Abstract
The molybdenum cofactor (Moco) forms part of the catalytic center in all eukaryotic molybdenum enzymes and is synthesized in a highly conserved pathway. Among eukaryotes, very little is known about the processes taking place subsequent to Moco biosynthesis, i.e. Moco transfer, allocation, and insertion into molybdenum enzymes. In the model plant Arabidopsis thaliana, we identified a novel protein family consisting of nine members that after recombinant expression are able to bind Moco with KD values in the low micromolar range and are therefore named Moco-binding proteins (MoBP). For two of the nine proteins atomic structures are available in the Protein Data Bank. Surprisingly, both crystal structures lack electron density for the C terminus, which may indicate a high flexibility of this part of the protein. C-terminal truncated MoBPs showed significantly decreased Moco binding stoichiometries. Experiments where the MoBP C termini were exchanged among MoBPs converted a weak Moco-binding MoBP into a strong binding MoBP, thus indicating that the MoBP C terminus, which is encoded by a separate exon, is involved in Moco binding. MoBPs were able to enhance Moco transfer to apo-nitrate reductase in the Moco-free Neurospora crassa mutant nit-1. Furthermore, we show that the MoBPs are localized in the cytosol and undergo protein-protein contact with both the Moco donor protein Cnx1 and the Moco acceptor protein nitrate reductase under in vivo conditions, thus indicating for the MoBPs a function in Arabidopsis cellular Moco distribution.
Keywords: Metals/Molybdenum, Protein/Metalloproteins, Protein/Protein-Protein Interactions, Metalloenzymes, Protein Cross-linking, Arabidopsis, Molybdenum Cofactor, Molybdenum Enzyme
Introduction
The molybdenum cofactor (Moco)2 is a prosthetic group highly conserved in all kingdoms of life and consists of a tricyclic pterin, referred to as molybdopterin or metal-binding pterin (MPT) and a molybdenum (Mo) atom covalently bound to the dithiolate moiety of MPT (1). Moco is required for the activity of all Mo-dependent enzymes with the exception of nitrogenase (2). Molybdenum enzymes (Mo-enzymes) are essential for a broad variety of metabolic processes such as nitrate assimilation and phytohormone synthesis in plants (3) and sulfur detoxification and purine catabolism in mammals (4).
Synthesis of Moco proceeds in a highly conserved multistep pathway, involving at least six proteins named Cnx in plants (3). Much is known about the final step of Moco biosynthesis where one Mo atom is ligated to the MPT dithiolate function, which is catalyzed by the two-domain protein Cnx1 (5, 6): the C-terminal Cnx1-G domain activates MPT by adenylation, which is handed over to the N-terminal Cnx1-E domain where it is converted to Moco by inserting Mo into MPT under simultaneous cleavage of the pyrophosphate bond.
After completion of biosynthesis, Moco has to be allocated and inserted into the apoMo-enzymes. In prokaryotes, a complex of proteins synthesizing the last steps of Moco biosynthesis donates the mature cofactor to apoenzymes assisted by enzyme-specific chaperones (7). In eukaryotes, however, no Mo-enzyme-specific chaperone has been found. As free Moco is extremely sensitive to oxidation it is also assumed that Moco occurs permanently protein-bound in the cell. Therefore, a cellular Moco distribution system should meet two demands: (i) it should bind Moco subsequent to its synthesis, and (ii) it should maintain a directed flow of Moco from the Moco donor Cnx1-E to the Mo-dependent enzymes. This is important to ensure the fast and efficient incorporation of Moco into apoMo-enzymes. In the alga Chlamydomonas reinhardtii a Moco carrier protein (MCP) was identified that was found to bind Moco and protect it against oxidation (8–10). Without any denaturing procedure, subsequent transfer of Moco from MCP to apo-nitrate reductase (NR) from Neurospora crassa mutant nit-1 was possible (10), thus indicating that MCP-bound Moco was readily transferable. These properties of Chlamydomonas MCP make it a promising candidate for being part of a cellular Moco delivery system. It is, however, unknown whether MCP is also able to donate Moco to Mo-enzymes other than NR.
Here we present the cloning and characterization of Moco-binding proteins (MoBP) from Arabidopsis thaliana that form a novel protein family consisting of nine members in A. thaliana with functional properties different from Chlamydomonas MCP. The recombinantly expressed proteins are able to bind Moco, enhance Moco transfer to apo-NR in the Moco-free N. crassa mutant nit-1, and undergo protein-protein interaction with both the cellular Moco donor protein Cnx1-E and the Moco acceptor protein NR, thus indicating for the MoBPs a function in Arabidopsis cellular Moco distribution but not in Moco storage.
EXPERIMENTAL PROCEDURES
Identification of Arabidopsis Moco-binding Proteins
Primary sequences of those two Arabidopsis proteins (AGI accession numbers At2g37210 and At5g11950) with structural homology to C. reinhardtii MCP (10) were used for BLAST searches in the TAIR data base. See supplemental Tables S1 and S2 for sequences and annotation.
Preparation of RNA and Reverse Transcription
Total RNA from Arabidopsis was prepared by using the NucleoSpin® RNA Plant kit (Macherey-Nagel, Germany). For construction of an Arabidopsis cDNA library, Arabidopsis mRNA was reverse transcribed with avian myeloblastosis virus-reverse transcriptase (Promega, Germany) using an oligo-d(T)18-BamHI primer.
Cloning of Moco-binding Proteins from Arabidopsis
The cDNAs of Arabidopsis MoBP proteins were cloned PCR-based into the expression vector pQE80 (Qiagen) at appropriate restriction sites, thus fusing the respective N terminus of the protein to a vector-encoded His6 tag. When appropriate, cDNAs were primarily cloned into the pGEM®-T Easy (Promega) cloning vector, using primers derived against the 3′- and 5′-untranslated regions of the corresponding cDNAs.
PCR were carried out by use of an Arabidopsis cDNA library, which was constructed as described above. Thus cDNAs coding for any of the identified Arabidopsis MoBPs have been successfully cloned. The gene-specific primers that have been used for this purpose are listed in supplemental Table S3. Construction of C-terminal-truncated variants as well as fusion of the MoBP3 C terminus to MoBP6 and accordingly replacement of MoBP2 C terminus with MoBP3 C terminus were carried out as PCR based, and the resulting altered cDNAs were subcloned into the expression vector pQE80 (Qiagen, Germany) at appropriate restriction sites.
Expression and Purification of Recombinant Proteins
For cofactor binding studies, MoBP proteins from Arabidopsis were expressed aerobically in Escherichia coli strain DL41 at 30 °C, using LB medium containing 100 μg/ml of ampicillin. After cell density reached an A600 = 0.5, cells were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside and allowed to grow for 20 h. Cell lyses was achieved by two passages through a French pressure cell followed by sonication for 1.5 min. Cell lyses was carried out at 4 °C. After centrifugation, His6-tagged proteins were purified at 4 °C under native conditions using Ni-Sepharose 6 Fast Flow (Amersham Biosciences) according to the manufacturer's manual. Cell lyses buffer contained 50 mm NaPO4, 300 mm NaCl, 10 mm imidazole, 10% glycerol, pH 8.0. Washing steps were carried out using washing buffer containing 50 mm NaPO4, 300 mm NaCl, 20 mm imidazole, 10% glycerol, pH 6.0. Proteins were eluted in elution buffer (50 mm NaPO4, 300 mm NaCl, 250 mm imidazole, 10% glycerol, pH 8.0). All buffers were degassed prior to use. Eluted fractions were separated electrophoretically on 15% SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue.
Pure protein fractions were concentrated (10 kDa molecular mass cut off, Vivaspin 15, Sartorius) up to 50 mg/ml and stored in 20-μl aliquots at −70 °C. For Moco transfer studies to apoNR, Arabidopsis MoBP proteins were expressed in E. coli strain RK5204 (11). Cells were grown aerobically at 25 °C in LB medium containing 100 μg/ml of ampicillin until cell density of the culture reached an A600 = 0.5. Protein synthesis was induced by addition of 0.5 mm isopropyl β-d-thiogalactopyranoside and cells were allowed to grow for 20 h. Proteins were purified and stored as described for proteins expressed in DL41. Detection of Moco/MPT bound to Arabidopsis MoBP proteins throughout the protein purification was carried out after protein expression in E. coli strains RK5206 (11) and TP1000 (12), respectively. To synchronize intracellular protein and cofactor synthesis, expression temperature and expression levels were adapted. Expression was carried out in LB medium containing 100 μg/ml of ampicillin and at 22 °C, using different isopropyl β-d-thiogalactopyranoside concentrations (10 nm, 100 nm, 10 μm, and 100 μm). For expression in TP1000 cells, additionally 0.1 mm sodium molybdate was added. Upon addition of a saturated overnight culture (5 ml/liter of expression culture), recombinant protein synthesis in the cells was induced directly and cells were allowed to grow aerobically for 48 h. Proteins were purified and stored as described for proteins expressed in DL41.
Size Exclusion Chromatography
Purified recombinant proteins were subjected to chromatography on an analytical Superdex 200 column (Amersham Biosciences), using 50 mm Tris-HCl, 150 mm NaCl, pH 7.4, as running buffer. Molecular weight standards (Amersham Biosciences) were used for calibration.
Determination of Lysine Decarboxylase Activity
Lysine decarboxylase activity was determined essentially as described by Phan et al. (13).
Quantification of Moco/MPT
The amount of total Moco/MPT was quantified by HPLC FormA analysis as described earlier (14).
Generation of Free Moco
The Moco carrier protein MCP from C. reinhardtii was chosen as source of Moco because this protein binds specifically Moco and not MPT (10). Expression and purification were performed as described (10). Preparation of Moco was carried out using 600 μl of a 200 μm C. reinhardtii MCP solution in degassed 50 mm Tris-HCl, 150 mm NaCl, 2% (w/v) glycerol, 5 mm sodium molybdate, pH 7.4. Moco was released from its protein environment by heat treatment at 82 °C for 5 min. After centrifugation at 21,000 × g for 2 min the supernatant was subjected to ultrafiltration, using a membrane with a molecular mass cut off of 10 kDa (Vivaspin 500, Sartorius). The protein-free flow-through was used as Moco source.
Binding of Moco to Arabidopsis MoBP Proteins
Moco binding was carried out using 2.5 μm protein and varying amounts of Moco. Protein and Moco were incubated anaerobically for 5 min at room temperature in degassed 50 mm Tris-HCl, 150 mm NaCl, 2% (w/v) glycerol, pH 7.4. Subsequently, unbound Moco was removed by gel filtration using Nick columns (GE Healthcare). The Moco amount in the protein fraction was determined by HPLC FormA analysis (14).
Determination of KD Values
KD values for Moco binding were determined using 2.5 μm of each Arabidopsis MoBP for coincubation with free Moco (0–20 μm). Coincubation was carried out at 20 °C for 5 min in degassed coincubation buffer (50 mm Tris-HCl, 150 mm NaCl, 2% (w/v) glycerol, pH 7.4). After coincubation, the samples were transferred to Vivaspin 500 concentrators (molecular mass cut off 10 kDa) and centrifuged at 11,000 × g for 5 min. As a control, samples containing only free Moco but no protein were used. The amount of Moco in the flow-through was quantified by HPLC FormA analysis (14). Afterward the amount of Moco bound per protein monomer was plotted against unbound Moco. The KD value was calculated according to Equation 1 and assuming one Moco binding site per MoBP monomer. At equilibrium, the population of MoBP was divided into free MoBP ([A]) and MoBP in complex with Moco ([AB]). Accordingly the population of Moco molecules ([B]) was divided into free Moco and Moco in complex with MoBP ([AB]). At saturating Moco concentrations, Moco binding stoichiometries for MoBP3 and MoBP5 were found to be significantly below 1.0 (approximately 0.8 molecules of Moco per MoBP monomer), the fmax variable was added to Equation 1, where fmax represents the binding stoichiometry at saturating Moco concentrations, thus making accurate KD measurements possible.
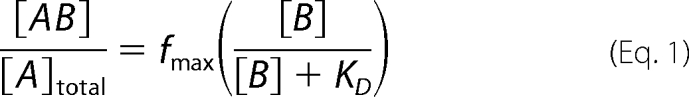 |
Monitoring of MoBP-mediated Moco Transfer
Monitoring of Moco transfer from MoBP to apoNR was carried out using the nit-1 reconstitution assay (15). Crude extracts from the N. crassa nit-1 mutant were prepared (15) and stored in aliquots at −70 °C. Reconstitutions were performed in the presence of 3 mm reduced glutathione and 5 mm sodium molybdate. Reconstitution assays were carried out in a 25-μl reaction volume containing either protein-free Moco or Moco bound to MoBP proteins. Protein-bound Moco was generated by coincubation of 15 μm Moco with 2.5 μm MoBP proteins. Coincubation was carried out for 5 min at 20 °C in degassed 50 mm Tris-HCl, 150 mm NaCl, 2% (w/v) glycerol, pH 7.4. Subsequently, unbound Moco was removed by gel filtration using Nick columns (GE Healthcare), and the Moco-containing protein fraction was concentrated using Vivaspin 500 concentrators (molecular mass cut off 10 kDa). Thereafter, aliquots of the protein fraction were added to the nit-1 extract. In parallel, increasing amounts of protein-free Moco were added. For calibration, nit-1 NR activity was plotted against the concentrations of protein-free Moco in the reconstitution assays, showing the linear dependence of nit-1 NR activity on the amount of protein-free Moco in the reconstitution assay. The amount of Moco in the reconstitution assays was quantified as described earlier (14). Upon addition of protein-bound Moco or free Moco to nit-1 extract, complementation was carried out anaerobically for 16 h at 4 °C. Reconstituted NADPH-NR activity was determined as described (15). Nitrite was quantified by absorbance at 540 nm using a 96-well plate reader (Versa max, Molecular Devices). One unit of Moco activity was defined as reconstituted nit-1 NR activity sufficient to produce an increase of 1.0 absorbance units at 540 nm/25 min of reaction time.
Cross-linking and Biotin Label Transfer
Cross-linking experiments were carried out with the trifunctional cross-linker sulfo-SBED-biotin (sulfo-N-hydroxysuccinimidyl-2-(6-iotinamido]-2-(p-azidobenzamido)-hexanoamido)ethyl-1,3′-dithioproprionate) (Pierce/Perbio) and affinity purified proteins. The amine-reactive NHS group was used to incorporate the cross-linker at the N terminus and side chain of lysine residues of the bait protein. Cross-linking with the prey protein was achieved by the UV light-activatable aryl azide group, reacting nonspecifically with the protein side chains and backbone upon UV-light exposure. After cross-linking and reduction of the disulfide bridge between linker and bait protein, the biotin label is transferred from the bait protein to the prey protein. Bait proteins were mixed with a 5-fold excess of cross-linker in a total volume of 300 to 500 μl (phosphate-buffered saline, pH 7.4) and a concentration of 7 mg/ml. After a 1-h incubation at room temperature unreacted cross-linker was removed by Zeba Desalt spin columns (Pierce/Perbio). For cross-linking, bait and prey protein were mixed equimolar at 10 μm in a volume of 12.5 μl. The reaction mixture was incubated for 30 min at 22 °C. Samples were stored on ice and the UV light-activatable aryl azide group was activated by UV light irradiation for 7 min at a distance of 10 cm. Up to this point all reactions were carried out in subdued light to prevent a light-induced reaction of the aryl azide group of the cross-linker. After irradiation, 6 μl of SDS-loading buffer containing β-mercaptoethanol were added. The samples were heated to 75 °C for 7 min and separated by denaturing gel electrophoresis in 15% polyacrylamide gels. For detection of biotin, proteins were blotted to a polyvinylidene difluoride membrane (GE Healthcare) and probed with horseradish peroxidase-conjugated NeutrAvidin (Pierce/Perbio).
Subcellular Localization
The cDNAs for A. thaliana MoBP1 (At2g28305), MoBP3 (At2g37210), MoBP4 (At4g35190), MoBP5 (At3g53450), Cnx1(E) (At5g20990), NR (At1g37130), CBL10 (At4g33000), and the cyan fluorescent protein (CFP) were amplified with a gene-specific attB primer by PCR and cloned into the donor vector pDONR/Zeo (Invitrogen) using the GATEWAY-BP reaction system to create entry clones, with and without the final stop codon. To fuse the respective MoBP, Cnx1(E), NR, and CBL10 sequence with the coding sequences for the yellow fluorescent protein (YFP) and/or CFP, the entry clones in combination with GATEWAY destination vectors pEarleyGate101, pEarleyGate102, and pEarleyGate104 (16), kindly provided by the Arabidopsis Biological Resource Center, Columbus, OH, were used to create the final YFP and/or CFP expression vectors by LR reaction3 (Invitrogen). Constructs for Bimolecular fluorescence complementation (BiFC) were cloned using the GATEWAY destination vectors pDEST-SCYCE(R)GW, pDEST-VYNE(R)GW, and pDEST-GWVYNE (17). CFP was introduced into pK7WGC2 and used as the cytoplasmic expression control. All GATEWAY expression constructs were introduced into Agrobacterium tumefaciens strain C58C1 (helper plasmid pMP90) and used for infiltration into leaves of Nicotiana benthamiana as described earlier (17). Transient fluorescence was visualized with the confocal laser scanning microscope cLSM-510META connected to an Axiovert 200M (Carl Zeiss) in the lower epidermis of leaf discs 2–3 days after infiltration with laser excitation of 488 nm and BP 505–530 for YFP/BiFC fluorescence and LP 650 for chlorophyll autofluorescence; laser excitation of 458 nm and BP 475–525 for CFP fluorescence. The λ mode was used to examine the spectral signature of the fluorochromes. All images were processed with LSM Image Browser Release 4.2 for two-dimensional pictures (Carl Zeiss) and Volocity 4.4 for three-dimensional pictures (Improvision).
RESULTS
Identification of Arabidopsis Putative Moco-binding Proteins
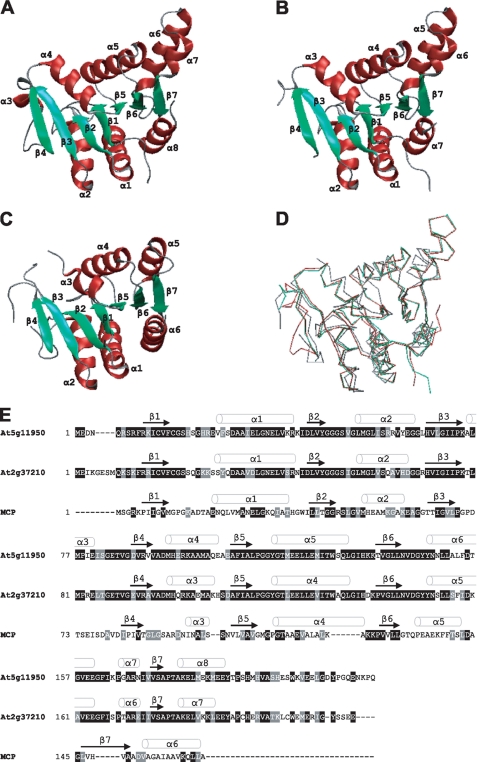
To identify putative Moco-binding proteins from Arabidopsis, we used the primary sequence of Chlamydomonas MCP as query for a homology search using BLAST (18), without retrieving any meaningful hits. However, a homology-based search using the atomic structure of Chlamydomonas MCP led to the identification of structurally closely related proteins from A. thaliana, Thermus thermophilus, and Bacillus subtilis (10). In Arabidopsis two proteins (Protein Data Bank codes 2A33 and 1YDH) with pronounced structural similarities to Chlamydomonas MCP were identified (Fig. 1). Regarding quarternary structure the Arabidopsis homologs, however, differ from Chlamydomonas MCP as they have been crystallized as dimers (19), whereas the latter forms stable homotetramers (9, 10). Superposition of Chlamydomonas MCP (Protein Data Bank Code 2IZ5) with the two proteins from Arabidopsis demonstrates a high degree of structural conservation (Fig. 1D) documented by an average root mean square deviation of 1.84 Å (2A33 and 2IZ5) and 1.74 Å (1YDH and 2IZ5) as determined by EBI-SSM (20). Consequently we assumed a function of both Arabidopsis proteins related to the function of the MCP from Chlamydomonas. When comparing the amino acid sequences of all three proteins and aligning them with the structural motifs (Fig. 1E) it becomes obvious that their close structural relationships are not reflected on the primary sequence level. The two Arabidopsis MoBP proteins 2A33 and 1YDH share about 20% primary sequence identity with Chlamydomonas MCP.
FIGURE 1.
Tertiary structure-based identification of putative MoBP proteins in Arabidopsis. Ribbon representations of Arabidopsis proteins At5g11950 (Protein Data Bank code 1YDH, chain B) (A) and At2g37210 (Protein Data Bank code 2A33, chain B) (B). C, ribbon representation of Chlamydomonas MCP (code 2IZ5, chain A). D, superposition of 1YDH chain B (red), 2A33 chain B (green), and 2IZ5 chain A (gray). Figures were generated by use of EBI-SSM (20) and rendered using the VMD Molecular Graphics Viewer (30). E, sequence alignment of Arabidopsis proteins At5g11950 and At2g37210 with Chlamydomonas MCP. The alignment was generated with ClustalW. Identical residues are shaded in black and similar residues are shaded in gray.
To identify more putative MoBP proteins in Arabidopsis, the primary sequences of the two Arabidopsis proteins 2A33 and 1YDH were used as queries in TAIR-BLAST searches. As a result, seven other proteins (At2g28305, At4g35190, At3g53450, At5g03270, At5g06300, At2g35990, and At5g26140) with high levels of sequence similarities have been identified (supplemental Fig. S1). Common to all but one (At5g26140) of them is the sequence motif PGGXGTXXE, which is attributed to members of a protein family including annotated lysine decarboxylases (21).
Characterization of Arabidopsis Putative MoBP Proteins
Cloning of Arabidopsis putative MoBPs was performed by using an Arabidopsis cDNA library. Recombinant overexpression in E. coli was possible for all proteins with the exception of At5g26140, and yielded highly pure proteins after nickel affinity chromatography. Gel filtration chromatography revealed the dimeric nature of the Arabidopsis putative MoBP proteins (data not shown). As both the eight identified Arabidopsis putative MoBP proteins and Chlamydomonas MCP have a sequence motif that is also found in lysine decarboxylases we tested all of them for this activity. No lysine decarboxylase activity was observed (data not shown). Therefore we exclude a function as lysine decarboxylases.
To study substrate binding properties of the eight Arabidopsis putative MoBP proteins, each protein was expressed in different E. coli strains. The strains used were RK5204 (11), which lacks MPT and Moco, RK5206, which accumulates MPT but has no Moco (11), and TP1000, which accumulates Moco (12). Yield and purity of all protein preparations obtained were similar. No co-purified MPT/Moco was detected upon expression in RK5204 and trace amounts of co-purified pterin were detected upon expression in RK5206 and TP1000, respectively (data not shown). Hence, neither Moco, nor the metal-free MPT are bound to Arabidopsis putative MoBP proteins throughout nickel affinity chromatography. Therefore we assumed that any protein-bound Moco was lost during the purification procedure. A similar observation was reported for the xanthine dehydrogenase-specific chaperone XdhC from Rhodobacter capsulatus where, however, in vitro Moco binding to this protein was possible subsequent to purification (22). Consequently we tested the eight Arabidopsis putative MoBPs for their ability to bind Moco in vitro.
In Vitro Moco Binding to Arabidopsis Putative MoBP Proteins
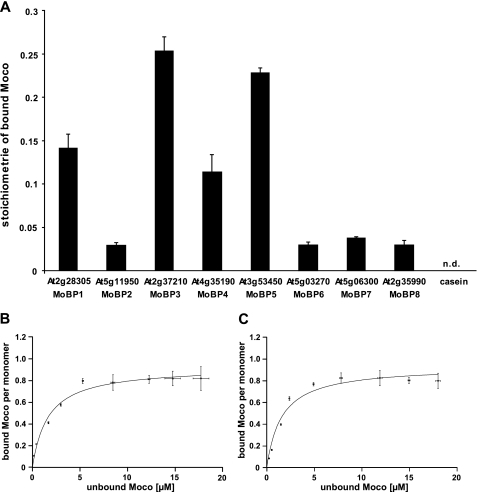
As source of Moco for in vitro binding studies we chose recombinantly expressed Chlamydomonas MCP, which can be purified in large quantities, containing up to 25% co-purified Moco. Furthermore, Chlamydomonas MCP was shown to bind exclusively Moco but not the metal-free MPT (10). Free Moco was obtained after heat treatment of 200 μm recombinant Chlamydomonas MCP. Immediately after release, extracted Moco was incubated anaerobically with each of the Arabidopsis putative MoBPs for 5 min at room temperature before unbound Moco was removed by gel filtration. The amount of Moco bound to the protein fraction was quantified by HPLC FormA analysis (14). In this way significant Moco binding to At2g28305, At2g37210, At4g35190, and At3g53450 was shown (Fig. 2A). No Moco was found in the protein fraction of the casein control, whereas the remaining Arabidopsis putative MoBPs showed Moco saturation levels in the lower percentage range. Therefore, Arabidopsis putative MoBPs are henceforth referred to as Arabidopsis MoBP proteins and are numbered MoBP1 to MoBP8. The corresponding GenBank codes are given in supplemental Table S1. Using a 2.8-fold excess of Moco over each monomer of Arabidopsis MoBP resulted in a 25% Moco saturation, as found for Arabidopsis MoBP3. Incubation with a higher molar excess of Moco gave Moco saturation levels up to 50% (data not shown). Among Arabidopsis MoBPs remarkable differences in Moco saturation levels were observed that could most likely be ascribed to varying Moco binding affinities. Based on these observations we conclude that the binding properties of Arabidopsis MoBP proteins do not permit their co-purification with Moco, whereas Moco binding throughout gel filtration is well possible.
FIGURE 2.
Moco binding properties of Arabidopsis MoBP proteins. A, Moco binding stoichiometries of Arabidopsis MoBP proteins. The coincubation reaction contained 2.5 μm concentrations of each protein and 7 μm Moco. Proteins and Moco have been incubated anaerobically for 5 min at room temperature. Unbound Moco was removed by gel filtration, and the protein-bound Moco amount was subsequently quantified by HPLC FormA analysis (14). The average values were obtained in three duplicate experiments. Casein was used as negative control. Moco binding to Arabidopsis MoBP3 (B) and MoBP5 (C): bound Moco per MoBP monomer is plotted against unbound Moco. Increasing amounts of Moco were coincubated with 2.5 μm concentrations of the respective protein. After 5 min of anaerobic incubation at room temperature, unbound Moco was separated from MoBP proteins by ultrafiltration. Average values were obtained by three duplicate experiments. Moco amounts were quantified by HPLC FormA analysis (14). Error bars, S.E.
Quantification of Moco Binding Properties
To determine the dissociation constants for Moco binding to Arabidopsis MoBPs, the proteins were incubated with increasing amounts of freshly prepared Moco derived from heat-treated Chlamydomonas MCP and processed as described under “Experimental Procedures.” Determination of Moco dissociation constants (KD) was possible for Arabidopsis MoBP1, MoBP3, MoBP4, and MoBP5. For the remaining proteins, no Moco saturation could be achieved, thus making the determination of KD values impossible. Slightly different Moco saturations ranging from 0.81 to 1.07 have been found for these MoBPs, pointing in each case toward a Moco/MoBP ratio of 1:1. The corresponding KD values have been determined, ascribing the highest affinities for Moco to MoBP3 (1.58 ± 0.26 μm) and MoBP5 (1.65 ± 0.32 μm) (Fig. 2, B and C).
Both proteins share >90% sequence identity, explaining their very similar affinities for Moco. Lower KD values were obtained for MoBP1 and MoBP4, corresponding to 4.03 ± 0.64 and 4.72 ± 0.64 μm, respectively. These binding efficiencies for Moco correlate well with the different Moco saturation levels found after gel filtration (compare Fig. 2A), showing that the latter result indeed from varying Moco binding affinities.
MoBP C Terminus Is Involved in Moco Binding
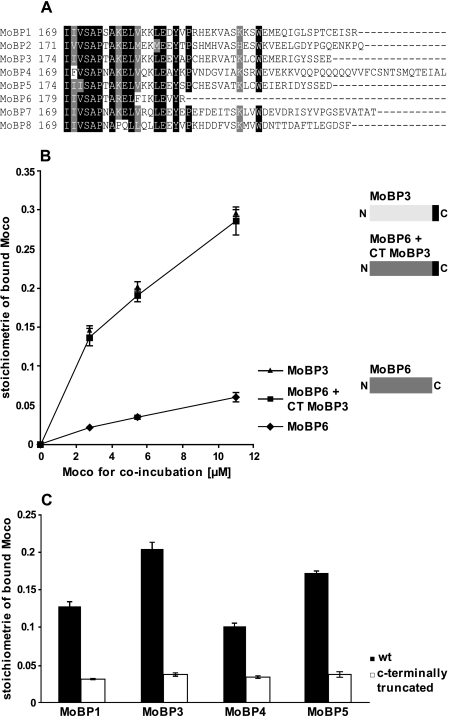
To identify amino acids responsible for the varying Moco binding stoichiometries among Arabidopsis MoBP proteins, we compared their primary sequences. The last 22–40 C-terminal amino acids were found to be highly variable in sequence (Fig. 3A), whereas the remaining protein core showed sequence identities above 70% (supplemental Fig. S1), which would argue for the C terminus to be responsible for the varying Moco binding affinities observed among the MoBP. To support this assumption the following experiment was carried out. MoBP6 is C-terminal shorter than the other MoBPs, i.e. it lacks a corresponding C terminus. In direct comparison to MoBP3, MoBP6 displayed only 5% saturation with Moco (Fig. 3B), whereas MoBP3 exhibited 30% saturation under identical experimental conditions. MoBP3 possesses a C terminus of 23 amino acids that does not occur in MoBP6. To determine whether or not these amino acids are causal for the higher Moco saturation levels observed we fused them to MoBP6 (Fig. 3B). The chimeric protein was assayed for its Moco binding properties and found to bind Moco in the elevated stoichiometries observed for MoBP3 (Fig. 3B). Likewise the poorly Moco-binding protein MoBP2 was substituted with the C terminus of MoBP3, thus again giving yield to a similar increase in the Moco binding stoichiometries (data not shown). Consequently, the enhancement of Moco binding stoichiometry can be ascribed to the last 23 amino acids of MoBP3. Next we deleted this sequence stretch of MoBP3.
FIGURE 3.
Moco binding properties of Arabidopsis MoBP proteins. A, sequence comparison of MoBP C termini. The alignment was generated with ClustalW. Identical residues are shaded in black, and similar residues are shaded in gray. B, Moco binding to MoBP6 and MoBP6 + C terminus (CT) from MoBP3 and MoBP3. For generation of the chimeric protein the last C-terminal 23 amino acids of MoBP3 were fused to MoBP6. 2.5 μm concentrations of each protein were incubated anaerobically with increasing amounts of Moco for 5 min at room temperature. Unbound Moco was removed by gel filtration and the amount of protein-bound Moco subsequently quantified by HPLC FormA analysis (14). Average values were obtained in three duplicate experiments. C, Moco binding to Arabidopsis MoBP wild type proteins and their respective C-terminal-truncated variants. Truncated variants lack the last 27 (MoBP1), 23 (MoBP3), 50 (MoBP4), and 23 (MoBP5) amino acids, respectively. The coincubation reaction contained 2.5 μm concentrations of each protein and 6.4 μm Moco. Proteins and Moco were incubated anaerobically for 5 min at room temperature. Unbound Moco was removed by gel filtration and the amount of protein-bound Moco was subsequently quantified by HPLC FormA analysis (14). Average values were obtained in three duplicate experiments. Error bars, S.E.
Because three other MoBP proteins (MoBP1, MoBP4, and MoBP5) from Arabidopsis also exhibited Moco binding properties comparable with MoBP3, we likewise truncated them and tested for Moco binding (Fig. 3C). After coincubation with Moco all truncated MoBP proteins revealed significantly decreased Moco binding stoichiometries as compared with the wild type proteins. Thus Moco binding stoichiometries of MoBP1, MoBP3, MoBP4, and MoBP5 indeed go back to their respective C-terminal part.
Arabidopsis MoBP Gene Structure
Supportive evidence for the role of MoBP C terminus in Moco binding comes from the gene structure. Analysis of the exon-intron structure of the corresponding genes revealed that for each MoBP the C terminus is encoded by a single exon. Besides the highly variable C-terminal part, three additional amino acids are encoded by this exon, which are in part conserved among Arabidopsis MoBP proteins (supplemental Fig. S2). Clearly, such a gene structure may have promoted the exchange of the C-terminal part during the course of evolution, giving rise to proteins with apparently different properties.
Moco Transfer to Aponitrate Reductase
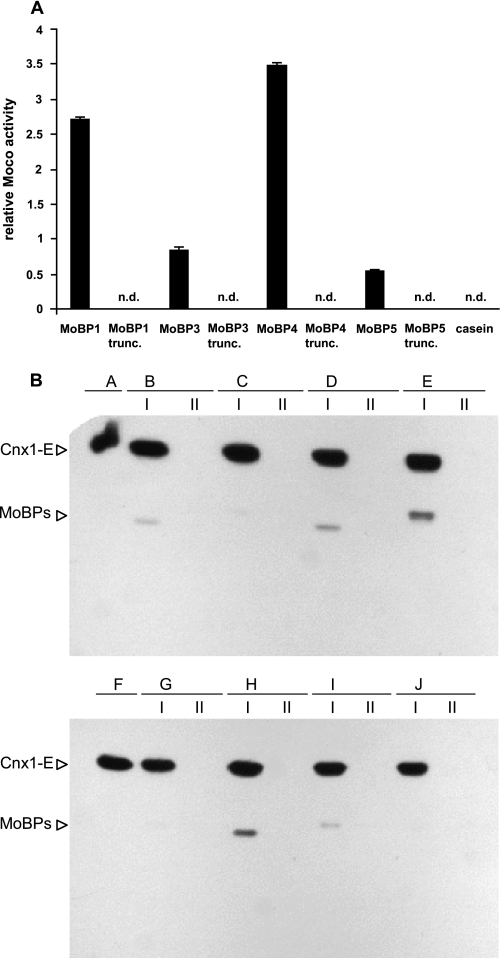
Protein-directed Moco flow to apoNR was investigated by use of the nit-1 reconstitution assay (15). Arabidopsis proteins MoBP1, -3, -4, and -5 were coincubated with 15 μm Moco. Subsequently, unbound Moco was removed by gel filtration, the Moco-loaded proteins were concentrated by ultrafiltration, and thereafter added to the nit-1 extract (see “Experimental Procedures”). In a parallel control experiment, increasing amounts of protein-free Moco were added to the nit-1 extract. Thus calibrated, we compared nit-1 NR reconstitutions based upon addition of MoBP-bound Moco with NR activities due to addition of protein-free Moco (Fig. 4A). No reconstitution activity was observed upon addition of casein after coincubation with 15 μm Moco, whereas for each of the MoBPs tested reconstitution activities became measurable. Moco bound to MoBP1 and MoBP4 was found to enhance nit-1 NR activity about 2.5- and 3.5-fold, respectively, as compared with the same amount of protein-free Moco added to the nit-1 extract. Surprisingly, unlike MoBP1 and MoBP4, the MoBP proteins with the highest affinity for Moco (MoBP3 and MoBP5) affected nit-1 NR activity adversely, namely 0.85- (MoBP3) and 0.55-fold (MoBP5) as compared with protein-free Moco added to the nit-1 extract. The four remaining MoBPs displayed nit-1 NR activities close to the detection limit. Coincident with the decreased Moco binding stoichiometries of C-terminal-truncated MoBP1, -3, -4, and -5, these proteins also showed merely no nit-1 reconstitution activity, and an effect on apoNR reconstitution was no longer assignable (Fig. 4A). Thus we conclude that the C terminus of MoBP1, MoBP3, MoBP4, and MoBP5 is involved in both, Moco binding and its delivery to apoNR.
FIGURE 4.
MoBP-mediated Moco transfer to apoNR and interaction of Arabidopsis MoBPs with Cnx1-E. A, Moco transfer to apoNR has been monitored by use of the nit-1 reconstitution assay. MoBP-bound Moco has been added to the nit-1 extract without cofactor releasing treatment. Casein was used as a control. For details see “Experimental Procedures.” The average values were obtained in three duplicate experiments. Error bars, S.E. B, cross-linked and immunoblotted samples with horseradish peroxidase-streptavidin conjugate (α-Biotin) and Cnx1-E as bait protein. Label transfer from Cnx1-E to the particular Arabidopsis MoBPs is shown in lane I for (B) MoBP1, (C) MoBP2, (D) MoBP3, (E) MoBP4, (G) MoBP5, (H) MoBP6, (I) MoBP7, and (J) MoBP8. As a control the respective Arabidopsis MoBP proteins were treated identically in the absence of Cnx1-E in the coincubation mixture (lane II). Biotin-labeled Cnx1-E is shown in A and F.
Cross-linking of Arabidopsis MoBP Proteins with the Moco Donor Cnx1
The results obtained point to a role of MoBP in the cellular flow of Moco from the site of its synthesis to the apoMo-enzymes. As, however, MoBPs do not bind Moco tightly it is difficult to unequivocally assign a more detailed role to them. Hence we used a different approach to address this question. We asked whether MoBPs are able to physically interact with the protein Cnx1, which catalyzes the last step of Moco biosynthesis (i.e. the insertion of Mo into MPT) and therefore serves as Moco donor for all Mo-enzymes. This assay has been carried out by a novel type of reversible cross-linking followed by label (=biotin) transfer from the labeled donor protein Cnx1 to a non-labeled acceptor protein. This label transfer only takes place when donor and acceptor get as close as 14 Å, and was monitored by subsequent immunoblot analysis. As the E-domain of Cnx1 catalyzes Mo insertion into MPT we used Cnx1-E as label donor, as described under “Experimental Procedures.” All eight Arabidopsis MoBP proteins were tested for interaction with labeled Cnx1-E (Fig. 4B): five proteins (MoBP1, MoBP3, MoBP4, MoBP6, and MoBP7) showed signals, and three proteins (MoBP2, MoBP5, and MoBP8) gave only very faint or no signals. No interaction was observed between Cnx1-E and bovine serum albumin (data not shown). These data demonstrate that the majority of MoBPs are able to undergo protein-protein contact with the Moco-donor protein Cnx1-E.
Subcellular Localization of Arabidopsis MoBP Proteins
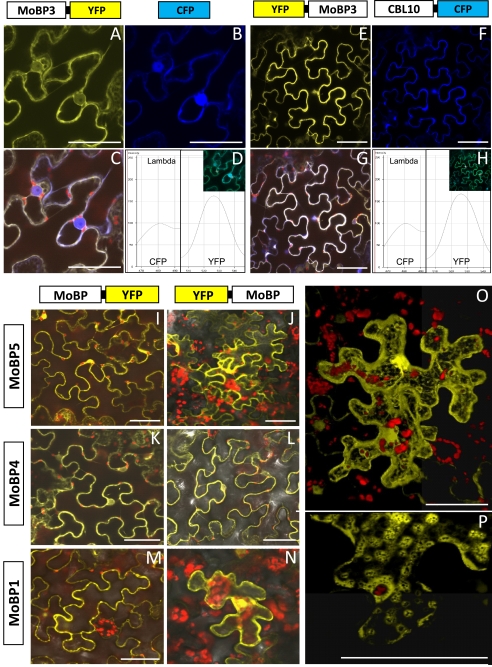
The results obtained demonstrate that Arabidopsis MoBP proteins interact with both Cnx1 and apoNR, which are known to be cytosolic proteins (23, 24). Therefore we assumed that likewise MoBPs are localized in the cytosol of the cell. To provide evidence for this assumption, MoBP1, MoBP3, MoBP4, and MoBP5, respectively, were fused to the YFP and the corresponding gene constructs were expressed in cells of N. benthamiana. The emerging fluorescence was then monitored by confocal laser scanning microscopy. For localization studies YFP was fused with the N and C terminus, respectively, of the characterized MoBPs. Fig. 5, A and E, show a typical cytoplasmic localization for MoBP3 fused to YFP within a cell in focus (which looks like a puzzle piece: the large vacuole presses the cytosol against the cell wall so that its fluorescence shows the contour of the cell). As the fusion protein MoBP3-YFP has a size of approximately 50 kDa, in addition to the evident cytoplasmic fluorescence a weak fluorescence is seen in the nucleus because the size of the fusion protein is just at the edge of the exclusion limit of nuclear pores (about 40–60 kDa (25)). To provide evidence for cytoplasmic localization of MoBP3-YFP we carried out colocalization studies with two proteins with known cytoplasmic localizations: CFP and CBL10. CBL10 (calcineurin B-like protein), is a calcium sensor known to localize to the cytoplasm (26). The C-terminal YFP fusion variant of MoBP3 and the control protein CFP were transferred and expressed in the same cell. Fig. 5B shows the fluorescence of the control protein CFP. The fluorescence was located in the cytoplasm but also in the nucleus as the size of the CFP (27 kDa) is far below the exclusion limit of nuclear pores. In Fig. 5C images of MoBP3-YFP (Fig. 5A) and CFP (Fig. 5B) localization are merged, showing that the YFP and CFP signals co-localize, thus verifying the cytoplasmic localization of MoBP3. Fig. 5D shows the spectra taken for both YFP and CFP in the yellow and blue channels, respectively, thus confirming the specificity of the fluorescence recorded.
FIGURE 5.
Cytoplasmic localization of MoBP-YFP fusion proteins. cDNAs of MoBP1, -3, -4, and -5 were fused to the N and C terminus of YFP, respectively, and transferred via Agrobacterium infiltration into N. benthamiana leaves. A–H show co-localization with cytoplasmic marker proteins (CFP and CBL10::CFP); A and E, YFP fusion with MoBP3; B and F, CFP channel; C and G, merged pictures of A and B and E and F, respectively (the red color shows autofluorescence of the chloroplasts); D and H, spectral signature of CFP/YFP (peak at 480 and 525 nm, respectively) as detected in the λ mode. I–N show pictures of YFP fluorescence merged with the chlorophyll autofluorescence and the transmitted light photomultiplier in the channel mode of the confocal laser scanning microscope for MoBP1, -4, and -5 in both orientations to the fluorochrome. O and P, all scanning images of a single cell transformed with the MoBP3::YFP fusion construct are combined (Z-stack), the picture was taken at higher magnification and calculated using Volocity 4.4. Bars represent 50 μm.
In a similar approach, localization of the N-terminal YFP fusion variant of MoBP3 was compared with the localization of the control protein CBL10 fused to CFP (Fig. 5, E–H). The results obtained corroborate the cytoplasmic localization of MoBP3-YFP.
Fig. 5, I–N, shows cytoplasmic localizations of MoBP1, MoBP4, and MoBP5 for both fusion variants, i.e. N- and C-terminal fused to YFP, respectively. When all scanning images of a cell (Z-stack) are combined (Fig. 5, O and P) it becomes clear that a given MoBP (here MoBP3) is distributed over the whole cytoplasm. As MoBPs are below the exclusion limit of nuclear pores they also entered the nucleus (Fig. 5O). However, they are not localized in the mitochondria, Fig. 5P shows mitochondria as little black spots that exhibit no fluorescence.
In Vivo Interaction of Arabidopsis MoBP Proteins with Moco Donor and Acceptor Proteins
The experiments demonstrate that MoBPs are cytoplasmic proteins that bind Moco, can be cross-linked in vitro to the Moco donor protein Cnx1, and are able to stimulate Moco transfer to the Moco-acceptor protein apoNR. However, these are in vitro experiments. If MoBPs have a physiological role in Moco distribution they should undergo protein interaction with both a Moco donor (=Cnx1) and a Moco acceptor protein (e.g. NR) in a living cell. For this purpose we used the novel approach of BiFC. BiFC is based on splitting a fluorescent protein (e.g. the yellow fluorescent protein YFP) into two halves and coupling the latter to proteins A and B, respectively. The genes for both fusion constructs are cotransferred into cells of the model plant N. benthamiana where they are transiently expressed. Only if proteins A and B undergo protein interaction the two non-functional fragments of split fluorescent proteins are brought into tight contact, refold together, and the fluorophore reconstitutes (27).
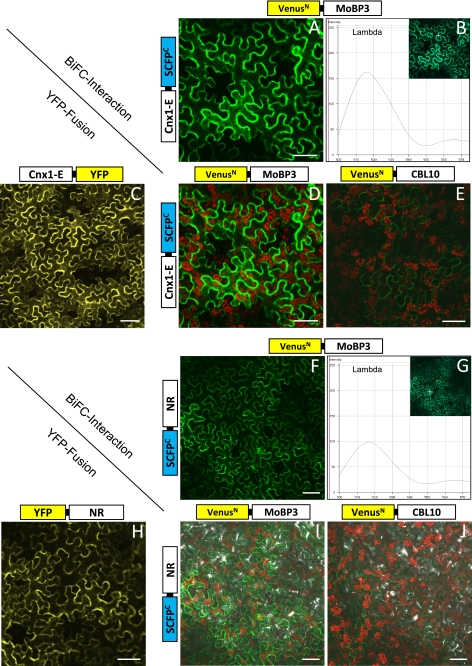
In a first set of experiments we wanted to see whether MoBP3 chosen as representative for the MoBP proteins does interact with the Moco donor Cnx1 in a living cell. Cnx1 is a two-domain protein that holds the final reaction product Moco on its E-domain. For BiFC experiments MoBP3 was N-terminal fused to half of the enhanced yellow fluorescent protein Venus, and Cnx1-E was fused to half of SCFP (SCFP is the enhanced version of CFP and reconstitutes in combination with Venus, resulting in a bright green fluorescence after excitation (17)). Fig. 6A leads to two conclusions: (i) the bright green fluorescence indicates that MoBP3 and Cnx1-E undergo tight protein interaction in vivo and (ii) the fluorescence localizes exclusively to the cytoplasm as the combined protein complex is above the exclusion limit of nuclear pores. In Fig. 6B, the spectral signature (λ mode) of the green BiFC fluorescence is recorded, as control Fig. 6C shows Cnx1-E fused to YFP. In Fig. 6D, the red channel is added to the green channel of the laser scanning microscope thus making the red autofluorescence of the chloroplasts visible. Fig. 6E shows the negative control that is Cnx1-E fused to half of SCFP that was tested for in vivo interaction with an unrelated protein, namely CBL10 fused to half of Venus. Here, only weak background fluorescence is visible, which is typical for this kind of in vivo approach (17, 27).
FIGURE 6.
BiFC of MoBP3 with Cnx1-E (A–E) and NR (F–J), respectively. In A and F the reconstitution of the fluorochrome is shown in the green channel mode, B and G give the spectral signature of the BiFC complex (peak at 515 nm) detected in the λ mode. C and H show the cytoplasmic fluorescence of Cnx1-E and NR with YFP fusion (expression control); D and I show the BiFC interaction signal of the protein partners studied plus chlorophyll autofluorescence; in E and J the negative interaction controls using cytoplasmic protein CBL10 in combination with MoBP3 and NR, respectively, are shown. Bars represent 50 μm.
The results obtained indicate an in vivo interaction occurring between MoBP3 and Cnx1-E. Following the assumption that MoBPs fulfill a function in cellular Moco trafficking between donor and acceptor proteins, in a second set of experiments we wanted to see whether MoBP3 is able to interact with the Moco-acceptor protein NR in a living cell. Thus MoBP3 was N-terminal fused to half of the enhanced yellow fluorescent protein Venus, and NR was fused to half of SCFP. Fig. 6, F, G, and I, show green fluorescence in the channel and λ mode, respectively, indicating that MoBP3 and NR undergo protein interaction in vivo, whereas the negative control (=NR fused to half of SCFP that was tested for in vivo interaction with the unrelated protein CBL10 fused to half of Venus) hardly showed any background fluorescence (Fig. 6J). Fig. 6H shows as control NR fused to YFP.
DISCUSSION
Research in the past years has shed much light on the Moco biosynthesis pathway both in bacteria (7) and eukaryotes (3). For bacteria, first insights also became available in the mechanism underlying Moco transfer and its incorporation into the appropriate apoenzymes (7). However, for eukaryotes little information is available about the processes taking place subsequent to Moco biosynthesis. Moco-binding proteins, i.e. Moco containing proteins without a Moco-dependent enzymatic activity, have been suggested to be involved in Moco transfer to apoenzymes (2), but only for the green alga C. reinhardtii are detailed data available. In this alga, the Moco carrier protein MCP binds Moco tightly and stabilizes it against oxidation (9, 10). Moreover Moco bound to this protein can be transferred without any denaturing procedure to apoNR from the N. crassa nit-1 mutant.
Using the tertiary structure of Chlamydomonas MCP (10) as a template, in silico analyses unveiled a family of nine MoBP proteins in the model plant A. thaliana. As cloning of any of the corresponding cDNAs was possible, it is obvious that the respective genes are expressed in the plant. However, only eight of them could be recombinantly expressed. Upon primary sequence analysis a possible lysine decarboxylase motif was identified both in the eight expressible Arabidopsis MoBP proteins and in Chlamydomonas MCP, but none of the recombinant proteins showed any lysine decarboxylase activity. Therefore we conclude that this motif has no function related to lysine decarboxylation.
Moco binding to Arabidopsis MoBP proteins was shown in a fully defined in vitro system and permitted to subdivide into two groups: one group (MoBP1, MoBP3, MoBP4, and MoBP5) bound Moco in a 1:1 ratio with KD values between 1.58 and 4.72 μm, whereas the other group showed significantly lower Moco binding stoichiometries that precluded determination of dissociation constants. Upon recombinant expression in the Moco accumulating E. coli strain TP1000, no copurification of Moco with any of the Arabidopsis MoBP proteins was possible. This behavior is markedly different from Chlamydomonas MCP that could be saturated up to 25% with Moco in TP1000 (10). In consequence we conclude that Arabidopsis MoBP proteins do not serve as Moco storage proteins and should therefore carry out another function in the cellular Moco flow.
Our truncation experiments indicated that the C terminus of Arabidopsis MoBP proteins seems to play a role in Moco binding. The C termini of the eight MoBPs are highly variable in sequence, whereas the protein cores exhibit a remarkable degree of identity. This is reflected by the crystal structures available for Arabidopsis MoBP2 (1YDH) and MoBP3 (2A33). Both crystal structures are highly similar to each other but lack electron density for the last 35 (2A33) and 37 (1YDH) amino acids, respectively, which is consistent with a high mobility and flexibility of this part of the proteins. Chlamydomonas MCP lacks this flexible C terminus comparable with Arabidopsis MoBPs, instead it binds Moco on a surface-exposed site (10). C-terminal-truncated MoBP proteins did not completely lose their Moco binding ability, rather they retained a basal level that would argue for part of the Moco binding site to be located to the protein body. Based on the high degree of sequence similarity among the MoBP cores, we propose that this part of the protein provides at least part of the Moco binding site in any of the eight MoBPs. Consistent with this, the assembly of a fully functional Moco binding site was possible by fusing the C terminus of MoBP3 (=protein with the best Moco binding properties) to the poorly Moco-binding proteins MoBP6 and MoBP2, respectively, that in turn adopted the Moco binding behavior of MoBP3. Most remarkably, the gene structure of Arabidopsis MoBP proteins points to a special role of the C termini as they are encoded by a separate exon in each case. This gene structure may have contributed to the observed C-terminal sequence diversity, accompanied by apparently different properties of the proteins. The C terminus of Arabidopsis MoBP proteins seems to be important from yet another point of view. In apoNR reconstitution assays, it was found to be crucial for enhancing NR reconstitution above those basal levels that were displayed by the C-terminal truncated protein versions.
Localization experiments with fluorescent fusion proteins showed that MoBPs are cytoplasmic proteins. Furthermore, our BiFC interaction studies revealed that in a living cell MoBP3 is able to interact with both the Moco donor Cnx1 and likewise with the Moco acceptor NR. Besides monitoring of protein interactions the BiFC approach has another advantage: it also shows where in the cell this interaction takes place. Interactions of MoBP3 with Cnx1 and NR take place in the cytoplasm, thus indicating that MoBP3 carries out its function in this cell compartment.
What is the function of Arabidopsis MoBP proteins in the cellular flow of Moco from the final step of its synthesis to the apoMo-enzymes? Unlike Chlamydomonas MCP, Arabidopsis MoBPs do not bind Moco tightly, which makes them unsuitable candidates to serve as storage proteins. However, their KD values for Moco binding (1.58–4.72 μm) turned out to be within the range previously determined (14) for the Moco donor protein Cnx1-E (KD = 1.6 μm). Our cross-linking experiments also revealed that five of the eight Arabidopsis MoBPs physically interact with Cnx1-E. Furthermore, Arabidopsis MoBPs are able to enhance Moco-dependent reconstitution of apoNR. Thus we come to the conclusion that Arabidopsis MoBP proteins are involved in cellular distribution of Moco.
The high number of eight expressed MoBP proteins in Arabidopsis let us assume that each of them may have a particular function for Moco distribution in the plant. Indeed, the first in silico analyses (28) revealed that for instance, MoBP1 and MoBP2 are expressed at elevated levels in the stem, and MoBP3 and MoBP4 are expressed at high levels in the apex during plant development. Data base searches (29) revealed that the high number of MoBPs is not a peculiarity of Arabidopsis: corn exhibits nine MoBP homologous sequences, rice (Oryza sativa, Indica Group) 10, and the evolutionary very old moss Physcomitrella nine. For Chlamydomonas a single MoBP homologous sequence has been identified (which is different from Chlamydomonas MCP), but nothing is known yet about the function of this protein.4
Another possible role for each of the eight Arabidopsis MoBPs may be found when one considers the number of different Mo-dependent enzymes in Arabidopsis. Altogether, Arabidopsis harbors four different Mo-dependent enzymes with several corresponding isoforms (3), and a fifth one has been identified very recently. Therefore one could assume that part of the eight Arabidopsis MoBPs may serve as private proteins being responsible for protein-directed Moco transfer to a certain type or group of Mo-enzymes. Arabidopsis MoBP proteins that were found to lack interaction with Cnx1-E may have a function as secondary acceptor proteins for Moco or may be involved in recognition of Mo-enzymes. As MoBPs form dimers it would be of interest to also consider the possible formation of heterodimers between different MoBPs with each monomer fulfilling a special task. Future experiments are needed to elucidate the detailed functions of Arabidopsis MoBPs.
Supplementary Material
Acknowledgments
We thank Günter Schwarz and Angel Llamas for contributions in the initial phase of this project.
This work was supported by the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2 and Tables S1–S3.
The LR reaction allows transfer of a gene sequence into expression vectors by recombining an Entry Clone (provided with attL sites) with Destination Vectors (provided with attR sites).
T. Kruse and R. R. Mendel, unpublished results.
- Moco
- molybdenum cofactor
- BiFC
- bimolecular fluorescence complementation
- MoBP
- Moco-binding protein
- Mo-enzyme
- molybdenum-enzyme
- NR
- nitrate reductase
- CBL
- calcineurin B-like protein
- MPT
- metal-binding pterin
- MCP
- Moco carrier protein
- HPLC
- high pressure liquid chromatography
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescence protein.
REFERENCES
- 1.Schwarz G., Mendel R. R., Ribbe M. W. (2009) Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan K. V., Johnson J. L. (1992) J. Biol. Chem. 267, 10199–10202 [PubMed] [Google Scholar]
- 3.Schwarz G., Mendel R. R. (2006) Annu. Rev. Plant Biol. 57, 623–647 [DOI] [PubMed] [Google Scholar]
- 4.Hille R. (2005) Arch. Biochem. Biophys. 433, 107–116 [DOI] [PubMed] [Google Scholar]
- 5.Llamas A., Otte T., Multhaup G., Mendel R. R., Schwarz G. (2006) J. Biol. Chem. 281, 18343–18350 [DOI] [PubMed] [Google Scholar]
- 6.Llamas A., Mendel R. R., Schwarz G. (2004) J. Biol. Chem. 279, 55241–55246 [DOI] [PubMed] [Google Scholar]
- 7.Vergnes A., Pommier J., Toci R., Blasco F., Giordano G., Magalon A. (2006) J. Biol. Chem. 281, 2170–2176 [DOI] [PubMed] [Google Scholar]
- 8.Witte C. P., Igeño M. I., Mendel R., Schwarz G., Fernández E. (1998) FEBS Lett. 431, 205–209 [DOI] [PubMed] [Google Scholar]
- 9.Ataya F. S., Witte C. P., Galván A., Igeño M. I., Fernández E. (2003) J. Biol. Chem. 278, 10885–10890 [DOI] [PubMed] [Google Scholar]
- 10.Fischer K., Llamas A., Tejada-Jimenez M., Schrader N., Kuper J., Ataya F. S., Galvan A., Mendel R. R., Fernandez E., Schwarz G. (2006) J. Biol. Chem. 281, 30186–30194 [DOI] [PubMed] [Google Scholar]
- 11.Stewart V., MacGregor C. H. (1982) J. Bacteriol. 151, 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer T., Santini C. L., Iobbi-Nivol C., Eaves D. J., Boxer D. H., Giordano G. (1996) Mol. Microbiol. 20, 875–884 [DOI] [PubMed] [Google Scholar]
- 13.Phan A. P., Ngo T. T., Lenhoff H. M. (1982) Anal. Biochem. 120, 193–197 [DOI] [PubMed] [Google Scholar]
- 14.Schwarz G., Boxer D. H., Mendel R. R. (1997) J. Biol. Chem. 272, 26811–26814 [DOI] [PubMed] [Google Scholar]
- 15.Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 3242–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. (2006) Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 17.Gehl C., Waadt R., Kudla J., Mendel R. R., Hänsch R. (2009) Mol. Plant 2, 1051–1058 [DOI] [PubMed] [Google Scholar]
- 18.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 19.Jeon W. B., Allard S. T., Bingman C. A., Bitto E., Han B. W., Wesenberg G. E., Phillips G. N., Jr. (2006) Proteins 65, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 20.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 21.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., Griffiths-Jones S., Howe K. L., Marshall M., Sonnhammer E. L. (2002) Nucleic Acids Res. 30, 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M., Schulte M., Jünemann N., Stöcklein W., Leimkühler S. (2006) J. Biol. Chem. 281, 15701–15708 [DOI] [PubMed] [Google Scholar]
- 23.Mendel R. R., Hänsch R. (2002) J. Exp. Bot. 53, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 24.Schwarz G., Schulze J., Bittner F., Eilers T., Kuper J., Bollmann G., Nerlich A., Brinkmann H., Mendel R. R. (2000) Plant Cell 12, 2455–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Görlich D., Mattaj I. W. (1996) Science 271, 1513–1518 [DOI] [PubMed] [Google Scholar]
- 26.Kim B. G., Waadt R., Cheong Y. H., Pandey G. K., Dominguez-Solis J. R., Schültke S., Lee S. C., Kudla J., Luan S. (2007) Plant J. 52, 473–484 [DOI] [PubMed] [Google Scholar]
- 27.Bhat R. A., Lahaye T., Panstruga R. (2006) Plant Methods 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J. U. (2005) Nat. Genet. 37, 501–506 [DOI] [PubMed] [Google Scholar]
- 29.Altschul S. F., Lipman D. J. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.