Abstract
Gpd1p is a cytosolic NAD+-dependent glycerol 3-phosphate dehydrogenase that also localizes to peroxisomes and plays an essential role in the cellular response to osmotic stress and a role in redox balance. Here, we show that Gpd1p is directed to peroxisomes by virtue of an N-terminal type 2 peroxisomal targeting signal (PTS2) in a Pex7p-dependent manner. Significantly, localization of Gpd1p to peroxisomes is dependent on the metabolic status of cells and the phosphorylation of aminoacyl residues adjacent to the targeting signal. Exposure of cells to osmotic stress induces changes in the subcellular distribution of Gpd1p to the cytosol and nucleus. This behavior is similar to Pnc1p, which is coordinately expressed with Gpd1p, and under conditions of cell stress changes its subcellular distribution from peroxisomes to the nucleus where it mediates chromatin silencing. Although peroxisomes are necessary for the β-oxidation of fatty acids in yeast, the localization of Gpd1p to peroxisomes is not. Rather, shifts in the distribution of Gpd1p to different cellular compartments in response to changing cellular status suggests a role for Gpd1p in the spatial regulation of redox potential, a process critical to cell survival, especially under the complex stress conditions expected to occur in the wild.
Keywords: Lipid/Fatty Acid, Subcellular Organelles/Peroxisomes, Chromatin Structure, Fatty Acid Oxidation, Nuclear Translocation, Gpd1p, Osmotic Stress, PTS2, Pnc1p, Subcellular Localization
Introduction
Glycerol 3-phosphate dehydrogenase (Gpd1p) is one of two NAD+-dependent glycerol 3-phosphate dehydrogenases in yeast (1, 2). It is classically defined as a cytosolic enzyme that catalyzes the conversion of dihydroxyacetone phosphate (DHAP)2 and NADH to glycerol 3-phosphate (Glycerol 3-phosphate) and NAD+ (2). In unstressed cells, this reaction prevents the accumulation of DHAP (3), which can otherwise be transformed into methyl glyoxylate (MG) (3, 4), a toxic compound that interacts with proteins (5–7). This reaction also allows for the reoxidation of NADH to NAD+, which can serve as a buffer for cytosolic redox balance, compensating for cellular reactions that produce NADH (2). Moreover, glycerol 3-phosphate is a key metabolite for the synthesis of glyceride lipids and phospholipids, as well as for the formation of glycerol (8–12).
Under hyperosmotic stress, Saccharomyces cerevisiae, as well as other yeasts, accumulates glycerol as a major solute (13, 14). This increased production of glycerol is caused mainly by an enhanced activity of Gpd1p, and accordingly, Gpd1p is essential for growth under osmotic stress (2, 15). In addition, GPD1 expression is altered by a wide variety of stresses, including heat, cold, and oxidative stress (16–18), suggesting its regulation is controlled by stress. Genome-wide monitoring of transcript changes in yeast under various stress conditions also showed that GPD1 belongs to a large group of common stress-response genes (19, 20); however, its regulation is complex and appears to be controlled by multiple signaling pathways (21). This likely reflects the multiple metabolic changes that yeast cells undergo in response to different stresses (19, 22, 23) and the function of Gpd1p at the interface of many metabolic pathways, including glycolysis, glycerolipid and phospholipid biosynthesis, and glycerol metabolism (8–12). Together, these data implicate Gpd1p in the coordination of multiple stress-responsive metabolic processes.
Gpd1p is generally considered a cytosolic protein, but quantitative mass spectrometry of the S. cerevisiae peroxisomal proteome and live cell fluorescence microscopy identified a pool of Gpd1p in peroxisomes (24, 25). Peroxisomes play major roles in oxidative stress in both the generation of, and protection of cells from, reactive oxygen species (26). This link to cellular stress has also implicated peroxisomes in aging; however, this relationship remains unclear. Although numerous studies have characterized important roles for Gpd1p in osmoregulation and other central metabolic processes, relatively little attention has been paid to its cellular localization and, in particular, its localization to peroxisomes.
Peroxisomal matrix proteins are imported into the organelle by one of two pathways (27, 28). Most matrix proteins harbor a C-terminal peroxisomal targeting signal type 1 (PTS1) having the consensus (S/A/C)(K/R/H)(L/M). This signal is recognized by Pex5p, which through interactions with membrane protein receptors mediates import into the peroxisome (29, 30). A second type of peroxisomal targeting signal, PTS2, is recognized by Pex7p (28, 30) and is usually located near the N terminus of proteins (31). The consensus sequence of PTS2 is defined as (R/K)(L/V/I)X5(Q/H)(L/A/I). Only two S. cerevisiae peroxisomal proteins, Pot1p (3-ketoacyl-CoA thiolase) and Pcd1p (peroxisomal nudix pyrophosphatase), are known to be dependent on PTS2 (32, 33).
We have investigated the localization of Gpd1p to peroxisomes. We demonstrate that Gpd1p localization to peroxisomes requires an N-terminal PTS2 and Pex7p. We also found that Gpd1p dynamically changes its subcellular distribution among the cytosol, peroxisome, and nucleus depending on the type of cellular stress, becoming primarily peroxisomal upon exposure to the fatty acid, oleic acid, and nucleus upon exposure to osmotic stress. Similar dynamics were investigated for Pnc1p, a peroxisomal protein also implicated in cellular stress and aging (34). Perturbations of normal distribution dynamics for Gpd1p suggest the controlled subcellular distribution of Gpd1p among these compartments is important for cells to respond appropriately to different environmental stresses.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
The yeast strains used in this study were derived from the parental strain BY4742 and corresponding deletion strain library (ResGen) or the GFP clone library (Invitrogen), unless otherwise indicated. For subtelomeric silencing assays, strains were derived from strain YTI249 (35). Yeast deletions were made by targeted PCR disruption. Genomically integrated GFP, red fluorescent protein, mCherry, and protein A fusions and pPOT1-RFP were made as described previously (24). pGPD1-GFP was constructed by ligating a PCR product of GPD1 (encompassing nucleotides −519 to +1771, where +1 is the A of the start codon) in-frame and upstream of the coding sequence for GFP into pRS315 (CEN and LEU2). pΔNGPD1-GFP lacking the 17 N-terminal amino acid residues after the start codon was generated by overlap extension PCR with pGPD1-GFP as a template, using internal primers to delete base pairs 4–54 from the start codon in pRS315. pGPD1-GFP-SKL was constructed by PCR-directed mutagenesis to yield GPD1-GFP containing a 3′-extension encoding a tripeptide PTS1 (Ser-Lys-Leu) using pGPD1-GFP as a template in pRS315. pGPD1-GFP-NLS was constructed by PCR-directed mutagenesis to yield GPD1-GFP containing a 3′-extension encoding an SV40 nuclear localization sequence (PKKKRKV) using pGPD1-GFP as a template in pRS316. pΔNGPD1-GFP-NLS was constructed by PCR-directed mutagenesis to yield ΔNGPD1-GFP containing a 3′-extension encoding an SV40 nuclear localization sequence (PKKKRKV) using pΔNGPD1-GFP as a template in pRS315. pGPD1(Ser to Ala)-GFP and pGPD1(Ser to Asp)-GFP were made by site-directed mutagenesis of pGPD1-GFP to convert amino acid residues 24 and 27 of Gpd1p to alanine or aspartic acid, respectively.
Culture Conditions
The following media were used for cell culture: YPD (1% yeast extract, 2% peptone, 2% glucose) and SCIM (0.7% yeast nitrogen base, 0.5% yeast extract, 0.5% peptone, 0.5% Tween 40, 0.79 g of complete synthetic medium/liter, 0.5% (NH4)2, and 0.15% oleic acid). When marker selection was required for each strain, defined synthetic medium (SM) supplemented with the necessary amino acids or nucleotides was also used; glucose at 2% or oleic acid (0.15%) plus Tween 40 (0.5%) was added as a carbon source. Agar was added to 2% in solid media. Cell densities of liquid cultures were determined from absorbance measurements at 600 nm using a BioPhotometer (Eppendorf). For growth assays, precultures (5 ml) were grown overnight at 30 °C on a rotator, inoculated to A600 0.1–0.2 in fresh medium, grown to mid-exponential growth phase (A600 0.8–1), and diluted 10-fold into glucose, oleic acid, or 1 m NaCl stress media. The growth of cells was recorded every 15 min using a Bioscreen (Oy Growth Curves AB Ltd.).
Fluorescence Microscopy
Confocal images were collected on an inverted microscope (model DM IRBE, Leica) with a TCS SP2 confocal system equipped with an HCX PL APO ×100/1.40 oil objective (Leica). Wide field images were obtained using a DeltaVision imaging system (Applied Precision) equipped with an Olympus IX-71 wide field microscope with a 250-watt xenon LED transillumination light source. All images were of cells harvested at the exponential phase of growth.
Subcellular Fractionation
Subcellular fractionation was performed as described previously (24, 36). Briefly, yeast cells were grown to mid-exponential growth phase (A600 ∼1) in SM-Leu. Cells were harvested, washed, and converted to spheroplasts with 1 mg of Zymolase 100T/g of cells for 1 h at 30 °C. Spheroplasts were lysed by homogenization in MES buffer (0.65 m sorbitol, 5 mm MES, pH 5.5) containing 1 mm KCl, 1 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, 0.4 μg of pepstatin A/ml, 1× SIGMAFASTTM protease inhibitor (Sigma). Cell debris and nuclei were pelleted from the homogenate by centrifugation for 10 min at 2,000 × g to generate a postnuclear supernatant, which was subjected to 20,000 × gmax for 30 min to yield a supernatant (20KgS) and a pellet (20KgP).
Western Blotting
For Western blotting, protein A was detected using affinity-purified rabbit IgG (Cappel), GFP using mouse anti-GFP, C-terminal SKL using an anti-SKL polyclonal antibody (37), and Gsp1p using rabbit polyclonal antibodies directed against Gsp1p (38). Antibody binding was visualized using secondary antibodies coupled to horseradish peroxidase (Amersham Biosciences) and ECL detection reagents (Pierce).
Analysis of Expression Profiles
The expression profiles for genes encoding peroxisomal proteins were log2-transformed and analyzed using Cluster and Treeview software (39). Genes were clustered hierarchically using default parameters and average linkage clustering. A Pearson correlation was used as the similarity metric. Genes in the entire yeast genome with the expression profiles most similar to that of GPD1 were identified using Pearson correlation.
Subtelomeric Silencing
The subtelomeric silencing assay was performed as described previously (40). Briefly, cells were pre-grown in synthetic media lacking uracil, collected, and resuspended in water. The cell mixtures were normalized to an absorbance of 1.0 and then serially diluted in 10-fold increments. Five μl of each dilution were spotted onto complete SM medium (CSM) and CSM with 1% 5-fluoroorotic acid. Plates were incubated at 30 °C for 2 days.
RESULTS
Gpd1p Is Both Peroxisomal and Cytosolic
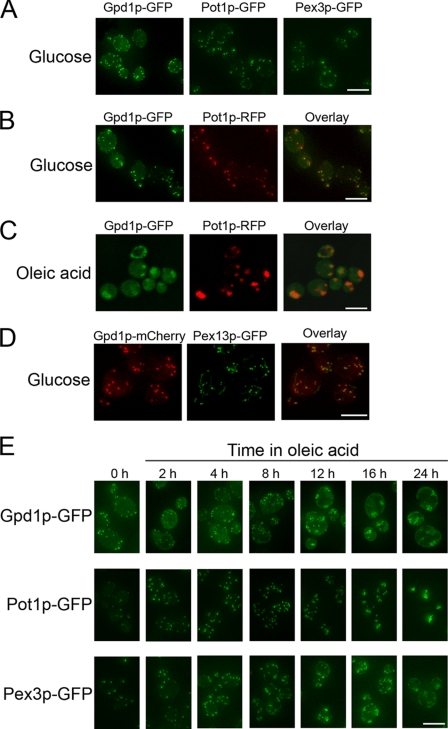
Gpd1p tagged with GFP (Gpd1p-GFP) was visualized by fluorescence microscopy of cells either metabolizing glucose, a condition in which peroxisomes are dispensable, or incubated in oleic acid, a condition in which cells require functional peroxisomal fatty acid β-oxidation for growth (41) and peroxisomes proliferate (42). In glucose medium, Gpd1p-GFP localized to punctate structures like the well characterized peroxisomal matrix protein, thiolase (Pot1p-GFP), or a peroxisomal membrane protein (Pex3p-GFP) (Fig. 1A). These structures were confirmed to be peroxisomes by double-labeling fluorescence microscopy with Pot1p-RFP and Gpd1p-GFP in cells grown in glucose (Fig. 1B) or oleic acid (Fig. 1C). Interestingly, in glucose-grown cells, the number of peroxisomes labeled with Gpd1p-GFP (∼10/cell) appeared to be greater than the number of peroxisomes labeled with either Pot1p-GFP or Pex3p-GFP (∼4/cell) (Fig. 1A, supplemental Video S1, and supplemental Table S1). Previous studies have reported ∼4 peroxisomes per glucose-grown cell (43). This difference in the number of peroxisomes depending on the type of GFP-tagged peroxisomal marker used is likely due to the inefficiency of labeling of peroxisomes with lowly expressed reporters; Pot1p expression is repressed in glucose medium (44), and Pex3p is a lowly abundant membrane protein (45). In contrast, Gpd1p is highly abundant in cells grown in glucose, facilitating its detection as a GFP-tagged protein. To further confirm that punctate structures of Gpd1p-GFP are peroxisomes, Gpd1p-mCherry was co-localized with Pex13p-GFP, an abundant peroxisomal membrane protein in cells grown in glucose (Fig. 1D). We also observed a diffuse cytosolic fluorescent signal for Gpd1p-GFP, whereas Pot1p-RFP was observed to be nearly exclusively peroxisomal, suggesting a dual localization of Gpd1p to peroxisomes and the cytosol.
FIGURE 1.
Cellular localization of Gpd1p. A, fluorescence micrographs depicting the subcellular localization of Gpd1p, Pot1p, and Pex3p genomically tagged with GFP in cells grown in glucose medium. B, double-labeling fluorescence confocal microscopy images of glucose-grown cells synthesizing genomically encoded Gpd1p-GFP and containing a plasmid coding for peroxisomal thiolase tagged with monomeric red fluorescent protein (Pot1p-RFP) showing co-localization of the two signals to punctate structures characteristic of peroxisomes (Overlay). C, fluorescence confocal microscopy images of doubly labeled cells as in B but incubated in oleic acid medium for 16 h. D, double-labeling fluorescence micrographs showing co-localization of Gpd1p-mCherry with Pex13p-GFP in glucose grown cells. E, localization of Gpd1p-GFP, Pot1p-GFP, and Pex3p-GFP in wild-type cells. The time in oleic acid medium is indicated. Bars, 10 μm.
When cells were shifted to and incubated in oleic acid medium, the number of peroxisomes per cell increased to ∼15, and they clustered over time (Fig. 1, C and E), as observed previously (43). As in cells grown in glucose, a significant portion of the Gpd1p-GFP signal remained diffuse in the cytosol in oleic acid-incubated cells. This agrees with previously published reports of both cytosolic and peroxisomal localizations for Gpd1p (24, 25) and suggests the existence of a mechanism regulating Gpd1p localization.
Gpd1p Is Targeted to Peroxisomes by an N-terminal PTS2
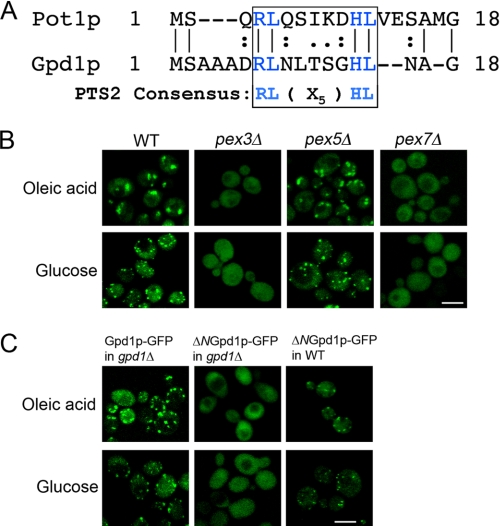
Examination of the amino acid sequence of Gpd1p shows that its N terminus contains a sequence of amino acids that conforms to the consensus sequence for the PTS2, (R/K)(L/V/I)X5(Q/H)(L/A) (Fig. 2A) (28, 31). To determine whether the peroxisomal localization of Gpd1p is dependent on this sequence, we studied the localization of Gpd1p-GFP in a pex3Δ mutant deficient in peroxisome formation, a pex5Δ mutant deficient in the import of matrix proteins targeted by a C-terminal PTS1, and a pex7Δ mutant deficient in PTS2-mediated import in both oleic acid and glucose media. The punctate localization of Gpd1p-GFP was lost in the absence of the PTS2 receptor, Pex7p, but not in the absence of the PTS1 receptor, Pex5p (Fig. 2B), suggesting that Gpd1p import is mediated by Pex7p and that its N terminus contains a bona fide PTS2. Support for the Pex7p-mediated import of Gpd1p came from the mass spectrometric identification of Pex7p in an immunopurification using Gpd1p tagged at its C terminus with protein A (data not shown). Moreover, deletion of the putative PTS2 of Gpd1p (ΔNGpd1p-GFP) led to a loss of the punctate Gpd1p-GFP signal, demonstrating that the peroxisomal targeting of Gpd1p is dependent on an N-terminal PTS2. Interestingly, this dependence on PTS2 for the import of Gpd1p into peroxisomes was observed only when the N-terminal deletion construct of Gpd1p was expressed in gpd1Δ cells (Fig. 2C). In wild-type haploid cells harboring an intact chromosomal copy of the GPD1 gene, ΔNGpd1p-GFP again localized to punctate structures characteristic of peroxisomes (Fig. 2C), suggesting that ΔNGpd1p-GFP dimerizes with Gpd1p to be imported into peroxisomes by a “piggyback” mechanism (32). Gpd1p has been reported to form a dimer (46).
FIGURE 2.
PTS2-dependent targeting of Gpd1p to peroxisomes. A, sequence comparison of the N termini of Pot1p and Gpd1p showing they contain the PTS2 consensus sequence. B, peroxisomal targeting of Gpd1p is mediated by Pex7p. Wild-type BY4742 cells or the respective pex3Δ, pex5Δ, and pex7Δ mutants synthesizing genomically encoded Gpd1p-GFP were incubated in oleic acid medium for 8 h at 30 °C or in glucose medium and observed by confocal microscopy. The Gpd1p-GFP chimera fails to localize to peroxisomes in pex3Δ cells lacking peroxisomes and in pex7Δ cells lacking the PTS2 receptor but targets efficiently to peroxisomes in pex5Δ cells lacking the PTS1 receptor. Bar, 10 μm. C, Gpd1p-GFP import into peroxisomes is mediated by a functional PTS2 at the N terminus of Gpd1p. GFP chimeras of Gpd1p or Gpd1p lacking its N-terminal 17 amino acids (ΔNGpd1p-GFP) were localized in WT and gpd1Δ cells incubated in oleic acid or glucose medium for 8 h at 30 °C. ΔNGpd1p-GFP failed to localize to peroxisomes in gpd1Δ but not in wild-type cells. Bar, 10 μm.
Gpd1p Localization to Peroxisomes Is Regulated by Phosphorylation
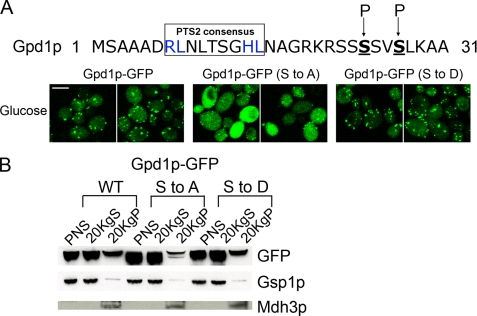
A global phosphoproteomic analysis in yeast (47) identified a phosphorylated peptide derived from Gpd1p in which two serines, Ser-24 and Ser-27, adjacent to the PTS2 sequence of Gpd1p were shown to be phosphorylated (Fig. 3A). This raised the possibility that Gpd1p import into peroxisomes could be regulated by phosphorylation. We therefore constructed plasmid-encoded site-directed mutants encoding chimeras that mimic constitutively phosphorylated (Gpd1p-GFP (Ser to Asp)) and dephosphorylated (Gpd1p-GFP (Ser to Ala)) forms of Gpd1p-GFP and introduced them into gpd1Δ cells. Gpd1p-GFP (Ser to Ala) failed to concentrate efficiently in peroxisomes compared with Gpd1p-GFP or Gpd1p-GFP (Ser to Asp) (Fig. 3A). Although small punctate structures were observed in cells expressing Gpd1p-GFP (Ser to Ala), our data suggest that the import of Gpd1p into peroxisomes is increased by phosphorylation adjacent to its PTS2 and that the cell exploits this post-translational modification to control the localization of Gpd1p. To confirm and extend the confocal data, subcellular fractions from cells expressing Gpd1p-GFP, Gpd1p-GFP (Ser to Ala), or Gpd1p-GFP (Ser to Asp) were prepared by differential centrifugation (24, 36). Consistent with the microscopy data, Western blot analysis showed that Gpd1p-GFP (Ser to Ala) is present in reduced amounts in the 20KgP fraction compared with Gpd1p-GFP or Gpd1p-GFP (Ser to Asp) (Fig. 3B). Taken together, these data provide strong evidence for a role of phosphorylation of the two serine residues of Gpd1p in efficient peroxisomal import of Gpd1p.
FIGURE 3.
Efficient peroxisomal import of Gpd1p by phosphorylation of two serine residues of Gpd1p. A, serines 24 and 27 (underlined and in boldface) of Gpd1p were changed to alanine (Ser to Ala) or aspartic acid (Ser to Asp), and the subcellular distributions of Gpd1p-GFP (Ser to Ala) and Gpd1p-GFP (Ser to Asp) were examined in glucose-grown cells by confocal microscopy. Bar, 10 μm. B, distribution of Gpd1p-GFP, Gpd1p-GFP (Ser to Ala), and Gpd1p-GFP (Ser to Asp) was analyzed by subcellular fractionation. Postnuclear supernatant (PNS), 20KgS fractions enriched for cytosol (loaded at 1 cell equivalent), and 20KgP fractions enriched for peroxisomes and mitochondria (loaded at 10 cell equivalents) were analyzed by Western blotting using anti-GFP antibodies. Gsp1p was used as a loading control for postnuclear supernatant and 20KgS. C-terminal SKL was used as a loading control for the 20KgP, and the molecular weight of the band corresponds to the molecular weight of Mdh3p.
Gpd1p Is Not Required for Peroxisomal β-Oxidation
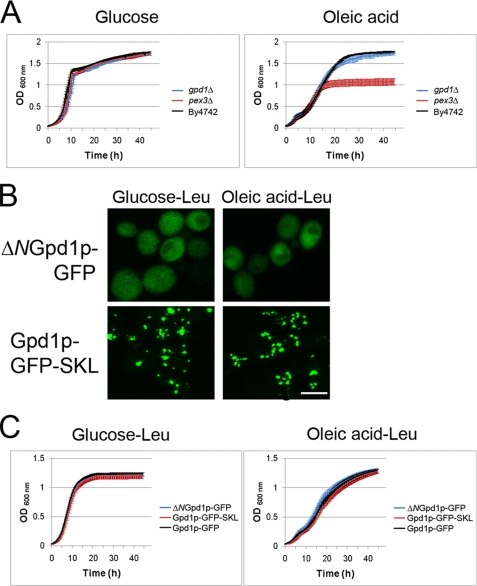
Because we and others have not detected differences between gpd1Δ and wild-type strains in large scale screens of growth in the presence of fatty acid (48, 49), we used more sensitive measures to determine whether Gpd1p is required for peroxisome biogenesis or peroxisomal β-oxidation. Liquid growth assays were done using a Bioscreen to compare the growth rates of wild-type, pex3Δ, and gpd1Δ cells in glucose medium and oleic acid medium (Fig. 4A). As expected, the peroxisome assembly mutant strain pex3Δ showed dramatically reduced growth in oleic acid medium (50). In contrast, the gpd1Δ strain grew essentially like the wild-type strain BY4742 in oleic acid medium. All three strains grew comparably in glucose medium.
FIGURE 4.
Gpd1p is not required for β-oxidation of fatty acids. A, cell growth assay of gpd1Δ, pex3Δ, and WT cells in media with glucose or oleic acid as the carbon source. Growth was monitored using a liquid growth assay in a Bioscreen automatic reader and plotted as the average of three independent experiments and three technical replicates of each. gpd1Δ cells grew at the same rate as WT cells in oleic acid and glucose. Error bars indicate standard deviation. B, subcellular localization of ΔNGpd1p-GFP and Gpd1p-GFP-SKL in SM-Leu media containing glucose or oleic acid (8 h). C, growth assays as in A, but the strains shown are gpd1Δ cells transformed with pGPD1-GFP, pΔNGPD1-GFP, or pGPD1-GFP-SKL.
To determine whether the peroxisomal pool of Gpd1p affects the growth of cells in oleic acid medium, we constructed pGPD1-GFP-SKL, which codes for a Gpd1p containing both a PTS1 and PTS2 and leads to almost exclusive peroxisomal localization of Gpd1p by both Pex5p- and Pex7p-mediated import (Fig. 4B), and tested the effect of shifting the localization of Gpd1p within the cell from peroxisome/cytosol to exclusively peroxisome. Liquid growth assays in glucose medium and oleic acid medium were done on gpd1Δ cells transformed with pGPD1-GFP (peroxisomal/cytosolic), pΔNGPD1-GFP (exclusively cytosolic), or pGPD1-GFP-SKL (exclusively peroxisomal). Regardless of which Gpd1p-chimera they expressed, gpd1Δ cells grew comparably in glucose medium or oleic acid medium (Fig. 4C), suggesting that a peroxisomal pool of Gpd1p is not necessary for normal peroxisomal β-oxidation activity and that Gpd1p does not play a primary role, or plays a redundant role, in peroxisomal β-oxidation in yeast.
Dynamic Changes in Subcellular Distribution of Gpd1p between the Nucleus and Peroxisomes under Stress Conditions
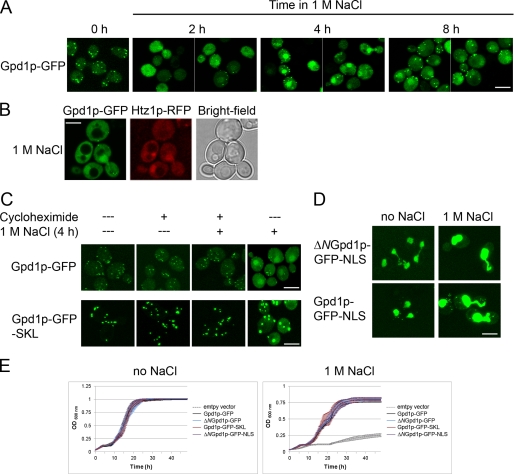
Given the previously established role for Gpd1p in response to osmotic stress, we investigated its localization over time in cells incubated under conditions of high osmolarity. Interestingly, Gpd1p-GFP accumulated in the nucleus after exposure of cells to osmotic stress in medium containing 1 m NaCl (Fig. 5A). We confirmed this nuclear localization of Gpd1p by double fluorescence microscopy co-localization of the GFP signal with Htz1p-RFP, a well characterized nuclear histone variant (Fig. 5B) (51). This shift of Gpd1p to the nucleus was common to exposure to a number of different stress conditions, including treatment with NaCl (0.3 and 0.6 m), 1 m sorbitol, 2.5 mm dithiothreitol, or 37 °C heat shock (data not shown).
FIGURE 5.
Gpd1p localizes to peroxisomes but redistributes to the nucleus under conditions of stress. A, localization of Gpd1p genomically tagged with GFP was analyzed by confocal microscopy at the different times upon exposure to 1 m NaCl. Bar, 10 μm. B, Gpd1p-GFP co-localizes in the nucleus with Htz1p-RFP as determined by confocal microscopy. Images were obtained after cells were exposed to 1 m NaCl for 4 h. Bar, 10 μm. C, peroxisomal Gpd1p does not exit peroxisomes upon exposure to 1 m NaCl. Cells expressing GFP chimeras of wild-type (pGPD1-GFP) or peroxisomal (pGPD1-GFP-SKL) Gpd1p were treated with 1 m NaCl for 4 h in the presence (+) or absence (−) of cycloheximide. D, subcellular localization of ΔNGpd1p-GFP-NLS and Gpd1p-GFP-NLS in glucose media in the absence or presence of 1 m NaCl. Cytoplasmic signals of both Gpd1p variants are increased by 1 m NaCl. E, cell growth assay of cells expressing GFP chimeras of wild-type (pGPD1-GFP), cytosolic (pΔNGPD1-GFP), peroxisomal (pGPD1-GFP-SKL), or nuclear (pΔNGPD1-GFP-NLS) Gpd1p. Cells were grown as in Fig. 4 but in the presence or absence of NaCl. All fusions were expressed from the plasmid pRS315 in gpd1Δ cells. Empty plasmid served as a control. The curves represent the average of three independent experiments and three technical replicates of each, and error bars represent standard deviation.
The increased cytosolic Gpd1p-GFP signals shortly after 1 m NaCl was concomitant with a relative decrease in the peroxisomal Gpd1p-GFP signal (Fig. 5A), which raised a question whether this phenomenon is due to exit of Gpd1p-GFP from peroxisomes or due to increased newly synthesized cytosolic Gpd1p-GFP, which fails to be efficiently imported into peroxisomes. To test this, the experiment was repeated in the presence of cycloheximide to inhibit new protein synthesis (52). In this case, there was no apparent shift in the localization of Gpd1p-GFP (or Gpd1p-GFP-SKL) (Fig. 5C), indicating that, upon shift to 1 m NaCl, the cytosolic accumulation of Gpd1p results from newly synthesized protein that fails to be imported into peroxisomes and not export of peroxisomal Gpd1p to the cytosol.
To address the potential role of subcellular localization of Gpd1p under conditions of osmotic stress, we asked whether altering the ability of Gpd1p to shift its localization between peroxisomes the nucleus affects the growth rate of cells exposed to 1 m NaCl. pΔNGPD1-GFP-NLS, which encodes Gpd1p lacking its PTS-2 and contains a classic nuclear localization signal (53), was constructed. This construct was localized predominantly to the nucleus under normal growth conditions with very little detectable cytosolic signal (Fig. 5D). Thus, liquid growth assays were conducted with cells expressing wild-type Gpd1p-GFP, cytosolic Gpd1p (ΔNGpd1p-GFP), peroxisomal Gpd1p (Gpd1p-GFP-SKL), or nuclear Gpd1p (ΔNGpd1p-GFP-NLS). All strains expressing each of the different versions of Gpd1p grew at rates similar to one another whether in YPD and/or in medium containing 1 m NaCl, suggesting a peroxisomal pool of Gpd1p is not necessary for the osmotic stress response (Fig. 5E). Moreover, these data suggest that Gpd1p localization to the peroxisome or the nucleus does not hinder the osmotic stress response. However, when the distributions of Gpd1p-GFP-SKL and Gpd1p-GFP-NLS were determined under these conditions, the salt stress led to an increased cytosolic signal for both chimeras (Fig. 5, C and D). Thus, despite the presence of the extra peroxisomal targeting signal or the classic nuclear localization signal, Gpd1p accumulated in the cytosol (albeit to lesser extents than WT Gpd1p), and the cells appear to generate sufficient cytosolic glycerol for osmoregulation.
Expression of Genes Encoding Peroxisomal Proteins under Various Stress Conditions
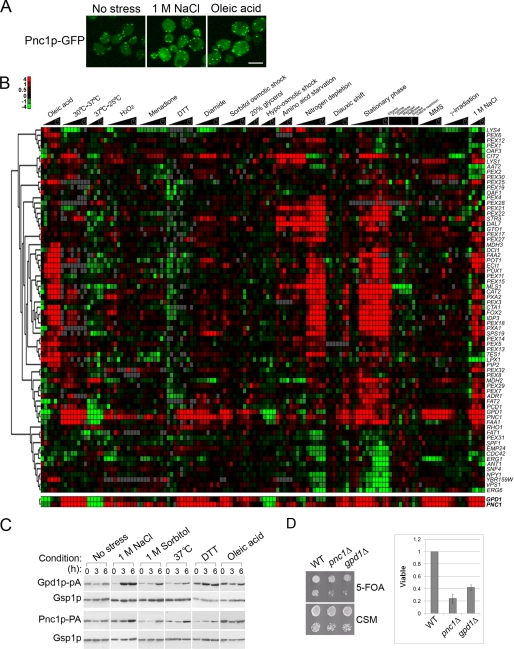
Pnc1p is a protein that undergoes similar shifts in localization in response to stress (34). It has been reported to localize to peroxisomes in a Pex7p-dependent manner, but it also localizes partially to the cytosol and nucleus (34). We confirmed this tripartite localization and observed a change in the localization of Pnc1p-GFP from peroxisomes to the cytosol and, to a lesser extent, the nucleoplasm in response to NaCl or oleic acid treatment (Fig. 6A). Likewise, PNC1 and GPD1 appear to be coordinately regulated. Examination of the transcriptional profiles of genes encoding peroxisomal proteins in S. cerevisiae cells exposed to a variety of different stress conditions (20, 36, 54–57) demonstrated that not all genes encoding peroxisomal proteins respond to these environmental cues in the same manner (Fig. 6B). Several genes encoding peroxisomal proteins were not induced in oleic acid but were induced in response to other stress conditions. Among this group of genes, the expression of GPD1 and PNC1 was strongly correlated, and among all genes in the yeast genome, the expression of PNC1 was the most closely correlated with that of GPD1 (Pearson correlation coefficient of 0.85). This coordinate control was confirmed by Western blotting (Fig. 6C), which showed increased levels of Gpd1p and Pnc1p in cells subjected to high osmolarity (1 m NaCl and 1 m sorbitol), heat shock (37 °C), or cell wall damage (2.5 mm dithiothreitol) but relatively constant levels in cells shifted from glucose to oleic acid.
FIGURE 6.
Similarities in expression and distribution of Pnc1p and Gpd1p. A, localization of Pnc1p-GFP chimera in glucose, 1 m NaCl (4 h), or oleic acid (4 h) as determined by confocal microscopy. Bar, 10 μm. B, hierarchical clustering of the transcriptional expression profiles of genes known to function in peroxisome biology over a broad range of stress conditions. GPD1 and PNC1 were among the most closely co-expressed genes within this dataset and within the whole yeast genome (correlation coefficient 0.85). C, co-synthesis of Gpd1p and Pnc1p across various stress conditions. Whole cell lysates of yeast strains subjected to various stress conditions were analyzed by Western blotting. Gsp1p was used as a loading control. D, strains lacking PNC1 or GPD1 failed to efficiently silence subtelomeric URA3. Wild-type, pnc1Δ, and gpd1Δ strains containing a subtelomeric URA3 gene were assayed for URA3 gene expression by spotting 10-fold dilutions onto CSM- and 5-fluoroorotic acid (5-FOA)-containing plates. The graph represents the silencing efficiency as the number of viable mutant colonies on 5-fluoroorotic acid normalized to wild type across nine independent experiments. Error bars represent the standard deviation.
Although its peroxisomal function is unclear, in the nucleus Pnc1p is thought to play a part in the NAD+ salvage pathway, which converts nicotinamide to nicotinic acid (58, 59). In this context, Pnc1p plays a significant role in Sir2p-mediated DNA silencing by reducing levels of nicotinamide, an inhibitor of the histone deacetylase Sir2p (34). The correlation of gene expression (39, 60), the Pex7p-dependent dynamic localization of Gpd1p and Pnc1p to peroxisomes, the nucleus, and the cytosol (34), and the links of these two proteins to NAD+ metabolism suggest the intriguing possibility that they function together. We therefore investigated whether Gpd1p plays a role in chromatin silencing. Our investigations identified a small but reproducible (n = 9) defect in subtelomeric silencing (35, 40) in gpd1Δ cells (Fig. 6D) but failed to reveal an effect on rDNA silencing, and we did not observe a correlation between silencing and Gpd1p localization to either the peroxisome or nucleus (data not shown).
Gpd1p Distribution Dynamics Are Important for Responses to Combined Stress
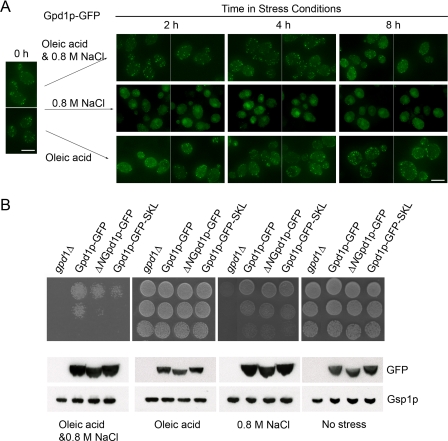
Gpd1p is localized predominantly in the nucleus and cytosol under conditions of osmotic stress. In oleic acid, Gpd1p is localized predominantly to peroxisomes. However, forced localization of Gpd1p to either location (by appending a PTS1 or by deleting its PTS2) did not compromise cells in either condition. We therefore asked whether its dynamics or its distribution balance is required under conditions of combined stress. We first examined the localization of Gpd1p-GFP in response to 0.8 m NaCl, oleic acid, or in a combined oleic acid-NaCl stress (Fig. 7A). As observed previously, Gpd1p remained peroxisomal in oleic acid and became cytosolic/nuclear in NaCl. In the combined stress, the localization was intermediate between the two extremes, and although the cytosolic fraction increased, there remained a significant peroxisomal signal. Assays for growth under these conditions demonstrated that this intermediate distribution is important (Fig. 7B). Cells lacking Gpd1p were unable to grow under the combined stress condition, whereas wild-type cells tolerated the condition (Fig. 7B). Cells expressing the nuclear/cytosolic version of Gpd1p (ΔN-Gpd1p-GFP) or the peroxisomal version of Gpd1p (Gpd1p-GFP-SKL) showed a significant growth defect under the combined stress (Fig. 7B). This suggests that the dynamic distribution of Gpd1p between the cytosolic, nuclear, and peroxisomal compartments is important under conditions of combined stress, a situation likely to dominate natural conditions.
FIGURE 7.
Balance of subcellular distribution of Gpd1p is required for efficient growth in the combined oleic acid and 0.8 m NaCl stress. A, localization of Gpd1p-GFP chimera in cells exposed to the combined stress of NaCl and oleic acid. Wild-type cells expressing Gpd1p-GFP were incubated in oleic acid, 0.8 m NaCl, or oleic acid and 0.8 m NaCl. Fluorescence images were captured at the times indicated using confocal microscopy. Bar, 10 μm. B, cell growth on plates containing oleic acid, 0.8 m NaCl, or oleic acid plus 0.8 m NaCl. Strains were grown to exponential growth phase in minimal medium, and the culture was serially diluted and applied to plates containing no stress, oleic acid, 0.8 m NaCl, or oleic acid plus 0.8 m NaCl. The plates were photographed following 2 days of incubation for control (no stress), 3 days incubation for oleic acid or 0.8 m NaCl, and 5 days incubation for both oleic acid and 0.8 m NaCl at 30 °C. Whole cell lysates of the strains subjected to each of the different growth conditions for 16 h were analyzed by Western blotting with the indicated antibodies. Gsp1p serves as a loading control.
DISCUSSION
Gpd1p is imported into peroxisomes by virtue of a PTS2 signal, and its import is dependent on Pex7p. There are two other peroxisomal proteins in S. cerevisiae transported by a cis-acting PTS2, Pot1p and Pcd1p. Unlike other PTS2-containing proteins, a significant portion of the cellular pool of Gpd1p remains cytosolic. This suggests that the localization of Gpd1p is regulated to maintain a cytosolic pool of Gpd1p. Indeed, we show that localization of Gpd1p is controlled by phosphorylation of two serine residues adjacent to its PTS2. Precisely how this imparts control over Gpd1p distribution is not yet clear. By analogy to other protein localization studies (61, 62), it is likely that PTS2 binding by Pex7p is altered by the phosphorylation of residues adjacent to the targeting signal. Alternatively, the mechanism could involve the cleavage of PTS2 to prevent peroxisomal import of this processed version. The cleavage of a signal sequence is a common mechanism by which cells regulate the localization of proteins, and in several organisms peroxisomal thiolase is proteolytically cleaved in the peroxisome to remove its PTS2 (63, 64). In support of this idea, proteolytic cleavage of the N terminus of Gpd1p, which removes the PTS2 signal sequence, has been reported previously (1). However, using SDS-PAGE, we could not detect a change in the mobility of Gpd1p under conditions where its distribution shifts (i.e. no stress versus osmotic stress; data not shown). In addition, mass spectrometric analyses of Gpd1p affinity purified from cytosolic fractions identified seven unique peptides derived from Gpd1p, one of which corresponded to its native N terminus, indicating that at least some of Gpd1p in the cytosolic pool is not proteolytically processed (data not shown).
Gpd1p changes its subcellular distribution among the cytosol, peroxisomes, and nucleus depending on environmental conditions. Gpd1p is primarily peroxisomal upon exposure to oleic acid. It has been suggested that Gpd1p may be part of a glycerol 3-phosphate shuttle to regenerate NAD+ from NADH produced by β-oxidation (25). This mechanism of NAD+ regeneration could be an alternative to the malate/oxaloacetate cycle. In the malate/oxaloacetate cycle, peroxisomal Mdh3p converts oxaloacetate to malate, consuming NADH and producing NAD+. Mdh2p, converts malate back to oxaloacetate, producing cytosolic NADH (electrons from which are shuttled to the mitochondrial electron transport chain by a mitochondrial glycerol phosphate shuttle) (65). However, the presence of Gpd1p is not sufficient to overcome the absence of MDH3 in cells grown on oleic acid (65), and cells lacking Gpd1p showed no growth defects on oleic acid, even when tested using sensitive, quantitative liquid growth assays. Thus, peroxisomal Gpd1p does not appear to play a primary role in peroxisomal NAD+ regeneration during fatty acid β-oxidation.
It therefore remains unclear why Gpd1p localizes to peroxisomes. It could reflect a mechanism to control activity of Gpd1p by simply sequestering the enzyme in peroxisomes. However, together with Gpd1p, two additional peroxisomal proteins, Pnc1p and Npy1p, implicate the organelle as playing an important role in NAD+ metabolism. Npy1p hydrolyzes the pyrophosphate linkage in NADH to produce nicotinamide mononucleotide and AMP (66). Gpd1p may play a role in regulating substrates levels for Npy1p, and as part of the NAD salvage pathway, Pnc1p provides a source of nicotinic acid through its nicotinamidase activity. It remains to be determined why these activities are localized to peroxisomes and how these functions are metabolically linked.
Under NaCl stress, Gpd1p becomes primarily cytosolic and nuclear. It is under this condition that cytosolic Gpd1p has been established to play an important role in cell survival. Yeast cells respond to osmotic stress by increasing cellular glycerol levels (13, 14), and Gpd1p catalyzes DHAP into glycerol 3-phosphate, a precursor of glycerol. It is less clear why Gpd1p accumulates in the nucleus under these conditions. One trivial possibility is that Gpd1p harbors a cryptic nuclear localization sequence, and the nuclear pool is relatively inconsequential to the cell. However, subcellular distribution of Gpd1p is intriguing given the example of Pnc1p (34). Although Pnc1p displays prominent peroxisomal localization, it regulates Sir2p-mediated chromatin silencing and life span extension under caloric restriction and stress conditions (34). Sir2p is exclusively nuclear (67), and it uses NAD+ as a substrate for its histone deactylase activity thereby inducing gene silencing (68, 69). During this reaction, nicotinamide is produced, which in turn it inhibits Sir2p function (34, 59). On the other hand, Pnc1p-mediated conversion of nicotinamide to nicotinic acid (58) relieves the inhibition of Sir2p, leading to increased histone deacetylation and increased silencing (34, 59).
As in the case of Pnc1p, shifts in the distribution of Gpd1p to the nucleus upon exposure to NaCl stress could lead to increased Sir2p activity, in this case by increasing the local NAD+ concentration. In our hands, subtelomeric silencing was modestly impaired in the absence of Gpd1p; however, overexpression of GPD1 did not lead to increased subtelomeric silencing (data not shown). Nevertheless, the increased longevity observed for cells exposed to osmotic stress depends on a coincident increase in GPD1 transcription (56).
Another possible role for increased Gpd1p in the nucleus is the protection from nuclear damage that results from the toxic molecule MG, which is derived from DHAP (4, 5, 7). The presence of Gpd1p may reduce the DHAP pool and thereby limit the production of MG. In fact, it has been shown that the level of MG is induced in cells upon exposure to osmotic stress and oxidative stress (70), and enzymes such as Gre3p and Glo1p involved in detoxification of MG (70) also localize to both the cytosol and nucleus (71).
The results we present here suggest that the dynamic distribution of Gpd1p among different cellular locations provides a mechanism for cells to respond to cellular stress. Indeed, when Gpd1p localization was altered, cell growth was dramatically compromised when cells were challenged with both oleic acid and osmotic stress. In this case, the cell is faced with the need to increase glycerol concentration through increased glycolysis but also must metabolize the oleic acid, which can be toxic otherwise (48). Both processes produce and consume NADH and are controlled in part by the NAD+/NADH ratio. It is unclear why altering Gpd1p localization compromises the cell growth under these conditions, but it is likely due to a need to maintain redox balance, in the context of spatially distributed and compartmentalized enzymes carrying out the multitude of metabolic reactions needed to appropriately respond to the combined stress. Ultimately, to understand this complexity, we will need to fully inventory the enzyme complement of peroxisomes and other organelles, but equally importantly, we will need to characterize and understand their dynamics in the face of different metabolic challenges.
Supplementary Material
Acknowledgments
We thank to Ramsey Saleem and Jennifer Smith and the Aitchison laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM075152, GM076547, and RR022220. This work was also supported by Canadian Institutes for Health Research Grant 53326.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Video S1.
- DHAP
- dihydroxyacetone phosphate
- MG
- methyl glyoxylate
- MES
- 4-morpholineethanesulfonic acid
- GFP
- green fluorescent protein
- WT
- wild type.
REFERENCES
- 1.Larsson K., Ansell R., Eriksson P., Adler L. (1993) Mol. Microbiol. 10, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 2.Larsson C., Påhlman I. L., Ansell R., Rigoulet M., Adler L., Gustafsson L. (1998) Yeast 14, 347–357 [DOI] [PubMed] [Google Scholar]
- 3.Aguilera J., Rodríguez-Vargas S., Prieto J. A. (2005) Mol. Microbiol. 56, 228–239 [DOI] [PubMed] [Google Scholar]
- 4.Phillips S. A., Thornalley P. J. (1993) Eur. J. Biochem. 212, 101–105 [DOI] [PubMed] [Google Scholar]
- 5.Vander Jagt D. L., Robinson B., Taylor K. K., Hunsaker L. A. (1992) J. Biol. Chem. 267, 4364–4369 [PubMed] [Google Scholar]
- 6.Martins A. M., Cordeiro C. A., Ponces Freire A. M. (2001) FEBS Lett. 499, 41–44 [DOI] [PubMed] [Google Scholar]
- 7.Lo T. W., Westwood M. E., McLellan A. C., Selwood T., Thornalley P. J. (1994) J. Biol. Chem. 269, 32299–32305 [PubMed] [Google Scholar]
- 8.Michnick S., Roustan J. L., Remize F., Barre P., Dequin S. (1997) Yeast 13, 783–793 [DOI] [PubMed] [Google Scholar]
- 9.Beopoulos A., Mrozova Z., Thevenieau F., Le Dall M. T., Hapala I., Papanikolaou S., Chardot T., Nicaud J. M. (2008) Appl. Environ. Microbiol. 74, 7779–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardocki M. E., Jani N., Lopes J. M. (2005) Biochim. Biophys. Acta 1735, 89–100 [DOI] [PubMed] [Google Scholar]
- 11.Athenstaedt K., Daum G. (1997) J. Bacteriol. 179, 7611–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nevoigt E., Stahl U. (1996) Yeast 12, 1331–1337 [DOI] [PubMed] [Google Scholar]
- 13.Blomberg A., Adler L. (1992) Adv. Microb. Physiol. 33, 145–212 [DOI] [PubMed] [Google Scholar]
- 14.Brown A. D. (1978) Adv. Microb. Physiol. 17, 181–242 [DOI] [PubMed] [Google Scholar]
- 15.Albertyn J., Hohmann S., Thevelein J. M., Prior B. A. (1994) Mol. Cell. Biol. 14, 4135–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boy-Marcotte E., Lagniel G., Perrot M., Bussereau F., Boudsocq A., Jacquet M., Labarre J. (1999) Mol. Microbiol. 33, 274–283 [DOI] [PubMed] [Google Scholar]
- 17.Panadero J., Pallotti C., Rodríguez-Vargas S., Randez-Gil F., Prieto J. A. (2006) J. Biol. Chem. 281, 4638–4645 [DOI] [PubMed] [Google Scholar]
- 18.Godon C., Lagniel G., Lee J., Buhler J. M., Kieffer S., Perrot M., Boucherie H., Toledano M. B., Labarre J. (1998) J. Biol. Chem. 273, 22480–22489 [DOI] [PubMed] [Google Scholar]
- 19.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. (2000) Mol. Biol. Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. (2001) Mol. Biol. Cell 12, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rep M., Albertyn J., Thevelein J. M., Prior B. A., Hohmann S. (1999) Microbiology 145, 715–727 [DOI] [PubMed] [Google Scholar]
- 22.Brauer M. J., Saldanha A. J., Dolinski K., Botstein D. (2005) Mol. Biol. Cell 16, 2503–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koerkamp M. G., Rep M., Bussemaker H. J., Hardy G. P., Mul A., Piekarska K., Szigyarto C. A., De Mattos J. M., Tabak H. F. (2002) Mol. Biol. Cell 13, 2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marelli M., Smith J. J., Jung S., Yi E., Nesvizhskii A. I., Christmas R. H., Saleem R. A., Tam Y. Y., Fagarasanu A., Goodlett D. R., Aebersold R., Rachubinski R. A., Aitchison J. D. (2004) J. Cell Biol. 167, 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valadi A., Granath K., Gustafsson L., Adler L. (2004) J. Biol. Chem. 279, 39677–39685 [DOI] [PubMed] [Google Scholar]
- 26.Schrader M., Fahimi H. D. (2006) Biochim. Biophys. Acta 1763, 1755–1766 [DOI] [PubMed] [Google Scholar]
- 27.Smith J. J., Aitchison J. D. (2009) Curr. Opin. Cell Biol. 21, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platta H. W., Erdmann R. (2007) FEBS Lett. 581, 2811–2819 [DOI] [PubMed] [Google Scholar]
- 29.Platta H. W., Erdmann R. (2007) Trends Cell Biol. 17, 474–484 [DOI] [PubMed] [Google Scholar]
- 30.Thoms S., Erdmann R. (2006) Biochim. Biophys. Acta 1763, 1620–1628 [DOI] [PubMed] [Google Scholar]
- 31.Petriv O. I., Tang L., Titorenko V. I., Rachubinski R. A. (2004) J. Mol. Biol. 341, 119–134 [DOI] [PubMed] [Google Scholar]
- 32.Glover J. R., Andrews D. W., Rachubinski R. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartwright J. L., Gasmi L., Spiller D. G., McLennan A. G. (2000) J. Biol. Chem. 275, 32925–32930 [DOI] [PubMed] [Google Scholar]
- 34.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iida T., Araki H. (2004) Mol. Cell. Biol. 24, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J. J., Marelli M., Christmas R. H., Vizeacoumar F. J., Dilworth D. J., Ideker T., Galitski T., Dimitrov K., Rachubinski R. A., Aitchison J. D. (2002) J. Cell Biol. 158, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szilard R. K., Titorenko V. I., Veenhuis M., Rachubinski R. A. (1995) J. Cell Biol. 131, 1453–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie D. M., Grill B., Rout M. P., Wozniak R. W., Aitchison J. D. (2002) Mol. Cell. Biol. 22, 2544–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., Gilson E., Olivo-Marin J. C., Scherthan H., Nehrbass U. (2002) Nat. Cell Biol. 4, 214–221 [DOI] [PubMed] [Google Scholar]
- 41.Erdmann R., Veenhuis M., Mertens D., Kunau W. H. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurvitz A., Rottensteiner H. (2006) Biochim. Biophys. Acta 1763, 1392–1402 [DOI] [PubMed] [Google Scholar]
- 43.Veenhuis M., Mateblowski M., Kunau W. H., Harder W. (1987) Yeast 3, 77–84 [DOI] [PubMed] [Google Scholar]
- 44.Einerhand A. W., Voorn-Brouwer T. M., Erdmann R., Kunau W. H., Tabak H. F. (1991) Eur. J. Biochem. 200, 113–122 [DOI] [PubMed] [Google Scholar]
- 45.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 46.Ou X., Ji C., Han X., Zhao X., Li X., Mao Y., Wong L. L., Bartlam M., Rao Z. (2006) J. Mol. Biol. 357, 858–869 [DOI] [PubMed] [Google Scholar]
- 47.Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 48.Lockshon D., Surface L. E., Kerr E. O., Kaeberlein M., Kennedy B. K. (2007) Genetics 175, 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J. J., Sydorskyy Y., Marelli M., Hwang D., Bolouri H., Rachubinski R. A., Aitchison J. D. (2006) Mol. Syst. Biol. 2, 2006.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Leij I., Van den Berg M., Boot R., Franse M., Distel B., Tabak H. F. (1992) J. Cell Biol. 119, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhillon N., Kamakaka R. T. (2000) Mol. Cell 6, 769–780 [DOI] [PubMed] [Google Scholar]
- 52.Lee D. C., Aitchison J. D. (1999) J. Biol. Chem. 274, 29031–29037 [DOI] [PubMed] [Google Scholar]
- 53.Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984) Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 54.Gasch A. P., Huang M., Metzner S., Botstein D., Elledge S. J., Brown P. O. (2001) Mol. Biol. Cell 12, 2987–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasch A. P., Werner-Washburne M. (2002) Funct. Integr. Genomics 2, 181–192 [DOI] [PubMed] [Google Scholar]
- 56.Kaeberlein M., Andalis A. A., Fink G. R., Guarente L. (2002) Mol. Cell. Biol. 22, 8056–8066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin S. J., Kaeberlein M., Andalis A. A., Sturtz L. A., Defossez P. A., Culotta V. C., Fink G. R., Guarente L. (2002) Nature 418, 344–348 [DOI] [PubMed] [Google Scholar]
- 58.Ghislain M., Talla E., François J. M. (2002) Yeast 19, 215–224 [DOI] [PubMed] [Google Scholar]
- 59.Gallo C. M., Smith D. L., Jr., Smith J. S. (2004) Mol. Cell. Biol. 24, 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcotte E. M., Pellegrini M., Thompson M. J., Yeates T. O., Eisenberg D. (1999) Nature 402, 83–86 [DOI] [PubMed] [Google Scholar]
- 61.Kaffman A., O'Shea E. K. (1999) Annu. Rev. Cell Dev. Biol. 15, 291–339 [DOI] [PubMed] [Google Scholar]
- 62.Jans D. A., Xiao C. Y., Lam M. H. (2000) BioEssays 22, 532–544 [DOI] [PubMed] [Google Scholar]
- 63.Osumi T., Tsukamoto T., Hata S., Yokota S., Miura S., Fujiki Y., Hijikata M., Miyazawa S., Hashimoto T. (1991) Biochem. Biophys. Res. Commun. 181, 947–954 [DOI] [PubMed] [Google Scholar]
- 64.Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. (1991) EMBO J. 10, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Roermund C. W., Elgersma Y., Singh N., Wanders R. J., Tabak H. F. (1995) EMBO J. 14, 3480–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.AbdelRaheim S. R., Cartwright J. L., Gasmi L., McLennan A. G. (2001) Arch. Biochem. Biophys. 388, 18–24 [DOI] [PubMed] [Google Scholar]
- 67.Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B. K., Grunstein M., Gasser S. M. (1997) EMBO J. 16, 3243–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gasser S. M., Cockell M. M. (2001) Gene 279, 1–16 [DOI] [PubMed] [Google Scholar]
- 69.Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 70.Aguilera J., Prieto J. A. (2001) Curr. Genet. 39, 273–283 [DOI] [PubMed] [Google Scholar]
- 71.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.