Abstract
N-Acetylglucosamine 6-O-sulfotransferase-2 (GlcNAc6ST-2) catalyzes the sulfation of mucin-like glycoproteins, which function as ligands for a lymphocyte homing receptor, L-selectin, in the lymph node high endothelial venules (HEVs). We previously showed that GlcNAc6ST-2 is expressed not only in lymph node HEVs but also in the colonic epithelial cells in mice. Here we investigated the regulatory mechanism and physiological significance of colonic expression of GlcNAc6ST-2 in mice. Treatment of a mouse colonic epithelial cell line with butyrate, a short-chain fatty acid produced by anaerobic bacteria, induced GlcNAc6ST-2 expression in the presence of epidermal growth factor. Administration of butyrate in the drinking water stimulated GlcNAc6ST-2 expression in the mouse intestine, indicating that butyrate could serve as a regulatory molecule for the GlcNAc6ST-2 expression in vivo. Immunohistochemical analysis indicated that the sulfation of colonic mucins was greatly diminished in GlcNAc6ST-2-deficient mice. Liquid chromatography coupled to electrospray ionization tandem mass spectrometry of the colonic-mucin O-glycans from wild-type and GlcNAc6ST-2-deficient mice showed that GlcNAc-6-O-sulfation was the predominant sulfate modification of these mucins, and it was exclusively mediated by GlcNAc6ST-2. After colitis induction by dextran sulfate sodium, significantly more leukocyte infiltration was observed in the colon of GlcNAc6ST-2-deficient mice than in that of wild-type mice, indicating that the sulfation of colonic mucins by GlcNAc6ST-2 has a protective function in experimental colitis. These findings indicate that GlcNAc6ST-2, whose expression is regulated by butyrate, is a major sulfotransferase in the biosynthesis of sulfomucins in the mouse colon, where they serve as a mucosal barrier against colonic inflammation.
Keywords: Carbohydrate, Carbohydrate/Biosynthesis, Carbohydrate/Function, Carbohydrate/Glycoconjugate, Carbohydrate/Glycoprotein, Carbohydrate/Structure
Introduction
Sulfated glycans have been shown to elicit diverse biological effects (1–4). The addition of sulfate groups to carbohydrate chains is catalyzed by sulfotransferases, which transfer a sulfate group from the sulfate donor, 3′-phosphoadenosine 5′-phosphosulfate, to a specific position on the acceptor oligosaccharide. N-Acetylglucosamine 6-O-sulfotransferases (GlcNAc6STs)2 catalyze the 6-O-sulfation of N-acetylglucosamine (GlcNAc) on the acceptor oligosaccharide. So far, five GlcNAc6STs in humans and four in mice have been identified (5, 6).
Among the GlcNAc6ST family members in mice, GlcNAc6ST-2 (also called HEC-GlcNAc6ST or L-selectin ligand sulfotransferase (LSST)) is known to be specifically expressed in lymph node high endothelial venules (HEVs), where L-selectin-mediated lymphocyte recruitment to the lymph nodes primarily occurs (7, 8). Another member of this sulfotransferase family, GlcNAc6ST-1, is also expressed in HEVs (9). Our group (10) and others (11) previously generated mice deficient in the two HEV-expressed sulfotransferases, and showed that GlcNAc-6-O-sulfation of the L-selectin ligand oligosaccharides plays a major role in lymphocyte recruitment to lymph nodes. The contact hypersensitivity responses are also significantly diminished in the double-null mice, due to a reduction in lymphocyte homing to the draining lymph nodes (10).
During the course of generating a transgenic mouse line expressing Cre recombinase under the transcriptional regulatory elements for the gene encoding GlcNAc6ST-2, we previously found that GlcNAc6ST-2 is strongly expressed not only in lymph node HEVs but also in the colonic villi (12). Further analysis indicated that the cells expressing GlcNAc6ST-2 are reactive with an antibody against Muc2, a major intestinal mucin produced by the goblet cells in the colon (13, 14), suggesting that GlcNAc6ST-2 catalyzes the sulfation of not only the L-selectin ligands in HEVs but also the colonic mucins in mice.
The gastrointestinal epithelium is covered by a protective mucus gel composed predominantly of mucins secreted by goblet cells (14). Muc2 is the most abundant intestinal mucin in the mucus gel produced by goblet cells (13). Muc2-deficient mice spontaneously develop colitis (15), and frequently develop adenomas in the small intestine that progress to invasive adenocarcinoma and rectal tumors (16), indicating that Muc2 is important for intestinal protection. Mice deficient in core 3 β1,3-N-acetylglucosaminyltransferase (C3GnT) display a colon-specific reduction in Muc2 protein and are highly susceptible to experimental triggers of colitis (17). In addition, mice deficient in core 2 β1,6-N-acetylglucosaminyltransferase-2 (C2GnT2) show an increased susceptibility to colitis (18). In the carbohydrate moieties of the mouse Muc2, fucosylated and sulfated oligosaccharides are abundant (19). However, it is unclear which sulfotransferases are responsible for the sulfation of Muc2, or whether the sulfation of Muc2 by those sulfotransferases affects the protective function of Muc2 against colitis.
Our previous study using the above-mentioned GlcNAc6ST-2-Cre transgenic mice also showed that the colonic expression of GlcNAc6ST-2 is regulated by commensal bacteria, as revealed by antibiotic administration (12), suggesting that bacterial components or their metabolites regulate the expression of GlcNAc6ST-2 in the mouse colon. In the present study, we first examined whether short-chain fatty acids (SCFAs), which are metabolites of anaerobic bacteria, might regulate the intestinal expression of GlcNAc6ST-2 in mice, because SCFAs regulate the expression of various genes in the colon (20). We show that one of the SCFAs, butyrate, induced GlcNAc6ST-2 in colonic epithelial cells. We also show that GlcNAc6ST-2 plays a major role in the biosynthesis of sulfomucins in the mouse colon, by immunohistochemical and biochemical analyses. Furthermore, we provide evidence that the sulfation of colonic mucins by GlcNAc6ST-2 in mice has a protective function against massive leukocyte infiltration in experimental colitis.
EXPERIMENTAL PROCEDURES
Mice
Mice deficient in GlcNAc6ST-1 and GlcNAc6ST-2 were backcrossed 5 generations to C57BL/6 mice and maintained as described previously (10, 21). GlcNAc6ST-2-Cre+/R26R mice were generated by mating GlcNAc6ST-2-Cre transgenic mice with ROSA26 reporter (R26R) mice (22) as described previously (12). p53-deficient mice (23) were generated by crossing C57BL-p53+/− mice (BRC No. 01361), provided by RIKEN BRC. Genomic DNA was isolated from the mouse tail and used for PCR genotyping. C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan). The mice were treated in accordance with the guidelines of the Animal Research Committee of the University of Shizuoka.
Establishment of a Mouse Colon Adherent Cell Line and Cell Culture
The colon was removed from p53-deficient mice and placed into sterile tubes containing the digestion buffer (RPMI 1640 containing 1 mg/ml collagenase A, 0.5 mg/ml dispase I, and 20 units/ml DNase I) at 37 °C for 1 h. The dissociated colonic cells were incubated in PBS containing 0.1% trypsin and 0.02% EDTA at 37 °C for 5 min, and passed through a 70-μm cell strainer. After centrifugation, the cell pellet was suspended in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum and 10 ng/ml recombinant human epidermal growth factor (EGF) (Invitrogen), and plated in a 100-mm cell culture dish at 37 °C in a humidified 5% CO2 atmosphere. The adherent cell line thus obtained was named CAdC1.
Immunofluorescence with Anti-Muc2 Antibody
CAdC1 cells were fixed with 0.5% glutaraldehyde, incubated with 3% BSA in PBS, and incubated with rabbit anti-Mucin 2 polyclonal antibody (pAb) (sc-15334, Santa Cruz Biotechnology, Inc.) or normal rabbit IgG (Zymed Laboratories Inc.). After washing, the cells were incubated with Texas Red-conjugated goat anti-rabbit IgG (H+L) (Vector Laboratories) together with 4′,6-diamidino-2-phenylindole (DAPI).
Treatment of CAdC1 Cells with Short-chain Fatty Acids
CAdC1 cells were treated with 4 mm each of sodium acetate (Kanto Chemicals Co., Inc.), sodium propionate (Wako Pure Chemicals Industries, Ltd.), or sodium butyrate (Sigma) in the cell culture medium in a 12-well plate for 24 h in the presence or absence of 10 ng/ml EGF. In some experiments, CAdC1 cells were treated with 0.5 μm trichostatin A (TSA) (Calbiochem) in the presence of 10 ng/ml EGF. The cells were subjected to RT-PCR analysis as described below, or Western blotting using a rabbit anti-acetylated-lysine pAb (Abcam) and horseradish peroxidase-conjugated goat anti-rabbit IgG (H+L) (Zymed Laboratories Inc.).
RT-PCR
The total RNA was purified from the ileum and colon of C57BL/6 mice, or from CAdC1 cells, and used for RT-PCR. The primers used were: 5′-TGTGGTCTGTGTGGGAACTTT-3′ and 5′-CATAGATGGGCCTGTCCTCAGG-3′ for mouse Muc2, 5′-ACCGCGTGTGCAAAAAG-3′ and 5′-CTGTTTGCAGCGTCTTGG-3′ for mouse GlcNAc6ST-1, 5′-TCCATACTAACGCCAGGAACG-3′ and 5′-TGGTGACTAAGGCTGGAACC-3′ for mouse GlcNAc6ST-2, 5′-TGCTGGTACTGTCCTCGTGG-3′ and 5′-TGATGTTGCCACGAGCGAAGG-3′ for mouse GlcNAc6ST-3, 5′-TCAACCTAAAGGTGGTGCAACT-3′ and 5′-GGTTAAGAAGAAATCAGCGCGT-3′ for mouse GlcNAc6ST-4, 5′-AAGCCCTACAACCTGGATGTG-3′ and 5′-GAGTTGCGCACTGTGCTGTAT-3′ for mouse keratan sulfate sulfotransferase (KSST), 5′-ATGACTCTGCTGCCAAAGAA-3′ and 5′-GTGCGTCTTCATGAACACAA-3′ for mouse galactose 3-O-sulfotransferase (Gal3ST)-1, 5′-ATGTCCATCCTGAGAGGCAC-3′ and 5′-GGTGGTTGCACATGATGTTA-3′ for mouse Gal3ST-2, 5′-ATGCCACCTATCCTCCAGCG-3′ and 5′-CGTGAGGTTGTGGCGTTCTG-3′ for mouse Gal3ST-3, 5′-TGCCCACTACCAGTTTGGCT-3′ and 5′-GTTACGGGCGTAGTGGTTGC-3′ for mouse Gal3ST-4, and 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′ for mouse β-actin. The PCR cycle (94 °C, 30 s; 62 °C 30 s; 72 °C 30 s) was repeated 38 times for GlcNAc6ST-1, GlcNAc6ST-3, GlcNAc6ST-4, and KSST; 32 times for Muc2, GlcNAc6ST-2, and β-actin; and 35 times for Gal3STs.
Histology and Immunostaining
LacZ staining was performed as described previously (12). For Alcian blue staining, frozen sections (7-μm thick) were fixed with 10% formaldehyde in PBS for 30 min. The fixed sections were stained with 1% Alcian blue 8GX (Sigma) in 0.1 n HCl (pH 1.0) for 2 h, and counterstained with Nuclear Fast Red solution (Sigma). For immunohistochemical analyses, the sections were fixed with 0.5% glutaraldehyde in PBS for 10 min, and the nonspecific-binding sites were blocked with 3% BSA in PBS for 30 min. The sections were then incubated with 4 μg/ml rabbit anti-Mucin 2 pAb (sc-15334, Santa Cruz Biotechnology, Inc.) for 2 h. After washing with PBS containing 0.1% BSA, the sections were incubated with 0.03% hydrogen peroxide in PBS containing 0.1% BSA to inactivate the endogenous peroxidase. After washing, the sections were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (H+L) (Zymed Laboratories Inc., diluted 1:400) for 2 h. After washing, the colored reaction product was developed using a metal-enhanced DAB substrate kit (Pierce Biotechnology, Inc.), and the sections were counterstained with Nuclear Fast Red solution.
Preparation of Muc2-enriched Fraction
A Muc2-enriched fraction was obtained according to a method described by Herrmann et al. (24) with some modifications. In brief, mucus from the colon was removed from the epithelial surface of the mouse colon by mechanical scraping. The mucus fraction thus obtained was solubilized by 6 m guanidinium chloride. After centrifugation at 12,500 × g for 60 min at 4 °C, the supernatant was decanted, and the gel-like pellet was re-extracted twice, as above. The pellet was finally suspended in water, dialyzed, and lyophilized.
SDS-PAGE and Lectin Blot Analysis
The Muc2-enriched fraction was solubilized in SDS-PAGE sample buffer containing 2-mercaptoethanol by heating at 95 °C for 5 min, and alkylated by incubation in 25 mm iodoacetamide for 30 min at room temperature. The samples were separated by SDS-PAGE using a 3% stacking and 4% running gel, and stained with 0.125% Alcian blue in solvent A (25% ethanol and 0.1 n hydrochloric acid, pH1.0) for 30 min. The gel was destained with solvent A for 10 min, 100% methanol for 20 min, and solvent A for 12 h. In some experiments, the samples separated by SDS-PAGE were transferred onto a polyvinylidene difluoride membrane, and lectin blot analysis was performed using biotinylated AAL (Vector Laboratories) and horseradish peroxidase-conjugated streptavidin (Zymed Laboratories Inc.).
Carbohydrate Structural Analysis
The lyophilized Muc2-enriched fraction (2 mg) was dissolved in 100 μl of water. To this was added 100 μl of 0.1 m KOH and 2.0 m NaBH4, and the mixture was incubated at 50 °C for 15 h. After cooling, 100 μl of 4.0 m acetic acid was added to the reaction mixture, and the sample was applied to a 1-ml column of AG50W-X8 resin (H+ form; Bio-Rad). The oligosaccharides were eluted from the column with 5 ml of water. The solvent was then evaporated, and the sample was dissolved in 200 μl of methanol with 20 μl of acetic acid, and evaporated. The last step was repeated three times, and the sample was stored in a desiccator at room temperature. The oligosaccharides thus obtained were dissolved in water and separated on a Hypercarb column (150 × 0.32 mm, 5-μm particles; Thermo Fisher Scientific) at a flow rate of 15 μl/min, with a 10 mm ammonium bicarbonate-acetonitrile gradient (0–40% acetonitrile) over 40 min. The column was coupled to a quadrupole orthogonal acceleration time-of-flight mass spectrometer (Q-TOF; Agilent Technologies), operated in negative or positive ion mode. For liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) operated in the negative ion mode, the electrospray voltage applied was −4.5 kV, and [M-H]− ions were collided with argon as the collision gas, with a collision energy from 5 to 90 eV for m/z 200–2000. For LC-ESI-MS/MS operated in the positive ion mode, the electrospray voltage applied was +3.5kV, and [M+H]+ ions were collided with a collision energy from 5 to 15 eV for m/z 200–2000.
Induction of Colitis by Dextran Sulfate Sodium (DSS)
Colitis was induced in mice by adding 5% DSS (M.W. = 36,000–50,000, MP Biomedicals) to their drinking water. The animals were allowed free access to the DSS-containing water for 7 days, then they were sacrificed, and their colon was dissected out, mounted in OCT compound, and stored at −80 °C until use. Frozen sections (7-μm thick) of the colons were stained with AlexaFluor 647-labeled anti-mouse CD45 monoclonal antibody (mAb) (BioLegend), AlexaFluor 647-labeled anti-mouse F4/80 mAb (BioLegend), or AlexaFluor 647-labeled anti-mouse Gr-1 mAb (BioLegend) together with DAPI. The CD45+ leukocyte infiltration and Gr-1+ granulocyte infiltration were assessed by counting the number of cells in a defined area using a BZ-9000 fluorescence microscope (Keyence, Co., Osaka, Japan). The F4/80+ macrophage infiltration was assessed by measuring the pixel area positively stained for F4/80 and dividing it by the total pixel area in the colon. Granulocyte infiltration into the colon was also quantified by measuring peroxidase activity. Briefly, a dissected colon was homogenized in the homogenizing buffer (20 mm HEPES-NaOH, 1.5 mm magnesium chloride, 400 mm sodium chloride, 1 mm EDTA, 1 mm DTT, 1× protease inhibitor mixture (Roche Applied Science), 1% nonidet-P40, and 20% glycerol, pH7.9) on ice using a pestle (As One, Tokyo, Japan). The homogenate was sonicated for 10 s and centrifuged at 12,500 × g for 15 min. To the supernatant 3,3′,5,5′-tetramentylbenzidine (Pierce) was added and incubated for 30 min, and the absorbance at 450 nm was measured to determine the peroxidase activity. One unit was defined as the activity of 1 mmol of peroxidase.
Statistical Analysis
The Student's t test was used for statistical analysis.
RESULTS
Regulation of GlcNAc6ST-2 Expression by Sodium Butyrate
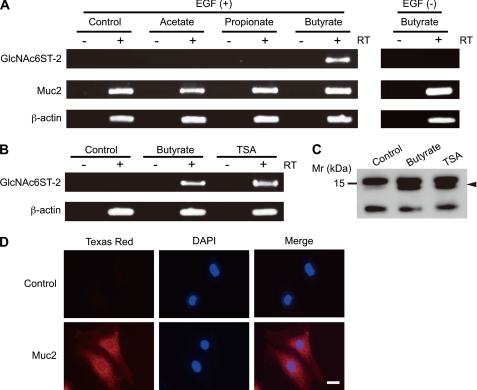
To test the effects of SCFAs on the expression of GlcNAc6ST-2 in the mouse colonic epithelium, we first prepared an immortalized colonic epithelial cell line, termed CAdC1, from the colon of p53-deficient mice, because cells from these mice become immortal at a high rate (23). The immunofluorescence study using the anti-Muc2 antibody indicated that all the CAdC1 cells expressed Muc2 (Fig. 1D), a major colonic mucin secreted from the goblet cells (13, 14), suggesting that CAdC1 cells were derived from goblet cells. The CAdC1 cells were treated with or without the sodium salt of one of the three major SCFAs: acetate, propionate, or butyrate. Only sodium butyrate strongly induced the expression of GlcNAc6ST-2, as revealed by RT-PCR analysis (Fig. 1A). The expression Muc2 was clearly detected in the CAdC1 cells without stimulation, and was slightly enhanced in the presence of sodium butyrate. The induction of GlcNAc6ST-2 was completely abolished in the absence of EGF, indicating that both sodium butyrate and EGF are required for the induction of GlcNAc6ST-2. Because sodium butyrate has an inhibitory effect on histone deacetylases (HDACs) (25), we next examined the effect of TSA (26), an HDAC-specific inhibitor, on the expression of GlcNAc6ST-2 in CAdC1 cells (Fig. 1B). TSA also clearly induced GlcNAc6ST-2 mRNA, indicating that the inhibition of HDACs in the presence of EGF induces GlcNAc6ST-2 expression. Western blot analysis showed that a 14-kDa band corresponding to the molecular size of histone H2A and H2B (27) became reactive with an anti-acetylated-lysine antibody after sodium butyrate or TSA treatment, confirming that sodium butyrate and TSA inhibited HDACs in this experiment (Fig. 1C).
FIGURE 1.
Induction of GlcNAc6ST-2 mRNA by sodium butyrate. CAdC1 cells were treated with or without (Control) 4 mm sodium acetate, sodium propionate, or sodium butyrate in the presence or absence of 10 ng/ml EGF, and subjected to RT-PCR analysis using primer pairs for GlcNAc6ST-2, Muc2, and β-actin (A). CAdC1 cells were treated with or without (Control) 4 mm sodium butyrate or 0.5 μm TSA in the presence of 10 ng/ml EGF, and subjected to RT-PCR analysis for GlcNAc6ST-2 or β-actin (B), or Western blotting using an anti-acetylated-lysine antibody (C). For PCR, single-stranded cDNAs prepared in the presence (+RT) or absence (−RT) of reverse transcriptase were used as templates. Arrowhead in C indicates a 14-kDa band corresponding to the molecular size of histone H2A and H2B (27). CAdC1 cells were stained with normal rabbit IgG (Control) or rabbit anti-Muc2 pAb (Muc2) together with Texas Red-conjugated secondary antibody and DAPI, and analyzed under fluorescence microscope BZ-9000 (D). Bar, 20 μm. Data are representative of four independent experiments.
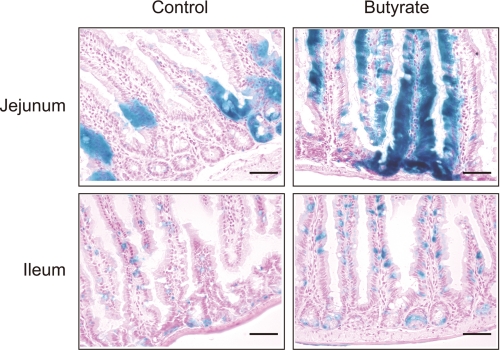
We previously generated a transgenic mouse line, GlcNAc6ST-2-Cre, which expresses Cre recombinase under the regulatory elements of the gene encoding GlcNAc6ST-2 (12). The GlcNAc6ST-2-Cre transgenic mice were crossed with ROSA26 reporter (R26R) mice (22), which express lacZ following Cre-mediated recombination. Because the expression of lacZ in the offspring (GlcNAc6ST-2-Cre+/R26R mice) recapitulates that of intrinsic GlcNAc6ST-2 (12), we next examined the effects of sodium butyrate on the expression of GlcNAc6ST-2 in vivo using these mice (Fig. 2). As described previously, a weak expression of GlcNAc6ST-2 was observed in the jejunum and ileum of GlcNAc6ST-2-Cre+/R26R mice that were given normal drinking water. The administration of sodium butyrate in the drinking water significantly enhanced the LacZ staining in the jejunum and ileum in these mice, indicating that sodium butyrate could induce the expression of GlcNAc6ST-2 in the mouse intestinal tissues in vivo. The reason why only some epithelial cells were positive for LacZ staining is probably because only goblet cells in the epithelium strongly express GlcNAc6ST-2 (12).
FIGURE 2.
Induction of LacZ by sodium butyrate in the small intestine of GlcNAc6ST-2-Cre+/R26R mice. GlcNAc6ST-2-Cre+/R26R mice (12) were given drinking water with or without (Control) 5 mm sodium butyrate for 24 h. Frozen sections of the jejunum and ileum were subjected to LacZ staining as described previously (12). Bar, 50 μm. Data are representative of four independent experiments.
Expression of Sulfotransferases in the Mouse Intestine
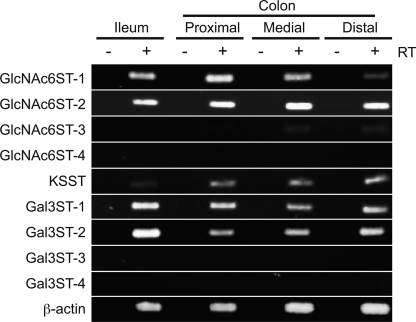
Although we detected the intestinal expression of GlcNAc6ST-2 in mice, other sulfotransferases might also be expressed in the mouse intestine. Thus, we next examined which types of carbohydrate sulfotransferases are expressed in the mouse intestine, by RT-PCR analysis (Fig. 3). Among the four GlcNAc 6-O-sulfotransferases in mice (6), only GlcNAc6ST-1 and GlcNAc6ST-2 were clearly detected in the ileum and colon in the RT-PCR analysis. In the medial and distal colon, only a very faint band of GlcNAc6ST-3 was detected, in contrast to the finding in humans that GlcNAc6ST-3, also called I-GlcNAc6ST, is highly expressed in the intestinal tissues including the colon (28), which may be due to species difference. Regarding galactose 6-O-sulfotransferase (Gal6ST), KSST expression was detected in the mouse colon. Among the Gal3STs, only the Gal3ST-1 and Gal3ST-2 expressions were detected. These results indicate that multiple carbohydrate sulfotransferases are expressed in the mouse intestine.
FIGURE 3.
Expression of sulfotransferases in the mouse intestine. RT-PCR analysis for various sulfotransferases was performed using mRNAs from the ileum, proximal colon, medial colon, and distal colon of the C57BL/6 WT mouse. For PCR, single-stranded cDNAs prepared in the presence (+RT) or absence (−RT) of reverse transcriptase were used as templates. Data are representative of three independent experiments.
Sulfation of the Colonic Mucins Is Largely Dependent on GlcNAc6ST-2
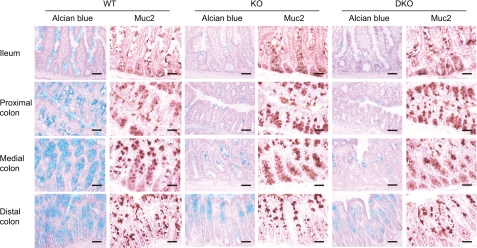
Because GlcNAc6ST-1 and GlcNAc6ST-2 were the two major sulfotransferases expressed in the mouse intestine among the GlcNAc6ST family members, we next sought to determine whether these sulfotransferases are involved in the sulfation of intestinal mucins, using GlcNAc6ST-2-deficient (KO) (21), and GlcNAc6ST-1 and GlcNAc6ST-2-doubly deficient (DKO) mice (10). To assess the mucin sulfation, serial sections were stained with Alcian blue and an antibody against Muc2, a major intestinal mucin (Fig. 4). The Alcian blue staining was performed at pH 1.0, because Alcian blue selectively binds to sulfated carbohydrates under this condition (29). In wild-type (WT) mice, both Alcian blue and the anti-Muc2 antibody clearly stained colonic villi, suggesting that the colonic mucins were highly sulfated. In the KO mice, the Alcian blue staining of the colon was significantly diminished, while the staining intensity with the anti-Muc2 antibody did not differ from that observed in the WT mice. No obvious further reduction of the staining intensity with Alcian blue was observed in the DKO mice, suggesting that GlcNAc6ST-1 is not very involved in the sulfation of colonic mucins, if at all. These results indicate that the sulfation of colonic mucins is largely mediated by GlcNAc6ST-2, although other sulfotransferases are also expressed in the mouse colon. In addition, a reduction in Alcian blue staining was also observed in the small intestine of KO and DKO mice, indicating that the sulfation of mucins in the small intestine is also largely mediated by GlcNAc6ST-2.
FIGURE 4.
Expression of sulfated carbohydrates and Muc2 in the intestine of WT, KO, and DKO mice. Frozen sections from WT, KO, and DKO mice were stained with Alcian blue (pH 1.0), or anti-Muc2 pAb. Bar, 50 μm. Data are representative of three (WT and KO), or two (DKO) independent experiments.
Sulfation of Colonic Mucins by GlcNAc6ST-2
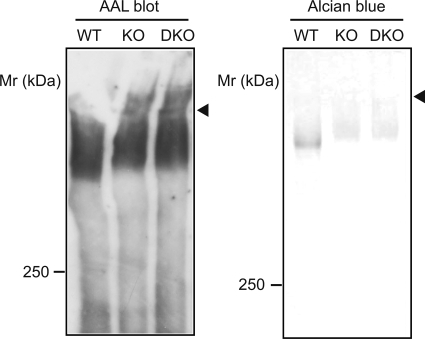
To elucidate the role of GlcNAc6ST-2 in the sulfation of colonic mucins, we next performed SDS-PAGE of the Muc2-enriched fraction from the WT, KO, and DKO mice, followed by lectin blot analysis and Alcian blue staining (Fig. 5). First, the mucus layer was prepared by scraping the luminal surface of the mouse colon, and solubilized by guanidinium chloride. After centrifugation, the Muc2-enriched fraction was recovered as the guanidinium chloride-insoluble pellet, according to a published method (24). After solubilization, the Muc2-enriched fraction was subjected to SDS-PAGE followed by lectin blot analysis using a fucose-binding lectin, AAL. An almost equivalent amount of colonic mucin reactive with AAL was present in the samples from WT, KO, and DKO mice. However, staining of the gel with Alcian blue at pH 1.0 showed that the sulfation of the colonic mucins was significantly decreased in the KO and DKO mice to a similar extent, suggesting that the GlcNAc-6-O-sulfation of colonic mucins is mediated by GlcNAc6ST-2, and not by GlcNAc6ST-1.
FIGURE 5.
Lectin blot and Alcian blue staining of colonic mucins. Muc2-enriched fractions from WT, KO, and DKO mice were separated by SDS-PAGE and subjected to lectin blotting using AAL, and to Alcian blue staining. Arrowheads indicate the boundary between the stacking and separating gel. Data are representative of three independent experiments.
Carbohydrate Structural Analysis of the Colonic Mucins
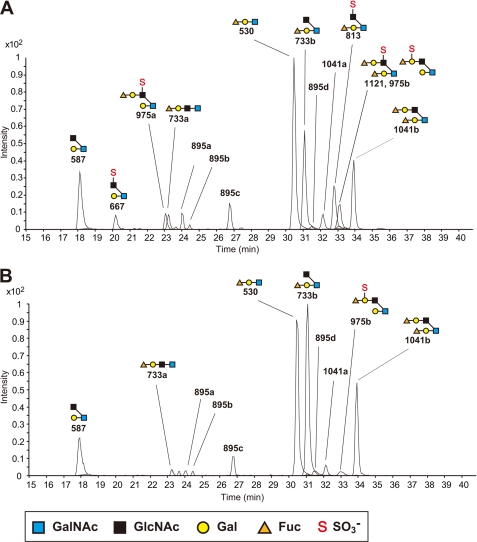
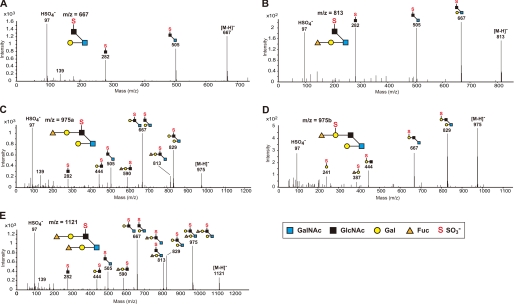
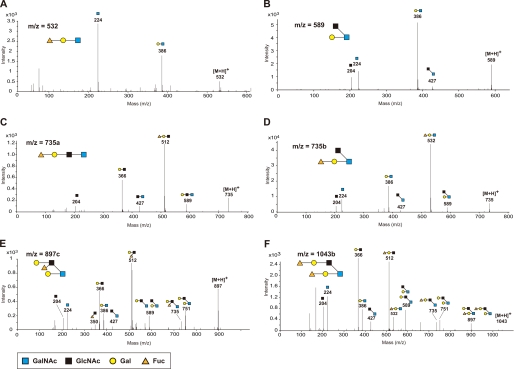
We next performed carbohydrate structural analysis of the O-glycans of the colonic mucins using LC-ESI-MS/MS spectrometry. The O-linked oligosaccharides were released from the colonic mucins in the Muc2-enriched fraction, by β-elimination and reduction with sodium borohydride, and separated by HPLC coupled to ESI-MS/MS. As shown in Fig. 6A, 15 types of oligosaccharides from the Muc2-enriched fraction of WT mice were detected as their [M-H]− ions in the LC-ESI-MS analysis. Oligosaccharides containing GlcNAc-6-O-sulfate were found on core 2-branched oligosaccharides from WT mice at m/z 667, 813, 975a, and 1121. In contrast, these sulfated oligosaccharides were completely absent from the oligosaccharide fraction obtained from the KO mice (Fig. 6B). The structures of these sulfated oligosaccharides were characterized by analyzing the daughter ion spectra obtained by the LC-ESI-MS/MS analysis (Fig. 7). Two isomeric sulfated oligosaccharides at m/z 975, namely 975a and 975b, were found in the LC-ESI-MS/MS analysis. The former, containing GlcNAc-6-O-sulfate, was completely absent in the oligosaccharide fraction from the KO mice, whereas the latter, containing Gal-3- or Gal-6-O-sulfate, was present in the KO mice at a comparable level as in the WT mice. We also analyzed the neutral oligosaccharides by LC-ESI-MS/MS operated at the positive ion mode, because the daughter ions of neutral oligosaccharides were less stable in LC-ESI-MS/MS operated at the negative ion mode. As shown in Fig. 8, [M+H]+ ions at m/z 532, 589, 735, 897, and 1043, which correspond to [M-H]− ions at m/z 530, 587, 733, 895, and 1041, respectively, were detected. Table 1 summarizes the carbohydrate structural analysis of the O-glycans of the colonic mucins from WT and KO mice. Collectively, these results indicate that GlcNAc-6-O-sulfation is the predominant sulfate modification of the colonic mucins, and that GlcNAc6ST-2 is essential for this carbohydrate modification.
FIGURE 6.
LC-ESI-MS analysis of the oligosaccharides on colonic mucins. LC-ESI-MS total ion chromatogram of the oligosaccharides in the Muc2-enriched fraction from the colon of WT (A) and KO (B) mice. LC-ESI-MS was operated in the negative ion mode. The annotations correspond to the [M-H]− ions listed in Table 1. Data are representative of five independent experiments.
FIGURE 7.
LC-ESI-MS/MS analysis of the sulfated oligosaccharides on colonic mucins. ESI tandem mass spectra of the [M-H]− ions obtained from m/z 667 (A), m/z 813 (B), m/z 975a (C), m/z 975b (D), and m/z 1121 (E). The collision energies applied were: for m/z 667, 50 eV; for m/z 813, 60 eV; for m/z 975a, 80eV; for m/z 975b, 70 eV; and for m/z 1121, 90 eV. LC-ESI-MS/MS was operated in the negative ion mode. Note the presence of m/z 97 (HSO4−) in all the ESI tandem mass spectra. Data are representative of three independent experiments.
FIGURE 8.
LC-ESI-MS/MS analysis of the neutral oligosaccharides on colonic mucins. ESI tandem mass spectra of [M+H]+ ions obtained from m/z 532 (A), m/z 589 (B), m/z 735a (C), m/z 735b (D), m/z 897c (E), and m/z 1043b (F), corresponding to the [M-H]− ions obtained from m/z 530, m/z 587, m/z 733a, m/z 733b, m/z 895c, and m/z 1041b in Table 1, respectively. LC-ESI-MS/MS was operated in the positive ion mode, and the collision energies applied were: for m/z 532, and 589, 5 eV; for m/z 735a, and 735b, 10 eV; and for m/z 897c and 1043b, 15 eV. Data are representative of three independent experiments.
TABLE 1.
Structural characterization and relative abundance of the O-glycans on the colonic mucins from WT and KO mice
| Molecular ion [M-H]− | Proposed sequence/compositiona | Relative abundance (%)b |
|
|---|---|---|---|
| WT | KO | ||
| 587 | Gal-3(GlcNAc-6)GalNAcol | 9.89 | 8.39 |
| 530 | Fuc-Gal-3GalNAcol | 30.43 | 30.50 |
| 733a | Fuc-Gal-GlcNAc-GalNAcol | 2.14 | 1.30 |
| 733b | Fuc-Gal-3(GlcNAc-6)GalNAcol | 18.08 | 34.98 |
| 895a, 895b, 895d | Fuc, 2Gal, GlcNAc, GalNAcol | 3.58 | 2.46 |
| 895c | Gal-3(Gal-(Fuc-)GlcNAc-6)GalNAcol | 3.68 | 3.41 |
| 1041a | 2Fuc, 2Gal, GlcNAc, GalNAcol | 2.41 | 1.70 |
| 1041b | Fuc-Gal-3(Fuc-Gal-GlcNAc-6)GalNAcol | 10.27 | 16.16 |
| 667 | Gal-3(SO3−-6GlcNAc-6)GalNAcol | 2.05 | –c |
| 813 | Fuc-Gal-3(SO3−-6GlcNAc-6)GalNAcol | 9.13 | – |
| 975a | Gal-3(Fuc-Gal-(SO3−-6)GlcNAc-6)GalNAcol | 2.13 | – |
| 975b | Gal-3(Fuc-(SO3−-3/6)Gal-GlcNAc-6)GalNAcol | 0.99 | 1.10 |
| 1121 | Fuc-Gal-3(Fuc-Gal-(SO3−-6)GlcNAc-6)GalNAcol | 5.21 | – |
a For structural characterization, the following assumptions were made based on the reported structures of O-glycans: the hexose and deoxyhexose are Gal and fucose, respectively, the N-acetylhexosamine residue linked to N-acetylhexosamininitol is GlcNAc linked to GalNAcol, and the core 2 branch is formed after the core 1 structure is formed (46).
b The relative abundance was calculated by dividing the area of each peak by the total area of the peaks annotated in Fig. 6, A and B. Peaks of impurities, which did not produce secondary ions of oligosaccharide fragments in the MS/MS analysis, were excluded from the calculation.
c Not detected.
Protective Function of GlcNAc6ST-2 in DSS-induced Colitis
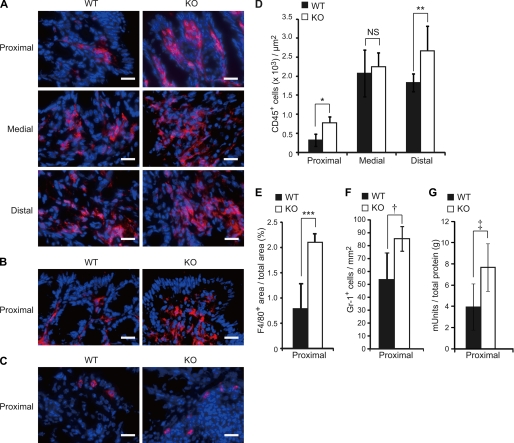
Colonic mucins are regarded as a mucosal barrier to infection (30). However, the contribution of the structures of the mucins' carbohydrate moieties to their barrier function is not well known. To examine the role of the colonic mucin sulfation by GlcNAc6ST-2 under pathological conditions, we measured the extent of inflammation in DSS-induced experimental colitis. Seven days after 5% DSS was administered to WT and KO mice in their drinking water, a significant increase in the CD45+ leukocyte infiltration into the colon was observed in the KO mice compared with the WT mice (Fig. 9, A and D). Staining of the sections with an anti-F4/80 mAb, which is specific for macrophages, showed that significantly more macrophages infiltrated the proximal colon of the KO mice than that of the WT mice (Fig. 9, B and E). Furthermore, staining of the sections with anti-Gr-1 mAb, which is specific for granulocytes, and the measurement of peroxidase activity, which reflects granulocyte numbers, showed that significantly more granulocytes infiltrated the proximal colon of the KO mice than that of the WT mice (Fig. 9, C, F, and G). These results indicate that the sulfation of colonic mucins by GlcNAc6ST-2 has a protective function against massive leukocyte infiltration in experimental colitis in mice.
FIGURE 9.
Accelerated leukocyte infiltration in KO mice after DSS treatment. WT and KO mice were given drinking water containing 5% DSS for 7 days. A, frozen sections of the proximal, medial, and distal colon were stained with AlexaFluor 647-labeled anti-CD45 mAb (red) and DAPI (blue), and analyzed by fluorescence microscopy (BZ-9000). Bar, 20 μm. B and C, frozen sections of the proximal colon were stained with AlexaFluor 647-labeled anti-F4/80 mAb (B) or anti-Gr-1 mAb (C) (red), and DAPI (blue). Bar, 20 μm. D, number of CD45+ leukocytes per 1 μm2 in the colon after DSS treatment. Each bar represents the mean ± S.D. of triplicate determinations. n = 3. *, p = 0.015; **, p = 0.054, and NS, not significant. E, percentage of the total area that was F4/80+ in the proximal colon. Each bar represents the mean ± S.D. of triplicate determinations. n = 3. ***, p = 0.006. F, number of Gr-1+ granulocutes per one mm2 in the proximal colon after DSS treatment. Each bar represents the mean ± S.D. of triplicate determinations. n = 3. †, p = 0.019. G, determination of peroxidase activity as an index of granulocyte infiltration into the proximal colon. Results are expressed as peroxidase milliunits per gram total protein. Each bar represents the mean ± S.D. of triplicate determinations. WT, n = 5; KO, n = 4. ‡, p = 0.020.
DISCUSSION
The questions addressed by the present study were whether GlcNAc6ST-2 catalyzes the sulfation of colonic mucins in mice, and if so, whether the sulfation affects the protective function of the mucins against colitis. Previous studies, including those from our group, indicated that GlcNAc6ST-2 plays a critical role in the biosynthesis of the 6-sulfo sialyl Lewis X structure, which functions as a major L-selectin ligand in lymph node HEVs (10, 11). We also previously showed that GlcNAc6ST-2 is expressed not only in lymph node HEVs but also in colonic epithelial cells in mice (12). In the present study, we demonstrated that GlcNAc6ST-2 plays a major role in the sulfation of colonic mucins in the mouse, and that the sulfation of colonic mucins has a protective function against massive leukocyte infiltration in the experimental colitis induced by DSS. We also showed that butyrate, a metabolite from anaerobic bacteria in the colon, induces the expression of GlcNAc6ST-2.
The expression of GlcNAc6ST-2 was thought to be specific for the HEVs in lymph nodes (31, 32). However, our recent study indicated that GlcNAc6ST-2 is also strongly expressed in the goblet cells in the mouse colon, and that its expression is inhibited by the elimination of commensal bacteria in the colon (12). These previous findings led us to investigate in the present study whether metabolites from commensal bacteria regulate the expression of GlcNAc6ST-2 in the colon. It was reported that the colonic lumen contains total SCFAs at a concentration of 80–130 mmol per kg of total intestinal contents (33). The three major SCFAs, acetate, propionate, and butyrate, constitute about 90% of the total SCFAs, with a molar ratio of 60:20:20 for acetate/propionate/butyrate (33). Our finding that 4 mm butyrate, which is lower than its physiological concentration, induced the expression of GlcNAc6ST-2 in an immortalized colonic epithelial cell line, suggested that butyrate induces GlcNAc6ST-2 expression in vivo. In support of this idea, the administration of sodium butyrate in the drinking water induced the expression of GlcNAc6ST-2 in vivo in the small intestines of mice that weakly expressed GlcNAc6ST-2 (Fig. 2).
Our finding that EGF is required for the butyrate-induced expression of GlcNAc6ST-2 suggested that multiple extracellular signals are required for its gene induction. Because butyrate functions as an HDAC inhibitor and enhances the accessibility of transcription factors to the enhancer/promoter region of various genes by inducing histone acetylation (25), it is plausible that transcription factors activated by the EGF-EGF receptor pathway are involved in the induction of the gene encoding GlcNAc6ST-2. It is reported that the normal human colon expresses little, if any, GlcNAc6ST-2 (34, 35). Because butyrate is produced by commensal bacteria in the human colon (33), signals equivalent to those transmitted from the EGF receptor might be lacking in the human colon. In this regard, it is noteworthy that some human colon adenocarcinoma cells express GlcNAc6ST-2 (34, 35), because those colon adenocarcinoma cells might be receiving certain growth signals from the environment, similar to those from EGF receptors. Further studies are required to determine the detailed molecular events leading to the induction of GlcNAc6ST-2 in the mouse colon.
Our carbohydrate structural analysis of mouse colonic mucins indicated that sulfation was preferential on the C-6 of GlcNAc residues on the core 2 branch of O-glycans. We also detected a small amount of galactose-sulfated oligosaccharides, consistent with the RT-PCR results that KSST and Gal3STs were expressed in the mouse colon. The extended core 1 structure modified with GlcNAc-6-O-sulfate that is reactive with the mAb MECA-79 (36) was not detected in our carbohydrate analysis of colonic mucins, consistent with our observation that the MECA-79 antibody does not bind to frozen sections of the mouse colon.3 In contrast to the abundant GlcNAc-6-O-sulfation of mouse colonic mucins, most of the sulfation of human MUC2 is found on galactose residues (37). Consistent with this finding, the mAb 91.9H, which was raised against human colonic sulfomucin, recognizes the HSO3-3Galβ1-3(Fucα1–4)GlcNAc structure (38). Interestingly, this mAb binds well to normal epithelial cells in the human colon, but minimally to the adjacent colon carcinoma cells in clinical specimens (39), and this finding was confirmed by biochemical studies (40). In addition, studies using rectal biopsies taken at colonoscopy from patients with ulcerative colitis indicated that the sulfation of the mucins is significantly reduced in these patients (41). These clinical data suggest that the sulfation of colonic mucins plays important roles in maintaining the normal physiological function of the colon. In this regard, it is notable that the colonic mucins in both humans and mice are highly sulfated, even though different monosaccharides are preferentially sulfated on the colonic mucins in humans and mice.
Our experiments using KO mice showed that the GlcNAc-6-O-sulfation of colonic mucins was exclusively mediated by GlcNAc6ST-2 in the mouse colon, even though GlcNAc6ST-1 was also expressed in the colon. Considering the previous finding that both of these sulfotransferases can transfer sulfate onto core 2-branched oligosaccharides (10, 35), and our present observation that sulfate residues were exclusively present on core 2 branched oligosaccharides of the colonic mucins, the preference for a particular acceptor oligosaccharide structure may not account for the differential activity of GlcNAc6ST-1 and GlcNAc6ST-2 for the colonic mucins. One possible explanation is that the subcellular distribution of GlcNAc6ST-2 is more suitable for transferring sulfate to the GlcNAc-terminated O-glycans of colonic mucins, because GlcNAc6ST-1 is confined to the trans-Golgi network, whereas GlcNAc6ST-2 is distributed throughout the Golgi apparatus (42). Another possibility is that differential preferences of GlcNAc6ST-1 and GlcNAc6ST-2 for the Muc2 protein, a major colonic mucin, accounts for the difference. Indeed, some glycosyltransferases preferentially act on specific core proteins and modulate the functions of such glycoproteins (43, 44).
The mucus layer of the intestinal tract functions as a barrier against pathogens and inflammatory stimuli (30). Spontaneous colitis (15), the frequent development of adenomas in the small intestine, and rectal tumors (16) are all found in Muc2-deficient mice, indicating the importance of Muc2 as a mucosal barrier. However, only a few reports have shown the consequences of structural changes in the carbohydrate moiety of Muc2. FUT2-deficient mice lack the terminal fucosylation of colonic mucin (19), but it is still unclear if the fucosylation of colonic mucins has functional significance in inflammation or other biological responses. C3GnT-deficient mice are highly susceptible to experimental triggers of colitis, which was attributed to a colon-specific reduction in Muc2 protein (17). C2GnT2-deficient mice also show an increased susceptibility to colitis without a reduction in Muc2 core proteins (18), indicating that the core 2-branched O-glycans are required to protect the animal against colitis. Because our results showed that GlcNAc-6-O-sulfation was exclusively present on core 2-branched O-glycans and that the KO mice did not show any reduction in the Muc2 core protein, we thought that our KO mice could serve as useful tools for examining the roles of GlcNAc-6-O-sulfation on the core 2 branch. Recently, Dawson et al. (45) reported that mice lacking a sulfate transporter, NaS1, show a decrease in mucin sulfation, accompanied by an enhanced susceptibility to experimental colitis. However, the NaS1-null mice show only a partial reduction in mucin sulfation, and no structural data for the O-glycans attached to the mucins were reported. To our knowledge, there are no previous reports describing the phenotype of mice lacking the GlcNAc-6-O-sulfation of O-glycans on the colonic mucins in experimental colitis. The results shown in the present study provide evidence that the GlcNAc-6-O-sulfation of colonic mucins was completely eliminated in GlcNAc6ST-2-deficient mice, and that this sulfation is important for the barrier function of colonic mucins to prevent the massive leukocyte infiltration induced by DSS administration, indicating a relationship between structure and function of the carbohydrate moieties of colonic mucins in mice.
In conclusion, the present study demonstrated that the expression of GlcNAc6ST-2 is regulated by a SCFA, butyrate, produced by anaerobic bacteria, and that GlcNAc6ST-2 functions as a major sulfotransferase in the sulfation of colonic mucins in mice, which serve as a mucosal barrier against inflammatory stimuli in the intestinal tract. These findings provide new insights into the importance of the structure of the carbohydrate moieties of mucins in health and disease.
Acknowledgments
We thank Dr. Junzhe Min (University of Shizuoka) for help with the LC-ESI-MS/MS analysis. We also thank Jotaro Hirakawa and Atsushi Takakura for animal care and technical support. The C57BL-p53+/− mouse strain (BRC No. 01361) was provided by RIKEN BRC with the support of the National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
This work was supported by National Institutes of Health Grant P01CA71932 (to M. F.), Grants-in-aid for Scientific Research, Category (B) and Grants-in-aid for Scientific Research on Priority Areas, Dynamics of Extracellular Environments, from the Ministry of Education, Culture, Sports, Science and Technology, Japan (21390023 and 20057022, respectively, to H. K.), and the Takeda Science Foundation (to H. K.).
H. Kawashima, unpublished observations.
- GlcNAc6ST
- N-acetylglucosamine 6-O-sulfotransferase
- GlcNAc
- N-acetylglucosamine
- HEVs
- high endothelial venules
- C3GnT
- core 3 β1,3-N-acetylglucosaminyltransferase
- C2GnT2
- core 2 β1,6-N-acetylglucosaminyltransferase-2
- SCFA
- short chain fatty acid
- EGF
- epidermal growth factor
- TSA
- trichostatin A
- pAb
- polyclonal antibody
- KSST
- keratan sulfate sulfotransferase
- Gal3ST
- galactose 3-O-sulfotransferase
- LC-ESI-MS/MS
- liquid chromatography coupled to electrospray ionization tandem mass spectrometry
- DSS
- dextran sulfate sodium
- mAb
- monoclonal antibody
- DAPI
- 4′,6-diamidino-2-phenylindole
- HDAC
- histone deacetylase
- KO
- GlcNAc6ST-2-deficient
- DKO
- GlcNAc6ST-1 and GlcNAc6ST-2-doubly deficient
- WT
- wild-type
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin.
REFERENCES
- 1.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 2.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honke K., Zhang Y., Cheng X., Kotani N., Taniguchi N. (2004) Glycoconj. J. 21, 59–62 [DOI] [PubMed] [Google Scholar]
- 4.Kawashima H. (2006) Biol. Pharm. Bull. 29, 2343–2349 [DOI] [PubMed] [Google Scholar]
- 5.Fukuda M., Hiraoka N., Akama T. O., Fukuda M. N. (2001) J. Biol. Chem. 276, 47747–47750 [DOI] [PubMed] [Google Scholar]
- 6.Hemmerich S., Lee J. K., Bhakta S., Bistrup A., Ruddle N. R., Rosen S. D. (2001) Glycobiology 11, 75–87 [DOI] [PubMed] [Google Scholar]
- 7.von Andrian U. H., Mempel T. R. (2003) Nat. Rev. Immunol. 3, 867–878 [DOI] [PubMed] [Google Scholar]
- 8.Girard J. P., Springer T. A. (1995) Immunol. Today 16, 449–457 [DOI] [PubMed] [Google Scholar]
- 9.Uchimura K., Kadomatsu K., El-Fasakhany F. M., Singer M. S., Izawa M., Kannagi R., Takeda N., Rosen S. D., Muramatsu T. (2004) J. Biol. Chem. 279, 35001–35008 [DOI] [PubMed] [Google Scholar]
- 10.Kawashima H., Petryniak B., Hiraoka N., Mitoma J., Huckaby V., Nakayama J., Uchimura K., Kadomatsu K., Muramatsu T., Lowe J. B., Fukuda M. (2005) Nat. Immunol. 6, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 11.Uchimura K., Gauguet J. M., Singer M. S., Tsay D., Kannagi R., Muramatsu T., von Andrian U. H., Rosen S. D. (2005) Nat. Immunol. 6, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 12.Kawashima H., Hirakawa J., Tobisawa Y., Fukuda M., Saga Y. (2009) J. Immunol. 182, 5461–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson M. E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deplancke B., Gaskins H. R. (2001) Am. J. Clin. Nutr. 73, 1131S–1141S [DOI] [PubMed] [Google Scholar]
- 15.Van der Sluis M., De Koning B. A., De Bruijn A. C., Velcich A., Meijerink J. P., Van Goudoever J. B., Büller H. A., Dekker J., Van Seuningen I., Renes I. B., Einerhand A. W. (2006) Gastroenterology 131, 117–129 [DOI] [PubMed] [Google Scholar]
- 16.Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. (2002) Science 295, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 17.An G., Wei B., Xia B., McDaniel J. M., Ju T., Cummings R. D., Braun J., Xia L. (2007) J. Exp. Med. 204, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone E. L., Ismail M. N., Lee S. H., Luu Y., Ramirez K., Haslam S. M., Ho S. B., Dell A., Fukuda M., Marth J. D. (2009) Mol. Cell. Biol. 29, 3770–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurd E. A., Holmén J. M., Hansson G. C., Domino S. E. (2005) Glycobiology 15, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanderson I. R. (2007) J. Nutr. 137, 2557S–2562S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiraoka N., Kawashima H., Petryniak B., Nakayama J., Mitoma J., Marth J. D., Lowe J. B., Fukuda M. (2004) J. Biol. Chem. 279, 3058–3067 [DOI] [PubMed] [Google Scholar]
- 22.Soriano P. (1999) Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 23.Tsukada T., Tomooka Y., Takai S., Ueda Y., Nishikawa S., Yagi T., Tokunaga T., Takeda N., Suda Y., Abe S., Matsuo I., Ikawa Y., Aizawa S. (1993) Oncogene 8, 3313–3322 [PubMed] [Google Scholar]
- 24.Herrmann A., Davies J. R., Lindell G., Mårtensson S., Packer N. H., Swallow D. M., Carlstedt I. (1999) J. Biol. Chem. 274, 15828–15836 [DOI] [PubMed] [Google Scholar]
- 25.Davie J. R. (2003) J. Nutr. 133, 2485S–2493S [DOI] [PubMed] [Google Scholar]
- 26.Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 27.Naldi M., Andrisano V., Fiori J., Calonghi N., Pagnotta E., Parolin C., Pieraccini G., Masotti L. (2006) J. Chromatogr. A 1129, 73–81 [DOI] [PubMed] [Google Scholar]
- 28.Lee J. K., Bhakta S., Rosen S. D., Hemmerich S. (1999) Biochem. Biophys. Res. Commun. 263, 543–549 [DOI] [PubMed] [Google Scholar]
- 29.Spicer S. S., Baron D. A., Sato A., Schulte B. A. (1981) J. Histochem. Cytochem. 29, 994–1002 [DOI] [PubMed] [Google Scholar]
- 30.Linden S. K., Sutton P., Karlsson N. G., Korolik V., McGuckin M. A. (2008) Mucosal Immunol. 1, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiraoka N., Petryniak B., Nakayama J., Tsuboi S., Suzuki M., Yeh J. C., Izawa D., Tanaka T., Miyasaka M., Lowe J. B., Fukuda M. (1999) Immunity 11, 79–89 [DOI] [PubMed] [Google Scholar]
- 32.Bistrup A., Bhakta S., Lee J. K., Belov Y. Y., Gunn M. D., Zuo F. R., Huang C. C., Kannagi R., Rosen S. D., Hemmerich S. (1999) J. Cell Biol. 145, 899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummings J. H., Pomare E. W., Branch W. J., Naylor C. P., Macfarlane G. T. (1987) Gut 28, 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seko A., Nagata K., Yonezawa S., Yamashita K. (2002) Glycobiology 12, 379–388 [DOI] [PubMed] [Google Scholar]
- 35.Uchimura K., El-Fasakhany F. M., Hori M., Hemmerich S., Blink S. E., Kansas G. S., Kanamori A., Kumamoto K., Kannagi R., Muramatsu T. (2002) J. Biol. Chem. 277, 3979–3984 [DOI] [PubMed] [Google Scholar]
- 36.Yeh J. C., Hiraoka N., Petryniak B., Nakayama J., Ellies L. G., Rabuka D., Hindsgaul O., Marth J. D., Lowe J. B., Fukuda M. (2001) Cell 105, 957–969 [DOI] [PubMed] [Google Scholar]
- 37.Larsson J. M., Karlsson H., Sjövall H., Hansson G. C. (2009) Glycobiology 19, 756–766 [DOI] [PubMed] [Google Scholar]
- 38.Tsuiji H., Hong J. C., Kim Y. S., Ikehara Y., Narimatsu H., Irimura T. (1998) Biochem. Biophys. Res. Commun. 253, 374–381 [DOI] [PubMed] [Google Scholar]
- 39.Irimura T., Wynn D. M., Hager L. G., Cleary K. R., Ota D. M. (1991) Cancer Res. 51, 5728–5735 [PubMed] [Google Scholar]
- 40.Yamori T., Kimura H., Stewart K., Ota D. M., Cleary K. R., Irimura T. (1987) Cancer Res. 47, 2741–2747 [PubMed] [Google Scholar]
- 41.Raouf A. H., Tsai H. H., Parker N., Hoffman J., Walker R. J., Rhodes J. M. (1992) Clin. Sci. 83, 623–626 [DOI] [PubMed] [Google Scholar]
- 42.de Graffenried C. L., Bertozzi C. R. (2003) J. Biol. Chem. 278, 40282–40295 [DOI] [PubMed] [Google Scholar]
- 43.Angata K., Chan D., Thibault J., Fukuda M. (2004) J. Biol. Chem. 279, 25883–25890 [DOI] [PubMed] [Google Scholar]
- 44.Taniguchi N., Miyoshi E., Gu J., Jianguo G., Honke K., Matsumoto A. (2006) Curr. Opin. Struct. Biol. 16, 561–566 [DOI] [PubMed] [Google Scholar]
- 45.Dawson P. A., Huxley S., Gardiner B., Tran T., McAuley J. L., Grimmond S., McGuckin M. A., Markovich D. (2009) Gut 58, 910–919 [DOI] [PubMed] [Google Scholar]
- 46.Fukuda M. (2002) Biochim. Biophys. Acta 1573, 394–405 [DOI] [PubMed] [Google Scholar]