Abstract
Hypoxia has been shown to promote tumor metastasis and lead to therapy resistance. Recent work has demonstrated that hypoxia represses E-cadherin expression, a hallmark of epithelial to mesenchymal transition, which is believed to amplify tumor aggressiveness. The molecular mechanism of E-cadherin repression is unknown, yet lysyl oxidases have been implicated to be involved. Gene expression of lysyl oxidase (LOX) and the related LOX-like 2 (LOXL2) is strongly induced by hypoxia. In addition to the previously demonstrated LOX, we characterize LOXL2 as a direct transcriptional target of HIF-1. We demonstrate that activation of lysyl oxidases is required and sufficient for hypoxic repression of E-cadherin, which mediates cellular transformation and takes effect in cellular invasion assays. Our data support a molecular pathway from hypoxia to cellular transformation. It includes up-regulation of HIF and subsequent transcriptional induction of LOX and LOXL2, which repress E-cadherin and induce epithelial to mesenchymal transition. Lysyl oxidases could be an attractive molecular target for cancers of epithelial origin, in particular because they are partly extracellular.
Keywords: Diseases/Cancer/Transformation, Oxygen/Hypoxia, Transcription/Target Genes, Gene Expression, Kidney, E-cadherin, Hypoxia-inducible Transcription Factor (HIF), Invasion, Lysyl Oxidase, von Hippel Lindau protein (VHL)
Introduction
Constant availability of molecular oxygen is crucial for the structure and function of any mammalian cell. Therefore, cellular responses to reduced oxygen tensions (hypoxia) play an important role in development and many aspects of physiological homeostasis. Many important disease processes, including ischemic vascular diseases and cancer, involve reduced tissue oxygenation, and cellular adaptation to this is implicated in disease progression and clinical outcome. The hypoxia-inducible transcription factor (HIF)2 is a central mechanism responding to low cellular oxygenation and mediates a variety of systemic and local adaptive responses, including the control of red cell production, regulation of angiogenesis, modulation of vascular tone, enhancement of glycolysis, and cellular glucose uptake (for a review, see Refs. 1–3).
HIF consists of a heterodimer of α- and β-subunits, both being basic helix-loop-helix-Per Arnt Sim domain proteins. Whereas HIFβ is constitutively expressed, HIFα subunits are unstable and inversely correlated to the availability of molecular oxygen. At least two oxygen-regulated isoforms of HIFα have been identified, HIF-1α and HIF-2α, sharing a high degree of sequence homology and a similar domain structure (4). Regulation of HIF is primarily governed by oxygen-dependent hydroxylation of its HIFα subunits, which influences protein stability and transcriptional activity.
Growth and behavior of tumor cells is strongly dependent on their microenvironment, where hypoxia is both a stress factor and an important signal (for a review, see Refs. 5 and 6). Dating back to 1927, Otto Warburg had already described that tumor cells have a much increased utilization of the glycolytic pathway (7). Since then, a number of studies have established that indeed HIF is necessary to activate glycolysis in tumor cells in order to maintain energy homeostasis (8, 9). Accordingly, most (but not all) experimental studies have demonstrated that HIF is a growth-promoting mechanism for tumors (10, 11). In human solid tumors, HIFα subunits are regionally stabilized in response to microenvironmental stimulation, of which hypoxia would presumably be the most important stimulus (first described in Refs. 12 and 13). In contrast, in the majority of renal cell carcinoma (RCC), HIFα subunits are globally stabilized (14, 15), which is the result of biallelic inactivation of the von Hippel Lindau (VHL) tumor suppressor gene. The VHL gene product has previously been shown to be part of an E3 ubiquitin ligase complex targeting HIFα subunits for degradation (16) in response to oxygen-dependent prolyl hydroxylation (17). Importantly, the VHL-associated HIF activation not only seems to be relevant for tumor growth in RCC but also plays a dominant role in tumorigenesis. HIFα is activated in the earliest renal lesions of the familial VHL syndrome (18) and seems to be the decisive factor for growth initiation of experimental RCCs, where HIF-2α plays the dominant role over HIF-1α (19–23).
Further to the unique role of the VHL/HIF axis, RCCs show a typical clinical feature, which is resistance to established forms of therapy with radiation and chemotherapy and a high rate of metastasis at diagnosis (24). To date, it can only be speculated whether these features are linked to each other. However, extensive data has accumulated which points toward hypoxia-driven dedifferentiation of tumor cells, which could lead to the process of epithelial to mesenchymal transition (EMT), leading to enhanced invasion and metastasis (25, 26). Clinically, this has been suspected for a long time, because severely hypoxic tumors have been described to be more aggressive and less responsive to therapy (27, 28).
The process of EMT is characterized by a complex dedifferentiation program, where epithelial cells lose their polarity and epithelial surface markers and induce the expression of mesenchymal markers, resulting in fibroblast-like and motile phenotypic cells (29, 30). In general, the invasive and metastatic phenotype is associated with down-regulation of E-cadherin expression, a hallmark of EMT (31), being a mediator and indicator of EMT, simultaneously. Several mechanisms like genetic, epigenetic, and transcriptional changes have been implicated in the regulation of E-cadherin expression during tumor progression (32). Importantly, recent studies have shown that E-cadherin expression can depend on the VHL status of cells, where hypoxic incubation was able to suppress E-cadherin protein expression (33–35), confirming a hypothesis that was previously formulated (36). These data indicate that VHL and/or HIF are capable of regulating E-cadherin, possibly influencing the process of EMT. The mechanism by which hypoxia and/or HIF represses E-cadherin expression is less clear, because a number of different pathways have been suggested.
The HIF target gene lysyl oxidase (LOX) has been implicated in involvement in E-cadherin regulation. LOX has been drawn as one of the highest regulated genes from numerous gene arrays searching for novel HIF-regulated genes, performed by us and others (37–39). Importantly, LOX has been shown to be a direct target of HIF-1, which is functionally required for hypoxia-induced metastasis (40). Furthermore, the lysyl oxidases LOX and LOX-like 2 (LOXL2) have been shown to physically interact with Snail, an important repressor of E-cadherin, which might influence Snail activity (41). Lysyl oxidases are a family of copper-dependent amine oxidases, catalyzing the covalent cross-link of the component side chains of collagen and those of elastin, thus stabilizing these proteins in the extracellular matrix (ECM). Four LOX-like (LOXL) proteins have been described so far, with varying degrees of similarity to LOX. Interestingly, the lysyl oxidases seem to carry additional functions beyond ECM stabilization, because they were found not only in the extracellular compartment but also in the cytoplasm and the nucleus, influencing proliferation and transformation between the normal and the malignant phenotype (for a review, see Refs. 42 and 43). LOX and LOXL2 have been demonstrated to be highly expressed in many human cancers, partly and adversely in correlation to clinical outcome (42).
In our study, we have investigated the role of LOX and LOXL2 in hypoxic repression of E-cadherin. We demonstrate that LOXL2 is also a direct HIF-1 target gene and that induction of lysyl oxidases is necessary and sufficient to repress E-cadherin in hypoxia. Our data provide experimental confirmation of a hypothesis that has been recently suggested, spanning a molecular pathway from hypoxia to HIF and lysyl oxidases, which is essential for E-cadherin repression (10) and which may be most relevant for development and progression of RCC.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Hep3B, HEK293, and HeLa cells were from the European Cell Collection. The following cell lines were kind gifts from different investigators: HKC-8 (L. Racusen, Baltimore, MD), A498 (P. Staller, Copenhagen, Denmark), RCC4 (P. Ratcliffe, Oxford, UK), RCC10 (T. Acker, Frankfurt, Germany). The last three were VHL-deficient RCC cell lines and were analyzed in parallel with stably reconstituted lines. All cell lines were cultured in Dulbecco's modified Eagle's medium, 10% fetal calf serum, 100 units/ml penicillin, 0.1 mg/ml streptomycin (all from PAA) to subconfluence and exposed to either normoxia or hypoxia (1% O2, 5% CO2, 94% N2) for the indicated time points. Hypoxic incubations were performed in an “invivo400” hypoxic workbench (Ruskinn Technology Ltd., West Yorkshire, UK). Reagents used to treat cells were 2,2′-dipyridyl (DP) (ICN, Costa Mesa, CA) and dimethyloxaloylglycine (Biomol, Hamburg, Germany). β-Aminopropionitrile (β-APN), bathocuproinedisulfonic acid (BCS), d-penicillamine, and cobalt chloride (CoCl2) were from Sigma.
Collection of Human Tissues
Tissues were from a historical collection (14), collected from radical tumor nephrectomies and fixed in 4% paraformaldehyde. Written informed consent was obtained from each patient before nephrectomy. The collection of human tissues was approved by the local ethics committee.
Knockdown with Small Interfering RNA (siRNA)
For mRNA knockdown, the following sequences were used at 100 nm (Eurogentec, Bruxelles, Belgium): HIF-1α, 5′-GCCACUUCGAAGUAGUGCUTT-3′; HIF-1α.2, 5′-CUGAUGACCAGCAACUUGATT-3′; HIF-2α, 5′-GCGACAGCUGGAGUAUGAATT-3′; HIF-2α.2, 5′-CAGCAUCUUUGAUAGCAGTT-3′; LOX, 5′-GACUGCCAGUGGAUUGAUATT-3′; LOXL2, 5′-GCGGGCUCUUAAACAACCATT-3′; LOXL2.1, 5′-GGCAAUGAGAAGUCCAUUATT-3′; LOXL2.2, 5′-GAAUCCGAUUACUCCAACATT-3′. As a negative control, we used an siRNA targeting green fluorescence protein, 5′-GGUGUGCUGUUUGGAGGUCTT-3′. Cells were transfected with the siRNAs at 50% confluence by the use of Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Protein Extraction and Immunoblot Analysis
Cells were homogenized into extraction buffer (7 m urea, 10% glycerol, 10 mm Tris-HCl, pH 6.8, 1% SDS, 5 mmol/liter dithiothreitol, 1 mm 4-(2-aminoethyl)-benzenesulfonylfluoride, Complete Mini EDTA-free) using a T8 Ultra-Turrax homogenizer (IKA, Staufen, Germany) for 10 s at full speed. Extracts were quantified using the DC protein assay (Bio-Rad). Proteins were resolved in 10% SDS-polyacrylamide gels and transferred to Immobilon P (Millipore, Bedford, MA) overnight in blotting buffer (10 mm Tris, 100 mm glycine, 10% methanol, 0.05% SDS). Membranes were blocked with 3% fat-free dried milk in PBS with 0.1% Tween 20 and probed with monoclonal antibodies against E-cadherin (0.2 μg/ml; clone HECD-1, Abcam (Cambridge, UK)), HIF-1α (1 μg/ml; clone 54 (Transduction Laboratories, Lexington, KY)), and β-actin (0.48 ng/ml; clone AC-15 (Sigma)) and horseradish peroxidase-conjugated secondary goat anti-mouse antibodies (0.5 ng/ml; DAKO (Glostrup, Denmark)). Signals were visualized by chemiluminescence (Pierce). The displayed results are representative data of independent experiments.
Luciferase Reporter Constructs
The pGL2/LOXL2 reporter construct, including a 502-bp fragment of the first intron of the LOXL2 gene (nucleotides +1758 to +2260; accession number NT_023666) was generated, amplified from human genomic DNA (forward primer, 5′-cgacgcgtaaattgagtctcacta-3′; reverse primer, 5′-ggaagatcttgtcagccaatgtgtg-3′ (generated MluI and BglII restriction sites are underlined) and cloned in the pGL2 promoter vector (Promega, Madison, WI). For sequential analysis of the three putative hypoxia response elements, site-directed mutagenesis was performed, generating the plasmids pGL2/LOXL2-mut1–3. For mutagenesis, the following primers were used: LOXL2-Bmut1, 5′-cgcTTTtttgtgtatgcatgtgtg-3′; LOXL2-Bmut1′, 5′-aaaAAAgcgcgcgtgcacacacag-3′; LOXL2-Bmut2, 5′-ggcTTTtgtgtgtgcacgcaat-3′; LOXL2-Bmut2′, 5′-acaAAAgcccgcgcacacacacc-3′; LOXL2-Bmut3, 5′-atgtgtgcTTTtgtgtatatgtatatgtgt-3′; LOXL2-Bmut3′, 5′-atatacacaAAAgcacacatgcgtgcaca-3′.
Transient Transfection Assays
For luciferase reporter assays cells were seeded in 24-well plates and transfected at a confluence of ∼50–60% with 0.3 μg of the LOXL2 luciferase reporter plasmids or 0.1 μg of the E-cadherin reporter plasmids and 0.1 μg of pCMV-β-galactosidase expression vector (Stratagene, La Jolla, CA) using jetPei transfection reagent (Polyplus, New York, NY) for HEK-293 cells or FuGene HD reagent (Roche Applied Science) for HKC-8, following the manufacturer's instructions. One day after transfection, cells were stimulated with hypoxia for 24 h. For HIF-1 overexpression, 50 ng of the expression plasmids HIF-1α and HIF-1β or an equimolar amount of the empty pcDNA3 vector was co-transfected. For RNA knockdown, cells were transfected with siRNAs as described above the day before reporter transfection. Luciferase activities were determined using luciferase assay reagent (Promega) and normalized to β-galactosidase expression. For 48-h overexpression of lysyl oxidases, cells were seeded in 10-cm dishes or on glass slides and transfected with FuGene HD (Roche Applied Science) with 10 μg of pcDNA-hLOX or pcDNA-hLOXL2-FLAG.
Immunofluorescence and Immunohistochemistry
RCC10 and RCC10/VHL cells were grown on glass slides to subconfluence and exposed to normoxia or hypoxia for 48 h. 3% paraformaldehyde-fixed cells were incubated with primary antibodies to E-cadherin (Abcam) (clone HECD-1, 0.2 μg/ml), FLAG tag (Stratagene) (clone M2, 4 μg/ml), and ZO-1 (Invitrogen) (2.5 μg/ml) overnight at 4 °C. Actin staining was performed with phalloidin-FluoroProbe 547 (0.164 nmol/ml; Interchim (Montlucon, France)). Cells were incubated with fluorescent dye-labeled secondary antibodies (AlexaFluor 488 or AlexaFluor 584; 0.4 μg/ml) (Molecular Probes/Invitrogen) at room temperature for 1 h. Nuclei were stained with DAPI (6.5 μg/ml; Invitrogen) for 1 h. Cells were mounted at 4 °C with Mowiol (Roth, Karlsruhe, Germany). Images of cells were acquired with a Nikon Eclipse 80i fluorescence microscope (Düsseldorf, Germany) and a CCD camera (SPOT RT KE/SE, Visitron Systems (Puchheim, Germany)), using the Spot Advanced 4.6 software (Visitron Systems).
RNase Protection Assay
For analysis of mRNA expression, cells were exposed to hypoxia or stimulated with hypoxia mimetics 16 h before total RNA was prepared using RNazol B (Biozol, Eching, Germany). 32P-Labeled antisense RNA probes were transcribed with SP6 or T7 polymerase from plasmids containing cDNA fragments of human HIF-1α, HIF-2α, and U6 small nuclear RNA (U6sn), as described (44). For lysyl oxidases, probes were generated in pcDNA3 (LOX, accession number NM_002317, nucleotides 805–990; LOXL2, accession number NM_002318, nucleotides 1049–1213). Displayed results are representative data of independent experiments.
Chromatin Immunoprecipitation
Two independent chromatin immunoprecipitation assays were performed using the Upstate protocol (Millipore). Cells were sonicated in 15-s on/off pulses for a total of 12 min (Bioruptor, Diagenode). Chromatin was immunoprecipitated using rabbit polyclonal antiserum to HIF-1α (PM14), which has previously been shown to perform well in chromatin immunoprecipitation assays and to be highly specific for HIF-1α (45). Preimmune serum was used as a negative control. Immunoprecipitated chromatin was amplified using the Sigma whole genome amplification kit according to the manufacturer's instructions. Real-time quantitative PCR for DNA quantification employed SYBR Green gene expression assays on a StepOne thermocycler (Applied Biosystems, Foster City, CA). Normalization was to β-actin DNA, and -fold enrichment at each locus was calculated using the ΔΔCt method. For the quantitative PCR, the following primers were used: β-actin-fw, 5′-accatggatgatgatatcgcc-3′; β-actin-rev, 5′-gccttgcacatgccgg-3′; LOXL2-fw, 5′-cacacatacacgtgcacaca-3′; LOXL2-rev, 5′-aggctctccccaaggaaat-3′.
Electrophoretic Mobility Shift Assay
Recombinant protein for HIF-1α and HIF-1β was generated by in vitro transcription and translation using the TNT® quick coupled transcription/translation system (Promega) and incubated with a 32P-labeled 24-bp LOXL2 HRE1 wild type (sense strand, 5′-gcacgcgcgcacgtttgtgtatgc-3′) and the LOXL2-mut1 (sense strand, 5′-gcatacacaaacgtgcgcgcgtgc-3′) oligonucleotide probe. For supershift assays, anti-HIF-1α antibody (clone 54, Transduction Laboratories; 750 ng) was added, and reactions were incubated for 20 min on ice before electrophoresis. An electrophoretic mobility shift assay was performed as described (16).
Invasion Assay
Collagen gel was prepared by mixing calf skin type I collagen G (Serva Electrophoresis GmbH, Heidelberg, Germany) and rat tail type I collagen R (Biochrom AG, Berlin, Germany) at a ratio of 1:1. 0.1 volume of sodium bicarbonate (22 mg/ml) and 0.1 volume of 10× Dulbecco's modified Eagle's medium were added, and the solution was neutralized with sodium hydroxide. Aliquots of 1.2 ml were allowed to gelatinize in 6-well culture dishes at 37 °C. 5 × 104 tumor cells were directly seeded in Dulbecco's modified Eagle's medium containing 2% fetal calf serum onto the collagen surfaces. Cells invading the collagen gel were detected by focusing down into the matrix and quantified by counting 20 optical fields/well (31) 3 days after seeding. For inhibition of lysyl oxidases, assays were performed in the presence of β-APN. For lysyl oxidase knockdown, cells were transfected with siRNAs as described above 24 h before seeding on collagen I gel. Statistical analysis was performed using the two-sided Student's t test. Statistical significance was presumed at a value of <0.05.
RESULTS
Expression of EMT Markers E-cadherin, F-actin, and ZO-1 Is Influenced by VHL and Hypoxia
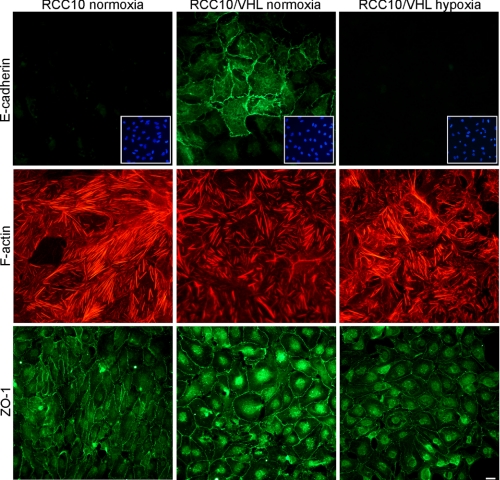
The cell line RCC10 (46) has been studied in the context of VHL-dependent E-cadherin regulation before (34). Fig. 1 shows the results of immunofluorescent staining for different markers of EMT in RCC10 cells, which are either defective (RCC10) or reconstituted for VHL (RCC10/VHL). Since the reconstituted cells show hypoxic inducibility for HIF and its target genes, we studied RCC10/VHL cells also in normoxia and hypoxia. As reported previously, the VHL status of the cells strongly determines the expression of E-cadherin, with no detectable signal in VHL-deficient cells and marked membranous expression in VHL-reconstituted cells. After 36 h of hypoxia (1% O2), E-cadherin expression in RCC10/VHL cells was completely abolished (Fig. 1, top panels). Similar results were found for other markers of EMT, actin stress fibers and ZO-1 (zonular occludens protein-1). RCC10/VHL cells showed only a moderate amount of actin stress fibers in normoxia, but a clear increase occurred after incubation in hypoxia (Fig. 1, middle panels). In contrast, VHL-deficient RCC10 cells displayed high levels of actin stress fibers already in normoxia. Similarly, ZO-1 expression, a marker for epithelial differentiation, showed a clear dependence for VHL and hypoxia (Fig. 1, lower panels). ZO-1 was strongly expressed in normoxic RCC10/VHL, where specific staining appeared as a continuous membranous pattern. In contrast, VHL-deficient RCC10 cells displayed weaker and irregular membranous expression. In hypoxic RCC10/VHL cells, the expression of ZO-1 could be significantly reduced, and discontinuous membranous staining appeared. Furthermore, RCC10 cells showed clear morphological differences between the VHL-deficient and VHL-reconstituted cell lines, which is best seen in the ZO-1 staining (Fig. 1, lower panels). Cells lacking VHL displayed a spindel-like, fibroblastoid cell shape. In contrast, VHL-reconstituted RCC10 cells showed an epitheloid cell morphology with a typical cobblestone appearance. Taken together, these findings are compatible with the process of EMT, induced by hypoxia and VHL deficiency. However, this feature is not complete because fibronectin and vimentin staining did not display these changes (data not shown).
FIGURE 1.
VHL and hypoxia influence markers of EMT. RCC10 cells were incubated under normoxia or hypoxia (1% O2) for 40 h on glass slides and analyzed for E-cadherin, F-actin, and ZO-1 expression by immunofluorescence staining. All images displayed for the different markers were photographed with identical exposition times. Insets in the E-cadherin panels depict nuclear staining (DAPI) of identical positions on the slide to confirm presence and density of cells where no E-cadherin was detected. Bar, 10 μm; representative for all panels.
LOX and LOXL2 Are Regulated by Hypoxia and Are Direct Targets of HIF-1
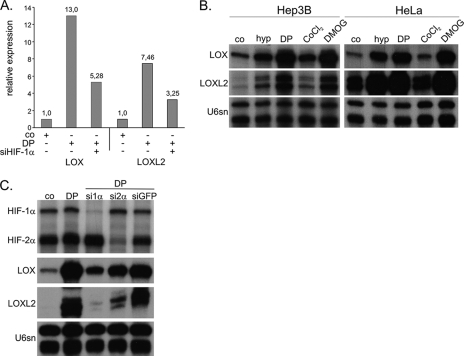
Because both VHL and hypoxia are dominant regulators of the transcription factor HIF, we were interested in identifying putative HIF target genes that could mediate cellular transformation by an Affymetrix gene array in Hep3B cells (39). This array identified two lysyl oxidases to be highly induced by HIF, LOX and a related protein, LOXL2 (LOX-like 2), which have been implicated in regulation of E-cadherin (see Introduction and Ref. 10). LOX and LOXL2 were induced 13- and 7.5-fold, respectively, in comparison with the untreated control. Importantly, HIF-1α siRNA reduced the expression of both lysyl oxidases significantly (Fig. 2A). We next aimed for verification of the array data by the specific and quantitative RNase protection assay for both genes. Samples from Hep3B and HeLa cells confirmed the regulation (Fig. 2B). Exposure of the cells to hypoxia (1% O2) or treatment with the hypoxia mimetic cobalt chloride (CoCl2), the prolylhydroxylase inhibitor dimethyloxaloglycine, and the iron chelator DP profoundly induced LOX and LOXL2 mRNA, where CoCl2 was the weakest stimulus. To investigate the dependence of the induction of LOX and LOXL2 expression on either HIF-α isoform, siRNA knockdown against HIF-1α and HIF-2α was performed with previously characterized siRNAs (39) (see Fig. 2C and supplemental Fig. 1 for independent siRNAs against HIF-1α and HIF-2α). Stimulated Hep3B cells transfected with HIF-1α siRNA markedly reduced LOX and LOXL2 mRNA expression in comparison with cells treated with siRNA against green fluorescent protein and untransfected cells. However, a slight reduction of both genes was also observed after HIF-2α knockdown but was rather low in comparison with the effect of HIF-1α knockdown.
FIGURE 2.
LOX and LOXL2 mRNA are induced under hypoxia and HIFα stabilizing conditions. A, Affymetrix gene array was performed with mRNA of normoxic or the iron chelator DP (100 μm, 16 h) stimulated Hep3B cells. B, LOX and LOXL2 mRNA expression was analyzed by RNase protection assay in Hep3B and HeLa cells after 16 h of hypoxic conditions or treatment with hypoxia mimetics (DP, 100 μm; CoCl2, 100 μm; dimethyloxaloylglycine (DMOG), 1 mm). Assays were internally controlled by U6 small nuclear RNA (U6sn). C, HIF dependence of LOX and LOXL2 expression was analyzed by knockdown of HIF-1α or HIF-2α under DP treatment in Hep3B cells by RNase protection. All samples were treated with transfection reagent. Control was treated with siRNA against green fluorescent protein (siGFP).
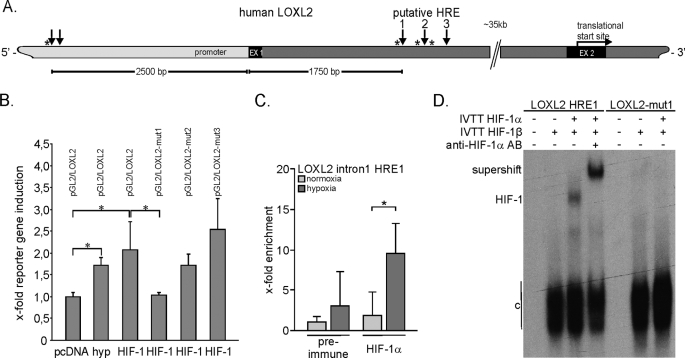
In order to further characterize the HIF-dependent transcriptional regulation of the lysyl oxidases, we searched the genomic sequences for putative hypoxia response elements (HREs; 5′-RCGTG-3′) (47) in silico, using the Genomatix MatInspector® software. While this work was in progress, Erler et al. (40) demonstrated a functional HIF binding site in the LOX gene, which corresponded well with our findings (as shown in supplemental Fig. 2). We therefore decided to focus our efforts on the regulation of the LOXL2 gene. In the genomic sequence of the LOXL2 gene, two separate regions were found, with two and three putative HREs in close proximity to each other (Fig. 3A). One region was located 2500 bp upstream of the transcriptional start site, containing two possible HIF binding sites. Three further putative response elements were found 1750 bp downstream of the transcriptional start site, in the first intron of the LOXL2 gene. Further analysis revealed that there were three HIF ancillary sequences (marked with asterisks in Fig. 3A) adjacent to these putative HREs, which were previously characterized to frequently lie in close proximity to functional HREs (such as erythropoietin, 5′-CACAG-3′ (48)). The more 5′ region displayed only one HIF ancillary sequence, close to the first putative HRE. Due to the lesser distance from the intronic HREs to the transcriptional start site and the appearance of the three HIF ancillary sequences close to the HRE consensus sequences, a 500-bp fragment containing these three HREs was cloned into a luciferase reporter vector. 16 h of hypoxic treatment induced luciferase activity up to 1.7-fold and HIF-1 overexpression up to 2.1-fold (Fig. 3B). Site-directed mutagenesis of each putative HRE sequence revealed the first HRE to be functional. Mutation of the putative HRE1 abolished inducibility, whereas mutation of sites 2 and 3 did not disable the reporter stimulation. To validate HIF-1 binding to HRE1 of LOXL2, chromatin immunoprecipitation with a HIF-1α antibody was performed. After hypoxic stimulation of HeLa cells, a 3.1-fold enrichment of HIF-1α binding to the LOXL2-HRE1 was detected, as compared with the normoxic control (Fig. 3C). A comparable result was observed in Hep3B cells after dimethyloxaloylglycine treatment (2 mm, 16 h; data not shown). To further validate the HIF regulation of the HRE1, electrophoretic mobility shift assays were performed (Fig. 3D). Using the combination of recombinant HIF-1α and HIF-1β proteins, generated by in vitro transcription and translation, we were able to show that HIF-1 could directly bind to the human LOXL2-HRE1 wild type oligonucleotide. Furthermore, the addition of a specific monoclonal antibody to HIF-1α further retarded the migration of the HIF-1-DNA complex (supershift). However, no binding of HIF-1 was present on a mutated oligonucleotide for the HRE1. In conclusion, these data together with the previously published date (40) clearly demonstrate that LOX and LOXL2 are direct transcriptional targets of HIF-1.
FIGURE 3.
LOXL2 is a direct transcriptional target of HIF-1. A, scheme of the partial genomic structure of the human LOXL2 gene indicating putative HREs in the vicinity of the transcriptional start site and the first intron (light gray, uncoding region; black (EX), exons; gray, introns; arrow indicates position of putative HREs; asterisk indicates ancillary sequence). The region with the three putative HREs located in the first intron was subcloned into a luciferase reporter vector (see below). B, LOXL2 reporter assays were performed in transiently transfected HEK 293 cells (pGL2/LOXL2). Luciferase activity was measured after hypoxic stimulation (1% O2) or transient HIF-1 overexpression with HIF-1α and HIF-1β expression plasmids (mean values of three independent experiments with the error bars representing S.D.; *, p < 0.05). For identification of the functional HRE, site-directed mutagenesis of each putative HRE (pGL2/LOXL2-mut1–3) was performed. Results represent mean values of three independent experiments, with the error bars representing S.D. C, for verification of HIF-1 binding activity on the HRE1 of LOXL2 after hypoxic stimulation (1% O2, 16 h) of HeLa cells, chromatin immunoprecipitation was performed using an HIF-1α antibody. HIF-1 enrichment was measured by quantitative PCR. Results show one representative of two independent experiments, depicted with mean values of quantitative PCR triplicate measurements, with S.D. *, p < 0.05. D, for validation of HIF-1 binding activity on the HRE1 of LOXL2, an electromobility shift assay was performed, using recombinant HIF-1α and HIFβ proteins generated by in vitro transcription and translation (IVTT) and the addition of a HIF-1α-specific antibody (AB) for retardation of the HIF-1α/HIFβ/LOXL2-HRE1 complex (supershift). c, constitutive binding activity.
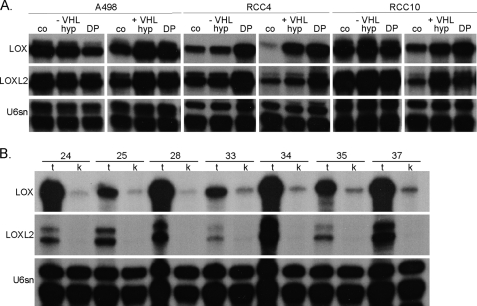
A clear HIF-dependent regulation should result in overexpression of target genes in cells lacking functional VHL. We therefore analyzed the expression of both lysyl oxidases in cells derived from RCCs deficient for VHL in comparison with their reconstituted clones by RNase protection assay (Fig. 4A). All three VHL-deficient cell lines, A498, RCC4, and RCC10, showed a high basal expression of both genes under normoxic control conditions, where neither hypoxia nor DP treatment considerably enhanced the expression. In the VHL-positive counterparts of these cell lines, normoxic expression of both lysyl oxidases was significantly repressed, showing strong inducibility by hypoxia or DP treatment. In addition to these in vitro data, we also investigated LOX and LOXL2 expression in vivo by analysis of tissues obtained from radical tumor nephrectomies and which have been characterized for their HIF status and target gene expression before (49). All of the samples investigated in this study were of clear cell histology and showed strong and global overexpression of HIFα isoforms and thus most likely were VHL-deficient (data not shown). RNA lysates from renal carcinomas in comparison with adjacent kidney tissues of the same patients were analyzed by an RNase protection assay (Fig. 4B). A low mRNA expression of LOX could be detected in every single kidney lysate, whereas profound up-regulation was detected in all analyzed tumor samples. In contrast, LOXL2 expression could not be detected in kidney extracts, but it was also highly expressed in the tumor tissues.
FIGURE 4.
LOX and LOXL2 regulation and expression is VHL-dependent. A, LOX and LOXL2 mRNA expression was analyzed by RNase protection assay in von Hippel Lindau-deficient (−VHL) and reconstituted (+VHL) renal cell carcinoma cell lines (A498, RCC4, and RCC10) after hypoxia or DP treatment (co, control/normoxia; hyp, hypoxia, 1% O2, 16 h; DP, 100 μm, 16 h). B, LOX and LOXL2 expression was analyzed in samples of radical nephrectomies, from patients with renal clear cell carcinomas with loss of VHL (14) (representative samples of 10 analyzed patients; t, tumor; k, kidney; numbers indicate individual patients).
E-cadherin Expression Is Lost under Hypoxic Conditions, Which Is Mediated by LOX/LOXL2
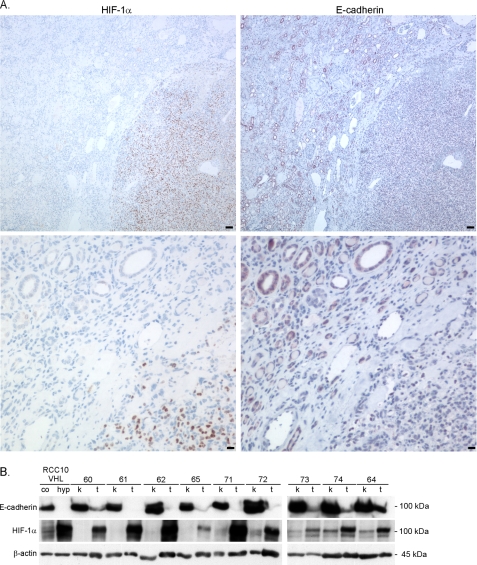
We next aimed to investigate the functional consequences that HIF and lysyl oxidase overexpression could have. Because HIF activity is negatively correlated with E-cadherin expression in cell cultures (Fig. 1), we studied the correlation of both proteins in RCC and kidney. Fig. 5A shows immunostaining for HIF-1α (left panels) and E-cadherin (right panels) on consecutive sections of human radical nephrectomy samples, where the border of tumor (lower right corners) with adjacent kidney tissue is displayed. In the kidney, E-cadherin is highly expressed in distal tubuli (50), whereas little expression can be detected in the adjacent tumor tissue. In contrast, no expression of HIF-1α can be detected in the kidney, whereas very strong and global expression is readily seen in the tumor tissue. Furthermore, we analyzed the E-cadherin and HIF-1α expression in protein lysates from renal carcinomas in comparison with adjacent kidney tissues from the same patient by immunoblotting (Fig. 5B). A broad E-cadherin protein expression was detected in all kidney samples, whereas the E-cadherin expression was strongly reduced in the tumor samples. In contrast, HIF-1α protein expression was clearly detectable in the tumor extracts, whereas no expression or only weak HIF-1α expression was detected in normal kidney extracts. These in vivo data are compatible with our expectation that strong activation of HIF would lead to a decrease of E-cadherin, possibly through activation of lysyl oxidases.
FIGURE 5.
E-cadherin expression in renal clear cell carcinomas is inversely associated with HIF-1 expression. A, serial sections of the border between renal tissue (upper left) to cancer (lower right corners) of samples from nephrectomies were analyzed by immunohistological staining for HIF-1α and E-cadherin expression. Bar, 100 μm (upper panels) and 10 μm (lower panels). B, E-cadherin and HIF-1α protein expression was analyzed in samples of radical nephrectomies, from nine patients with renal clear cell carcinomas with loss of VHL (14) (t, tumor; k, kidney; numbers indicate individual patients; co, control/normoxia; hyp, hypoxia, 1% O2, 72 h).
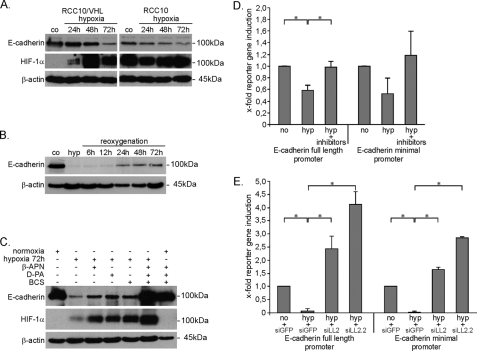
Pursuing this hypothesis, we investigated whether lysyl oxidases are necessary to suppress E-cadherin in hypoxia. As reported previously by others (34, 35, 51), prolonged hypoxia up to 72 h led to a strong decrease of E-cadherin protein in the presence of functional VHL (Fig. 6, A–C). In contrast, VHL-deficient cells show lower base-line E-cadherin expression and only marginal decreases of E-cadherin protein under hypoxia. Of note, protein levels of E-cadherin in the VHL-deficient cells were higher as measured by immunoblotting than could have been expected from immunofluorescence, where no detectable signal could be seen (compare Fig. 6A with Fig. 1). We were concerned that the repression of E-cadherin could partially be caused by cellular damage in hypoxia. However, light microscopy did not reveal obvious changes to the cellular morphology and confirmed confluent cell layers at the end of the experiments (data not shown). Furthermore, the high levels of HIF-1α protein in these cultures even after 72 h of continuous hypoxia implicate vital cells. Nevertheless, to ensure vitality of our cultures and investigate whether E-cadherin regulation is a dynamic cellular response, we tested whether E-cadherin expression could be restored, after 72 h of hypoxic incubation. Again, E-cadherin was strongly repressed in RCC10/VHL cells after 72 h of hypoxia (Fig. 6B). However, E-cadherin protein could be restored, which started at 24 h of reoxygenation and confirmed the vitality of our cultures.
FIGURE 6.
Hypoxia leads to loss of E-cadherin expression, which requires LOX/LOXL2. A, E-cadherin expression was analyzed by immunoblotting in RCC10 cells after 72 h of hypoxia (1% O2). HIF-1α was analyzed in parallel to verify hypoxic induction. co, normoxic control. B, in RCC10/VHL cells E-cadherin expression was analyzed after reoxygenation, from 72 h of hypoxia. C, E-cadherin expression was analyzed in RCC10/VHL cells exposed to hypoxia for 72 h with or without treatment with pharmacological inhibitors of lysyl oxidases by immunoblotting. Hypoxia, 1% O2. For β-APN, d-penicillamine, and BCS, a 300 μm concentration of one was used or 100 μm each in combination. D and E, HKC-8 human renal tubular cells were transiently transfected with luciferase reporter constructs containing either the full-length E-cadherin promoter or a minimal promoter fragment containing two E-boxes of E-cadherin. D, 24 h after transfection, cells were stimulated with hypoxia (hyp), with or without the addition of lysyl oxidase inhibitors for a further 48 h (β-APN, d-penicillamine (d-PA), and BCS, each 100 μm). E, the day before reporter transfection, knockdown for LOXL2 with the two independent siRNAs siLOXL2 (siLL2) and siLOXL2.2 (siLL2.2) was performed. Controls were treated with siRNA against green fluorescent protein. Cells were exposed to hypoxia 24 h after reporter transfection. Results in D and E represent mean values of two independent experiments, with the error bars representing S.D. *, p < 0.05.
We next aimed to interfere with the function of lysyl oxidases to address the question of lysyl oxidase involvement in hypoxic E-cadherin down-regulation. Unfortunately, the experimental requirements of the procedures, which are confluent cultures at the start of the experiment and a duration of at least 72 h of hypoxic incubation, excluded siRNA from analysis of endogenous E-cadherin because of low transfection efficiencies of the siRNA. For this reason, we referred to treatment with established pharmacological inhibitors of lysyl oxidases (40, 52) (Fig. 6, C and D). β-APN, d-penicillamine, and BCS could partially inhibit hypoxic repression of E-cadherin (compare lane 2 with lanes 3–5). However, only the mixture of all three inhibitors could completely restore E-cadherin expression to the same extent as normoxic control, but it had no influence on the basal E-cadherin expression under normoxic conditions. As stated, the possibilities to study and modify endogenous E-cadherin in cell culture were restricted because of the requirement of confluent cultures. However, E-cadherin promoter constructs gave us the possibility to experimentally interfere with and measure E-cadherin regulation in cell cultures that could be more easily treated by transfection. For this purpose, HKC-8 proximal renal tubular cells, which show excellent transfection efficacies, were transfected with the full-length E-cadherin promoter and a minimal promoter fragment, containing only E-box binding sites for E-cadherin repressor proteins. Hypoxic incubation decreased the reporter gene activity of both E-cadherin promoter constructs to ∼50%. Using the combination of the three inhibitors to block lysyl oxidase function, complete restoration of the reporter gene activity could be achieved (Fig. 6D). Under these experimental conditions, we were able to study the effects of LOX and LOXL2 siRNA, which worked at satisfactory efficiency, as demonstrated in RNase protection assays (supplemental Fig. 3). Knockdown of LOXL2 was consistently able to restore reporter gene activity to levels of normoxic activity (Fig. 6E). However, LOX knockdown produced weaker and more variable results (data not shown).
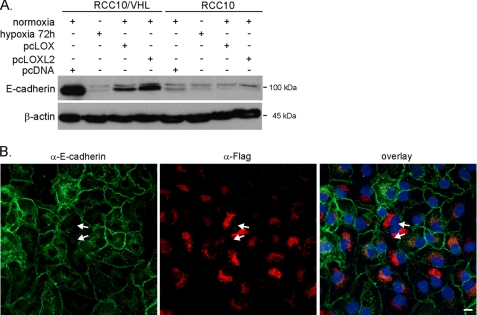
These data clearly show that lysyl oxidases are involved in E-cadherin regulation under hypoxia. To further investigate whether the lysyl oxidases have the ability to regulate E-cadherin on their own, we referred to overexpression. With the reagent FuGene HD®, we were able to transfect a sufficient number of cells to high levels in order to study functional consequences. Forced expression of either LOX or LOXL2 reduced E-cadherin levels in RCC10/VHL cells and marginally in RCC10 cells (Fig. 7A), where already high levels of constitutive lysyl oxidases exist. However, the level of E-cadherin repression was below the hypoxic effect, which may be the result of insufficient transfection efficacy. We therefore next looked at the effect of LOXL2 overexpression on single cell basis by immunofluorescence, where the transfected protein could be visualized by an antibody against the FLAG tag. Individual RCC/VHL cells that displayed overexpressed FLAG-tagged LOXL2 (Fig. 7B, red) showed a markedly reduced E-cadherin (green) expression. These cells lose E-cadherin at their site of contact. Interestingly, the LOXL2 transgene shows a distinct perinuclear expression pattern, which has been reported before (41).
FIGURE 7.
Ectopic expression of LOX and LOXL2 alone reduces E-cadherin expression in RCC10/VHL cells. A, RCC10/VHL cells were transiently transfected with expression plasmids bearing either LOX (pcLOX) or LOXL2 (pcLOXL2) cDNA 72 h prior to harvesting the cells and analyzed by immunoblotting. Equimolar amounts of empty vector (pcDNA) were transfected as a control. B, transient transfection of LOXL2 cDNA, which is tagged with a FLAG tag. Transfection was purposely performed less efficiently than in A, in order to be able to compare negative cells with positively transfected cells. Cells were fixed 48 h after transfection and analyzed by immunofluorescence for E-cadherin, LOXL2-FLAG tag, and DAPI nuclear stain. The arrows indicate adjacent RCC10/VHL cells, which have lost E-cadherin at their site of contact. Green, α-E-cadherin; red, α-FLAG staining; blue, DAPI; bar, 10 μm.
Modulation of Cellular Invasiveness by Hypoxia and VHL, Which Is Dependent on Lysyl Oxidases
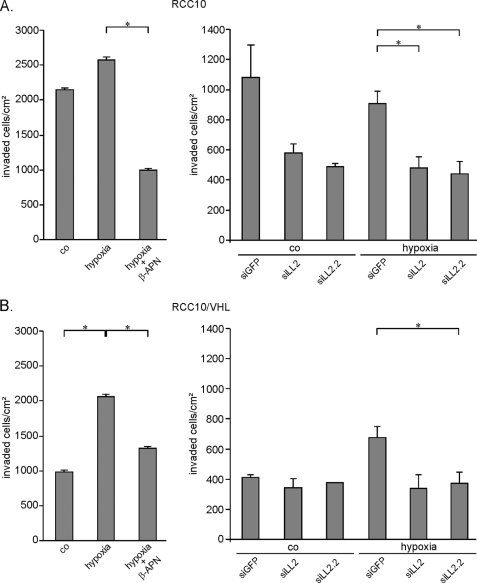
Given the hypoxic reduction of E-cadherin as one main feature of the transitional process shifting an epithelial cell to a motile and invasive phenotype, we were interested in whether hypoxic effects would be reflected in an in vitro invasion model and if lysyl oxidases play a role in these effects. To address this, we analyzed the invasive potential of RCC10 (Fig. 8A) and RCC10/VHL (Fig. 8B) cells into collagen I gels under normoxic and hypoxic conditions. RCC10 cells strongly invaded into the collagen matrix, whereas RCC10/VHL cells were nearly non-invasive under normoxia. Invasiveness of RCC10/VHL cells could be strongly induced by hypoxia up to the same level as normoxic VHL-deficient RCC10 cells. Also, the invasion of the RCC10 cells could be further promoted by hypoxia but to a much lesser degree than it was in RCC10/VHL cells. Treatment of the RCC10/VHL cells with the lysyl oxidase inhibitor β-APN significantly reduced the invasiveness of these cells under hypoxia in both cell lines (Fig. 8, A and B, left). The results were comparable at two different time points assayed, 1 day (data not shown) and 3 days after seeding. The knockdown of LOXL2 was able to reduce the invasiveness of these cells under hypoxia in both cell lines (Fig. 8, A and B, right). Furthermore, lysyl oxidase knockdown equally reduced the invasiveness of VHL-deficient RCC10 cell under normoxic conditions (data not shown). These results implicate that lysyl oxidases, under the influence of hypoxia, have the ability to influence the invasive potential of cells, a phenomenon that could be of relevance in metastasis of human cancers.
FIGURE 8.
Invasiveness of RCC cells in collagen gels is influenced by hypoxia and is partly LOX/LOXL2-dependent. The invasive potential of VHL-deficient (A) and VHL reconstituted (B) RCC10 cells into collagen I gels was analyzed under normoxic and hypoxic conditions. Invaded cells were counted after 3 days of seeding. Lysyl oxidase activity was blocked by treatment with the lysyl oxidase inhibitor β-APN under hypoxia (A and B, left-hand panels; co, normoxic control; hypoxia, 1% O2; β-APN, 300 μm; one representative experiment of three independent experiments, depicted with S.D.; *, p < 0.05). Additionally, knockdown for LOXL2 was performed with the two independent siRNAs siLOXL2 (siLL2) and siLOXL2.2 (siLL2.2) under normoxia and hypoxia (A and B, right-hand panels). Results are mean values of two independent experiments with the error bars representing S.D.; *, p < 0.05).
DISCUSSION
Based on the well known clinical context that hypoxic tumors show an aggressive phenotype and are less responsive to classical therapy strategies, we were interested in putative underlying mechanisms that could mediate these effects. We and others have identified two lysyl oxidases in gene arrays searching for hypoxic targets, which have been previously implicated in tumor progression and metastasis. We show here that the lysyl oxidases LOX and LOXL2 are highly regulated by hypoxia and HIF, are strongly expressed in RCC, and are responsible for E-cadherin repression, which is a hallmark of EMT. These data are compatible with an important role of lysyl oxidases in tumor progression and raise the question of whether inhibition of lysyl oxidases could be a potent strategy for tumor control.
A number of studies have shown that HIF has the potential of inducing EMT (53–57), but the detailed mechanism leading from hypoxia and HIF stabilization to EMT remains incompletely understood. Our study is the first to show that LOXL2 is a direct target of HIF-1, where the same has previously been shown for LOX (40). Our studies confirm and extend the work of previous groups in that E-cadherin negatively correlates with VHL function of a cell and that hypoxia is able to repress E-cadherin in VHL-competent cells (Figs. 1 and 6). Furthermore, we show for the first time that LOX and LOXL2 are functionally necessary and sufficient for the hypoxic down-regulation of E-cadherin (Figs. 6 and 7), which has merely been a hypothesis so far. Our data demonstrate a functional role of lysyl oxidases for E-cadherin repression utilizing inhibitory strategies (pharmacological and knockdown) as well as overexpression in two different settings. These experiments are complementary and do indicate a role of lysyl oxidases in E-cadherin expression. However, the detailed molecular mechanism of lysyl oxidase-mediated hypoxic E-cadherin repression remains unclear. A transcriptional regulation has been suspected (34) and would be compatible with our results of the E-cadherin reporter assays (Fig. 6). Transcriptional repression of E-cadherin is believed to be largely mediated by binding of repressors to E-box elements in the E-cadherin promoter. Because the reporter plasmid containing the minimal E-cadherin promoter (only the E-cadherin E-boxes) displayed a similar repression under hypoxia as the full-length promoter (Fig. 6), these findings are in agreement with dominant regulation by such transcriptional repressors. Snail was described as such a direct and potent E-cadherin repressor in epithelial cells and could induce EMT and migration/invasion (58, 59). Furthermore, Snail was reported to be a direct binding partner to the catalytic domain of LOX and LOXL2. Thus, it could have been a good candidate in this pathway (41). Although the classical functions of lysyl oxidases focus on ECM stabilization, other functions have been reported. Interestingly, LOX-dependent alterations in chromatin structure have been observed, and LOX could be partially localized in the nucleus (60–63). An involvement in transcriptional regulation of the collagen IIIα1 gene has been demonstrated, by binding of LOX to the Ku antigen (64). Peinado et al. (41) suggested a model for the LOXL2-dependent regulation of Snail, in which the oxidation of Snail at Lys-198 and/or Lys-137 by LOXL2 could lead to intramolecular linkage, inducing conformational changes and thereby influencing Snail function or Snail sensitivity to other regulating interaction partners. In RCC10 cells, we were not able to observe hypoxic induction of Snail; nor did knockdown with Snail siRNA produce reproducible effects in an E-cadherin reporter assay (data not shown). A large number of other transcriptional repressors of E-cadherin have been shown to be regulated by VHL, hypoxia, and/or HIF: SIP1 (also known as ZFHX1B and ZEB2), ZEB1 (also known as ZFHX1A), Twist, and TCF3 (also known as E47) (33, 35, 51, 55). Intriguingly, Twist has been shown recently to be a direct transcriptional target of HIF (55, 65), which could indicate a direct molecular link. Importantly, a recent study has shown that the intracellular domain of Notch may be involved in LOX-dependent effects on hypoxia-induced EMT and invasion, adding a further regulating factor to the list (56). Considering the large number of putatively involved proteins, this pathway is inclined to be biologically important. However, it is not known which, if any, of these repressors exert dominant functions over others, which may well be cell type- and context-dependent. Therefore, a better description of the molecular mechanism leading from lysyl oxidases on to E-cadherin repression will be required in future studies.
In addition to mediating E-cadherin repression and thus initiating the EMT program, as demonstrated in our study, LOX has been shown to activate the Src kinase and the focal adhesion kinase, which are both key proteins enabling cells to migrate by forming stable adhesions to the ECM (66). Interestingly, it has recently been shown that hypoxia strongly stimulates LOX gene expression, but enzymatic activity requires subsequent reoxygenation, which results in focal adhesion kinase/Src activation (67). The first study that elegantly showed that hypoxia-induced LOX has the potential to promote experimental invasion and metastasis was by Erler et al. (40). The same study demonstrated that LOX expression significantly correlates with clinical outcome in breast and head and neck cancer. LOX and its related proteins have previously been shown to be overexpressed in a large number of human tumors, shown by numerous investigators (reviewed in Ref. 42). Recently, LOXL2 has been demonstrated to promote migration of breast cancer cells (68). Together with the clinical observations that hypoxic tumors show an adverse clinical outcome (reviewed in the Introduction), extensive data have been gathered showing that hypoxia has the potential to promote metastasis in experimental models (69). Whether these processes are partly or largely dependent on the function of lysyl oxidases will have to be revisited. In this context, it is of particular interest that pharmacological inhibition of lysyl oxidases results in diminished metastatic potential in experimental models (70).
Most of the data concerning expression and function of lysyl oxidases exist for LOX. In our study, we show, that the related protein LOXL2 is also highly expressed in human RCC (Fig. 4) and, similarly to LOX, has the ability to repress E-cadherin on its own (Fig. 7). Considering that the C-terminal catalytic domain of all lysyl oxidases is highly conserved, similarities in their biological function could be suspected. However, gene regulation of the different lysyl oxidases may differ, since the Affymetrix array that we undertook to screen for HIF-regulated genes did not yield any lysyl oxidase other than LOX and LOXL2, although LOXL, LOXL3, and LOXL4 were contained in the array. It can therefore be assumed that the described effects in hypoxia are unique to LOX and LOXL2. It is of particular interest that the inhibitor β-APN has been shown not to influence LOXL2 function (71). If true, the effects observed for E-cadherin repression under β-APN (Figs. 6C and 8) would be solely confined to LOX.
Considering the profound effects that the lysyl oxidases are suspected to have for tumorigenesis and disease progression, these enzymes should be considered as therapeutic targets. A molecular target is particularly attractive if extracellular pharmacological approaches can be used. Although the function and effectors of lysyl oxidases that were of interest in our study would be suspected to be intracellular, extracellular targeting of the enzyme would still be possible. LOX is synthesized as a 46-kDa preproprotein, which is cleaved and glycosylated intracellularly. The 50-kDa large pro-LOX is then secreted into the extracellular space, where it is activated by cleavage by the bone morphogenic protein-1 to the active 32-kDa protein. The active protein is then able to reenter the cell, which would then exert its effects on intracellular substrates (for a review, see Refs. 42 and 43). Thus, during the extracellular cycle, targeting LOX could be feasible with the appropriate tools. Because BCS cannot be taken up into the cell and thus merely takes effect in the extracellular space (40, 54), partial E-cadherin restoration in hypoxia by BCS alone (Fig. 6C) may affirm this theory.
Considering inhibition of lysyl oxidases as a therapeutic strategy to prevent tumor progression and metastasis, clear cell RCC may well turn out to be the most effective tumor to do so. First, the majority of clear cell RCCs have a constitutive activation of the HIF pathway because of biallelic inactivation of the tumor suppressor VHL (as reviewed in the Introduction). This leads to particularly strong activation of target genes, such as LOX and LOXL2, which is not only dependent on the microenvironment but is genetically caused by the VHL mutation. Second, high levels of lysyl oxidase expression in well oxygenated cells may be specifically relevant for these enzymes because it has been shown that lysyl oxidases require oxygen to be fully active (67). Third, RCCs are clinically aggressive and not responsive to classical tumor therapy. The prognosis of these patients is usually not determined by tumor growth at the primary site but by the extent of metastasis, which is frequently present already at the time of diagnosis (24). Therefore, early up-regulation and activation of lysyl oxidases are likely to contribute to the bad prognosis of RCC. Furthermore, since increasing evidence has reported an important involvement of the enzymes in the process of metastasis, this may well be particularly relevant in RCC. In summary, lysyl oxidases could qualify as an interesting anti-tumor target. We show that the hypoxic E-cadherin repression, as an indicator of EMT, is dependent on lysyl oxidases and that a protein related to the previously demonstrated LOX is also competent in mediating these effects, LOXL2. Based on our results, we therefore postulate that any strategy designed to target lysyl oxidase activity in tumors will have to include both enzymes, LOX and LOXL2, to be effective.
Supplementary Material
Acknowledgments
E-cadherin full-length promoter and the E-cadherin minimal promoter constructs were a kind gift from G. Berx (University Gent, Belgium). pcDNA-hLOXL2-FLAG was provided by H. Peinado (University of Madrid, Spain), and pTRE2hyg-LOX was provided by T. Ushijiama (National Cancer Center, Tokyo, Japan) and subcloned into pcDNA3. Excellent technical assistance of M. Reutelshoefer and Tina Herter-Kermann is greatly acknowledged.
This work was supported by grants from the Interdisciplinary Centre for Clinical Research (to M. W. and J. B.), the European Commission Sixth Framework Programme (Euroxy) (to K. U. E.), the Wellcome Trust (to J. S.), and the German Research Foundation (SFB 423) (to M. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- HIF
- hypoxia-inducible transcription factor
- HRE
- hypoxia response element
- RCC
- renal cell carcinoma
- EMT
- epithelial to mesenchymal transition
- ECM
- extracellular matrix
- DP
- 2,2′-dipyridyl
- β-APN
- β-aminopropionitrile
- BCS
- bathocuproinedisulfonic acid
- siRNA
- small interfering RNA
- DAPI
- 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Semenza G. L. (2001) Cell 107, 1–3 [DOI] [PubMed] [Google Scholar]
- 2.Maxwell P. H., Ratcliffe P. J. (2002) Semin. Cell Dev. Biol. 13, 29–37 [DOI] [PubMed] [Google Scholar]
- 3.Wenger R. H. (2002) FASEB J. 16, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 4.O'Rourke J. F., Tian Y. M., Ratcliffe P. J., Pugh C. W. (1999) J. Biol. Chem. 274, 2060–2071 [DOI] [PubMed] [Google Scholar]
- 5.Semenza G. L. (2003) Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 6.Harris A. L. (2002) Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 7.Warburg O., Wind F., Negelein E. (1927) J. Gen. Physiol. 8, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seagroves T. N., Ryan H. E., Lu H., Wouters B. G., Knapp M., Thibault P., Laderoute K., Johnson R. S. (2001) Mol. Cell Biol. 21, 3436–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza G. L. (2007) J. Bioenerg. Biomembr. 39, 231–234 [DOI] [PubMed] [Google Scholar]
- 10.Pouysségur J., Dayan F., Mazure N. M. (2006) Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 11.Pugh C. W., Ratcliffe P. J. (2003) Semin. Cancer Biol. 13, 83–89 [DOI] [PubMed] [Google Scholar]
- 12.Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 13.Talks K. L., Turley H., Gatter K. C., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Am. J. Pathol. 157, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesener M. S., Münchenhagen P. M., Berger I., Morgan N. V., Roigas J., Schwiertz A., Jürgensen J. S., Gruber G., Maxwell P. H., Löning S. A., Frei U., Maher E. R., Gröne H. J., Eckardt K. U. (2001) Cancer Res. 61, 5215–5222 [PubMed] [Google Scholar]
- 15.Turner K. J., Moore J. W., Jones A., Taylor C. F., Cuthbert-Heavens D., Han C., Leek R. D., Gatter K. C., Maxwell P. H., Ratcliffe P. J., Cranston D., Harris A. L. (2002) Cancer Res. 62, 2957–2961 [PubMed] [Google Scholar]
- 16.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 17.Kaelin W. G., Jr. (2007) Clin. Cancer Res. 13, 680s–684s [DOI] [PubMed] [Google Scholar]
- 18.Mandriota S. J., Turner K. J., Davies D. R., Murray P. G., Morgan N. V., Sowter H. M., Wykoff C. C., Maher E. R., Harris A. L., Ratcliffe P. J., Maxwell P. H. (2002) Cancer Cell 1, 459–468 [DOI] [PubMed] [Google Scholar]
- 19.Maranchie J. K., Vasselli J. R., Riss J., Bonifacino J. S., Linehan W. M., Klausner R. D. (2002) Cancer Cell 1, 247–255 [DOI] [PubMed] [Google Scholar]
- 20.Kondo K., Klco J., Nakamura E., Lechpammer M., Kaelin W. G., Jr. (2002) Cancer Cell 1, 237–246 [DOI] [PubMed] [Google Scholar]
- 21.Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G., Jr. (2003) PLoS Biol. 1, E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer M., Doucette D., Siddiqui N., Iliopoulos O. (2004) Mol. Cancer Res. 2, 89–95 [PubMed] [Google Scholar]
- 23.Raval R. R., Lau K. W., Tran M. G., Sowter H. M., Mandriota S. J., Li J. L., Pugh C. W., Maxwell P. H., Harris A. L., Ratcliffe P. J. (2005) Mol. Cell Biol. 25, 5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen R. B. (2005) in Cancer and the Kidney (Cohen E. P. ed) pp. 201–226, Oxford University Press, Oxford, UK [Google Scholar]
- 25.Sullivan R., Graham C. H. (2007) Cancer Metastasis Rev. 26, 319–331 [DOI] [PubMed] [Google Scholar]
- 26.Axelson H., Fredlund E., Ovenberger M., Landberg G., Påhlman S. (2005) Semin. Cell Dev. Biol. 16, 554–563 [DOI] [PubMed] [Google Scholar]
- 27.Vaupel P., Mayer A. (2007) Cancer Metastasis Rev. 26, 225–239 [DOI] [PubMed] [Google Scholar]
- 28.Brown J. M., Giaccia A. J. (1998) Cancer Res. 58, 1408–1416 [PubMed] [Google Scholar]
- 29.Thiery J. P. (2002) Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 30.Grünert S., Jechlinger M., Beug H. (2003) Nat. Rev. Mol. Cell Biol. 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 31.Behrens J., Mareel M. M., Van Roy F. M., Birchmeier W. (1989) J. Cell Biol. 108, 2435–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peinado H., Olmeda D., Cano A. (2007) Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 33.Imai T., Horiuchi A., Wang C., Oka K., Ohira S., Nikaido T., Konishi I. (2003) Am. J. Pathol. 163, 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteban M. A., Tran M. G., Harten S. K., Hill P., Castellanos M. C., Chandra A., Raval R., O'brien T. S., Maxwell P. H. (2006) Cancer Res. 66, 3567–3575 [DOI] [PubMed] [Google Scholar]
- 35.Krishnamachary B., Zagzag D., Nagasawa H., Rainey K., Okuyama H., Baek J. H., Semenza G. L. (2006) Cancer Res. 66, 2725–2731 [DOI] [PubMed] [Google Scholar]
- 36.Beavon I. R. (1999) Mol. Pathol. 52, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang V., Davis D. A., Haque M., Huang L. E., Yarchoan R. (2005) Cancer Res. 65, 3299–3306 [DOI] [PubMed] [Google Scholar]
- 38.Elvidge G. P., Glenny L., Appelhoff R. J., Ratcliffe P. J., Ragoussis J., Gleadle J. M. (2006) J. Biol. Chem. 281, 15215–15226 [DOI] [PubMed] [Google Scholar]
- 39.Warnecke C., Weidemann A., Volke M., Schietke R., Wu X., Knaup K. X., Hackenbeck T., Bernhardt W., Willam C., Eckardt K. U., Wiesener M. S. (2008) Exp. Cell Res. 314, 2016–2027 [DOI] [PubMed] [Google Scholar]
- 40.Erler J. T., Bennewith K. L., Nicolau M., Dornhöfer N., Kong C., Le Q. T., Chi J. T., Jeffrey S. S., Giaccia A. J. (2006) Nature 440, 1222–1226 [DOI] [PubMed] [Google Scholar]
- 41.Peinado H., Del Carmen Iglesias-de la Cruz M., Olmeda D., Csiszar K., Fong K. S., Vega S., Nieto M. A., Cano A., Portillo F. (2005) EMBO J. 24, 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Payne S. L., Hendrix M. J., Kirschmann D. A. (2007) J. Cell Biochem. 101, 1338–1354 [DOI] [PubMed] [Google Scholar]
- 43.Lucero H. A., Kagan H. M. (2006) Cell Mol. Life Sci. 63, 2304–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiesener M. S., Turley H., Allen W. E., Willam C., Eckardt K. U., Talks K. L., Wood S. M., Gatter K. C., Harris A. L., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (1998) Blood 92, 2260–2268 [PubMed] [Google Scholar]
- 45.Lau K. W., Tian Y. M., Raval R. R., Ratcliffe P. J., Pugh C. W. (2007) Br. J. Cancer 96, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krieg M., Haas R., Brauch H., Acker T., Flamme I., Plate K. H. (2000) Oncogene 19, 5435–5443 [DOI] [PubMed] [Google Scholar]
- 47.Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 48.Kimura H., Weisz A., Ogura T., Hitomi Y., Kurashima Y., Hashimoto K., D'Acquisto F., Makuuchi M., Esumi H. (2001) J. Biol. Chem. 276, 2292–2298 [DOI] [PubMed] [Google Scholar]
- 49.Wiesener M. S., Münchenhagen P., Gläser M., Sobottka B. A., Knaup K. X., Jozefowski K., Jürgensen J. S., Roigas J., Warnecke C., Gröne H. J., Maxwell P. H., Willam C., Eckardt K. U. (2007) Int. J. Cancer 121, 2434–2442 [DOI] [PubMed] [Google Scholar]
- 50.Nouwen E. J., Dauwe S., van der Biest I., De Broe M. E. (1993) Kidney Int. 44, 147–158 [DOI] [PubMed] [Google Scholar]
- 51.Evans A. J., Russell R. C., Roche O., Burry T. N., Fish J. E., Chow V. W., Kim W. Y., Saravanan A., Maynard M. A., Gervais M. L., Sufan R. I., Roberts A. M., Wilson L. A., Betten M., Vandewalle C., Berx G., Marsden P. A., Irwin M. S., Teh B. T., Jewett M. A., Ohh M. (2007) Mol. Cell Biol. 27, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasquali Ronchetti I., Fornieri C., Baccarani Contri M., Quaglino D., Jr., Caselgrandi E. (1986) Am. J. Pathol. 124, 436–447 [PMC free article] [PubMed] [Google Scholar]
- 53.Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., Semenza G. L. (2003) Cancer Res. 63, 1138–1143 [PubMed] [Google Scholar]
- 54.Higgins D. F., Kimura K., Bernhardt W. M., Shrimanker N., Akai Y., Hohenstein B., Saito Y., Johnson R. S., Kretzler M., Cohen C. D., Eckardt K. U., Iwano M., Haase V. H. (2007) J. Clin. Invest. 117, 3810–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M. H., Wu M. Z., Chiou S. H., Chen P. M., Chang S. Y., Liu C. J., Teng S. C., Wu K. J. (2008) Nat. Cell Biol. 10, 295–305 [DOI] [PubMed] [Google Scholar]
- 56.Sahlgren C., Gustafsson M. V., Jin S., Poellinger L., Lendahl U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6392–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun S., Ning X., Zhang Y., Lu Y., Nie Y., Han S., Liu L., Du R., Xia L., He L., Fan D. (2009) Kidney Int. 75, 1278–1287 [DOI] [PubMed] [Google Scholar]
- 58.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 59.Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 60.Mello M. L., Contente S., Vidal B. C., Planding W., Schenck U. (1995) Exp. Cell Res. 220, 374–382 [DOI] [PubMed] [Google Scholar]
- 61.Li W., Nellaiappan K., Strassmaier T., Graham L., Thomas K. M., Kagan H. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12817–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nellaiappan K., Risitano A., Liu G., Nicklas G., Kagan H. M. (2000) J. Cell Biochem. 79, 576–582 [DOI] [PubMed] [Google Scholar]
- 63.Jansen M. K., Csiszar K. (2007) Matrix Biol. 26, 136–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giampuzzi M., Botti G., Di Duca M., Arata L., Ghiggeri G., Gusmano R., Ravazzolo R., Di Donato A. (2000) J. Biol. Chem. 275, 36341–36349 [DOI] [PubMed] [Google Scholar]
- 65.Gort E. H., van Haaften G., Verlaan I., Groot A. J., Plasterk R. H., Shvarts A., Suijkerbuijk K. P., van Laar T., van der Wall E., Raman V., van Diest P. J., Tijsterman M., Vooijs M. (2008) Oncogene 27, 1501–1510 [DOI] [PubMed] [Google Scholar]
- 66.Payne S. L., Fogelgren B., Hess A. R., Seftor E. A., Wiley E. L., Fong S. F., Csiszar K., Hendrix M. J., Kirschmann D. A. (2005) Cancer Res. 65, 11429–11436 [DOI] [PubMed] [Google Scholar]
- 67.Postovit L. M., Abbott D. E., Payne S. L., Wheaton W. W., Margaryan N. V., Sullivan R., Jansen M. K., Csiszar K., Hendrix M. J., Kirschmann D. A. (2008) J. Cell Biochem. 103, 1369–1378 [DOI] [PubMed] [Google Scholar]
- 68.Hollosi P., Yakushiji J. K., Fong K. S., Csiszar K., Fong S. F. (2009) Int. J. Cancer 125, 318–327 [DOI] [PubMed] [Google Scholar]
- 69.Rofstad E. K. (2000) Int. J. Radiat. Biol. 76, 589–605 [DOI] [PubMed] [Google Scholar]
- 70.Bondareva A., Downey C. M., Ayres F., Liu W., Boyd S. K., Hallgrimsson B., Jirik F. R. (2009) PLoS One 4, e5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vadasz Z., Kessler O., Akiri G., Gengrinovitch S., Kagan H. M., Baruch Y., Izhak O. B., Neufeld G. (2005) J. Hepatol. 43, 499–507 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.