Sp8 | 37,809 | 18 |
| 157 | GlcNAcα1-6Galβ1-4GlcNAcβ-Sp8 | 44,395 | 11 |
| 176 | GlcNAcβ1-6Galβ1-4GlcNAcβ-Sp8 | 41,449 | 18 |
| 165 | GlcNAcβ1-3Galβ1-4GlcNAcβ-Sp8 | 2,951 | 64 |
| Group L-3 | |||
| 151 | Galβ1-4GlcNAcβ1-6GalNAcα-Sp8 | 35,929 | 11 |
| 144 | Galβ1-4GlcNAcβ1-3GalNAcα-Sp8 | 472 | 78 |
a Average relative fluorescence of four replicates.
b Coefficient of variation expressed as %.
c Spacer arm code: Sp0, CH2CH2NH2; Sp8, CH2CH2 CH2NH2.
In agreement with previous dot-blot and competition analyses (16, 17, 33), our SPR experiments show that M-ficolin binds specifically to acetylated compounds, including the L-ficolin ligand GlcNAc. However, the main GlcNAc binding sites of M- and L-ficolins are clearly different, as indicated by the recognition properties of M-ficolin Y271F in which Tyr271 is replaced by Phe, its counterpart in L-ficolin. Indeed, the inability of this mutant to bind acetylated and sialylated ligands demonstrates that the OH group of Tyr271 in site S1 is essential in the recognition specificity of M-ficolin, in accordance with former studies (33, 45).
The glycan array analysis of M-ficolin reveals that this protein is a sialic acid-binding lectin, able to recognize gangliosides and sialylated biantennary N-linked type glycans, two types of ligands that had never been identified before (see supplemental Fig. S10 and Table S2 for binding data to all sialylated glycans of the Version 3.2 array). Moreover, M-ficolin exhibits strong binding to 9-O-acetylated sialic acid, defining a novel recognition specificity. Remarkably, binding to 9-O-acetylated sialylated LacNAc occurs exclusively when sialic acid is connected to galactose through an α2-6 linkage (glycan 49). In contrast, binding to the unmodified sialic acid moiety is restricted to the α2-3 linkage as illustrated when the underlying structure is a biantennary N-linked glycan (Table 3). In addition, our site-directed mutagenesis studies demonstrate that two small residues, Gly221 and Ala256, located in the vicinity of the acetyl binding pocket, condition the specificity of M-ficolin for 9-O-acetylated sialic acid. Indeed, replacement of either residue by its bulkier counterpart in L-ficolin (Phe and Val, respectively) virtually abolished binding to this ligand. In line with these data, we have previously shown that, based on the superposition of the S1 binding sites of L-ficolin and of the M-ficolin-sialic acid complex, the Val and Phe residues are positioned within 2.3–2.8 Å of the OH group of the C9 atom of sialic acid (33). Grafting a 9-O-linked acetyl group would, therefore, clearly prevent binding of the sialic acid derivative by steric hindrance. Further structural analysis of the FBG domain of M-ficolin in complex with 9-O-acetylated sialic acid derivatives will be needed to gain further insights into the determinants of this particular binding specificity. Interestingly, mutations G221F and A256V maintained the interaction of M-ficolin with the conserved GalNAcβ1-4(Neu5Acα2-3)Gal motif present in all ganglioside-related structures of groups M-2 and M-3. The fact that the G221F mutation inhibits binding to the biantennary N-linked type glycan 142 likely results from a less favorable positioning of the sialic acid residue in the context of this particular glycan, as also reflected by the lower binding efficiency of the A256V mutant (Fig. 5). In line with their preserved capacity to recognize acetyl groups, the G221F and A256V mutants retained the ability to activate the lectin complement pathway on immobilized acetylated BSA, although less efficiently than wild-type M-ficolin, the Y271F mutant exhibiting no complement activation ability.
TABLE 3.
Binding of M-ficolin to selected carbohydrate structures; effect of sialic acid linkage of glycans of groups M-1 and M-4
The values listed are those obtained at a concentration of M-ficolin of 120 μg/ml using the v3.2 version of the array. CV, coefficient of variation. The Neu5Ac linkages are indicated in bold characters.
| Glycan no. | Structure | Averagea | CVb |
|---|---|---|---|
| RFU | % | ||
| Group M-1a | |||
| 47 | Neu5Ac(9OAc)α2-6Galβ1-4GlcNAcβ-Sp8c | 28,171 | 13 |
| 244 | Neu5Acα2-6Galβ1-4GlcNAcβ-Sp8 | 684 | 62 |
| 322 | Neu5Ac(9OAc)α2-3Galβ1-4GlcNAcβ-Sp0 | 1,201 | 28 |
| 236 | Neu5Acα2-3Galβ1-4GlcNAcβ-Sp8 | 626 | 17 |
| Group M-4c | |||
| 142 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 22,846 | 6 |
| 52 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp-12 | 70 | 63 |
| 53 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp-13 | 89 | 65 |
| 51 | Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6) Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp-13 | 57 | 49 |
a Average relative fluorescence of four replicates.
b Coefficient of variation expressed as %.
c Spacer arm code: Sp0, CH2CH2NH2; Sp8, CH2CH2 CH2NH2; Sp12, Asn; Sp13, Gly.
Although previous studies have shown binding of L- and H-ficolins to apoptotic cells and binding of the three ficolins to certain pathogens as stated above, we lack precise information about the molecular nature of their physiological ligands, and the present study brings new information in this respect. The capacity of L-ficolin to bind sulfated glycans such as heparin is unlikely to be involved in pathogen recognition as sulfation of microbial glycosaminoglycan chains has not been described (46). However, heparin is known to interact with several complement proteins and to regulate several steps in the complement cascade (47). In this respect, it has been proposed recently that properdin, a component of the alternative complement pathway, may interact with apoptotic cells through surface-sulfated glycosaminoglycans chains (48). It will be interesting to investigate whether L-ficolin fulfils the same function and what would be its functional consequences.
The capacity of M-ficolin to recognize sialic acid as found in human gangliosides and in common N-linked polysaccharide chains of mammalian glycoproteins likely accounts for its reported localization on the surface of host cells such as monocytes (49). The physiological relevance of ganglioside binding is difficult to assess as no preferential localization of M-ficolin in the nervous system has been reported yet. However the presence of surface polysaccharides with ganglioside GM1 and GD1a mimicry has been reported in bacterial pathogens such as Campylobacter jejuni and Haemophilus influenzae (50). It should also be kept in mind that sialic acids and their 9-O-acetylated derivatives are found at the surface of many pathogens, including capsulated bacteria (51), protozoa (52, 53) and fungi (54, 55). In addition, sialylation changes in host glycoconjugates have been associated with malignant transformation and tumor progression, and 9-O-acetylation of sialic acid connected to GalNAc through an α2-6 linkage has been proposed recently to be a prognostic marker in acute lymphoblastic leukemia (56, 57). Further investigations will be needed to assess the functional relevance of the potential interactions of M-ficolin with sialic acid-bearing pathogens and malignant cells.
In conclusion glycan array screening of the three human ficolins shows that L- and M-ficolin have different recognition specificities and confirms the poor lectin activity of H-ficolin. Novel ligands are revealed, such as sulfated N-acetyllactosamine for L-ficolin and gangliosides for M-ficolin. Moreover, we show that M-ficolin has high specificity for 9-O-acetylated sialic acid in an α2-6 linkage to galactose, which is a relatively rare specificity. Indeed, although sialic acids are recognized by a variety of animal, plant, or viral lectins, only a few sialic acid-binding proteins show preferential binding to 9-O-acetylated sialic acid (58). The physiological relevance of these observations to pathogen and/or altered self cell recognition by L- and M-ficolin awaits further investigation, whereas specific ligands for H-ficolin remain to be identified.
Supplementary Material
Acknowledgments
We are indebted to Pr. Teizo Fujita (Fukushima Medical University, Japan) for the gift of the pMT/Bip/V5-His A plasmid encoding M-ficolin. We acknowledge Core H of The Consortium for Functional Glycomics for the glycan array analyzes. We thank Izabel Berard for mass spectrometry measurements and Dr. Véronique Rossi for preparing the Biacore sensor chip with immobilized heparin. We acknowledge access to beamlines at the European Synchrotron Radiation Facility in Grenoble and to facilities of the Partnership for Structural Biology (Grenoble). The resources provided by the Consortium for Functional Glycomics were funded by National Institutes of Health Grant GM62116 (NIGMS).
This work was supported by the Commissariat à l'Energie Atomique, the CNRS, the Université Joseph Fourier, Grenoble, and Agence Nationale de la Recherche Grant ANR-05-MIME-023-01.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S10.
The atomic coordinates and structure factors (code 2WNP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GlcNAc
- N-acetyl-d-glucosamine
- FBG
- fibrinogen
- GalNAc
- N-acetylgalactosamine
- LacNAc
- N-acetyllactosamine
- Neu5Ac
- N-acetylneuraminic acid
- SPR
- surface plasmon resonance
- RU
- resonance units
- RFU
- relative fluorescence units
- BSA
- bovine serum albumin
- GM
- monosialoganglioside
- GD
- disialoganglioside.
REFERENCES
- 1.Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 2.Fearon D. T., Locksley R. M. (1996) Science 272, 50–53 [DOI] [PubMed] [Google Scholar]
- 3.Fraser D. A., Tenner A. J. (2008) Curr. Drug Targets 9, 113–122 [DOI] [PubMed] [Google Scholar]
- 4.Matsushita M., Fujita T. (2002) Immunobiology 205, 490–497 [DOI] [PubMed] [Google Scholar]
- 5.Ohashi T., Erickson H. P. (2004) J. Biol. Chem. 279, 6534–6539 [DOI] [PubMed] [Google Scholar]
- 6.Lu J., Tay P. N., Kon O. L., Reid K. B. (1996) Biochem. J. 313, 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harumiya S., Takeda K., Sugiura T., Fukumoto Y., Tachikawa H., Miyazono K., Fujimoto D., Ichijo H. (1996) J. Biochem. 120, 745–751 [DOI] [PubMed] [Google Scholar]
- 8.Matsushita M., Endo Y., Taira S., Sato Y., Fujita T., Ichikawa N., Nakata M., Mizuochi T. (1996) J. Biol. Chem. 271, 2448–2454 [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto R., Yae Y., Akaiwa M., Kitajima S., Shibata Y., Sato H., Hirata J., Okochi K., Izuhara K., Hamasaki N. (1998) J. Biol. Chem. 273, 20721–20727 [DOI] [PubMed] [Google Scholar]
- 10.Ohashi T., Erickson H. P. (1998) Arch. Biochem. Biophys. 360, 223–232 [DOI] [PubMed] [Google Scholar]
- 11.Le Y., Lee S. H., Kon O. L., Lu J. (1998) FEBS Lett. 425, 367–370 [DOI] [PubMed] [Google Scholar]
- 12.Kilpatrick D. C., Fujita T., Matsushita M. (1999) Immunol. Lett. 67, 109–112 [DOI] [PubMed] [Google Scholar]
- 13.Krarup A., Sørensen U. B., Matsushita M., Jensenius J. C., Thiel S. (2005) Infect. Immun. 73, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munthe-Fog L., Hummelshøj T., Hansen B. E., Koch C., Madsen H. O., Skjødt K., Garred P. (2007) Scand. J. Immunol. 65, 383–392 [DOI] [PubMed] [Google Scholar]
- 15.Teh C., Le Y., Lee S. H., Lu J. (2000) Immunology 101, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frederiksen P. D., Thiel S., Larsen C. B., Jensenius J. C. (2005) Scand. J. Immunol. 62, 462–473 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Endo Y., Iwaki D., Nakata M., Matsushita M., Wada I., Inoue K., Munakata M., Fujita T. (2005) J. Immunol. 175, 3150–3156 [DOI] [PubMed] [Google Scholar]
- 18.Honoré C., Rørvig S., Munthe-Fog L., Hummelshøj T., Madsen H. O., Borregaard N., Garred P. (2008) Mol. Immunol. 45, 2782–2789 [DOI] [PubMed] [Google Scholar]
- 19.Wittenborn T., Thiel S., Jensen L., Nielsen H. J., Jensenius J. C. (2010) J. Innate Immun., in press [DOI] [PubMed] [Google Scholar]
- 20.Kuraya M., Ming Z., Liu X., Matsushita M., Fujita T. (2005) Immunobiology 209, 689–697 [DOI] [PubMed] [Google Scholar]
- 21.Jensen M. L., Honoré C., Hummelshøj T., Hansen B. E., Madsen H. O., Garred P. (2007) Mol. Immunol. 44, 856–865 [DOI] [PubMed] [Google Scholar]
- 22.Krarup A., Thiel S., Hansen A., Fujita T., Jensenius J. C. (2004) J. Biol. Chem. 279, 47513–47519 [DOI] [PubMed] [Google Scholar]
- 23.Faro J., Chen Y., Jhaveri P., Oza P., Spear G. T., Lint T. F., Gewurz H. (2008) Clin. Exp. Immunol. 151, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y. G., Cho M. Y., Zhao M., Park J. W., Matsushita M., Fujita T., Lee B. L. (2004) J. Biol. Chem. 279, 25307–25312 [DOI] [PubMed] [Google Scholar]
- 25.Lynch N. J., Roscher S., Hartung T., Morath S., Matsushita M., Maennel D. N., Kuraya M., Fujita T., Schwaeble W. J. (2004) J. Immunol. 172, 1198–1202 [DOI] [PubMed] [Google Scholar]
- 26.Aoyagi Y., Adderson E. E., Min J. G., Matsushita M., Fujita T., Takahashi S., Okuwaki Y., Bohnsack J. F. (2005) J. Immunol. 174, 418–425 [DOI] [PubMed] [Google Scholar]
- 27.Aoyagi Y., Adderson E. E., Rubens C. E., Bohnsack J. F., Min J. G., Matsushita M., Fujita T., Okuwaki Y., Takahashi S. (2008) Infect. Immun. 76, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsujimura M., Ishida C., Sagara Y., Miyazaki T., Murakami K., Shiraki H., Okochi K., Maeda Y. (2001) Clin. Diagn. Lab. Immunol. 8, 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honoré C., Hummelshoj T., Hansen B. E., Madsen H. O., Eggleton P., Garred P. (2007) Arthritis Rheum. 56, 1598–1607 [DOI] [PubMed] [Google Scholar]
- 30.Garlatti V., Belloy N., Martin L., Lacroix M., Matsushita M., Endo Y., Fujita T., Fontecilla-Camps J. C., Arlaud G. J., Thielens N. M., Gaboriaud C. (2007) EMBO J. 26, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munk Zacho R., Thiel S., Jensenius J. C. (2008) Mol. Immunol. 45, 4135–4135 [Google Scholar]
- 32.Lacroix M., Dumestre-Pérard C., Schoehn G., Houen G., Cesbron J. Y., Arlaud G. J., Thielens N. M. (2009) J. Immunol. 182, 456–465 [DOI] [PubMed] [Google Scholar]
- 33.Garlatti V., Martin L., Gout E., Reiser J. B., Fujita T., Arlaud G. J., Thielens N. M., Gaboriaud C. (2007) J. Biol. Chem. 282, 35814–35820 [DOI] [PubMed] [Google Scholar]
- 34.Tanio M., Kondo S., Sugio S., Kohno T. (2007) J. Biol. Chem. 282, 3889–3895 [DOI] [PubMed] [Google Scholar]
- 35.Kairies N., Beisel H. G., Fuentes-Prior P., Tsuda R., Muta T., Iwanaga S., Bode W., Huber R., Kawabata S. I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teillet F., Gaboriaud C., Lacroix M., Martin L., Arlaud G. J., Thielens N. M. (2008) J. Biol. Chem. 283, 25715–25724 [DOI] [PubMed] [Google Scholar]
- 38.Vivès R. R., Sadir R., Imberty A., Rencurosi A., Lortat-Jacob H. (2002) Biochemistry 41, 14779–14789 [DOI] [PubMed] [Google Scholar]
- 39.Kabsch. W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 40.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallog. 40, 654–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43.Krarup A., Mitchell D. A., Sim R. B. (2008) Immunol. Lett. 118, 152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garlatti V., Martin L., Lacroix M., Gout E., Arlaud G. J., Thielens N. M., Gaboriaud C. (2010) J. Innate Immun. 2, 17–23 [DOI] [PubMed] [Google Scholar]
- 45.Tanio M., Kohno T. (2009) Biochem. J. 417, 485–491 [DOI] [PubMed] [Google Scholar]
- 46.DeAngelis P. L. (2002) Anat. Rec. 268, 317–326 [DOI] [PubMed] [Google Scholar]
- 47.Sahu A., Pangburn M. K. (1993) Mol. Immunol. 30, 679–684 [DOI] [PubMed] [Google Scholar]
- 48.Kemper C., Mitchell L. M., Zhang L., Hourcade D. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9023–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honoré C., Hummelshoj T., Garred P. (2008) Mol. Immunol. 45, 4134–4135 [Google Scholar]
- 50.Yu R. K., Usuki S., Ariga T. (2006) Infect. Immun. 74, 6517–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Severi E., Hood D. W., Thomas G. H. (2007) Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 52.Ghoshal A., Mukhopadhyay S., Chava A. K., Gerwig G. J., Kamerling J. P., Chatterjee M., Mandal C. (2009) Parasitology 136, 159–173 [DOI] [PubMed] [Google Scholar]
- 53.Monteiro V. G., Lobato C. S., Silva A. R., Medina D. V., de Oliveira M. A., Seabra S. H., de Souza W., DaMatta R. A. (2005) Parasitol. Res. 97, 380–385 [DOI] [PubMed] [Google Scholar]
- 54.Soares R. M., de A Soares R. M., Alviano D. S., Angluster J., Alviano C. S., Travassos L. R. (2000) Biochim. Biophys. Acta 1474, 262–268 [DOI] [PubMed] [Google Scholar]
- 55.Rodrigues M. L., Rozental S., Couceiro J. N., Angluster J., Alviano C. S., Travassos L. R. (1997) Infect. Immun. 65, 4937–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandal C., Chatterjee M., Sinha D. (2000) Br. J. Haematol. 110, 801–812 [DOI] [PubMed] [Google Scholar]
- 57.Varki A. (2008) Trends Mol. Med. 14, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinha D., Chatterjee M., Mandal C. (2000) Trends Glycosci. Glycotechnol. 12, 17–33 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.