Abstract
Cell cycle progression is dependent upon coordinate regulation of kinase and proteolytic pathways. Inhibitors of cell cycle transitions are degraded to allow progression into the subsequent cell cycle phase. For example, the tyrosine kinase and Cdk1 inhibitor Wee1 is degraded during G2 and mitosis to allow mitotic progression. Previous studies suggested that the N terminus of Wee1 directs Wee1 destruction. Using a chemical mutagenesis strategy, we report that multiple regions of Wee1 control its destruction. Most notably, we find that the activation domain of the Wee1 kinase is also required for its degradation. Mutations in this domain inhibit Wee1 degradation in somatic cell extracts and in cells without affecting the overall Wee1 structure or kinase activity. More broadly, these findings suggest that kinase activation domains may be previously unappreciated sites of recognition by the ubiquitin proteasome pathway.
Keywords: Cdk (cyclin-dependent kinase), Cell Cycle, Mitosis, Protein Turnover, Ubiquitin
Introduction
The tyrosine kinase Wee1 inhibits mitotic entry, and its degradation is essential for exit from the G2 phase of the cell cycle (1). Here, Wee1 inactivates the mitosis-specific cyclin-dependent kinase Cdk1-cyclin B complex during the S and G2 phases by phosphorylating Cdk1 at tyrosine 15. However, Wee1 activity is opposed by the phosphatase Cdc25, which removes phosphate from tyrosine 15 on Cdk1, thereby activating Cdk1-cyclin B at the G2-M transition (2). A major mechanism that tips the balance toward Cdc25 and mitotic entry is Wee1 degradation during G2, and as this occurs active Cdc25 and Cdk1-cyclin B form a positive feedback loop, which ensures that mitotic entry is unidirectional (3, 4). Thus, Wee1 degradation is an essential component of the Cdk1 activation circuit.
Wee1 degradation has been observed in Saccharomyces cerevisiae, Xenopus egg extracts, and somatic cells (5–11). In Xenopus, the nuclei are required for proper Wee1 degradation, and the completion of DNA replication is required to achieve maximal rates of Wee1 degradation (6). Thus, Wee1 degradation is part of a sensing mechanism that signals the completion of the DNA replication phase, ensuring proper timing of entry into mitosis. For example, when DNA replication stalls, this fail-safe mechanism allows defects to be corrected before cells enter mitosis. Indeed, current theories suggest that many cancer cells have ineffective checkpoint pathways that cause them to divide with incompletely replicated DNA, which leads to genomic instability (12). Consistent with this notion are the findings that wee1 knock-out mice are not viable and that wee1−/− cells undergo mitotic catastrophe (13).
Wee1 phosphorylation is required for its degradation and is regulated by the DNA replication checkpoint (6, 10). Here in the presence of the DNA replication checkpoint, Wee1 phosphorylation is negligible, whereas when DNA replication proceeds normally, Wee1 phosphorylation and degradation and mitotic entry are normal.
One aspect of Wee1 degradation conserved between embryonic and somatic cell cycles is the requirement for SCF3 ubiquitin ligases that target Wee1 for destruction by the proteasome. Our prior studies uncovered a novel Wee1-interacting protein, Tome-1 (trigger of mitotic entry), which is required for Wee1 degradation during G2 and binds to Wee1 in a phosphorylation-dependent manner, as does the well characterized F-box protein β-trcp-1 (10, 14). Redundant mechanisms regulate Wee1 degradation, because either β-trcp or Tome-1 depletion stabilizes Wee1 during the S and G2 phases of the cell cycle (11). However, it is still unclear which sequence elements target Wee1 for destruction via SCFβ-trcp versus Tome-1. For example, phosphorylation of serine 38 is required for degradation of Xenopus Wee1 in vitro and in vivo, yet this site is not conserved in human Wee1 (10, 15). Similarly, the domain containing human Wee1 serine 53 and 123, which are required for association with β-trcp and degradation, is not conserved in Xenopus Wee1A (14).
Prior studies that have attempted to identify the sequences of SCF substrates required for degradation have relied on mass spectrometry to identify the phosphorylation sites necessary for interactions with SCF ubiquitin ligases. However, analyses are sometimes difficult because phosphorylated SCF substrates are efficiently targeted for degradation. Here we report a method for identifying sites required for substrate turnover, which relies on cell-based screens to detect stabilized protein targets following chemical mutagenesis. Using this method we show that the kinase activation domain of human somatic Wee1 is required for proper degradation during G2 and mitosis. Further, the Wee1 kinase activation domain and the newly identified sites required for its destruction are conserved, suggesting that this is an evolutionarily important mechanism that controls Wee1 proteolysis.
EXPERIMENTAL PROCEDURES
Wee1 Mutagenesis Screen
Mutagenesis of the PGL3 vector-Wee1 was performed by treating 1-μg aliquots of plasmid DNA with 1 m hydroxylamine (hydroxylamine hydrochloride; Sigma), pH 6.7, for 70 min at 70 °C. The DNA was washed with the Qiagen Miniprep kit PB buffer with three extra washes with PE buffer to remove hydroxylamine. The washed Wee1-luciferase plasmids were transformed into MACH1 competent cells and plated as previously described for site-directed mutagenesis (16). Approximately 1,100 colonies were picked from these plates. 96 deep-well plates filled with Terrific Broth and carbenecillin (Sigma) from the NucleoSpin Robot-96 plasmid purification kit (Macherey-Nagel) were inoculated with one colony for each well and grown overnight at 37 °C in a shaker at 230 rpm along with transformed colonies with control constructs of unaltered pGL3Wee1-luciferase and pGL3 vector only. The resulting bacterial cultures were pelleted, and the plasmid was purified using the NucleoSpin Robot-96 plasmid purification kit (Macherey-Nagel) using the Microlab Star Robot (Hamilton). The resulting purified DNA clones were loaded into 96-well UV spectrometry plates, and the DNA concentrations were determined. The DNA was normalized using the Microlab Star (Hamilton) via dilution and spotted in quadruplicate onto 384-well plates (Corning) with the Minitrak from PerkinElmer Life Sciences, resulting in 40 ng of DNA/well.
Asynchronous HeLa cells were harvested with trypsin, resuspended in Dulbecco's modified Eagle's medium with 20% fetal bovine serum (Invitrogen) and 2% penicillin-streptomycin (Invitrogen) at a concentration of 5 × 105 cells/ml. TransIT LT-1 (Mirus) transfection reagent was diluted in OptiMEM medium along with vector only DNA such that 85 ng of plasmid DNA and 125 μg of Mirus reagent were present per 20 μl of final mixture. The Mirus transfection reagent mixture was spotted with the Multidrop 384 (Titan) onto each 384-well plate in aliquots of 20 μl/well and incubated for 30 min. The HeLa cell/media mixture was spotted with the Multidrop 384 (Titan) onto each plate after transfection reagent DNA mixing in aliquots of 20 μl/well. The plates were covered with sterile metal lids with aerating holes and incubated at 37 °C in 10% CO2 for 20 h. A Brite-lite luminescence reporter gene assay system (PerkinElmer Life Science) was spotted with the Multidrop 384 (Titan) onto the 384-well plates in aliquots of 40 μl/well, and each plate was incubated for 1 min at room temperature. Each plate was read using the Analyst GT from Molecular Devices, and the relative luminescence values were determined. The relative luminescence values were taken for each well. The relative luminescence values were loaded and analyzed in Microsoft Excel.
Wee1 Constructs
The Wee1 gene purchased from Invitrogen is the same as the NCBI (Wee1 tyrosine kinase; Homo sapiens; NP 003381), with the exception of base pair 256, which is an A, not a C. DNA purifications were performed using Qiagen Mini or Maxi prep kits according to the manufacturer's protocols. PCR purification and agarose gel extractions were performed using the Qiagen QIAquick kit. Wee1 was cloned using EcoRI and XhoI restriction enzyme sites using standard protocols into the Myc-tagged pCS2 mammalian expression vector. The Wee1-luciferase construct was made through standard cloning of Wee1 from the pCS2 plasmid into the pGL3 luciferase expression vector (Promega) using EcoRI and XhoI. Wild-type and mutant Wee1 constructs were cloned into a modified pCS2-FLAG expression plasmid using EcoRI and NotI. The GC codon optimized opti-Wee1 construct was made by GenScript and was cloned into the cs2 Myc-tagged mammalian expression vector. PCR-based site-directed mutagenesis of Wee1 was performed using the following primers: FL483H, GGGCGATAGTCGTTTTCATGCAAATGAAGTTTTAC; RL483H, GTAAAACTTCATTTGCATGAAAACGACTATCGCCC; FS480A, CAAGTTGAAGAGGGCGATGCTCGTTTTCTTGCAAATG; RS480A, CATTTGCAAGAAAACGAGCATCGCCCTCTTCAACTTG; FS471A, GGCATGTAACAAGGATCGCAAGTCCACAAGTTGAAGAG; RS471A, CTCTTCAACTTGTGGACTTGCGATCCTTGTTACATGCC; FS472A, CATGTAACAAGGATCTCCGCTCCACAAGTTGAAGAGG; RS472A, CCTCTTCAACTTGTGGAGCGGAGATCCTTGTTACATG; FL483F, GAGGGCGATAGTCGTTTTTTCGCAAATGAAGTTTTACAG; and RL483F, CTGTAAAACTTCATTTGCGAAAAAACGACTATCGCCCTC.
PCR was performed using the opti-Wee1 construct as a template. Site-directed mutagenesis PCR primers were used at a concentration of 125 nmol and opti Wee1 at 50 ng using KOD Hotstart DNA polymerase (EMD Biosciences) according to manufacturer's protocol, with the cycling program of one initial min at 95 °C, followed by 16 cycles of 30 s at 95 °C, 1 min at 55 °C, and 6 min at 68 °C. The PCR products were digested with DpnI (Promega) according to the manufacturer's protocol for 3 h, and then the restriction enzyme mixtures were heat-inactivated. These digests were ligated overnight with T4 DNA ligase at 15 °C. The ligations were transformed into MACH1 competent cells with a 10-min incubation on ice, a 1-min incubation at 37 °C, a 2-min incubation on ice, and 15 min of incubation shaking at 37 °C in 200 μl of SOC medium at 230 rpm. Transformations were plated onto LB plates containing 100 μg/liter carbenicillin (Sigma) and incubated at 37 °C overnight.
Mass Spectrometry Analysis
Immunoprecipitations were made from transfected 293 Freestyle cells (Invitrogen) and then were silver-stained using Pierce silver SNAP stain for mass spectrometry kit. Protein bands of Wee1 were excised, digested, and analyzed using liquid chromatography mass spectrometry. Briefly, the excised bands were reduced and alkylated prior to trypsin digestion. Following digestion the peptides were extracted and taken to dryness. The digestion mixtures were reconstituted in 0.1 m acetic acid, loaded onto precolumns (360 mm outer diameter × 100 mm inner diameter fused silica; Polymicro Technologies, Phoenix, AZ) packed with 3-cm irregular C18 (5–15 μm nonspherical; YMC, Inc., Wilmington, NC) and washed with 0.1 m acetic acid for 5 min before switching in-line with the resolving column (7-cm spherical C18; 360-mm outer diameter × 100-mm inner diameter fused silica). Once the columns were in-line, the peptides were eluted along a gradient from 2% to 80% acetonitrile in 0.1 m acetic acid at an approximate flow rate of 350 nl/min. All of the samples were analyzed using a Thermo Electron LTQ (San Jose, CA). Electrospray was accomplished using a pulled fused silica emitter tip of ∼5 μm with a voltage of 1.7 kV. The mass spectrometer was operated in the data-dependent mode with the top five most abundant ions in each spectrum selected for sequential MS/MS experiments. The exclusion list was used (1 repeat, 180-s return time) to increase dynamic range. All of the MS/MS spectra were searched with Sequest (version 2.7) using sample-dependent data bases. The searches were performed with a fixed carbamidomethylation of cysteine and variable oxidation of methionine and phosphorylation of serine and threonine. The data base search results were tabulated and visually inspected for correct assignment using Scaffold version 1.7 (Proteome Software, Portland, OR).
In Vitro Degradation Assays
In vitro degradation assays were performed as previously described (17). HeLa cells were grown and synchronized as previously described (17). Five hours after the final thymidine release, HeLa cells were harvested, washed, and resuspended in swelling buffer (20 mm Hepes, pH 7.7, 10 mm MgCl2, 5 mm KCl, 1 mm dithiothreitol, 3 mm creatine phosphate, 0.4 mm ATP, pH 7.4, 0.04 mm EGTA, pH7.7, and protease inhibitor mixture (P8340; Sigma)) in a ratio of 1:0.75 of pellet volume to buffer volume and allowed to incubate on ice for 20 min. A small volume of unlysed cells were added to −20 °C 70% ethanol for fluorescence-activated cell sorter analysis to determine cell cycle state (mitotic index was calculated by measuring phosphohistone H3 staining). Cell lysis was performed by Dounce homogenization, and complete cell lysis was monitored by trypan blue exclusion. The resulting cell lysates were passed through a 26.5-gauge needle and spun at top speed in a 4 °C tabletop centrifuge (∼21,000 × g) for 20 min. The resulting pellet was discarded, and the cell extract supernatant was saved for in vitro degradation assays. Radiolabeled in vitro translated proteins of Wee1 mutants were made using the Promega TnT SP6 coupled reticulate lysate system according to the manufacturer's directions for in vitro translation with 1 μg of Myc-tagged cs2 plasmid constructs. Degradation assays were assembled on ice with 20 μl of cell extracts, 1 μl of radiolabeled protein, and 1 μl of reaction mixture energy mix (150 mm creatine phosphate, 20 mm ATP, pH 7.4, 2 mm EGTA, pH 7.7), cycloheximide (0.1 mg/ml; Sigma), and 0.1 mg/ml ubiquitin (Boston BioChem) in a 1:1:1 ratio). The reactions were moved from ice to room temperature, and the time points were taken by adding 4 μl of reaction mixture to 20 μl of Laemmli sample buffer (Bio-Rad) supplemented with 2 mm dithiothreitol.
Pulse-Chase Analysis
Pulse-chase analysis was performed essentially as described (11). The cells were transfected with Myc-wild-type Wee1, Myc-S53A/123A-Wee1, Myc-S472A/L483F Wee1, Myc-K328M-Wee1 (9, 14), Myc-K328M-S472A-Wee1, or Myc-K328M-L483F-Wee1 and analyzed by pulse-chase analysis 48 h after transfection. An identical protocol was utilized for pulse-chase performed in supplemental Fig. S3.
Mitotic Entry Assay of Transfected HeLa Cells
HeLa cells were plated at 40% confluency in 10-cm dishes with 10 ml of 1× HeLa medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin-streptomycin) containing 2 mm thymidine and grown in a humidified 37 °C incubator with 10% CO2 for 18–20 h. The cells were released from the thymidine block by washing twice in phosphate-buffered saline and adding 9 ml of Optimem to the plate for transfection. Lipofectamine 2000 (Invitrogen) transfection was done according to the manufacturer's instructions with 7 μg of pCS2+MT-HsOptiWee1, S472A mutant of Wee1, L483F mutant of Wee1, or vector alone. 1 μg of pCS2+eGFP was added to determine transfection efficiency (greater than 90% in all experiments). The cells were transfected shortly after release from the thymidine block, and the complexes were allowed to incubate with cells for 4–6 h. Each 10-cm transfected plate was then split into a 6-well dish and plated in 1× HeLa medium containing 2 mm thymidine and allowed to incubate for 18–20 h. The cells were released from the thymidine block, and the time points were collected every 2 h for 10 h by scraping cells in growth medium, washing once in 10 ml of phosphate-buffered saline, and snap freezing pellets in liquid nitrogen. The pellets were resuspended in 1× SDS-containing sample buffer and processed for phosphohistone, Myc, or Skp-1 immunoreactivity.
Immunoblotting
Anti-Myc (A-14; Santa Cruz), anti-Wee1(H-300; Santa Cruz), anti-skp1(H-163; Santa Cruz), anti-p-histone (H3) (Ser 10-R; Santa Cruz), ant-Cdk1 (9112; Cell Signaling), phospho-Y15-Cdk1 (9111S; Cell Signaling), and anti-green fluorescent protein (FL; Santa Cruz) rabbit polyclonal IgG were used as primary antibodies. Anti-rabbit IgG donkey IgG (GE Healthcare) was used as secondary antibody. Imaging of Western blots was performed using the GE Healthcare Amersham Biosciences ECL Plus Western blotting detection system.
RESULTS
A Chemical Mutagenesis Strategy Identifies Essential Wee1 Proteolysis Sites
We established a cell-based screening method to identify sites that control Wee1 degradation based on the studies of Sato et al. (16), using a human Wee1-luciferase (Wee1-Luc) fusion as a surrogate to measure Wee1 turnover. C-terminal fusions of proteins with luciferase to measure degradation have been validated for a number of unstable proteins including cyclin B1 and p27Kip1 (18, 19). Indeed, we established that Wee1 is a very unstable protein relative to luciferase (supplemental Fig. S1) and that turnover of Wee1-Luc was very similar to that of Wee1 (supplemental Fig. S1).
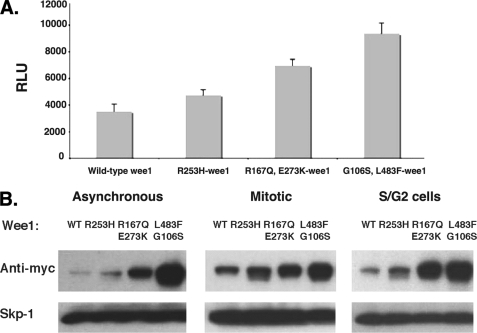
To identify mutations that might affect Wee1 turnover, we treated Wee1-Luc plasmid DNA with hydroxylamine and transformed Escherichia coli to isolate individual mutated plasmids whose concentration was normalized and spotted robotically on 384-well plates. Each clone (1,100 total) was then individually transfected into HeLa cells, which were cultured for 24 h before adding Brite-lite reagent (PerkinElmer Life Science) to simultaneously lyse cells and determine luminescence. We then retested any clone that gave a signal 1.5 times greater than that generated by wild-type Wee1-Luc. Once we identified the most stable Wee1-Luc constructs (those having the highest signal relative to wild-type Wee1 luciferase, shown in Fig. 1A), we sequenced the Wee1 coding region to identify potential mutations (supplemental Fig. S2). These mutations were then shuttled into Myc epitope-tagged Wee1 and assessed for their steady-state levels relative to wild-type, Myc-tagged Wee1 following transfection in HeLa cells. In asynchronous, S phase/G2 cells or mitotic cells, the three Wee1 mutants (R253H, R167Q/E273K, and the G106S/L483F) were more highly expressed than wild-type Wee1 (Fig. 1B), and here increased levels of protein directly correlated with increased luciferase activity of Wee1-Luc fusions, confirming that Wee1-Luc was an accurate surrogate of Wee1.
FIGURE 1.
Steady-state levels of wild-type and mutant Wee1 proteins. A, wild-type Wee1 or the various mutant Wee1 constructs were transfected into HeLa cells using a reverse transfection procedure. 24 h after transfection, the cells were lysed using Brite-lite reagent. The relative luciferase units (RLU) are indicated. The experiments were performed in quadruplicate. The averages and standard deviations are shown. B, various mutants were generated in a Myc-Wee1 fusion plasmid and transfected into HeLa cells. After synchronization in the S/G2 or mitotic phases of the cell cycle, anti-Myc Western analysis was performed to detect the various proteins. A representative assay of three independent experiments is shown. WT, wild type.
Identified Wee1 Mutations Inhibit Wee1 Turnover in Vitro
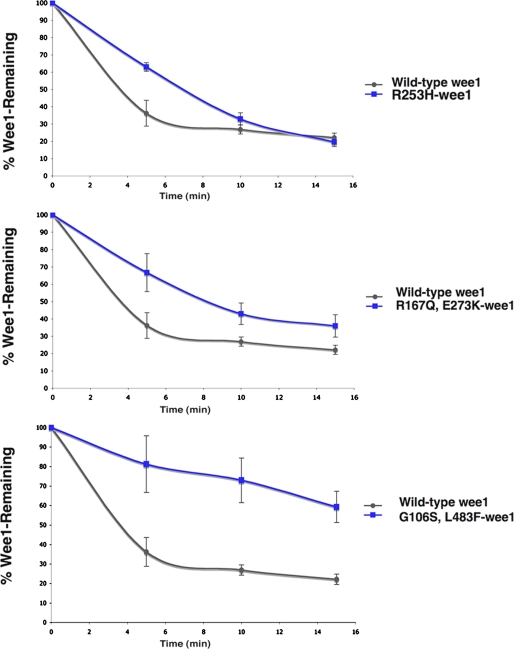
To demonstrate that Wee1 degradation was indeed inhibited by these mutations, we measured the turnover of either Myc-wild-type Wee1 or the corresponding Wee1 mutant proteins in somatic cell extracts (Fig. 2). Extracts from S phase/G2-arrested HeLa cells were incubated with either in vitro translated 35S-labeled wild-type Wee1, R253H-Wee1, R167Q/E273K-Wee1, or G106S/L483F-Wee1. At specific intervals, the reactions were terminated, and their in vitro degradation rates were determined following SDS-PAGE and PhosphorImager analysis. Indeed all three of these mutants had reduced rates of turnover compared with wild-type Wee1 (Fig. 2), and their relative rates of degradation were inversely proportional to their steady-state levels (Fig. 1), with the G106S/L483F-Wee1 mutant having the slowest degradation rate.
FIGURE 2.
In vitro degradation assays of 3 Wee1 mutants relative to wild-type Wee1. 35S-Labeled Wee1 or Wee1 mutants were incubated in somatic cell extracts, and the amount of Wee1 remaining after incubation was analyzed after SDS-PAGE. The results are the averages of three independent experiments.
Serines in the Kinase Activation Domain Direct Wee1 Turnover
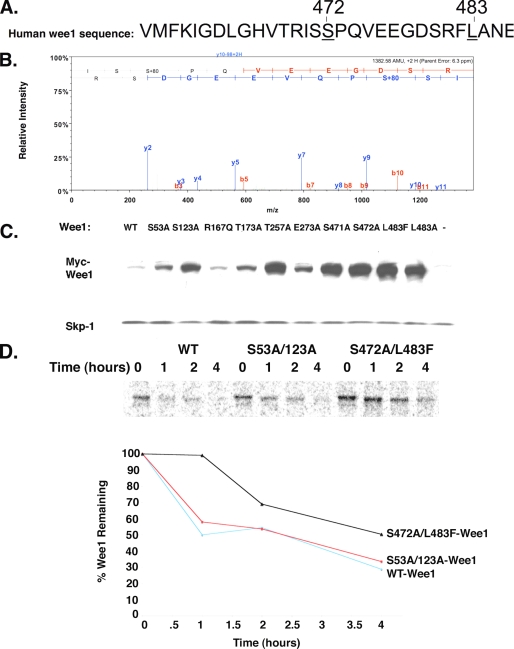
Because the G106S/L483F Wee1 mutant was the most stable (Fig. 1 and supplemental Figs. S2 and S3), we tested the contribution of glycine 106 or leucine 483 to Wee1 turnover. Mutating glycine 106 to serine did not affect the steady-state levels of Wee1, suggesting that changes in turnover were due to changing leucine 483. Indeed, mutation of leucine 483 to alanine, phenylalanine, or even histidine was sufficient to stabilize Wee1 (Fig. 3C and supplemental Figs. S2 and S3).
FIGURE 3.
Mutation of serine residues near the L483 site increases the steady-state levels of Wee1. A, sequence of the region of human Wee1 identified as required for turnover. Leucine 483 and serine 472 are underlined. B, human Wee1 is phosphorylated at serine 472. FLAG-tagged Wee1 was immunostained from transfected 293T cells, and the resulting immunoprecipitate is resolved by SDS-PAGE. The bands were excised, and liquid chromatography-MS/MS was performed on Wee1 band after trypsin digest. Scan indicated +80 AMU on serine 472, suggesting that it was phosphorylated C. Mutation of activation domain residues 471, 472, or 483 stabilizes Wee1. HeLa cells were transfected with either wild-type (WT) Wee1 or the indicated mutants. The known sites required for Wee1 destruction are serines 53 and 123. C, pulse-chase analysis of Myc-wild-type Wee1, Myc-S53A/123A-Wee1, or Myc-S472A/L483F. A representative assay is shown.
Phosphorylation of the N-terminal Wee1 residues serines 53 and 123 are important for its turnover (9, 14). Given the locale of leucine 483 within the Wee1 kinase activation domain (residues 461–488) (20), we postulated that this domain was necessary for Wee1 destruction and that degradation might be mediated by phosphorylation of serines 471, 472, and/or 480 that reside within this domain. To test this notion, we initially performed liquid chromatography-MS/MS analysis of immunopurified FLAG-tagged Wee1 expressed in 293F cells, which allowed us to express a sufficient quantity of protein for analysis. We demonstrated that serine 472 was indeed phosphorylated (Fig. 3B; phosphorylation of endogenous Wee1 has been recently reported by another group to be induced during mitosis) (21). Moreover, mutation of serine 472 to alanine led to marked increases in steady-state levels of the protein, and the same effects were also observed in S471A- and S480A-Wee1 mutants (Fig. 3C and supplemental Fig. S3). When directly compared with the known S53A and S123A stabilizing mutations, the kinase activation domain mutants S471A, L483A (F, H), S472A, or S480A had much more profound effects on the steady-state levels of Wee1, and this was also the case for T257A (Fig. 3C and supplemental Fig. S3; for comparison we also mutated residue Thr257 to alanine, which was predicted to be phosphorylated by Prosite). Notably, pulse-chase analysis demonstrated that Wee1 turnover was severely impaired when we mutated serine 472 to alanine and leucine 483 to phenylalanine (Fig. 3D). Whereas ∼50% of wild-type Wee1 was turned over after 1 h, the S472A/L483F mutant remained stable during that time frame and continued to be more stable than wild-type Wee1 over 4 h. By contrast, the S53A/S123A mutant was only slightly more stable than wild-type Wee1. Therefore, serines within the kinase activation domain of Wee1, along with leucine 483, are required for proper Wee1 degradation in somatic cells.
Activation Domain Mutations Do Not Affect Wee1 Structure or Activity
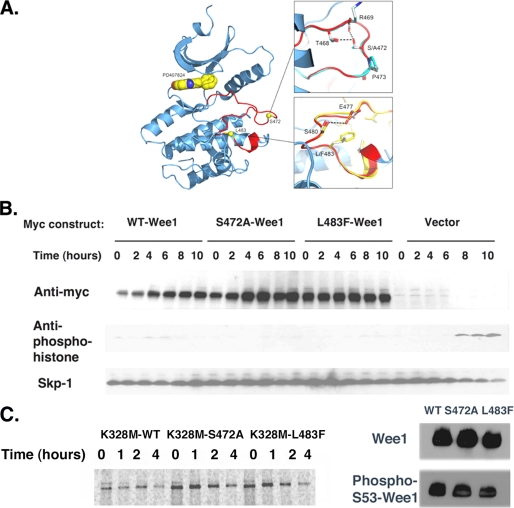
Possible explanations for the increased stability of the kinase activation domain mutants were that they disturbed Wee1 structure or kinase activity. To assess the structural consequences of these mutations, modeling and energy minimization of the Wee1 structure was performed and compared with that of wild-type Wee1 (20). Importantly, the L483F or S472A Wee1 mutant structures were very similar to wild-type Wee1 (Fig. 4A). Because the Wee1 N terminus is lacking in the Wee1 crystal structure, the affects of these mutations on structure and folding of the Wee1 N terminus could not be assessed. However, the S472A and L483F mutants did not affect the phosphorylation of the N-terminal serine residue, serine 53, which is also required for Wee1 turnover in somatic cells (Fig. 4C).
FIGURE 4.
S472A-Wee1 or L483F-Wee1 mutation does not affect Wee1 structure or kinase activity. A, X-ray crystal structure of the kinase domain of human Wee1 kinase highlighting the ATP-binding site and activation segment. The small molecule inhibitor PD407824 is drawn in the active site as a CPK model (space filling model by Corey et al. (29)). The insets show overlays of phenylalanine for leucine at position 483 and alanine for serine at position 472. The Wee1 activation domain is colored in red. B, HeLa cells were synchronized in the S phase using thymidine, released for 4 h, and transfected with Myc-WT-Wee1, Myc-S472A-Wee1, Myc-L483F-Wee1, or vector alone. Thymidine was then added for another 18 h, at which time the cells were released and isolated at 0, 2, 4, 6, 8, and 10 h and processed for Myc, anti-phosphohistone, and Skp1 immunoreactivity. C, left panel, pulse-chase analysis of Myc-K328M-Wee1, Myc-K328M-S472A-Wee1, or Myc-K328M-L483F-Wee1 in HeLa cells. Right panel, anti-Wee1 or phosphoserine 53-Wee1 immunoreactivity for wild-type (WT) Wee1, S472A-Wee1, or L483F-Wee1. A representative assay of three independent experiments is shown.
To test whether the S472 and L483 played critical roles in mitotic entry, HeLa cells expressing wild-type, S472A, or L483F Wee1 were synchronized at the G1/S transition by a double-thymidine block and were then released. Serine 10 phosphorylation of histone H3 was then monitored to measure mitotic entry. As expected, control HeLa cells transfected with empty vector entered mitosis 6 h after thymidine release, and this was blocked in cells expressing wild-type Wee1 (Fig. 4B). Further, cells expressing the S472A or L483 Wee1 also failed to enter mitosis, and indeed here the block was slightly more complete than wild-type Wee1 (Fig. 4B). Therefore the mutations in the kinase activation domain that stabilize Wee1 do not affect its overall structure or function.
To test the effects of these activation domain mutations on Wee1 kinase activity, FLAG-tagged wild-type, S472A, or L483F Wee1 were transfected into HeLa cells, and phosphorylation of endogenous Cdk1 by Wee1 was detected using a commercially available phospho-Y15-Cdk1 antibody. As shown in supplemental Fig. S4, wild-type Wee1 increased phosphorylation of Cdk1 to the same extent as the S472A or L483F mutant. Therefore, the effects of activation domain mutations on Wee1 turnover are independent of effects on kinase activity. Consistent with this notion, mutating serine 472 or leucine 483 in a kinase-inactive version of Wee1, K328M-Wee1 (9, 14), impaired turnover as judged by pulse-chase analysis (Fig. 4C).
DISCUSSION
Prior studies have suggested key roles for N-terminal serine residues of Wee1 (serines 53 and 123) in directing Wee1 destruction, yet the unbiased screen reported herein has established that the kinase activation domain also contributes to Wee1 turnover. Consistent with this notion, we find that an N-terminal deletion mutant lacking serines 53 and 123 is still rapidly turned over (supplemental Fig. S5). Thus, other domains of Wee1 contribute to harness its levels and may suggest that multiple ubiquitin ligases and kinases control Wee1 turnover. Such differential regulation may explain how Wee1 is controlled during mitotic entry, apoptosis, and development (22). Thus, the activation domain, like the Wee1 N terminus, is a major site of Wee1 turnover.
The precise mechanisms that control Wee1 remain unresolved. Both the N terminus and the activation domain of Wee1 contain or are proximal to putative PEST regions, which often contribute to degradation and are consistent with the observation that two or more substrate degrons are necessary to promote efficient binding of ubiquitin ligases (supplemental Fig. S6) (23, 24). Thus, one would predict that the binding of β-trcp and/or Tome-1 to Wee1 would be compromised by stabilizing mutations in the activation domain. However, we only detected slight defects in binding of β-trcp or Tome-1 to S472A or L483F Wee1, at least in vitro.4 This is perhaps due to the presence of N-terminal residues that provide an alternate docking site for these ubiquitin ligases. Alternatively, the S472A and L483F mutations may disrupt binding to another protein required for Wee1 turnover. Indeed, a recent paper demonstrates that cyclin A1/Cdk2 binds somatic Wee1 kinase at multiple sites including Leu483 and Arg611 (25). These two sites were identified via our mutagenesis approach as required for Wee1 turnover (Fig. 1 and supplemental Figs. S2 and S7). Thus, these data suggest that cyclin A1/Cdk2 may be an important regulator of somatic Wee1 turnover. Cyclin A1/Cdk2 could control Wee1 degradation either by inducing Wee1 recognition by ubiquitin ligases or by inducing Wee1 transport to the cytoplasm where it is degraded. Consistent with the former possibility, we find that Cyclin A1/Cdk2 increases Wee1 recognition by Tome-1 and β-trcp (supplemental Fig. S8).
Whether cyclin A1/Cdk2-dependent mechanisms contribute to the increased stability of the S472A and L483F mutants remains to be determined. However, the Wee1 activation domain is clearly required for efficient turnover of Wee1 and is linked to phosphorylation of at least one serine residue, serine 472. It will be important to determine whether such requirements are important in Wee1-directed control of embryonic cell mitotic entry and gastrulation (22).
The unbiased hydroxylamine-directed mutagenesis and cell-based screens described herein to assess Wee1 turnover can easily be applied to resolve the control of other cell cycle regulators. For instance, although phosphorylation of p27Kip1 on Thr187 was thought to direct recognition by an E3 ligase containing the ubiquitin ligase Skp-2, subsequent studies using a mouse knock-in model demonstrated that phosphorylation of this site was dispensable for control of p27Kip1 turnover (26). Although this may indicate that ubiquitin ligases other than Skp-2 such as Kip1 ubiquitination-promoting complex (KPC) regulate p27Kip1 destruction in vivo, Skp-2 mediated destruction of p27Kip1 may also require sites that have not been identified (27). Our cell-based means of identifying sites required for protein destruction can be easily implemented to identify these sites as well as those of virtually every unstable protein. Although hydroxylamine-mediated mutagenesis is ideally suited for cDNAs such as Wee1 that are GC rich because it specifically targets GC pairs (28), we have also utilized error-prone PCR, which is more efficient than hydroxylamine at introducing random mutations, and have identified putative new degrons for p21Cip1 and p27Kip1.5
Supplementary Material
Acknowledgments
We thank all members of the cell-based screening core and Cancer Biology Dept. at Scripps Florida for helpful discussions. We also thank Dr. Howard Petrie and Dr. John Cleveland for critical review of the manuscript. We thank Dr. Ning Zheng for providing purified GST-β-trcp.
This work was supported, in whole or in part, by National Institutes of Health Grant 1 R21 NS056991-01. This work was also supported by a Landenberger Foundation grant (to N. G. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
L. Owens and N. G. Ayad, unpublished observations.
S. Simanski and N. G. Ayad, unpublished observations.
- SCF
- ubiquitin ligase containing Skp1, cul1, and F box protein
- β-trcp
- β-transducin repeat containing protein
- MS/MS
- tandem mass spectrometry
- E3
- ubiquitin-protein isopeptide ligase.
REFERENCES
- 1.Reed S. I. (2003) Nat. Rev. Mol. Cell Biol. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 2.Gautier J., Solomon M. J., Booher R. N., Bazan J. F., Kirschner M. W. (1991) Cell 67, 197–211 [DOI] [PubMed] [Google Scholar]
- 3.Pomerening J. R., Sontag E. D., Ferrell J. E., Jr. (2003) Nat. Cell Biol. 5, 346–351 [DOI] [PubMed] [Google Scholar]
- 4.Pomerening J. R., Kim S. Y., Ferrell J. E., Jr. (2005) Cell 122, 565–578 [DOI] [PubMed] [Google Scholar]
- 5.McMillan J. N., Theesfeld C. L., Harrison J. C., Bardes E. S., Lew D. J. (2002) Mol. Biol. Cell 13, 3560–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michael W. M., Newport J. (1998) Science 282, 1886–1889 [DOI] [PubMed] [Google Scholar]
- 7.Watanabe N., Broome M., Hunter T. (1995) EMBO J. 14, 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Vugt M. A., Brás A., Medema R. H. (2004) Mol Cell 15, 799–811 [DOI] [PubMed] [Google Scholar]
- 9.Watanabe N., Arai H., Iwasaki J., Shiina M., Ogata K., Hunter T., Osada H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11663–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayad N. G., Rankin S., Murakami M., Jebanathirajah J., Gygi S., Kirschner M. W. (2003) Cell 113, 101–113 [DOI] [PubMed] [Google Scholar]
- 11.Smith A., Simanski S., Fallahi M., Ayad N. G. (2007) Cell Cycle 6, 2795–2799 [DOI] [PubMed] [Google Scholar]
- 12.Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Jr., Kastrinakis N. G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T. D. (2005) Nature 434, 907–913 [DOI] [PubMed] [Google Scholar]
- 13.Tominaga Y., Li C., Wang R. H., Deng C. X. (2006) Int J Biol Sci 2, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe N., Arai H., Nishihara Y., Taniguchi M., Watanabe N., Hunter T., Osada H.(2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S. Y., Song E. J., Lee K. J., Ferrell J. E., Jr. (2005) Mol. Cell. Biol. 25, 10580–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A., Hogenesch J. B. (2004) Neuron 43, 527–537 [DOI] [PubMed] [Google Scholar]
- 17.Ayad N. G., Rankin S., Ooi D., Rape M., Kirschner M. W. (2005) Methods Enzymol. 399, 404–414 [DOI] [PubMed] [Google Scholar]
- 18.Verma R., Peters N. R., D'Onofrio M., Tochtrop G. P., Sakamoto K. M., Varadan R., Zhang M., Coffino P., Fushman D., Deshaies R. J., King R. W. (2004) Science 306, 117–120 [DOI] [PubMed] [Google Scholar]
- 19.Zhang G. J., Safran M., Wei W., Sorensen E., Lassota P., Zhelev N., Neuberg D. S., Shapiro G., Kaelin W. G., Jr. (2004) Nat Med 10, 643–648 [DOI] [PubMed] [Google Scholar]
- 20.Squire C. J., Dickson J. M., Ivanovic I., Baker E. N. (2005) Structure 13, 541–550 [DOI] [PubMed] [Google Scholar]
- 21.Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 22.Murakami M. S., Moody S. A., Daar I. O., Morrison D. K. (2004) Development 131, 571–580 [DOI] [PubMed] [Google Scholar]
- 23.Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 24.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Mol Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 25.Li C., Andrake M., Dunbrack R., Enders G. H. (2009) Mol. Cell. Biol. 1, 116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malek N. P., Sundberg H., McGrew S., Nakayama K., Kyriakides T. R., Roberts J. M., Kyriakidis T. R. (2001) Nature 413, 323–327 [DOI] [PubMed] [Google Scholar]
- 27.Kamura T., Hara T., Matsumoto M., Ishida N., Okumura F., Hatakeyama S., Yoshida M., Nakayama K., Nakayama K. I. (2004) Nat Cell Biol. 6, 1229–1235 [DOI] [PubMed] [Google Scholar]
- 28.Tessman I., Poddar R. K., Kumar S. (1964) J. Mol. Biol. 9, 352–363 [DOI] [PubMed] [Google Scholar]
- 29.Corey R., Pauling L. (1953) Rev. Sci. Instruments 24, 621–627 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.