Abstract
The Notch signaling pathway is a cell-cell communication network giving rise to cell differentiation during metazoan development. Activation of the pathway releases the intracellular portion of the Notch receptor to translocate to the nucleus, where it is able to interact with the effector transcription factor CSL, converting CSL from a transcriptional repressor to an activator. This conversion is dependent upon the high affinity binding of the RAM region of the Notch receptor to the β-trefoil domain (BTD) of CSL. Here we probe the energetics of binding to BTD of each conserved residue of RAM through the use of isothermal titration calorimetry and single residue substitution. We find that although the highly conserved ΦWΦP motif is the largest determinant of binding, energetically significant interactions are contributed by N-terminal residues, including a conserved Arg/Lys-rich region. Additionally, we present a thermodynamic analysis of the interaction between the Epstein-Barr virus protein EBNA2 with BTD and explore the extent to which the EBNA2- and RAM-binding sites on BTD are nonoverlapping, as proposed by Fuchs et al. (Fuchs, K. P., Bommer, G., Dumont, E., Christoph, B., Vidal, M., Kremmer, E., and Kempkes, B. (2001) Eur. J. Biochem. 268, 4639–4646). Combining these results with displacement isothermal titration calorimetry, we propose a mechanism by which the ΦWΦP motif of RAM and EBNA2 compete with one another for binding at the hydrophobic pocket of BTD using overlapping but specific interactions that are unique to each BTD ligand.
Keywords: Gene Transcription, Notch Pathway, Peptide Interactions, Signal Transduction, Viral Protein, CSL, EBNA2, Epstein Barr, RAM, Isothermal Titration Calorimetry
Introduction
The Notch signaling pathway is a highly conserved network of interactions by which adjacent cells communicate, giving rise to cell-specific differentiation during embryogenesis and stem cell homeostasis in adulthood (1, 2). Both mutations and viral infection cause misactivation of Notch signaling, resulting in vasculature deformations, gross developmental defects, and numerous cancers (3–5).
The central component of the signaling pathway is the Notch receptor, a 300-kDa single-pass transmembrane receptor protein located at the cell surface. Binding of protein ligands from the DSL (Delta, Serrate, Lag2 for the mammalian, Drosophila melanogaster, and Caenorhabditis elegans orthologs, respectively) class to Notch family receptors on an adjacent cell causes ligand-activated proteolytic processing of the receptor. Following an initial cleavage of Notch by an ADAM (a disintegrin and metalloprotease) protease just outside the plasma membrane (6), a second cleavage by the γ-secretase complex within the transmembrane region of the receptor releases the Notch intracellular domain (NICD)2 from the plasma membrane (7). NICD then translocates to the nucleus, activating transcription of target genes. NICD is composed of an unstructured, membrane-proximal region denoted RAM (RBP-Jκ-associated molecule), followed by seven ankyrin repeats (ANK), a nuclear localization sequence, and a C-terminal PEST degradation sequence (8–11).
The major target of NICD, the transcription factor CSL (CBF1/RBP-Jκ, Su(H), Lag1) is believed to be bound to specific DNA sequences at the promoters of Notch-sensitive genes. Genetic, biochemical, and structural studies have elucidated two primary sites of interaction between CSL and NICD. High affinity binding occurs between the β-trefoil domain (BTD) of CSL and the N-terminal 25 residues of the RAM region. This interaction is centered on an absolutely conserved ΦWΦP motif, where Φ is any hydrophobic residue (9, 12, 13). The ΦWΦP motif interacts with a hydrophobic pocket on the surface of BTD, and mutation of the W and P renders the RAM region binding-incompetent (see Fig. 1A) (9, 12, 14). The C-terminal domain of CSL interacts with ANK, albeit with much lower affinity than the BTD·RAM interaction, and is instead stabilized by interaction with MAM (Mastermind), a third activating protein (10, 15, 16).
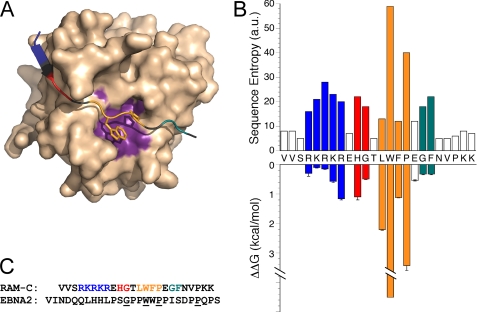
FIGURE 1.
Structure of the BTD·RAM complex. A, Molecular representation of BTD (wheat) in complex with the N terminus of RAM, with the four regions of conservation colored in blue (basic region), red (HG), orange (ΦWΦP motif), and teal (GF). The absolutely conserved Trp and Pro side chains are displayed as sticks. The ΦWΦP motif of RAM interacts with the hydrophobic pocket (purple) on BTD, composed of residues Phe210, Leu259, Leu262, Ile264, Leu298, and Ile305 (Protein Data Bank code 3brd). B, sequence of the RAM consensus peptide, along with the sequence entropy (upper bar plot) and the free energy change for single-residue alanine substitution from ITC measurements, color-coded as described above (lower bar plot; Table 1). For Trp14, the ΔΔG° estimate is an upper limit because we cannot detect binding. For Gly18 and Phe19, we partitioned the modest energy change for the doubly substituted G18A/F19A peptide evenly between the two positions. C, sequence alignment of RAM-C with CR6 of EBNA2. Only four residues of EBNA2 (underlined) match the conserved residues of RAM-C, but both have been observed to interact with CSL via their ΦWΦP motif. Molecular representations were generated using PyMOL (45).
Upon activation of Notch, displacement of co-repressor proteins is coupled to the high affinity binding between RAM and BTD (17). This tight binding event may be regarded as an anchoring point, bringing ANK into close proximity to its binding site on the C-terminal domain, possibly increasing the effective concentration of ANK to promote this otherwise weak binding reaction (11, 16). The C-terminal domain·ANK interaction creates an elongated groove in which the co-activator MAM is able to bind, further recruiting histone acetyltransferase (p300) and converting the chromatin to an active conformation (12, 15, 18–20). In addition to recruiting the co-activators necessary to activate transcription, MAM also recruits the kinase cdk8, such that the activated transcription complex is short-lived, and the target gene is returned to the repressed state quickly (21).
In addition to the normal mechanism of activation described above, several herpesviruses have adopted mechanisms by which the Notch signaling pathway is misactivated. This misactivation is essential for the viral life cycle (22). For example, the Epstein-Barr virus transcriptional transactivator protein EBNA2 (Esptein-Barr nuclear antigen 2) activates CSL-dependent genes through direct interaction with CSL (22). Although in some regards EBNA2 substitutes for NICD, it lacks an ANK domain, bypassing the recruitment of MAM and the associated kinase (23). In doing so, EBNA2-responsive genes can be activated without the rapid turnover back to the repressed state, although it is expected that the absence of the ANK·MAM interaction with CSL may weaken the EBNA2·CSL interaction.
Given the importance of the interaction between BTD and RAM as an energetically crucial binding event in switching from gene repression to activation, we determined the energetic contribution made by each of the conserved residues in binding RAM to BTD. Additionally, we report the in vitro binding behavior between BTD and conserved region 6 (CR6) of EBNA2, which also has a ΦWΦP motif (PPWWPP). To better understand how RAM and EBNA2 engage BTD and to gain insight to potential cross-talk between these activators, we explore the extent to which binding of the RAM and EBNA2 sequences to BTD are exclusive, both by using displacement isothermal titration calorimetry (ITC) and by quantifying the effects on binding of BTD point substitutions reported to specifically and exclusively inhibit interactions of one or the other ligands (24).
EXPERIMENTAL PROCEDURES
Mutagenesis, Protein Expression, and Purification
The BTD construct studied here contains residues 161–349 of human CSL, as previously described (13). BTDF235L and BTDK249M were made using the QuikChange® mutagenesis kit (Stratagene); inverse PCR was used to make BTDQ307L utilizing a primer spanning the AflII restriction site and the Q307L substitution. All of the protein constructs were expressed in an Escherichia coli BL21*(DE3) Rosetta2 cell line (Novagen) in TB. The cells were grown at 37 °C in 1 liter of medium/3-liter culture flask to an A600 of 1.5, plunged into an ice bath for 30 min, and subsequently treated to a final concentration of 2% ethanol and 1 mm isopropyl β-d-thiogalactopyranoside. Induction proceeded for 18 h at 18 °C, after which cell pellets were stored at −80 °C. Cell lysis and BTD purification were carried out as previously described (13). Purified protein was dialyzed overnight against 1 liter of buffer containing 200 mm NaCl, 25 mm Tris (pH 8.0), 1 mm EDTA, 5% (v/v) ethylene glycol, and 0.1 mm tris(2-carboxyethyl)phosphine hydrochloride (TCEP) before being flash frozen and stored at −80 °C.
Peptide Synthesis and Purification
All of the peptides used in these experiments were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on a Protein Technologies Symphony Quartet. The peptides were removed from the resin via trifluoroacetic acid cleavage (95% trifluoroacetic acid, 2.5% anisole, 2.5% water) for 2 h with agitation and were then precipitated with cold ether. Crude peptides were purified using reverse-phase high pressure liquid chromatography with a C18 column and a linear acetonitrile gradient (0–50%) over 10 column volumes. Purified peptides were frozen at −80 °C, lyophilized, and stored at −20 °C. The peptide masses were verified using a Finnigan LCQ Deca ion trap electrospray mass spectrometer (Thermo Finnigan, San Jose, CA).
Isothermal Titration Calorimetry
BTD was dialyzed for 24 h against 4 liters of buffer containing 200 mm NaCl, 20 mm HEPES (pH 7.5), 1 mm EDTA, 5% (v/v) ethylene glycol, and 0.25 mm TCEP. RAM and EBNA2 peptides were resuspended in buffer of identical composition. After dialysis and resuspension, BTD and RAM peptides were filtered through a 0.2-μm pore diameter filter, and the concentrations were determined using UV absorbance. All of the titrations were carried out using 10 μm BTD and 100 μm titrant (RAM-C) at 25 °C on a VP-ITC microcalorimeter from MicroCal (Northhampton, MA); the data were analyzed using the provided Origin 5.0 software with a one-site binding model.
For displacement ITC, 10 μm BTD was incubated overnight at 4 °C in the presence of EBNA2 (0–200 μm), loaded into the ITC cell, and titrated with 100 μm RAM-C. To deconvolute the apparent binding affinity into that of RAM-C (fixed) and EBNA2 (floated), we used the DISPLACE algorithm by Sigurskjold (25) using Equations 2 and 3 in the Appendix (25). The Keq and ΔH° for the BTD·RAM binding reaction were fixed at 1.38 × 107 m−1 and −19.8 kcal·mol−1, respectively, to better constrain the model during the fitting process. The reported value for displacement-derived EBNA2 binding affinity is the mean ± standard deviation of the means for six experiments, each performed at a different concentration of EBNA2 in complex with BTD.
Protein Stability Measurements
Ultrapure guanidine HCl was purchased from Invitrogen and dissolved in buffer to a final composition of 7.48 m guanidine HCl, 200 mm NaCl, 20 mm HEPES (pH 7.5), 1 mm EDTA, 5% (v/v) ethylene glycol, and 0.25 mm TCEP. The protein was denatured by titrating native BTD in the above buffer with guanidine HCl buffer using a Hamilton Microlab 500 titrator at a constant volume. The samples were equilibrated for 300 s at each guanidine HCl step (0.16 m) at 25 °C prior to measurement of tryptophan fluorescence on an ATF-105 automated titrating differential/ratio spectrofluorometer (Aviv Biomedical, Lakewood, NJ). Tryptophan fluorescence was excited at 295 nm (4.0-nm bandwidth); emission was detected at 331 nm (6.4-nm bandwidth). All equilibrium unfolding experiments were performed with an initial concentration of 3 μm BTD and subsequently corrected for the dilution during the titration. The data were fitted with a two-state model, using the linear extrapolation method (26–28), with the nonlinear least squares tool in KaleidaGraph (Synergy Software).
ΔCp Calculation
Protein Data Bank files 1ttu (DNA·CSL) and 3brd (DNA·CSL·RAM) were modified to contain only those residues corresponding to BTD. Residues that are missing from 3brd (residues 374–378 and 433–436 in worm CSL) were deleted from 1ttu to calculate surface area changes between the free and bound states. The changes in solvent-accessible surface area (ΔASA) were calculated by rolling a ball with a radius of 1.4 Å (approximately a water molecule) over the surface (29). The ΔASA for the binding reaction of BTD and RAM was calculated as follows: ASA(BTD·RAM) − ASA(BTD) − ASA(RAM). The ΔASA obtained from the calculation was separated into polar and nonpolar components, which were used to calculate ΔCp from the equation ΔCp = 0.28(ΔASAnp) − 0.09(ΔASApol) (30).
Co-immunoprecipitation and Peptide Competition
HEK293 cells were grown in Dulbecco's modified Eagle's medium + 10% fetal bovine serum at 37 °C and 5% CO2 and were transfected with either pcDNA3.1+hNICD1 (coding the intracellular domain of human Notch1) or pcDNA3.1+hCSLmyc/his (coding the three structured domains of human CSL, with a C-terminal Myc/His tag) or co-transfected with both expression vectors using FuGENE (Roche Applied Science). At 48 h post-transfection, the cells were harvested and lysed in cold co-immunoprecipitation buffer as previously described (13). Cleared cell lysates from cells expressing either hNICD1 or hCSLmyc were pooled and aliquoted into microcentrifuge tubes containing varying concentrations of different RAM and EBNA2 synthetic peptides. In a separate experiment, cleared cell lysates from cells co-expressing hNICD1 and hCSLmyc were pooled and aliquoted into microcentrifuge tubes containing identical peptide conditions to those described above. To monitor the effects of the various RAM and EBNA2 peptides on the NICD·CSL complex, hCSLmyc was immunoprecipitated using 9E10 anti-Myc ascites and Protein A beads (Sigma), and the NICD bound to CSL was assessed by Western blot analysis. NICD and CSL were detected with AN1 and anti-Myc (Cell Signaling) primary antibodies, and anti-mouse and anti-rabbit horseradish peroxidase secondary antibodies, respectively.
RESULTS
Thermodynamic Contribution by Conserved Residues in RAM to BTD Binding
Previous studies of the interaction of the ∼140-residue RAM region with CSL show the interaction to be localized to the N-terminal 25 residues of RAM (Fig. 1A) (9, 13, 14, 31). Sequence alignment of this segment of the RAM region from diverse Notch homologs, centered on the absolutely conserved ΦWΦP motif (supplemental Fig. S1), reveals three additional regions of significant conservation: (i) an N-terminal Arg/Lys-rich region, herein described as the “basic” region; (ii) an HG motif; and (iii) a C-terminal GF dipeptide (Fig. 1B). There is no detectable sequence conservation C-terminal to region III. To probe the relative contributions of these regions to binding, we synthesized a RAM consensus peptide from this region as a reference and monitored its binding to BTD using ITC. We then substituted the conserved residues with alanine, either as small blocks or as single substitutions, and measured the effects on binding.
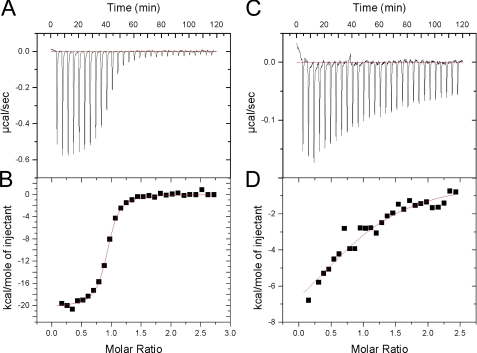
Fig. 2 shows base line-corrected heat peaks and integrated peaks for injection of the consensus peptide into BTD, fitted with a single-site model. The consensus peptide (herein referred to as RAM-C) binds BTD with high affinity (Kd = ∼70 nm, ΔG° = −9.75 kcal·mol−1), favorable enthalpy (ΔH° = −19.8 kcal·mol−1), and unfavorable entropy (TΔS° = −10.1 kcal·mol−1; Table 1). The measured binding affinity for RAM-C is consistent with previously published values for native Notch RAM sequences, especially considering the differences in protein constructs, buffer composition, and temperature (13, 32). Although the consensus sequence is not identical to any of the native ligands of human BTD (Notch 1–4), the differences are, for the most part, at residues of low conservation, largely C-terminal to the conserved GF dipeptide (region III).
FIGURE 2.
Isothermal titration calorimetry of peptide binding to BTD. A, the heat peaks resulting from the enthalpically favorable (exothermic) binding reaction of the RAM consensus peptide with BTD. B, integrated heat peaks (data points) with the associated single-site fit (red line). C and D, the heat peaks and fitted integrated heat peaks for the binding reaction of EBNA2 CR6 with BTD. Note the y axis deflection is different for RAM-C (A and B) than that of EBNA2 (C and D).
TABLE 1.
ITC data for RAM consensus peptide variants and EBNA2 binding to the β-trefoil domain of CSL
The reported values are the means ± standard deviation of the means of three independent experiments, excepting RAM-C where n = 6. Uncertainty in ΔΔG° was calculated by standard propagation of errors.
| Ligand | Kd | ΔG° | ΔH° | TΔS° | ΔΔG° |
|---|---|---|---|---|---|
| μm | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | |
| RAM consensus | 0.072 ± 0.004 | −9.75 ± 0.02 | −19.8 ± 0.5 | −10.1 ± 0.5 | |
| R4A/K5A/R6A/K7A/R8A | 5.4 ± 0.3 | −7.19 ± 0.04 | −11.7 ± 0.8 | −4.5 ± 0.8 | 2.56 ± 0.04 |
| H10A/G11A | 1.0 ± 0.1 | −8.18 ± 0.07 | −13.6 ± 0.6 | −5.4 ± 0.6 | 1.57 ± 0.07 |
| W14A/P16Aa | NBD | ||||
| G18A/F19A | 0.20 ± 0.02 | −9.11 ± 0.05 | −16.5 ± 0.4 | −7.4 ± 0.5 | 0.64 ± 0.05 |
| Δ1–3 | 0.10 ± 0.01 | −9.54 ± 0.02 | −19.6 ± 0.02 | −10.0 ± 0.1 | 0.21 ± 0.03 |
| R4A | 0.12 ± 0.02 | −9.45 ± 0.10 | −18.7 ± 0.3 | −9.2 ± 0.5 | 0.30 ± 0.10 |
| K5A | 0.084 ± 0.001 | −9.65 ± 0.01 | −20.1 ± 0.1 | −10.4 ± 0.1 | 0.10 ± 0.02 |
| R6A | 0.092 ± 0.002 | −9.60 ± 0.01 | −21.5 ± 0.2 | −11.9 ± 0.2 | 0.15 ± 0.02 |
| K7A | 0.19 ± 0.01 | −9.18 ± 0.03 | −19.8 ± 0.1 | −10.6 ± 0.2 | 0.57 ± 0.04 |
| R8A | 0.50 ± 0.03 | −8.59 ± 0.03 | −20.1 ± 0.6 | −11.5 ± 0.7 | 1.16 ± 0.04 |
| H10A | 0.47 ± 0.08 | −8.65 ± 0.10 | −17.7 ± 0.8 | −9.0 ± 0.8 | 1.10 ± 0.10 |
| G11A | 0.16 ± 0.01 | −9.26 ± 0.02 | −14.6 ± 0.2 | −5.4 ± 0.2 | 0.49 ± 0.02 |
| L13A | 3.0 ± 0.2 | −7.54 ± 0.05 | −19.6 ± 1.2 | −12.1 ± 1.2 | 2.21 ± 0.05 |
| W14Aa | NBD | ||||
| W14Yb | >10 | −6.79 ± 0.11 | −9.93 ± 0.6 | −3.12 ± 0.7 | 2.96 ± 0.11 |
| F15A | 0.474 ± 0.004 | −8.63 ± 0.01 | −17.3 ± 0.02 | −8.67 ± 0.01 | 1.12 ± 0.02 |
| P16Ab | >10 | −6.32 ± 0.17 | −16.0 ± 3.4 | −9.7 ± 3.6 | 3.43 ± 0.17 |
| E17A | 0.18 ± 0.04 | −9.21 ± 0.01 | −18.5 ± 0.01 | −9.3 ± 0.01 | 0.54 ± 0.02 |
| EBNA2 CR6 | 4.6 ± 1.3 | −7.33 ± 0.18 | −10.2 ± 3.1 | −2.8 ± 1.2 | 2.42 ± 0.18 |
a Neither W14A/P16A nor W14A peptide variants produced heats of binding above the background heats of injection; hence no binding was detected (NBD).
b For W14Y and P16A, binding to BTD could be detected, though the c value (product of Keq·Mtot) was below the range considered statistically significant for ITC (1 < c < 1000) (46).
To probe the contribution of each residue to the binding free energy, we made a series of RAM consensus peptides with alanine substitutions at residues of greatest conservation. To obtain an initial coarse picture of the contribution by conserved regions to binding affinity, we made multiple alanine substitutions through each conserved block and measured the effect on the energetics of association to BTD. Block alanine substitution at regions I and II (basic and HG) significantly decreased the binding affinity for BTD, increasing the free energy of association by 2.56 and 1.57 kcal·mol−1, respectively (Table 1). Changing the tryptophan and proline in the ΦWΦP motif to alanines completely abolished binding to BTD; no heat of binding was detected above the background heat of injection. This effect is consistent with previously published reports that underscore the importance of these two residues (9, 13, 14). Block alanine substitution of region III (GF) modestly decreased affinity, increasing the free energy of association by 0.64 kcal·mol−1. To obtain a more detailed map of the large effects of block alanine substitution of the residues in regions I and II and the ΦWΦP motif and to gain insight into the relationship between binding energy and conservation, we made single alanine variants and measured binding to BTD.
Residues in the ΦWΦP Motif
Like the doubly substituted W14A/P16A, no binding was detected between BTD and the W14A peptide variant. Weak binding was detected between BTD and the W14Y variant, corresponding to a 150-fold decrease in affinity. Likewise, only weak binding was detected between BTD and the P16A peptide variant, corresponding to a 350-fold decrease in affinity. Substitution of the Φ residues within the ΦWΦP motif decreased affinity, albeit less severely than either Trp or Pro. The L13A (Φ1) variant destabilized the BTD·RAM complex by 2.21 kcal·mol−1, whereas the F15A (Φ2) variant destabilized the complex by 1.12 kcal·mol−1 (Table 1 and Fig. 1B). Thus, all four of these residues contribute significant binding energy to the BTD·RAM binding reaction, although as assessed by alanine substitution, the absolutely conserved Trp and Pro contribute significantly more to the binding free energy than the more variable (Φ1 and Φ2) positions.
Residues in the N-terminal HG Motif (Region II)
Dissection of region II into residue-specific contributions revealed that alanine substitution of His10 and Gly11 increased the binding free energy by 1.1 and 0.49 kcal·mol−1, respectively. The sum of these free energy changes is nearly identical to that for the H10A/G11A double variant, which displayed a ΔΔG° value of 1.57 kcal·mol−1; thus, contributions appear to be additive despite the close proximity of these two residues.
Residues of the N-terminal Basic Region (Region I)
Block alanine substitution of the five-residue N-terminal basic region resulted in significant (ΔΔG° = 2.56 kcal·mol−1) destabilization of the BTD·RAM complex. To explore the extent to which individual residues contribute to this interaction, we measured the binding of singly alanine-substituted RAM-C peptides within this block. The R8A variant had the largest ΔΔG° value of 1.16 kcal·mol−1. Substitution of the adjacent lysine (K7A) had the next largest ΔΔG° value of 0.57 kcal·mol−1. Substitution of the arginine at position six (R6A) only modestly weakened binding (ΔΔG° = 0.15 kcal·mol−1). Surprisingly, the relative change in binding energy for these three residues is opposite to the extent of conservation (33); Arg6 is most conserved, whereas Arg8 is least conserved. Substitution of the arginine and lysine at positions four and five (R4A and K5A), the two least conserved residues within the basic region, modestly weakened the interaction, with ΔΔG° values of 0.3 and 0.1 kcal·mol−1, respectively. As with block II above, the contributions made by each of the five basic residues, as probed by alanine substitution, appears to be approximately additive. The sum of the ΔΔG° values for the five single-alanine substitutions is 2.28 ± 0.12 kcal·mol−1, compared with that of the five-site substitution with a ΔΔG° value of 2.56 ± 0.04 kcal·mol−1 (Table 1).
In addition, we probed the contribution of the three N-terminal RAM residues (VVS) by N-terminal truncation but found only a very modest change in affinity (Table 1). These results, when taken together, indicate that the six N-terminal residues of RAM (starting at the major site of γ-secretase cleavage) contribute little to the binding affinity of RAM to BTD (ΔΔG° = 0.76 kcal·mol−1). This finding is consistent with the lack of electron density for five of these residues in the crystal structures of Wilson and Kovall (12) and Friedmann et al. (32).
Heat Capacity Change Associated with Binding of RAM to BTD
One of the most structurally interpretable thermodynamic parameters related to conformational transitions and binding reactions is the heat capacity change, or ΔCp. This parameter is highly sensitive to solvation of nonpolar groups by water, as well as to other sources of enthalpy fluctuation in the unbound and bound states (34). The contribution of solvation to ΔCp can be estimated from changes in nonpolar surface area, although the contribution from water-independent enthalpy fluctuations remains a significant challenge. We determined the value of ΔCp for binding of RAM-C to BTD by monitoring the enthalpy of binding over the temperature range of 288–308 K. In this temperature range, ΔH° decreases linearly with increasing temperature, indicating that ΔCp for the BTD·RAM binding reaction is negative. Linear regression gives a ΔCp value of −0.49 kcal·mol−1·K−1 with r = 0.99 (Fig. 3A; see Equation 1 in the Appendix). We did not observe any conformational changes in the unligated BTD in the temperature range explored here, as assessed by far-UV CD (not shown).
FIGURE 3.
Determination of the heat capacity change upon binding of the RAM consensus peptide to BTD. A, plot of enthalpy of binding versus temperature. The slope of the line (−0.49 kcal·mol−1·K−1) is equal to ΔCp. B, structural alignment of BTD in the free (blue) and RAM-bound (wheat) states. Phe235 is represented by a space-filling model to highlight the conformational change in BTD upon binding RAM (orange). The Protein Data Bank codes were 1ttu (worm CSL, blue) and 3brd (worm CSL·RAM complex, wheat).
Several crystal structures of CSL have been solved in recent years, including worm CSL bound to DNA, worm and human CSL·DNA bound to NICD and MAM, and most recently, worm CSL·DNA bound only to the RAM region of NICD (12, 15, 31, 32). Alignment of the CSL·DNA structure with that of RAM·CSL·DNA reveals a conformational change in BTD that may be associated with RAM binding (Fig. 3B) (32). Using these structures, we were able to calculate a ΔCp value based on the change in solvent-accessible surface area (ΔASA, Å2) of polar and nonpolar side chains (ΔCp = 0.28; (ΔASAnp) − 0.09(ΔASApol), both as a result of binding and as a result of the observed conformational change (30). The calculated ΔCp value for the BTD·RAM association is −0.44 kcal·mol−1·K−1, remarkably close to the −0.49 kcal·mol−1·K−1 obtained experimentally. The agreement between the experimentally derived and calculated ΔCp values suggests that the conformational change observed in the crystal structure is representative of the binding reaction in solution (32).
Thermodynamic Parameters of Binding EBNA2 to BTD
In previous studies using a variety of cell culture assays and glutathione S-transferase pulldowns, CR6 of EBNA2 has been implicated in binding CSL. Moreover, when the two tryptophans of CR6 (PPWWPP) are replaced with serine and arginine, EBNA2 loses its ability to interact with CSL (33, 34). Using a peptide corresponding to CR6, we quantified the thermodynamics of binding to BTD using ITC (Table 1). We find that EBNA2 has a 60-fold weaker affinity for BTD than RAM-C, corresponding to an increase in free energy of association of 2.4 kcal·mol−1. The Kd measured here for binding of EBNA2 to BTD (4.6 μm) is remarkably close to an IC50 measured by Farrell et al. (37) using cell culture inhibition assays (1–10 μm).
One possible explanation for this lower affinity is that EBNA2 lacks the conserved basic region that our alanine substitution studies show to be important in RAM binding BTD. The ΔG° value for RAM lacking the basic region is nearly identical to that of EBNA2, suggesting that complex formation between EBNA2 and BTD is largely driven by the PPWWPP sequence.
Mutational Analysis of BTD and the Resulting Effects on Ligand Binding
Previous functional and biochemical assays have implicated residues on CSL that, when substituted, perturb EBNA2 binding but not RAM binding (E−/R+) and others that perturb RAM binding but not EBNA2 binding (R−/E+) (24, 38, 39). This may suggest distinct EBNA2 and RAM sites on CSL (as proposed by Fuchs et al. (24)) or may reflect a common (overlapping) site where different features of the two ligands are recognized. The possibility that the two ligands may engage distinct sites could provide potential therapeutic manipulation of the EBNA2 interaction without affecting the RAM binding necessary for native Notch signaling.
The residues of BTD reported to block RAM (but not EBNA2) interaction are located on a region that contacts the N terminus of RAM (Fig. 4A, red). These R−/E+ residues may help to directly stabilize the N terminus of RAM or may help drive the conformational change associated with RAM binding. One of the residues that blocked EBNA2 (but not RAM) binding is located at the edge of the hydrophobic pocket on BTD (Gln307; Fig. 4A, black) and is hydrogen-bonded to the proline carbonyl within the RAM ΦWΦP motif in the worm CSL·RAM crystal structure. To obtain a quantitative, mechanistic explanation for the selective interaction previously observed, we made three BTD variants reported to selectively perturb binding of RAM and EBNA2 (24), with two in the N-terminal-complementary region (F235L and K249M; R−/E+) and one at the edge of the hydrophobic pocket (Q307L; E−/R+), and determined the effects of these substitutions on the structural integrity of BTD and its thermodynamics of binding to RAM and EBNA2.
FIGURE 4.
Residues in BTD identified as selective for RAM and EBNA2. A, surface representation of the BTD·RAM complex. Gln307 (black) is located at the edge of the hydrophobic pocket. Substituting Gln307 with leucine has been implicated to disrupt binding of EBNA2, but not RAM, despite the structural proximity. Phe235 and Lys249 (red) are in a region of BTD complementary to the N terminus of RAM. Substitution of Phe235 and Lys249 with Leu and Met, respectively, have been implicated to block RAM binding without affecting the BTD·EBNA2 interaction. B, high resolution view of the hydrogen bonding interaction between the Gln307 side chain amide and the proline carbonyl of the ΦWΦP motif. Elemental coloring was blue for nitrogen and red for oxygen.
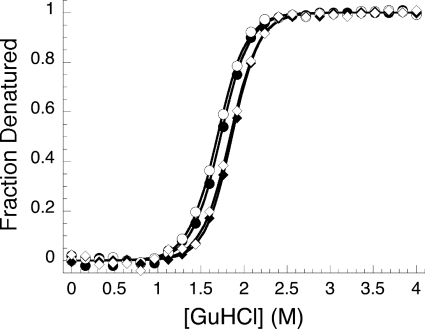
The three surface mutations did not adversely affect the structure or stability of BTD, as assessed by their far-UV CD spectra (not shown) and guanidine-induced unfolding (Fig. 5 and Table 2). BTDF235L, with a reported R−/E+ phenotype, decreased the affinity for RAM by 250-fold, increasing the Kd from 70 nm to >10 μm (Table 3). The same substitution had a modest effect on the binding of EBNA2, decreasing the affinity by 4-fold. The corresponding changes in free energy of association for RAM and EBNA2 were determined to be 3.27 and 0.82 kcal·mol−1, respectively. Similarly, BTDK249M, with a reported R−/E+ phenotype, reduced RAM affinity from 70 nm to >10 μm. Again, the effect on EBNA2 binding was modest, reducing the affinity 4-fold, from 4.6 to >10 μm. The changes in free energy of association for RAM and EBNA2 were determined to be 3.50 and 0.81 kcal·mol−1, respectively (Table 3).
FIGURE 5.
Guanidine HCl melts of BTD variants show minor effects on conformational stability. Unfolding transitions were monitored by tryptophan fluorescence and were normalized to facilitate comparison of slopes and midpoints. The solid lines represent fits using the two-state model. ●, wild type BTD; ○, BTDF235L; ♦, BTDK249M; ◇, BTDQ307L.
TABLE 2.
Stability of BTD variants
The reported values are the means ± standard deviation of the means for three independent experiments.
| BTD varianta | ΔG° | m value | Cm |
|---|---|---|---|
| kcal·mol−1 | kcal·mol−1·m−1 | m | |
| Wild type | 6.10 ± 0.05 | 3.51 ± 0.06 | 1.74 ± 0.02 |
| F235L (R−/E+) | 6.02 ± 0.30 | 3.52 ± 0.17 | 1.71 ± 0.01 |
| K249M (R−/E+) | 6.67 ± 0.36 | 3.58 ± 0.16 | 1.86 ± 0.02 |
| Q307L (E−/R+) | 6.46 ± 0.13 | 3.41 ± 0.05 | 1.90 ± 0.04 |
a Adopted from Ref. 24.
TABLE 3.
Calorimetric binding data of RAM and EBNA2 peptides to BTD variants
The reported values are the means ± standard deviation of the means of three independent experiments.
| BTD variant | Ligand | Kd | ΔG° | ΔH° | TΔS° |
|---|---|---|---|---|---|
| μm | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | ||
| Wild type | RAM-C | 0.072 ± 0.004 | −9.75 ± 0.02 | −19.8 ± 0.4 | −10.1 ± 0.4 |
| EBNA2 | 4.6 ± 1.3 | −7.33 ± 0.18 | −10.2 ± 3.1 | −2.8 ± 1.1 | |
| F235L | RAM-Ca | >10 | −6.48 ± 0.02 | −12.7 ± 0.2 | −6.3 ± 0.2 |
| RAM (A4–8)b,c | NBD | ||||
| RAM H10Ab | NBD | ||||
| EBNA2a | >10 | −6.51 ± 0.08 | −12.3 ± 0.7 | −5.8 ± 1.0 | |
| K249M | RAM-Ca | >10 | −6.25 ± 0.08 | −15.3 ± 1.8 | −9.0 ± 1.9 |
| RAM (A4–8)b,c | NBD | ||||
| RAM H10Ab | NBD | ||||
| EBNA2a | >10 | −6.52 ± 0.12 | −13.4 ± 1.0 | −6.9 ± 1.1 | |
| Q307L | RAM-C | 0.06 ± 0.01 | −9.85 ± 0.10 | −16.5 ± 0.4 | −6.7 ± 0.5 |
| EBNA2b | NBD |
a The c value is less than 1.
b No binding was detected (NBD) between ligand and the BTD variant.
c RAM (A4–8) is the consensus peptide with an alanine-substituted basic region (region I).
The large effect of these substitutions on the affinity for RAM suggests that a significant portion of RAM is unable to interact with BTD. The most likely region of RAM-C to be perturbed is the N-terminal basic region, given its proximity to the site of substitution and given the detectable enthalpy of binding suggesting that the ΦWΦP motif of RAM-C is still interacting with the hydrophobic pocket of BTDF235L and BTDK249M. To test this, we measured the binding of RAM-C lacking the basic region (RAM A4–8) to BTDF235L and BTDK249M. The loss of these basic residues within RAM further decreases binding affinity below our detection limit for BTDF235L and BTDK249M, indicating that the interaction with the basic region of RAM-C with these BTD variants remains at least partially intact. Similarly, the RAM H10A variant shows no detectable binding to BTDF235L or BTDK249M, suggesting the HG motif of RAM-C is contributing to the binding affinity to these BTD variants (Table 3).
The subtle, yet measurable effects of these substitutions on the affinity for EBNA2 suggest that EBNA2 has some residual interaction with this region of BTD. The significantly smaller effect on EBNA2 binding, compared with that of RAM binding, indicates that the interaction between this region of BTD and EBNA2 is weaker, possibly because of the absence of a basic region that we show here to contribute significantly to RAM binding. In summary, the N-terminal complementary region clearly has some interaction with both EBNA2 and RAM, although the specific contacts and binding determinants differ for each ligand.
BTDQ307L has a reported E−/R+ phenotype despite the observed hydrogen bond to the proline carbonyl of the RAM ΦWΦP motif. Given the importance of this sequence in binding of both ligands to BTD, we would expect this particular substitution to similarly disrupt the binding of both RAM and EBNA2. We measured the binding of RAM and EBNA2 to BTDQ307L, and in agreement with Fuchs et al. (24), substitution of Gln with Leu weakens the binding of EBNA2 to BTD, reducing it to a level undetectable by ITC experiments. In contrast, BTDQ307L binds RAM-C with wild type (or slightly tighter, ΔΔG° = −0.1 kcal·mol−1) affinity (Table 3).
In summary, all of the effects of the substitutions are consistent with the selectivity identified by Fuchs et al. (24). The R−/E+ variants selectively decrease the affinity of RAM, whereas the E−/R+ variant selectively decreases the affinity of EBNA2 binding.
Exclusive Binding of EBNA2 and RAM
The importance of the ΦWΦP motif in interacting with CSL, in both functional (9, 35, 36, 38, 40) and biophysical studies (13, 14), suggests that RAM and EBNA2 may share a common mode of interaction (and binding site) with CSL (Fig. 6A). However, Fuchs et al. (24) have shown that the two interactions are mediated by two different sets of structurally nonoverlapping residues on BTD, which is quantitatively confirmed by our ITC data using BTD variants targeting these residues. This specificity is consistent with separate binding sites (Fig. 6B).
FIGURE 6.
Models for two-ligand binding: overlapping versus nonoverlapping sites. A, schematic representation of two-ligand binding to overlapping (mutually exclusive) sites. B, schematic representation of two-ligand binding to two nonoverlapping sites. For nonoverlapping sites, two subclasses exist: independent and coupled (cooperative and anticooperative) sites. For independent two-ligand binding, the binding of one ligand has no effect on the binding of the second ligand, such that ΔGA (the free energy of binding ligand A in the absence of ligand B, purple) is equal to ΔGA(B) (the free energy of binding ligand A when ligand B (orange) is already bound). From the properties of thermodynamic cycles, the same relationship holds true for ligand B (ΔGB = ΔGB(A)). For coupled sites, binding of one ligand either promotes (positive coupling) or weakens (negative coupling, i.e. anticooperative) the affinity for the second ligand, such that ΔGA ≠ ΔGA(B).
To resolve whether this selectivity reflects binding to two separate sites, we used ITC to examine the effects of these two ligands on each other's binding. We pre-equilibrated BTD with varying concentrations of EBNA2 (weakly binding ligand) and then injected RAM-C (tightly binding ligand) into the BTD·EBNA2 complex. If the two peptides bind to independent sites (Fig. 6B), the presence of excess EBNA2 should have no effect on RAM binding, as schematized in Fig. 7A. In stark contrast, we see that the presence of EBNA2 greatly perturbs the ITC profile when RAM-C is titrated into BTD (Fig. 7B). The presence of EBNA2 decreases the enthalpy of RAM binding to BTD and lowers the apparent binding constant (Kapp; Fig. 7C), demonstrating that the two ligands do not bind independently. Decreasing the initial concentration of EBNA2 in complex with BTD gives rise to an increase in the apparent enthalpy of the BTD·RAM binding reaction, consistent with the larger (more negative) binding enthalpy of RAM-C compared with EBNA2 (Fig. 7C). Using a model for purely exclusive binding, we were able to extract from each titration experiment an equilibrium constant and enthalpy of binding for EBNA2 to BTD (25). The average ΔG° value derived from these displacement studies was determined to be −7.33 ± 0.06 kcal·mol−1, which is identical to the value obtained from direct measurement of injecting EBNA2 into BTD (ΔG° = −7.33 ± 0.18 kcal·mol−1). Moreover, the same displacement-derived ΔG° value is obtained regardless of the concentration of EBNA2 in complex with BTD. Although we cannot formally exclude the possibility that these two ligands bind with complete anticooperativity to separate sites, the agreement between these numbers, as well as the goodness of fit of the purely exclusive model (see legend for Fig. 7), is strong evidence that RAM-C and EBNA2 are indeed binding a common region of BTD in vitro and that binding of each ligand excludes the other.
FIGURE 7.
Displacement ITC with EBNA2 and RAM. A, simulated values of Kapp (solid lines) and ΔHapp (dashed lines) for three different modes of two-ligand binding: (i) independent sites (green, α = 1 in Equations 4 and 5), (ii) exclusive binding of two ligands at a single site (red, Equations 2 and 3), and (iii) negatively coupled binding (purple, Equations 4 and 5, α = 0.1 and Δh = 0). B, integrated heat peaks and fits for the injection of RAM-C into BTD·EBNA2 complex, analyzed with a purely exclusive model (25). Initial EBNA2 concentrations are 201.2 μm (red), 100.6 μm (orange), 28.7 μm (green), 14.4 μm (light blue), 7.1 μm (dark blue), 3.6 μm (purple), and 0.0 μm (black). C, plot of experimentally determined ΔHapp and Kapp as a function of EBNA2 concentration. The exclusive model simulated in A closely resembles the experimental data (C) and can be well fitted to the experimentally determined values of Kapp (solid purple line) and ΔHapp (dashed red line).
Peptide Competition with Cellular NICD·CSL Complexes
To test whether the synthetic peptide series exerts the same effects on full-length CSL as on BTD and to test whether the EBNA2 CR6 peptide directly competes with Notch binding to CSL, we monitored the ability of different peptides to block the interaction between NICD and CSL using cell lysates. This experiment was performed in two formats. In one, lysates from HEK293 cells separately expressing hNICD1 or hCSLmyc were mixed together; in the other, lysates from cells co-expressing both constructs were used directly. The lysates were mixed with varying concentrations of different peptides, hCSLmyc was immunoprecipitated, and the amount of hNICD bound to hCSL was monitored by co-immunoprecipitation and Western blot analysis.
In agreement with our ITC-derived binding affinities, the RAM-C peptide effectively inhibited binary complex (NICD·CSL) formation at all of the concentrations tested here (0.5–5 μm; Fig. 8A). In contrast, both EBNA2 CR6 and our RAM peptide lacking the basic region partially inhibited binary complex formation only at the highest concentration of peptide tested (5 μm), consistent with the similarly decreased affinities of both peptides as assessed by ITC. Our RAM peptide with Trp and Pro substituted for Ala could not inhibit binary complex formation at even the highest concentration tested, consistent with our inability to detect binding in the ITC experiments and the notion that the Kd is significantly weaker than 10 μm (Fig. 8A). In addition to inhibiting binary complex formation, the RAM and EBNA2 peptides could also effectively break apart preformed NICD·CSL complexes (Fig. 8B) and show the same efficacy in disrupting NICD·CSL complexes, regardless of whether or not the NICD·CSL complex is preformed.
FIGURE 8.
RAM-C and EBNA2 peptides are able to block hNICD1·hCSLmyc association from cellular lysates. A, lysates from cells separately expressing hNICD1 and hCSLmyc were combined and subsequently challenged with different RAM peptides or the EBNA2 CR6 peptide (5, 1.5, and 0.5 μm), and the hNICD1 bound to hCSLmyc was determined by co-immunoprecipitation and Western blot (WB) analysis. RAM-C was able to block the interaction at all concentrations tested here (lanes 3–5), whereas RAM A(4–8), which lacks the basic resides in region I, was only able to block NICD·CSL association at the highest concentration (lanes 6–8). RAM A(14/16), where the Trp and Pro of the ΦWΦP motif have been substituted to Ala, was unable to block NICD·CSL association at any concentration (lanes 9–11). Similar to RAM A(4–8), EBNA2 CR6 was only able to perturb NICD·CSL interaction at the highest concentration (lanes 12–14), consistent with the similar affinities of these two peptides for BTD. Top panel, hNICD1. Bottom panel, hCSLmyc. B, lysates from cells co-expressing hNICD1 and hCSLmyc were similarly challenged with the same peptides as those in A. Top panel, hNICD1. Bottom panel, hCSLmyc. Lane 1 is hNICD1 only, and lane 2 is hNICD1+hCSLmyc in the absence of peptide.
DISCUSSION
As the nuclear effector of Notch signaling, it is crucial that CSL be able to efficiently and reproducibly make the switch from a repressor to an activator of gene transcription. In making this switch, it is postulated that the dissociation of co-repressor proteins is coupled to the high affinity BTD·RAM binding event. It is well documented from genetic and biochemical studies that, of the 1000 residues of NICD, the N-terminal 25 residues of RAM encode high affinity binding to BTD and that the ΦWΦP motif is critical for interaction (9, 13, 14). In the current work, we have dissected the energetic contribution of the conserved residues of RAM binding to BTD, the overall thermodynamic signature of EBNA2 binding to BTD, and the mutual interactions between these two ligands on BTD.
Sequence Determinants of RAM Binding Affinity
Although consensus sequences are composed of the most frequent residues in an alignment, they do not always represent the most stably folded or strongest binding sequences. Consensus sequences do not account for specific pair-wise interactions that result in thermodynamic nonadditivity (41). The high affinity of a consensus RAM peptide binding to BTD, equal to or exceeding previously measured affinities of peptides derived from native (i.e. nonconsensus) Notch receptors, suggests that the sequence variations are additive and that variable positions contribute little to binding. As assessed by alanine substitution, the ΦWΦP motif is the largest energetic determinant of binding, because substitutions of W and P greatly reduce the interaction with BTD. N-terminal to the ΦWΦP motif, we find substantial energetic contributions to binding that are unevenly distributed throughout the first 11 residues. The three residues preceding the basic region contribute minimally to binding BTD, although they may serve an indirect role in stabilizing NICD in vivo, potentially maintaining Notch activity by slowing the rate at which N-end rule degradation occurs (42). The five residues within the basic region encode a significant portion of the energetics of BTD binding, ∼2.5 kcal·mol−1, with 70% being contributed by only two C-terminal residues (Lys7 and Arg8). BTD is still able to bind a RAM peptide lacking these basic residues, albeit with an 80-fold decrease in affinity. Lastly, we have determined that a highly conserved HG motif in between the basic region and ΦWΦP motif contributes ∼1.6 kcal·mol−1 to the binding reaction.
The residue-specific energetic contributions determined here are consistent with the measured affinities of RAM peptides derived from the four mammalian Notch receptors (13). These four peptides lack the N-terminal VVS sequence (ΔΔG° = 0.2 kcal·mol−1; Table 1) and also lack the first three basic residues. Approximating the interaction energy of these three residues, using the combined ΔΔG° values for the Arg/Lys to Ala substitutions (0.55 kcal·mol−1), the six residue N-terminal truncations should lead to an increase in binding energy of 0.76 ± 0.11 kcal·mol−1 or a free energy of binding in the range of −8.88 to −9.10 kcal·mol−1. The observed values for Notch1 (N1), N2, and N4 each lie within the range of −8.5 to −8.7 kcal·mol−1. Each of these three RAM sequences possesses the conserved residues probed in the current work. N3 was observed to bind with a modestly decreased affinity compared with the other three paralogs, possibly resulting from a substitution of the conserved Gly11 with serine. Our measurements indicate that the G11A substitution increases the binding free energy by 0.5 kcal·mol−1, which is roughly the energetic difference between N3 (Ser11, ΔG° = −8.0 kcal·mol−1) and N1, N2, and N4 (Gly11).
Conservation, Energetic Contribution, and Structural Implications
To quantitatively examine how the degree of conservation at each position of the RAM region is correlated with interaction energy, we calculated the sequence entropy at each position from our multiple sequence alignment. Sequence entropy combines the observed frequencies of each residue at a given position in the same way that probabilities combine in statistical calculations of entropy, correcting for general coding sequence biases. The sequence entropies of the absolutely conserved Trp and Pro within the ΦWΦP motif correlate well with the substantial contribution to binding energy made by each residue. However, the correlation between sequence entropy and binding energy contributions does not apply at other positions in the RAM sequence. Within the basic region, Arg6 is the most highly conserved residue, yet the free energy of association is only increased by 0.15 kcal·mol−1 for the R6A variant, compared with 0.57 and 1.16 kcal·mol−1 for K7A and R8A, respectively. Instead of following the degree of conservation, the residue-specific energetic contribution roughly decreases with increasing distance from the ΦWΦP motif. Overall, we did not find a strong correlation between conservation and energetic contribution; the large bias for Arg at position six does not appear to result from a strong contribution toward BTD binding affinity but rather from some other requirement of Notch signaling, perhaps in functioning as a “stop translocation” signal sequence (43).
Despite the weak correlation between conservation and binding affinity, we observed a robust trend between the residues with the most significant contribution and those displaying electron density in the crystal structures in which RAM is present (Protein Data Bank codes 2FO1 and 3brd) (12, 32). The worm crystal structures only display electron density for those residues corresponding to Arg6–Asn20. The first five residues, those which were determined to contribute very little to the binding reaction (0.61 kcal·mol−1, collectively), lack electron density. No electron density is observed downstream of position 20, immediately following the third region of conservation (Gly18/Phe19).
Observed and Calculated Heat Capacity of Binding RAM to BTD
The enthalpically driven binding of RAM to BTD displays a significant decrease in heat capacity over the temperature range explored here (Fig. 3A), closely matching ΔCp values calculated from surface area changes (−0.49 and −0.44 kcal·mol−1·K−1, respectively). Alignment of the structures of BTD in the free (Protein Data Bank code 1ttu) and RAM-bound (Protein Data Bank code 3brd) states reveals a local rearrangement and folding event in BTD at the site of RAM binding. In the free form, this region of BTD is in an open, extended conformation lacking regular secondary structure. Upon binding RAM, this open loop adopts β-stranded secondary structure that H-bonds to the backbone of RAM (Fig. 3B). Omitting this conformational change from calculation of ΔCp (using the bound state of BTD as a model for the unbound state by omitting RAM from the structure) gives a value of −0.36 kcal·mol−1·K−1. Including the conformational change brings the calculated ΔCp to −0.44 kcal·mol−1·K−1, a value very close to the experimentally derived value of −0.49 kcal·mol−1·K−1.
Binding Behavior of RAM and EBNA2 to BTD
The binding data presented here, along with numerous functional studies, highlight the importance of the ΦWΦP motif in BTD binding. Given that this is the only sequence similarity between the BTD-binding region of EBNA2 and RAM, it seems likely that both sequences bind BTD through a common mode of interaction and thus would show exclusive binding. However, this model is at odds with an interpretation based on a two-hybrid analysis by Fuchs et al. (24), in which separate binding sites were proposed. In the two-hybrid study, point substitutions in BTD were found to selectively block the interaction of one or the other ligand. To determine whether this selectivity is reflected in the thermodynamics of binding, we measured the energetic consequences of these selective point substitutions on RAM and EBNA2 binding. Consistent with the two-hybrid analysis, each BTD variant significantly destabilized binding of one ligand, but produced a much smaller destabilization of the second ligand. Most notably, BTDQ307L weakened binding of EBNA2 to an undetectable level. This result supports the idea that EBNA2 binds in close proximity to Gln307. Because Gln307 is at the rim of the hydrophobic pocket, to which the ΦWΦP motif of RAM binds, and is hydrogen-bonded to the proline carbonyl of RAM in the CSL·RAM crystal structure (Fig. 4B), it seems likely that RAM and EBNA2 bind to overlapping sites. However, it is quite surprising that RAM-C binding is unaffected by the Q307L substitution and suggests that the energetic contribution of proximal BTD residues is unique for each ligand.
With the notable exception of the ΦWΦP motif, there is very little sequence similarity between RAM and EBNA2. Motivated by the numerous reports demonstrating that substitution of the ΦWΦP motif of either ligand disrupts binding, we set out to test a model whereby both ligands bind similar or overlapping sites on BTD. Utilizing displacement ITC, which is normally used to determine the thermodynamic parameters of tightly binding (Kd < 10−10 m) competitive inhibitors, we examined whether EBNA2 and RAM bind unique or overlapping sites on BTD. By titrating a pre-equilibrated BTD·EBNA2 complex with RAM-C, we observed that the enthalpy of the reaction was directly related to the amount of EBNA2 in complex with BTD (Fig. 7B), clearly demonstrating that the ligands do not bind independently. If the two sites were unique and isolated, then the injection of RAM-C into BTD·EBNA2 would give rise to the same enthalpy and binding constant as the injection of RAM-C into BTD (Fig. 7A, green lines), which was not the case.
The displacement ITC model, which describes purely exclusive ligand binding, can be used to evaluate the equilibrium constant and enthalpy of binding of one ligand, given prior knowledge of the same parameters for the second ligand. To test whether binding of RAM and EBNA2 are exclusive, we fitted our displacement ITC data with a model for exclusive binding (Fig. 6A; Equations 2 and 3 in the Appendix). The agreement in the extracted quantities for EBNA2 with those from the direct binding of EBNA2 to BTD (ΔG° = −7.33 kcal·mol−1 for both methods) is strong evidence that the two ligands are binding exclusively. Because exclusive binding could result from even partial overlap of ligand-binding sites, our data suggest that EBNA2 and RAM are, at a minimum, binding overlapping sites on BTD and behave in a manner similar to that of competitive inhibitors, despite previous reports suggesting separate binding sites.
Combining the data from displacement ITC and the mutational analysis of BTD, we propose a model whereby EBNA2 and RAM overlap at a common site on BTD (likely involving the hydrophobic pocket that binds the ΦWΦP motif) but also make unique and specific stabilizing contacts with BTD. Partial overlap is consistent with our results from displacement ITC and from disruption of NICD·CSL complexes from cell lysates by the EBNA2 CR6 peptide. Unique modes of interaction by RAM versus EBNA2 CR6 are consistent with the results from ITC experiments on the R−/E+ and E−/R+ point substitutions of BTD, which show ligand-selective effects on binding free energy. This selectivity may result either from the substantial sequence variation between RAM and EBNA2 or potentially from structural differences in arrangements of the two ligands distant from their common overlapping sites.
Implications for Epstein-Barr Virus Therapeutics
The work presented here demonstrates that although there are some unique, nonoverlapping interactions in the RAM and EBNA2 complexes with BTD, there is substantial overlap between the ligand-binding sites. Thus, peptide therapeutics that block the interaction between endogenous EBNA2 and CSL in an effort to halt Epstein-Barr virus infection may also sterically block the BTD·RAM interaction necessary for native Notch signaling, which is known to play a critical role in the maintenance and homeostasis of stem cells in adults. Although direct competition makes a peptide therapeutic approach more challenging, the parameters resulting from our direct binding and displacement studies may provide a quantitative (albeit narrow) range through which selective disruption of the weaker EBNA2 interaction may be achieved.
Appendix
By monitoring the binding enthalpy over a range of temperatures, one can extract the heat capacity change ΔCp from the following relationship.
 |
For the simulation of two-ligand binding data in Fig. 7A, exclusive binding is defined by the apparent equilibrium constant and enthalpy defined as follows,
 |
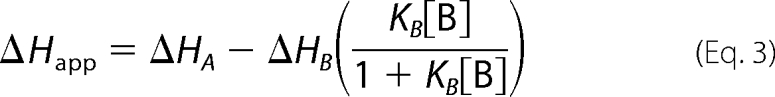 |
where KA is the equilibrium constant for the tightly binding ligand, KB is the equilibrium constant for the weakly binding ligand, ΔHA is the enthalpy of the binding reaction for the tightly binding ligand, ΔHB is the enthalpy of the binding reaction for the weakly binding ligand, and [B] is the concentration of free weakly binding ligand (25).
The simulation of cooperative (or in this case potentially anticooperative) two-ligand binding interactions is very similar to that above for exclusive binding, with additional parameters for the degree of cooperativity, α, and its enthalpic contribution, Δh. For positive cooperativity, α > 1. For negative cooperativity, α < 1. For independent binding, α = 1 (44). These two parameters can modify Equations 2 and 3 to give Equations 4 and 5.
 |
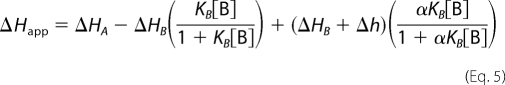 |
When α = 0, Equations 4 and 5 become identical to those for exclusive binding (Equations 2 and 3).
Supplementary Material
Acknowledgments
We thank Dr. Tamara Hendrickson and Megan Ehrenwerth for help with peptide synthesis. We also thank Dr. Joel Schildbach for instrument use, Bent Sigurskjold for the Displace fitting function, and Mohan Bolisetty for site-directed mutagenesis and construction of BTDF235L and BTDK249M.
This work was supported, in whole or in part, by National Institutes of Health Grants GM060001 (to D. B.) and P50 CA094056. This work was also supported by Instrumentation Development Grant 0500580 from the National Science Foundation Division of Biological Infrastructure for Peptide Synthesis Instrumentation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NICD
- Notch intracellular domain
- BTD
- β-trefoil domain
- ITC
- isothermal titration calorimetry
- ANK
- ankyrin repeat(s)
- CR
- conserved region
- RAM
- RBP-JK-associated molecule
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride.
REFERENCES
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Bonde J., Hess D. A., Nolta J. A. (2004) Curr. Opin. Hematol. 11, 392–398 [DOI] [PubMed] [Google Scholar]
- 3.Gridley T. (2003) Hum. Mol. Genet. 12, R9–R13 [DOI] [PubMed] [Google Scholar]
- 4.Weng A. P., Ferrando A. A., Lee W., Morris J. P, Silverman L. B., Sanchez-Irizarry C., Blacklow S. C., Look A. T., Aster J. C. (2004). Science 306, 269–271 [DOI] [PubMed] [Google Scholar]
- 5.Macsween K. F., Crawford D. H. (2003) Lancet Infect. Dis. 3, 131–140 [DOI] [PubMed] [Google Scholar]
- 6.Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 7.Ray W. J., Yao M., Mumm J., Schroeter E. H., Saftig P., Wolfe M., Selkoe D. J., Kopan R., Goate A. M. (1999) J. Biol. Chem. 274, 36801–36807 [DOI] [PubMed] [Google Scholar]
- 8.Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. (1985) Cell 43, 567–581 [DOI] [PubMed] [Google Scholar]
- 9.Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., Furukawa T., Honjo T. (1995) Curr. Biol. 5, 1416–1423 [DOI] [PubMed] [Google Scholar]
- 10.Nam Y., Weng A. P., Aster J. C., Blacklow S. C. (2003) J. Biol. Chem. 278, 21232–21239 [DOI] [PubMed] [Google Scholar]
- 11.Bertagna A., Toptygin D., Brand L., Barrick D. (2008) Biochem. Soc. Trans. 36, 157–166 [DOI] [PubMed] [Google Scholar]
- 12.Wilson J. J., Kovall R. A. (2006) Cell 124, 985–996 [DOI] [PubMed] [Google Scholar]
- 13.Lubman O. Y., Ilagan M. X., Kopan R., Barrick D. (2007) J. Mol. Biol. 365, 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Gall M., Giniger E. (2004) J. Biol. Chem. 279, 29418–29426 [DOI] [PubMed] [Google Scholar]
- 15.Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. (2006) Cell 124, 973–983 [DOI] [PubMed] [Google Scholar]
- 16.Del Bianco C., Aster J. C., Blacklow S. C. (2008) J. Mol. Biol. 376, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubman O. Y., Korolev S. V., Kopan R. (2004) Mol. Cell 13, 619–626 [DOI] [PubMed] [Google Scholar]
- 18.Petcherski A. G., Kimble J. (2000) Nature 405, 364–368 [DOI] [PubMed] [Google Scholar]
- 19.Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G. (2002) Mol. Cell Biol. 22, 7812–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L., Sun T., Kobayashi K., Gao P., Griffin J. D. (2002) Mol. Cell Biol. 22, 7688–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fryer C. J., White J. B., Jones K. A. (2004) Mol. Cell 16, 509–520 [DOI] [PubMed] [Google Scholar]
- 22.Hayward S. D. (2004) Semin. Cancer Biol. 14, 387–396 [DOI] [PubMed] [Google Scholar]
- 23.Sakai T., Taniguchi Y., Tamura K., Minoguchi S., Fukuhara T., Strobl L. J., Zimber-Strobl U., Bornkamm G. W., Honjo T. (1998) J. Virol. 72, 6034–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs K. P., Bommer G., Dumont E., Christoph B., Vidal M., Kremmer E., Kempkes B. (2001) Eur. J. Biochem. 268, 4639–4646 [DOI] [PubMed] [Google Scholar]
- 25.Sigurskjold B. W. (2000) Anal. Biochem. 277, 260–266 [DOI] [PubMed] [Google Scholar]
- 26.Pace C. N. (1986) Methods Enzymol. 131, 266–280 [DOI] [PubMed] [Google Scholar]
- 27.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 28.Street T. O., Courtemanche N., Barrick D. (2008) Methods Cell Biol. 84, 295–325 [DOI] [PubMed] [Google Scholar]
- 29.Fraczkiewicz R., Braun W. (1998) J. Comp. Chem. 19, 319–333 [Google Scholar]
- 30.Myers J. K., Pace C. N., Scholtz J. M. (1995) Protein Sci. 4, 2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovall R. A, Hendrickson W. A. (2004) EMBO J. 23, 3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedmann D. R., Wilson J. J., Kovall R. A. (2008) J. Biol. Chem. 283, 14781–14791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Samudrala R. (2006) BMC Bioinformatics 7, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhu N. V., Sharp K. A. (2005) Annu. Rev. Phys. Chem. 56, 521–548 [DOI] [PubMed] [Google Scholar]
- 35.Ling P. D., Rawlins D. R., Hayward S. D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9237–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling P. D., Hayward S. D. (1995) J. Virol. 69, 1944–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farrell C. J., Lee J. M., Shin E. C., Cebrat M., Cole P. A., Hayward S. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4625–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh J. J., Hayward S. D. (1995) Science 268, 560–563 [DOI] [PubMed] [Google Scholar]
- 39.Hsieh J. J., Nofziger D. E., Weinmaster G., Hayward S. D. (1997) J. Virol. 71, 1938–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Gall M., De Mattei C., Giniger E. (2008) Dev. Biol. 313, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells J. A. (1990) Biochemistry 29, 8509–8517 [DOI] [PubMed] [Google Scholar]
- 42.Tagami S., Okochi M., Yanagida K., Ikuta A., Fukumori A., Matsumoto N., Ishizuka-Katsura Y., Nakayama T., Itoh N., Jiang J., Nishitomi K., Kamino K., Morihara T., Hashimoto R., Tanaka T., Kudo T., Chiba S., Takeda M. (2008) Mol. Cell. Biol. 28, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velazquez-Campoy A., Goñi G., Peregrina J. R., Medina M. (2006) Biophys. J. 91, 1887–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano W. L. (2008) PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 46.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 47.Schuster-Böckler B., Schultz J., Rahmann S. (2004) BMC Bioinformatics 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.