Abstract
Obesity and insulin resistance are associated with deposition of triglycerides in tissues other than adipose tissue. Previously, we showed that a missense mutation (I148M) in PNPLA3 (patatin-like phospholipase domain-containing 3 protein) is associated with increased hepatic triglyceride content in humans. Here we examined the effect of the I148M substitution on the enzymatic activity and cellular location of PNPLA3. Structural modeling predicted that the substitution of methionine for isoleucine at residue 148 would restrict access of substrate to the catalytic serine at residue 47. In vitro assays using recombinant PNPLA3 partially purified from Sf9 cells confirmed that the wild type enzyme hydrolyzes emulsified triglyceride and that the I148M substitution abolishes this activity. Expression of PNPLA3-I148M, but not wild type PNPLA3, in cultured hepatocytes or in the livers of mice increased cellular triglyceride content. Cell fractionation studies revealed that ∼90% of wild type PNPLA3 partitioned between membranes and lipid droplets; substitution of isoleucine for methionine at position 148 did not alter the subcellular distribution of the protein. These data are consistent with PNPLA3-I148M promoting triglyceride accumulation by limiting triglyceride hydrolysis.
Keywords: Cell/Hepatocyte, Lipid, Lipid/Lipase, Lipid/Triacylglycerol, Membrane/Lipids, Metabolism/Fatty Acid, Metabolism/Lipid, Metabolism/Lipogenesis, Lipase, Lipolysis
Introduction
Triglycerides are the major energy storage molecules in eukaryotes. In mammals, triglycerides are stored primarily in adipose tissue, which serves as an energy buffer during long term caloric depletion. Under conditions of sustained caloric excess, the deposition of triglycerides in non-adipose tissues may increase substantially, particularly in the liver. The extent of triglyceride accumulation varies markedly among individuals, ranging from less than 1% to more than 50% of liver weight in the general population (1). A variety of factors are associated with deposition of fat in the liver, including obesity, diabetes, insulin resistance, and alcohol ingestion, but the factors responsible for the wide individual differences in susceptibility to hepatic steatosis are not known (2).
Recently, we identified a nonsynonymous polymorphism in PNPLA3 (I148M) that is strongly associated with hepatic fat content and with elevated serum levels of alanine aminotransferase and aspartate aminotransferase, which are markers of liver inflammation. Subsequent studies have confirmed the association of this allele with hepatic fat content, elevated circulating liver enzyme levels, and liver injury (3–7). Taken together, these studies support the notion that sequence differences in PNPLA3 lead to both fat deposition and inflammation in the liver.
The mechanism by which variation in PNPLA3 affects liver triglyceride content is not known. PNPLA3, alternatively referred to as adiponutrin, encodes a 481-amino acid protein of unknown function that belongs to the patatin-like phospholipase domain-containing family (8). The progenitor of the family, patatin, is a major protein of potato tubers and has nonspecific lipid acyl hydrolase activity (9). PNPLA3 is most closely related to PNPLA2 (adipose triglyceride lipase), which encodes the major triglyceride hydrolase of adipose tissue (10, 11). Human PNPLA3 was shown previously to hydrolyze triglycerides in vitro (12, 13), but overexpression of PNPLA3 in human embryonic kidney cells is not associated with any change in cellular triglycerides (13), and small interfering RNA-mediated knockdown of PNPLA3 does not affect the triglyceride content of 3T3-L1 cells (14). Thus, it remains unclear if PNPLA3 functions as a triglyceride hydrolase in vivo.
The tissue distribution and subcellular localization of PNPLA3 also have not been fully defined. Initially identified as a transcript restricted to the adipose lineage (8); subsequent studies indicated that PNPLA3 is also expressed in other tissues, including the liver (13) and adrenal (14). Baulande et al. (8) reported that recombinant PNPLA3 pelleted completely with the membrane fraction after ultracentrifugation at 150,000 × g. Immunolocalization studies using confocal microscopy localized epitope-tagged recombinant PNPLA3 to membranes (but not lipid droplets) in cultured adipocytes (8). These results were interpreted as being consistent with the prediction that PNPLA3 is an integral membrane protein with four transmembrane domains (8).
To examine the role of PNPLA3 in triglyceride metabolism and elucidate the mechanism by which the I148M substitution affects liver fat content, we expressed wild type and mutant forms of the protein in Sf9 cells, in cultured hepatocytes, and in livers of mice. Our data indicate that PNPLA3 is a lipid droplet protein that can catalyze the hydrolysis of triglycerides in vitro. Overexpression of wild type PNPLA3 had little effect on triglyceride concentration in hepatocytes, but the I148M mutation promoted triglyceride accumulation when fatty acid acylation was inhibited, as did expression of an isoform of PNPLA3 containing a substitution in the catalytic site (S47A). These findings are consistent with the I148M substitution interfering with hepatic triglyceride hydrolysis and thus promoting hepatic steatosis.
EXPERIMENTAL PROCEDURES
Materials
Rabbit anti-PNPLA3 polyclonal antibodies were raised against a synthetic peptide corresponding to the C-terminal 20 amino acids of human PNPLA3 (VPAGAEGLSTFPSFSLEKSL). Mouse monoclonal antibodies against adipose differentiation-related protein (ADRP)3 (catalog number 10R-All7a) and flotillin (catalog number 610821) were obtained from Fitzgerald Industries International (Acton, MA) and BD Biosciences, respectively. Mouse monoclonal anti-V5 antibody (catalog number R960-25) was purchased from Invitrogen, rabbit polyclonal anti-calnexin antibody (catalog number SPA860F) from Stressgen (Ann Arbor, MI), and anti-FLAG M2 antibody (catalog number F1804) from Sigma.
Molecular Modeling of PNPLA3
The sequences of patatin family members found in iterative PSI-BLAST searches (15) were aligned using PROMALS3D (16). The computer program SWISS-MODEL (17) was used to construct a homology model of the patatin-like domain of human PNPLA3 from this alignment, with the x-ray structure of heartleaf horsenettle patatin (Protein Data Bank code 1oxw) (18) as a template. The model was visualized in PyMOL (19).
In Vitro Assays of Recombinant PNPLA3 in Sf9 Cells
The cDNA for human PNPLA3 was inserted upstream of the cytomegalovirus promoter/enhancer elements in pcDNA3.1- TOPO-V5-His (Invitrogen). Single nucleotide changes were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by Sanger sequencing. A V5-epitope tag (GKPIPNPLLGLDST) was placed at the C terminus of each plasmid construct except for PNPLA3-S47A. To produce recombinant baculoviruses, the human PNPLA3 cDNA was inserted into a pFASTBac1 vector (Invitrogen). A FLAG tag (DYKDDDDK) and a tandem array of 10 histidines were inserted at the C terminus of PNPLA3 using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). All constructs were confirmed by DNA sequencing. Baculoviruses were prepared using the Bac-to-Bac baculovirus expression system protocol (Invitrogen) for Sf9 cells. Conditioned medium from baculovirus-infected Sf9 cells was used as a viral stock.
Sf9 cells obtained from Invitrogen were grown to a density of 1 × 106 cells/ml (Grace's medium, Sigma) and infected (multiplicity of infection of 2) with recombinant baculovirus. Partially purified PNPLA3 was prepared as described by Jenkins et al. (12). Seventy-two hours after infection, cells were centrifuged at 2600 × g for 15 min and resuspended in 30 ml of lysis buffer (25 mm Na2HPO4, pH 7.8, containing 20% (v/v) glycerol and 2 mm 2-mercaptoethanol). Cells were lysed by sonication (1-s bursts, 40% power, 30 times with a Branson Digital Sonifier 450 (Danbury, CT)) and centrifuged at 100,000 × g for 1 h to separate cytosolic and membrane fractions. The cytosolic fraction (2 mg/ml) was mixed with 3 ml of cobalt-resin (Pierce) for 2 h at 4 °C and then loaded into an Econo-column (1.5 × 10 cm; Bio-Rad). The column was washed with 10 column volumes of lysis buffer plus 500 mm NaCl. PNPLA3 was eluted using an imidazole gradient (0–200 mm) in lysis buffer, and fractions containing the highest concentration of PNPLA3, as determined by Western blotting (see below), were used in the enzymatic assays. Triolein emulsions were prepared by sonicating 1.5 mg of unlabeled triolein (Sigma), 20 × 106 cpm of [9,10-3H]triolein (60 μCi/μmol), and 40 μg of egg yolk phosphatidylcholine (Sigma) in 1 ml of 170 mm potassium phosphate (K2HPO4), pH 7.0, containing 200 μm sodium taurocholate. After sonication, disodium EDTA and dithiothreitol were added to the emulsion to a final concentration of 4 mm. For each assay, 100 μl of the substrate, [9,10-3H]triolein, was mixed with partially purified PNPLA3 (20–80 μg οf protein) in a glass tube to a final concentration of 60 μm [9,10-3H]triolein. The mixture was incubated at 37 °C for 15 min before the addition of 100 μl of butanol. The mixture was vortexed and then separated by centrifugation at 400 × g for 10 min. A total of 20 μl of the extracted lipids was loaded onto TLC plates and developed using chloroform/methanol/30% NH4OH (65/25/5) as the mobile phase. The lipids were visualized with iodine vapor, and the free fatty acid bands were scraped from the plates and quantitated by scintillation counting.
SDS-PAGE and Immunoblot Analysis
Protein concentrations were determined using the Bradford assay according to the manufacturer's protocol (Bio-Rad). An equivalent proportion (by volume) of each cell fraction was added to 5× sample loading buffer (0.313 M Tris-HCl, pH 6.8, 10% SDS, 0.05% bromphenol blue, 50% glycerol, 0.4 m dithiothreitol) to a final concentration of 1×. After heating to 95 °C for 5 min, the proteins were size-fractionated by 10% SDS-PAGE at 150 V and then transferred to a nitrocellulose membrane (Amersham Biosciences) at 100 V for 1 h (20). The membranes were incubated in TBST buffer (0.05 m Tris, 0.138 m NaCl, 2.7 μm KCl, 0.1% Tween 20, pH 8.0) with 5% dry nonfat milk (Nestle) at 4 °C overnight before adding the primary antibodies. Monoclonal anti-V5 antibody (Invitrogen) and a polyclonal anti-calnexin antibody (Stressgen, Ann Arbor, MI) were diluted 1:5,000 in TBST buffer with 5% dry nonfat milk and incubated with membranes for 60 min. Membranes were washed three times for 10 min each in TBST buffer. Horseradish peroxidase-conjugated donkey anti-rabbit IgG or sheep anti-mouse IgG (Amersham Biosciences) was diluted (1:10,000) in TBST buffer plus 5% dry milk and incubated with membranes for 60 min. Membranes were subject to three 10-min washes in TBST and visualized using SuperSignal-enhanced chemiluminescence (Pierce).
Expression of PNPLA3 in Cultured Cells
To generate cells that stably express PNPLA3, a human hepatoma cell line (HuH-7) was transfected with plasmids expressing wild type or mutant PNPLA3-V5. Three days after transfection, cells were passaged at low density in DMEM with 10% FCS containing 1 mg/ml G418 (Invitrogen). Medium was changed every 2 days. After 7 days, cells were suspended with trypsin, counted, and seeded in 96-well plates at a density of 2 cells/well in DMEM with 10% FCS containing 1 mg/ml G418. Wells were inspected with a light microscope. Those wells containing a single colony were treated with trypsin, and the cells were transferred to 48-well plates. Once cells reached 80% confluence, they were split into 6-well plates, and expression of PNPLA3 was confirmed by immunoblotting.
HuH-7 cells were grown to 90% confluence in DMEM with 10% FCS plus 100 IU/ml penicillin and 100 μg/ml streptomycin and infected with recombinant adenoviruses (pShuttle, Clontech). After 48 h the medium was replaced with serum-free DMEM with 1 μCi of [14C]palmitic acid. After 4 h, the medium was changed to DMEM with 10% FCS, and 20 and 44 h later, the lipids were extracted from cells with chloroform/methanol using the method of Bligh and Dyer (21). The lipids were separated by TLC using hexanes/diethyl ether/acetic acid (80/20/2) and visualized with iodine vapor and with also with phosphorimaging (Storm 820 PhosphorImager, Amersham Biosciences). Bands were also scraped from the TLC plate, and the radioactivity was counted using a scintillation counter.
In another set of experiments, HuH-7 cells were infected with recombinant adenoviruses for 48 h and then treated with 100 μm oleate and 1 μCi of [14C]palmitic acid and allowed to grow for 24 h. After 24 h, the medium was changed to DMEM and 10% FCS plus 5 μm triacsin C (Sigma), an inhibitor of long-chain acyl-CoA synthetase (22). At timed intervals, the lipids were extracted from cells and analyzed by TLC as described above. The relative intensities of the bands on the TLC plate were quantitated using phosphorimaging. The signal from cells expressing wild type PNPLA3 at the zero time point was assigned a value of 1.
Adenovirus-mediated Expression of PNPLA3 in Mice
Male C57Bl/6J mice purchased from Jackson Laboratory (Bar Harbor, ME) were housed in colony cages (≤5 mice/cage) maintained on a 12-h light/12-h dark daily cycle and fed standard chow diets (catalog number 7001; 4% fat, Harlan Teklad, Madison, WI). Five groups of six 12-week-old mice each were injected with 1.25 × 1011 recombinant adenoviral particles. Three days after injection, mice were sacrificed after a 4-h fast, and the liver and blood were collected. Lipids were extracted from 100–200 mg of frozen liver samples using the Folch and Lees method (23). Triglyceride, free fatty acid, total cholesterol, cholesteryl ester, free cholesterol, and phosphatidylcholine were measured using enzymatic assays (Infinity, Thermo Electron Corp. (Louisville, CO) and Wako Inc. (Richmond, VA)) and normalized to sample weight. Serum levels of alanine aminotransferase, aspartate aminotransferase, triglyceride, cholesterol, and glucose were measured using the Vitros 250 system (GMI, Inc.). All research protocols were reviewed and approved by the Institutional Animal Care and Use Committee of University of Texas Southwestern Medical Center at Dallas.
For Oil Red O staining, the liver was fixed in 4% paraformaldehyde for 2 days and equilibrated in 10% sucrose for 12 h and then in 18% sucrose for 12 h at 4 °C prior to cryosectioning. Cryosections of liver stored at −80 °C were brought to room temperature and air-dried for at least 2 h and then fixed in methanol-free 4% paraformaldehyde. Slides were washed with distilled water three times and then incubated for 10 min in 0.18% Oil Red O (Sigma) prepared in 60% isopropyl alcohol (24). The slides were washed in distilled water five times. Nuclei were counterstained with hematoxylin, and coverslips were affixed with aqueous mounting medium (Vector Laboratories, Inc., Burlingame, CA).
Subcellular Fractionation
For cell fractionation studies, transfected cells were disrupted by passage through a 27-gauge needle in 450 μl of Buffer A (50 mm Tris-Cl, pH 7.0, 250 mm sucrose, and 1 mm EDTA with protease inhibitors (Roche Applied Science)). Cell lysates were centrifuged (1000 × g), and membranes were isolated by centrifugation at 100,000 × g for 45 min at 4 °C. The pellet was resuspended in 200 μl of Buffer A, and equal proportions of the supernatant and pellet were analyzed by immunoblotting. For membrane extraction experiments, the membrane fractions prepared as described above were divided into four equal parts and centrifuged at 100,000 × g at 4 °C for 1 h. The membrane pellets were resuspended in 10 mm Tris-HCl (pH 7.4) and incubated on ice for 1 h in buffer containing either 1 m NaCl, 0.2 m Na2CO3 (pH 11.3), 1% Triton X-100, or 2% SDS (pH 7.4). Soluble and insoluble fractions were separated by centrifugation at 100,000 × g at 4 °C for 1 h.
Lipid droplets were isolated from HuH-7 cell lines that stably expressed wild type PNPLA3 and PNPLA3-I148M as described (25), except that cells were disrupted by passage through a 21-gauge needle 25 times, and the homogenates were centrifuged at 1,000 × g for 10 min at 4 °C to pellet nuclei and unbroken cells. The supernatant was removed, and 100 μl was retained for immunoblot analysis. The remaining postnuclear supernatant was adjusted to a concentration of 20% sucrose using Buffer C (20 mm Tris-HCl, pH 7.4, 1 mm EDTA) plus 60% sucrose and added to the bottom of an ultracentrifuge tube. A step gradient was constructed by overlaying the postnuclear supernatant with 5 ml of Buffer C plus 5% sucrose followed by Buffer C alone. The tube was clamped and centrifuged at 28,000 × g for 30 min at 4 °C. After centrifugation, the lipid layer was isolated by slicing the tube, combined with 20 volumes of −80 °C acetone, and stored overnight at −20 °C. The precipitated proteins were pelleted by centrifugation at 4,300 × g for 1 h at 4 °C and then resuspended in Buffer C plus 1% SDS. The lipid droplet proteins and postnuclear supernatant were subjected to immunoblotting using an anti-V5 antibody and monoclonal anti-ADRP antibody (Fitzgerald Industries International, Concord, MA). Experiments were also conducted using a modification of this protocol in which lipid droplets were isolated by centrifugation at 100,000 × g.
Immunofluorescence Microscopy
HuH-7 cells were grown on glass coverslips. The cells were washed with PBS and then incubated for 4 h at 37 °C in serum-free medium with 6 × 109 recombinant adenoviral particles. The cells were refed with DMEM with 10% FCS in the presence or absence of oleate-conjugated albumin (400 μm). The following day, cells were washed with PBS, fixed in 4% (w/v) paraformaldehyde in PBS for 15 min, quenched in 1% (w/v) l-glycine in PBS for 10 min, and permeabilized in 0.05% (w/v) Triton X-100 (Pierce) in PBS for 10 min at room temperature. Fixed/permeabilized cells were then blocked for 30 min in PBS plus 5% (w/v) bovine serum albumin and 0.025% (w/v) Triton X-100. Cells were stained with the anti-V5 antibody (dilution 1:500) overnight at 4 °C followed by a 1-h incubation at room temperature with Alexa Fluor 568-conjugated goat anti-mouse IgG (Invitrogen) (2 μg/ml) and 1 μg/ml of boron dipyrromethene (Invitrogen). Both incubations with the primary and secondary antibodies were followed by three 5-min washes in PBS with 0.025% Triton X-100. The coverslips were rinsed in PBS prior to mounting with a 4′,6′-diamino-2-phenylindole-containing mounting medium (Vectashield, Vector Laboratories, Burlingame, CA) and imaged on a Leica TCS SP5 confocal microscope using an HCX PL APO ×63/1.40–0.60 oil objective.
RESULTS
PNPLA3-I148M Interferes with Triglyceride Hydrolysis in Vitro
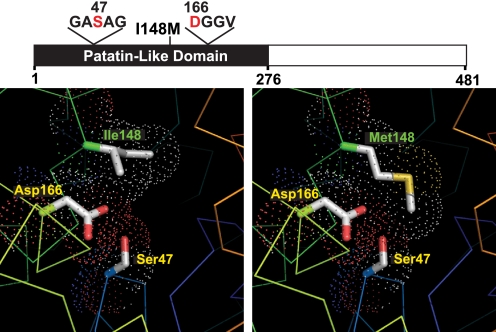
A structural model of the patatin-like domain of PNPLA3 was developed to examine the effect of substituting methionine for isoleucine at residue 148. In this model, the side chain of the isoleucine at residue 148 is located adjacent to the Ser47-Asp166 catalytic dyad (Fig. 1) and comprises part of a hydrophobic substrate-binding groove in the active site. Substitution of methionine for isoleucine is not predicted to alter the orientation of the catalytic dyad, but the longer side chain of methionine restricts access of substrate to the catalytic serine, thus inactivating PNPLA3.
FIGURE 1.
Structural model of wild type and mutant (I148M) PNPLA3. The domain structure of PNPLA3, showing the patatin-like domain (black) and locations of the catalytic dyad (Ser47 and Asp166) and the I148M substitution associated with increased hepatic triglyceride content (26), is shown. Structure models of normal (Ile148) and mutant (Met148) PNPLA3 are shown in the left and right panels, respectively. Protein traces are rainbow-colored from N to C terminus (blue to red) with side chains of catalytic dyad residues (positions 47 and 166) shown. The dots indicate a space-filling model corresponding to van der Waals atomic radii. Oxygen and sulfur atoms are colored red and yellow, respectively. The model was built using heartleaf horsenettle (Solanum cardiophllum) patatin (Protein Data Bank code 1oxw) as a template. Images were prepared in PyMOL (19).
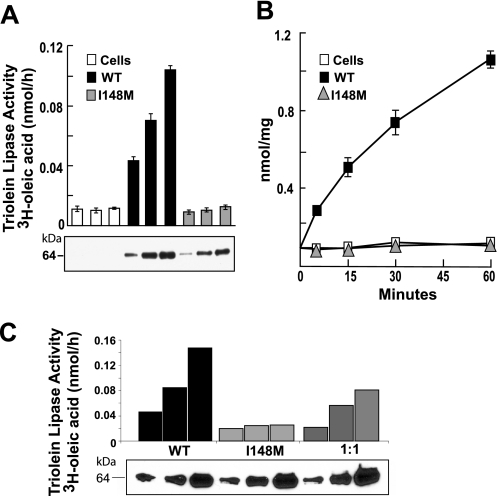
To test this prediction, we expressed both the wild type and the mutant forms of the enzyme in Sf9 cells and determined the effect of the substitution on the ability of the enzyme to hydrolyze triglyceride, as described previously by Jenkins et al. (12). Wild type PNPLA3 hydrolyzed emulsified triolein (Fig. 2A) in both a dose-dependent and time-dependent manner (Fig. 2, A and B). In contrast, PNPLA3-I148M did not catalyze triglyceride hydrolysis at any enzyme level or time point examined (Fig. 2B). To determine if the mutant isoform of PNPLA3 inhibited the activity of the wild type protein, equal amounts of partially purified wild type and mutant PNPLA3 were added to the substrate. The triglyceride hydrolase activity was intermediate between the wild type and mutant preparations (Fig. 2C). Thus, the mutant protein did not inhibit the activity of the wild type enzyme, at least in vitro. These findings are consistent with the prediction that the substitution of methionine for isoleucine at residue 148 of PNPLA3 limits access of substrate to the catalytic residues.
FIGURE 2.
Effect of I148M substitution on triglyceride hydrolysis in vitro. FLAG-tagged human wild type and mutant (I148M) PNPLA3 were partially purified from Sf9 cells using nickel affinity chromatography as described under “Experimental Procedures.” A, a total of 20, 40, or 80 μg of protein was incubated at 37 °C for 15 min with 3H-triolein emulsions (60 μm of [9,10-3H]triolein). Lipids were extracted with butanol and separated by TLC, and the free fatty acid bands were excised and quantitated by scintillation counting. B, emulsions of radiolabeled triolein were incubated with 40 μg of partially purified recombinant PNPLA3 for the times indicated. The free fatty acid release was quantitated as described in A. C, partially purified wild type PNPLA3, PNPLA3-I148M and a 1:1 mixture of wild type and mutant PNPLA3 were incubated with [3H]triolein emulsions, and free fatty acid release was measured as described in A. Proteins were examined by immunoblotting using an anti-FLAG epitope antibody (Sigma). Each experiment was repeated twice, and similar results were obtained. WT, wild type.
Functional Analysis of the I148M Substitution in PNPLA3 in Vivo
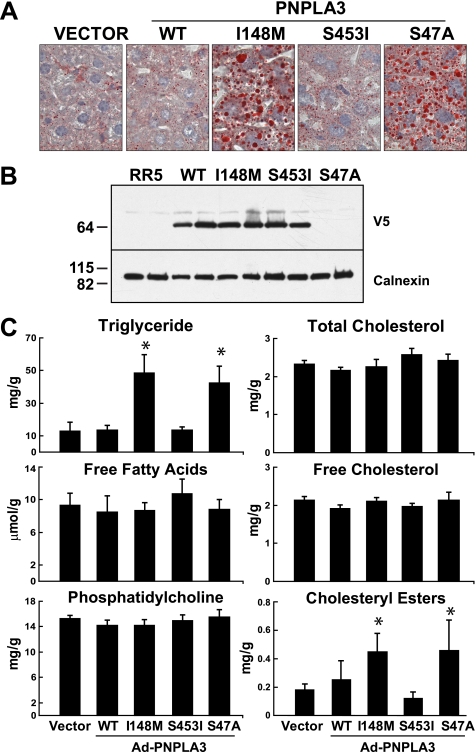
To determine the effect of the I148M substitution in vivo, we used recombinant adenoviruses to overexpress wild type and mutant forms of human PNPLA3 in the livers of mice (Fig. 3). Oil Red O staining of the liver sections revealed that overexpression of wild type PNPLA3 had no discernable effect on liver triglyceride content or distribution. Both the number and size of the lipid droplets in mice expressing wild type PNPLA3 were similar to those observed in animals infected with vector alone (Fig. 3A). Similarly, infection with virus expressing the S453I allele, a sequence variation associated with lower hepatic triglyceride content in humans (26), did not affect the pattern or amount of Oil Red O staining. Enzymatic measurements of tissue triglyceride levels confirmed that there was no change in hepatic triglyceride content in these mice (Fig. 3C). In contrast to these results, administration of viruses expressing PNPLA3-I148M or a catalytically dead form of the enzyme (S47A) was associated with a dramatic increase in the number of lipid droplets (Fig. 3A) and in the tissue levels of triglycerides and cholesterol esters (Fig. 3C). The size of the lipid droplets tended to be larger in the mice expressing either PNPLA3-I148M or PNPLA3-S47A. Similar levels of recombinant protein were expressed in the livers of mice receiving wild type and I148M viruses (Fig. 3B). (No V5 tag was present on the PNPLA3-S47A construct, so the protein was not detected by immunoblotting in this experiment). No significant differences were found in the levels of hepatic free fatty acids, phospholipids, or free cholesterol in any of the mice in this study (Fig. 3C and Table 1). Similar results were obtained in ob/ob mice, which have high levels of liver triglycerides. Hepatic steatosis in these animals was not altered by overexpression of wild type PNPLA3 and was exacerbated by the mutant proteins (I148M and S47A) (supplemental Fig. 1).
FIGURE 3.
Adenovirus-mediated expression of PNPLA3 in the livers of mice. A, 12-week-old male C57BL/6J mice (n = 6 mice/group) were injected with 1.25 × 1011 recombinant adenovirus particles expressing no insert (Vector) or V5-tagged versions of wild type PNPLA3 (WT) PNPLA3-I148M, PNPLA3-S453I, or untagged PNPLA3-S47A. Three days after injection, Oil Red O staining was performed on liver sections as described under “Experimental Procedures.” Pictures were taken by a Leica microscope at ×40 magnification. B, immunoblot analysis of PNPLA3-V5 expression in lysates from livers of mice injected with recombinant adenoviruses. Representative blots from two mice in each group are shown. C, lipids were extracted from the livers and assayed using enzymatic kits as described under “Experimental Procedures.” Values are means ± S.D. p values were calculated using analysis of variance and corrected for multiple testing using the Bonferroni procedure. *, p < 0.0001 for the triglyceride and cholesterol ester (vector versus S47A and versus I148M). This experiment was repeated three times, and the results were similar.
TABLE 1.
Characterization of liver and serum lipids and enzymes in mice infected with recombinant adenoviruses expressing wild type or mutant PNPLA3
| Empty adenovirus | PNPLA3 adenovirus |
||||
|---|---|---|---|---|---|
| Wild type | I148M | S453I | S47A | ||
| Body weight (g) | 27.5 ± 2.5 | 28.2 ± 1.0 | 27.2 ± 1.6 | 27.4 ± 1.0 | 28.1 ± 2.4 |
| Liver/body weight (×100) | 5.2 ± 0.2 | 5.4 ± 0.4 | 5.63 ± 0.3 | 5.28 ± 0.1 | 5.49 ± 0.3 |
| Liver | |||||
| Triglyceride (mg/g) | 12.7 ± 5.6 | 13.5 ± 2.9 | 48.4 ± 11.3 | 13.5 ± 2.0 | 42.5 ± 10.1 |
| Phosphotidylcholine (mg/g) | 15.3 ± 0.5 | 14.2 ± 0.8 | 14.2 ± 0.9 | 15.5 ± 1.2 | 14.9 ± 0.9 |
| Free fatty acids (μmol/g) | 9.3 ± 1.5 | 8.5 ± 1.9 | 8.7 ± 0.9 | 8.8 ± 1.2 | 10.7 ± 1.8 |
| Cholesterol esters (mg/g) | 0.2 ± 0.0 | 0.3 ± 0.0.1 | 0.5 ± 0.1 | 0.1 ± 0.0 | 0.5 ± 0.2 |
| Free cholesterol (mg/g) | 2.1 ± 0.1 | 1.9 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.2 | 2.1 ± 0.1 |
| Cholesterol (mg/g) | 2.3 ± 0.1 | 2.2 ± 0.1 | 2.4 ± 0.2 | 2.3 ± 0.2 | 2.6 ± 0.2 |
| Blood | |||||
| Alanine aminotransferase (units/liter) | 53.3 ± 13.0 | 102.8 ± 42.5 | 92.2 ± 29.4 | 128.7 ± 63.1 | 85.5 ± 30.8 |
| Aspartate aminotransferase (units/liter) | 65.3 ± 15.6 | 142.0 ± 61.5 | 115.2 ± 28.5 | 146.5 ± 56.6 | 119.0 ± 30.8 |
| Cholesterol (mg/dl) | 103.8 ± 12.0 | 94.3 ± 17.5 | 98.5 ± 10.1 | 101.2 ± 8.3 | 101.0 ± 10.5 |
| Trigylceride (mg/dl) | 123.0 ± 13.9 | 119.3 ± 24.4 | 107.7 ± 12.8 | 108.0 ± 14.9 | 127.0 ± 19.2 |
| Glucose (mg/dl) | 190.8 ± 30.7 | 128.8 ± 24.5 | 134.2 ± 12.9 | 129.2 ± 44.8 | 160.5 ± 45.7 |
These results indicate that the PNPLA3-I148M mutation promotes the accumulation of triglycerides and cholesterol esters in mouse liver, either by inhibiting hydrolysis of the neutral lipids or by promoting re-esterification of glycerides and cholesterol. To distinguish between these possibilities, we used recombinant adenoviruses to express PNPLA3 in human hepatoma cells (HuH-7).
Functional Analysis of the I148M Substitution in PNPLA3 in Cultured Hepatoma (HuH-7) Cells
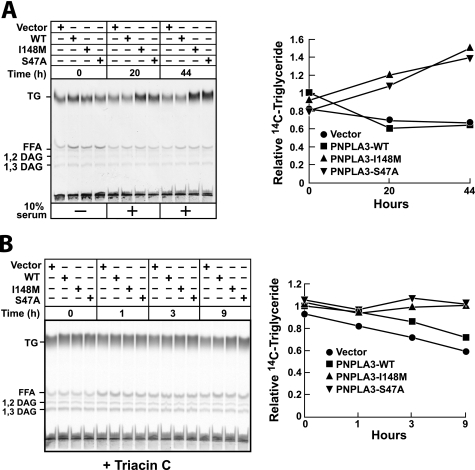
To determine the effect of PNPLA3 on cellular triglyceride content, HuH-7 cells expressing wild type or mutant forms of PNPLA3 were grown in serum-free medium plus [14C]palmitate for 4 h. The medium was then changed, 10% FCS was added, and the cells were incubated for 20 and 44 h. Cellular lipids were separated by TLC and visualized by phosphorimaging. At time 0, expression of wild type and mutant PNPLA3 did not affect cellular triglyceride content (Fig. 4A). After 20 h, the amount of triglyceride was similar in cells expressing the wild type protein and in cells expressing the vector alone. A ∼2-fold increase in triglyceride level was seen in cells expressing PNPLA3-I148M or an isoform of PNPLA3 in which alanine was substituted for the catalytic serine (S47A).
FIGURE 4.
Triglyceride hydrolase activity of PNPLA3 in HuH-7 cells. A, HuH-7 cells were infected with adenoviruses encoding wild type or mutant forms of human PNPLA3. After 48 h, the medium was changed to DMEM plus 1 μCi [14C]palmitate. After 4 h, the medium was changed to DMEM plus 10% FCS, and the cells were harvested after the indicated time intervals. Lipids were extracted from the cells, fractionated by TLC, and visualized using a Storm 820 PhosphorImager (Amersham Biosciences). The activity of each band was quantitated using ImageQuant TL analysis software (Molecular Dynamics). The relative amount of labeled triglyceride was expressed as a fraction of the value obtained at the zero time point from cells expressing wild type PNPLA3. B, HuH-7 cells were infected with adenoviruses encoding wild type or mutant forms of human PNPLA3. After 48 h, the medium was changed to DMEM plus 10% FCS plus 1 μCi of [14C]palmitate. After 24 h, the medium was changed to DMEM plus 10% FCS plus triacsin C (5 μm). Cells were harvested after the indicated time intervals, and lipids were analyzed by TLC as described above. These experiments were repeated twice, and the results were similar.
To determine if the increased triglyceride level associated with expression of either mutant protein was due to a decrease in hydrolysis or an increase in fatty acid esterification, oleate-loaded cells were treated with triacsin C (5 μm). A modest but consistent decrease in cellular triglyceride content was seen in hepatocytes infected with empty adenovirus, and a similar decrease was observed in cells expressing wild type PNPLA3 (Fig. 4B). In contrast, no change or, in some experiments, a modest increase in triglyceride content was seen in cells expressing the I148M and S47A forms of the virus. These results suggest that the I148M mutation promotes accumulation of triglyceride by inhibiting the hydrolysis of triglycerides.
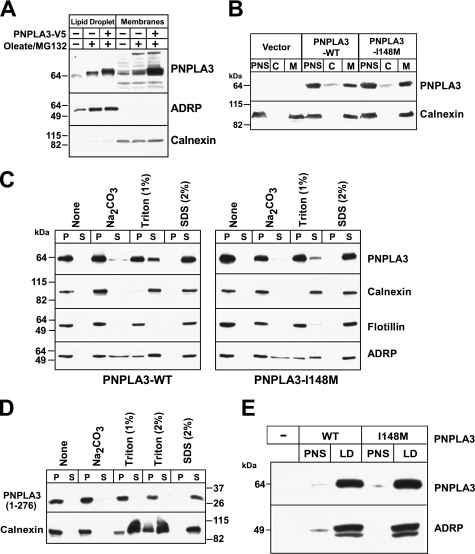
PNPLA3 Is Membrane-bound and Is Present in Lipid Droplets
PNPLA2, the PNPLA family member most similar to PNPLA3, is tightly associated with the ER membrane and traffics from the ER to lipid droplets (27). To determine if PNPLA3 follows a similar itinerary, we examined the distribution of endogenous PNPLA3 in Huh7 cells. In these cells, the protein was equally distributed between the membrane and lipid droplet fractions (Fig. 5A), and only trace amounts were detected in the cytoplasmic fraction (data not shown). Treatment of cells with oleate and MG-132, a proteasome inhibitor, increased the cellular content of endogenous immunodetectable PNPLA3, which was present both in the lipid droplets and in the membrane fraction. The distribution of an epitope-tagged recombinant protein was similar to that of the endogenous protein (Fig. 5A).
FIGURE 5.
Subcellular localization of PNPLA3 in cultured hepatoma (HuH-7) cells. A, HuH-7 cells were grown in the absence and presence of 400 μm oleate for 24 h and MG132 (2.5 μm) for 12 h. Membranes and lipid droplets were isolated by density gradient ultracentrifugation as described under “Experimental Procedures.” One-twentieth of the total volume of each fraction was analyzed by SDS-PAGE and immunoblotted for PNPLA3, ADRP, and calnexin. A rabbit polyclonal antibody to the last 20 amino acids of human PNPLA3 was used to detect endogenous PNPLA3. B, postnuclear supernatants (PNS) prepared from HuH-7 cells stably expressing PNPLA3-V5 were subjected to ultracentrifugation at 100,000 × g to separate the cytoplasm (C) and membranes (M) as described under “Experimental Procedures.” One-twentieth of the total volume of each fraction was analyzed by SDS-PAGE, and immunoblotting using a V5 monoclonal antibody and rabbit anti-calnexin polyclonal antibody. C and D, membrane fractions from HuH-7 cells expressing wild type PNPLA3, PNPLA3-I148M (C), or truncated PNPLA3 (D) were suspended in 450 μl of 10 mm Tris, pH 7.4. Membranes were repelleted by centrifugation at 100,000 × g for 1 h at 4 °C and resuspended in the indicated buffers as described under “Experimental Procedures.” Pellet (P) and supernatant (S) fractions were subjected to 10% SDS-PAGE and analyzed by immunoblotting. E, lipid droplets were isolated from HuH-7 cells stably expressing recombinant PNPLA3 and PNPLA3-I148M. The postnuclear supernatant was adjusted to a sucrose concentration of 20%, applied to the bottom of a discontinuous sucrose gradient, and centrifuged at 28,000 × g as described under “Experimental Procedures.” A total of 20 μg of protein from the PNS and 17 μg from the lipid droplet fractions were subjected to immunoblotting using antibodies against V5 and ADRP, a lipid droplet marker. There experiments were repeated at least twice, and the results were similar.
To determine if the I148M mutation alters the association of PNPLA3 with membranes, we used stably transfected HuH-7 cell lines expressing wild type or mutant PNPLA3 to compare the partitioning of PNPLA3 between the cytosolic and membrane fractions. Immunoblot analysis of equal proportions of membranes and cytosol revealed that the major portion of wild type PNPLA3 was in the membrane fraction (Fig. 5B). Wild type PNPLA3 was not dislodged from membranes by incubation in high salt (1 m NaCl) (data not shown), and only a small fraction of the protein entered the soluble fraction after the addition of sodium carbonate (0.2 m, pH 11.3) (Fig. 5C). The protein was poorly extracted with 1% Triton X-100 at 4 °C but was completely solubilized with 2% SDS. Substitution of methionine for isoleucine at residue 148 did not affect the localization or elution of the protein from membranes by salt, pH, or detergent (Fig. 5C). Calnexin, a membrane-bound ER protein (28); flotillin, a protein located in lipid rafts (29); and ADRP, a resident protein of lipid droplets, served as controls in this experiment.
We repeated this experiment using a construct expressing only the patatin domain of the protein (residues 1–276). The truncated version of the protein was also tightly bound to membranes, as reflected in the failure to dislodge the protein from the membrane with high salt, high pH, or 2% Triton X-100 (Fig. 5D). Thus, the patatin domain contains the residues required for membrane association.
To determine if the I148M substitution affects the association of PNPLA3 with lipid droplets, we supplemented cells stably expressing the wild type and mutant protein with oleate and then isolated the lipid droplets as described (25). Substitution of methionine for isoleucine at residue 148 did not affect the relative amount of PNPLA3 associated with lipid droplets (Fig. 5E). Taken together, these findings indicate that PNPLA3 is strongly associated with membranes and with lipid droplets and that the I148M mutation does not alter the partitioning of the protein between these fractions.
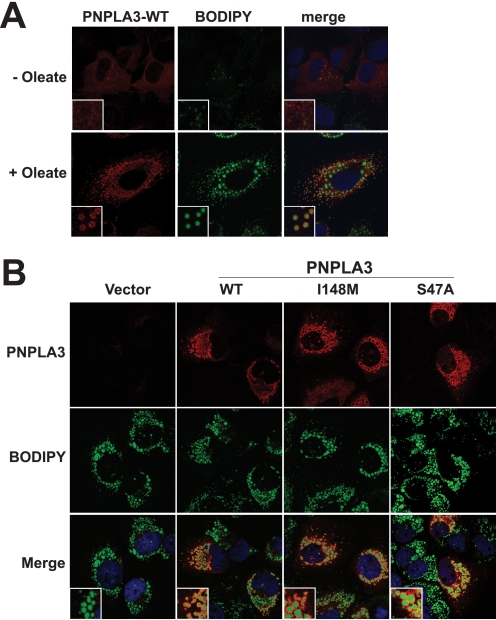
Immunolocalization of Wild Type and Mutant PNPLA3
Because hydrophobic proteins can adhere nonspecifically to lipids droplets that are fragmented during isolation (30), we confirmed the localization of PNPLA3 to lipid droplets by immunofluorescence microscopy (Fig. 6A). In cells grown in 10% FCS without the addition of oleate, PNPLA3 had a diffuse staining pattern consistent with an ER and cytoplasmic localization. After the addition of oleate to cells, the protein clustered into larger puncta that encircled the neutral lipid droplets, which were stained using a fluorescent lipid dye, boron dipyrromethene (Fig. 6A). Substitution of methionine for isoleucine at residue 148 or of alanine for serine at residue 47 did not affect the localization of PNPLA3 to lipid droplets (Fig. 6B). However, it appeared that the lipid droplets were larger in size in cells expressing the mutant proteins.
FIGURE 6.
Immunolocalization of recombinant human PNPLA3 to lipid droplets in oleate-treated HuH-7 cells. A, HuH-7 cells grown on glass coverslips were infected with a recombinant adenovirus encoding wild type PNPLA3. Cells were cultured for 16 h in DMEM plus 10% FCS with or without oleate-conjugated albumin (400 μm), fixed with 4% paraformaldehyde, permeabilized with 0.05% Triton X-100, and stained with an anti-V5 antibody and a goat anti-mouse antibody conjugated to Alexa Fluor 568. Lipid droplets were visualized using 1 μg/ml boron dippyrromethane (BODIPY). B, HuH-7 cells were grown on glass coverslips and infected with a control recombinant adenovirus (Vector) or with adenoviruses encoding either wild type PNPLA3 or PNPLA3-I148M or transfected with a plasmid encoding PNPLA3-S47A. All constructs contained a V5 epitope tag at the C terminus. The cells were cultured in medium containing oleate-conjugated albumin (400 μm) for 16 h and processed for immunofluorescence as described under “Experimental Procedures.”
DISCUSSION
The results of this study show that the amino acid substitution in PNPLA3 that confers susceptibility to nonalcoholic fatty liver disease (I148M) inhibits catalytic activity of the enzyme. Structural modeling indicated that the substitution does not perturb the position or orientation of the catalytic dyad residues (Ser-47 and Asp-166). Rather, the side chain of the methionine at residue 148 extends into the catalytic site, shielding the serine side chain from access to substrate. In vitro assays using recombinant PNPLA3 were consistent with the predictions of the structural model; whereas the wild type enzyme hydrolyzed emulsified triglycerides, the mutant enzyme was inactive against this substrate. Studies in mice indicated that the accumulation of triglyceride associated with the Met148 isoform is due to expression of the mutant protein, rather than loss of the wild type enzyme activity. Overexpression of wild type human PNPLA3 did not affect liver triglyceride content in mice, suggesting that PNPLA3 may not be a major triglyceride hydrolase in this organ. In contrast, expression of the I148M isoform in the liver of mice increased liver triglyceride content. These findings were recapitulated in cultured hepatoma cells (HuH-7), even when de novo triglyceride formation from acyl-CoAs was inhibited by triacsin C. These data are consistent with the notion that expression of the mutant protein promotes triglyceride accumulation by inhibiting triglyceride hydrolysis in the cell.
The predicted structure of the patain domain of PNPLA3 is highly congruent with the crystal structure of heartleaf horsenettle patatin (18) (Fig. 1). Patatins differ from the classical lipases in two key respects. First, patatins use a catalytic dyad (Ser-Asp), rather than the catalytic triad (Ser-His-Asp) usually present in lipases. The predicted catalytic serine of PNPLA3 (Ser47) lies in a highly conserved hydrolase motif (Gly-X-Ser-X-Gly) at a hairpin turn between a β-strand and an α-helix. The patatin fold brings this serine into close apposition to Asp166 at the edge of a putative substrate-binding groove formed by the side chains of several hydrophobic residues, including Ile148. In the wild type protein, the side chains of Ser47 and Asp166 project into the groove. When methionine is substituted for isoleucine at residue 148, the longer side chain of methionine is predicted to occlude access of substrates to the catalytic dyad (Fig. 1). These structural predictions are consistent with the observation that the I148M substitution abolishes cleavage of emulsified triglycerides (Fig. 2).
Second, the patatin structure does not contain a lid domain, which mediates interfacial activation in classical lipases. Therefore, it has been proposed that patatins act on solubilized lipids rather than on micelles like the lid domain-containing lipases (18). In contrast to plant patatins, which are soluble proteins (9, 18), PNPLA3 is tightly associated with membranes and with lipid droplets. Constructs comprising the patatin domain alone were also firmly anchored to the membrane (Fig. 5). Whereas these experiments do not preclude additional sites of interaction between PNPLA3 and membranes, they indicate that the patatin domain of PNPLA3 interacts with the membrane. Previous studies proposed that PNPLA3 contains membrane spanning domains on the basis of its predicted secondary structure (8). However, our model of the tertiary structure of the enzyme indicates that all of the helices form part of the globular structure of the protein and do not span the membrane. The active site of PNPLA3 is surrounded by hydrophobic and lysine residues that are exposed on the protein surface and probably mediate interaction with the membrane. The close proximity of the active site to the membrane suggests that PNPLA3 is active on lipids in membranes and/or lipid droplets rather than solubilized lipids. Experiments are in progress to define the site of attachment of PNPLA3 to membranes.
The association of PNPLA3 with lipid droplets, together with the finding that the I148M substitution inactivates triglyceride hydrolysis by PNPLA3, suggests that the association between the I148M allele and hepatic triglyceride content in humans (26) may be explained by a simple model in which PNPLA3 normally serves to hydrolyze hepatic triglycerides. In this model, the loss of enzymatic activity associated with the I148M substitution leads to triglyceride accumulation in this organ. However unlike PNPLA2 and hormone-sensitive lipase, which reduced liver triglycerides when overexpressed in this organ (31), overexpression of wild type PNPLA3 in mouse liver failed to lower triglyceride content, whereas overexpression of the mutant PNPLA3 actually increased hepatic triglyceride levels. These data suggest that PNPLA3 is not usually rate-limiting for triglyceride hydrolysis in the liver and that the increased liver triglyceride content associated with the I148M allele is due to the presence of the mutant protein rather than the absence of the wild type enzyme activity.
The lack of reduction in hepatic triglyceride content in the livers of mice expressing high levels of the wild type human PNPLA3 may reflect interspecies differences in the role of PNPLA3. This explanation seems unlikely because high level expression of PNPLA3 did not alter triglyceride levels in cultured human hepatocytes. An alternative explanation is that PNPLA3 stimulates fatty acid re-esterification, either directly, as proposed by Jenkins et al. (12), or indirectly by activating a signaling molecule that promotes triglyceride accumulation. If PNPLA3 promotes both hydrolysis and transacylation of triglycerides, then inactivation of the catalytic site may disrupt hydrolytic activity but spare the transacylase function of the enzyme, promoting triglyceride formation. In preliminary studies using recombinant PNPLA3, we failed to show appreciable transesterification of mono- and diglycerides by either the wild type or the mutant enzyme (data not shown). Additional studies will be required to clarify the role of PNPLA3 in hepatic triglyceride metabolism relative to the other lipases.
It remains possible that the inactive PNPLA3 isoforms sequester a cofactor required for hydrolysis or restrict access of the active allele (or another lipase) to the substrate, thus causing triglyceride accumulation. Many triglyceride hydrolases, including PNPLA2, the major triglyceride hydrolase in adipose tissue, require protein co-factors for activity (32), but a similar co-factor for PNPLA3 has not been identified. The mutant enzyme did not interfere with the wild type protein in our in vitro assay (Fig. 2C), but access to the substrate or to essential co-factors may be limited in vivo.
The localization of PNPLA3 to both the membrane and lipid droplet fractions in cultured hepatocytes mirrors the distribution reported previously for PNPLA2, the major triglyceride hydrolase of adipose tissue (27), and may represent partitioning of the protein between a membrane reservoir and an active pool on the surfaces of lipid droplets. Substitution of isoleucine 148 with methionine did not affect the partitioning of the protein between the membrane and cytosol or its localization into lipid droplets (Fig. 5E). To exclude the possibility that PNPLA3 associates nonspecifically with lipid droplets during the isolation procedure, a recognized artifact of lipid droplet isolation (30), we confirmed the localization of both the wild type and mutant proteins using immunofluorescence microscopy. Thus, it is unlikely that the mutation results in mistargeting the protein away from its normal sites of action.
Despite the apparent absence of a membrane-spanning domain, PNPLA3 is tightly associated with membranes; harsh treatment with high salt, high pH, or Triton X-100 failed to elute the protein from the membrane fraction. PNPLA3 may be targeted specifically to regions of the ER membrane that are destined to become nascent lipid droplets. Alternatively, PNPLA3 may be trafficked from the ER membrane to preexisting lipid droplets by components of the ER-Golgi transport machinery, as has recently been described for PNPLA2 (27). It is also possible that PNPLA3 performs distinct functions in membranes and lipid droplets. Elucidating the physiological substrate(s) of the enzyme will be essential to unraveling its role in lipid metabolism and in the pathogenesis of fatty liver disease.
Supplementary Material
Acknowledgments
We thank Christina Zhao, Sijing Niu, Liangcai Nie, Stephanie Blankenship, and Michele Alkalay for excellent technical assistance. We especially thank Richard W. Gross (Washington University, St. Louis, MO) for helpful discussions. We also thank James Richardson, Guosheng Liang, Jay Horton, and David Russell (University of Texas Southwestern) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL092550, UL1DE019684, PL1DK081182, and HL20948.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ADRP
- adipose differentiation-related protein
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- PBS
- phosphate-buffered saline
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. (2004) Hepatology 40, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 2.Browning J. D., Horton J. D. (2004) J. Clin. Invest. 114, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan B., Neuman R., Duan S. H., Weber J. L., Kwok P. Y., Saccone N. L., Wu J. S., Liu K. Y., Schonfeld G. (2000) Am. J. Hum. Genet. 66, 1699–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotronen A., Johansson L. E., Johansson L. M., Roos C., Westerbacka J., Hamsten A., Bergholm R., Arkkila P., Arola J., Kiviluoto T., Fisher R. M., Ehrenborg E., Orho-Melander M., Ridderstråle M., Groop L., Yki-Järvinen H. (2009) Diabetologia 52, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 5.Sookoian S., Castaño G. O., Burgueño A. L., Gianotti T. F., Risselli M. S., Pirola C. J. (2009) J. Lipid Res. 52, 2111–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romeo S., Sentinelli F., Dash S., Yeo G. S., Savage D. B., Leonetti F., Capoccia D., Incani M., Maglio C., Iacovino M., O'Rahilly S., Baroni M. G. (2010) Int. J. Obes. (Lond.) 34, 190–194 [DOI] [PubMed] [Google Scholar]
- 7.Kantartzis K., Peter A., Machicao F., Machann J., Wagner S., Königsrainer I., Königsrainer A., Schick F., Fritsche A., Häring H. U., Stefan N. (2009) Diabetes 58, 2616–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baulande S., Lasnier F., Lucas M., Pairault J. (2001) J. Biol. Chem. 276, 33336–33344 [DOI] [PubMed] [Google Scholar]
- 9.Galliard T. (1971) Biochem. J. 121, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kienesberger P. C., Oberer M., Lass A., Zechner R. (2009) J. Lipid Res. 50, S63–S68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson P. A., Gardner S. D., Lambie N. M., Commans S. A., Crowther D. J. (2006) J. Lipid Res. 47, 1940–1949 [DOI] [PubMed] [Google Scholar]
- 12.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 13.Lake A. C., Sun Y., Li J. L., Kim J. E., Johnson J. W., Li D., Revett T., Shih H. H., Liu W., Paulsen J. E., Gimeno R. E. (2005) J. Lipid Res. 46, 2477–2487 [DOI] [PubMed] [Google Scholar]
- 14.Kershaw E. E., Hamm J. K., Verhagen L. A., Peroni O., Katic M., Flier J. S. (2006) Diabetes 55, 148–157 [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei J., Kim B. H., Grishin N. V. (2008) Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 18.Rydel T. J., Williams J. M., Krieger E., Moshiri F., Stallings W. C., Brown S. M., Pershing J. C., Purcell J. P., Alibhai M. F. (2003) Biochemistry 42, 6696–6708 [DOI] [PubMed] [Google Scholar]
- 19.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, LLC, Palo Alto, CA [Google Scholar]
- 20.Romeo S., Yin W., Kozlitina J., Pennacchio L. A., Boerwinkle E., Hobbs H. H., Cohen J. C. (2009) J. Clin. Invest. 119, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 22.Omura S., Tomoda H., Xu Q. M., Takahashi Y., Iwai Y. (1986) J. Antibiot. 39, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 24.Sheehan D. C., Hrapchak B. B. (1980) Theory and Practice of Histotechnology, 2nd Ed., p. 205, Mosby, St. Louis [Google Scholar]
- 25.Brasaemle D. L., Wolins N. E. (2006) Current Protocols in Cell Biology, pp. 3.15.1–3.15.12, Wiley Interscience, New York [Google Scholar]
- 26.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., Hobbs H. H. (2008) Nat. Genet. 40, 1461–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soni K. G., Mardones G. A., Sougrat R., Smirnova E., Jackson C. L., Bonifacino J. S. (2009) J. Cell Sci. 122, 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada I., Rindress D., Cameron P. H., Ou W. J., Doherty J. J., 2nd, Louvard D., Bell A. W., Dignard D., Thomas D. Y., Bergeron J. J. (1991) J. Biol. Chem. 266, 19599–19610 [PubMed] [Google Scholar]
- 29.Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. (1997) J. Biol. Chem. 272, 13793–13802 [DOI] [PubMed] [Google Scholar]
- 30.Fujimoto T., Ohsaki Y. (2006) Ann. N.Y. Acad. Sci. 1086, 104–115 [DOI] [PubMed] [Google Scholar]
- 31.Reid B. N., Ables G. P., Otlivanchik O. A., Schoiswohl G., Zechner R., Blaner W. S., Goldberg I. J., Schwabe R. F., Chua S. C., Jr., Huang L. S. (2008) J. Biol. Chem. 283, 13087–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Cell Metabolism 3, 309–319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.