Abstract
Nedd4 (Nedd4-1) is a Hect domain E3 ubiquitin ligase that also contains a C2 domain and three WW domains. Despite numerous in vitro studies, its biological function in vivo is not well understood. Here we show that disruption of Nedd4-1 in mice (leaving Nedd4-2 intact) caused embryonic lethality at mid gestation, with pronounced heart defects (double-outlet right ventricle and atrioventricular cushion defects) and vasculature abnormalities. Quantitative mass spectrometry and immunoblot analyses of lysates from the wild type and knock-out mouse embryonic fibroblasts to identify Nedd4-1 in vivo targets revealed dramatically increased amounts of thrombospondin-1 (Tsp-1) in the knock-out mouse embryonic fibroblasts and embryos. Tsp-1 is an inhibitor of angiogenesis, and its elevated level was mediated primarily by enhanced transcription. Interestingly, the administration of aspirin (an inhibitor of Tsp-1) to the pregnant heterozygote mothers led to a reduction in Tsp-1 levels and a substantial rescue of the embryonic lethality. These results suggest that Nedd4-1 is a suppressor of Tsp1 and that increased levels of Tsp-1 in the Nedd4-1 knock-out mice may have contributed to the developmental defect observed in the embryos.
Keywords: Enzymes/Proteolytic, Extracellular Matrix/Thrombospondin, Organisms/Mouse, Proteases/Ubiquitin, Proteases/Ubiquitination, Protein/Post-translational Modification, Protein/Turnover, Tissue/Organ Systems/Heart
Introduction
Ubiquitination regulates stability of many cellular proteins (mainly cytosolic) by tagging them for degradation by the 26 S proteasome (1). It also controls other cellular fates such as endocytosis and vesicular sorting of transmembrane proteins (2–4). The ubiquitination reaction is carried out by the sequential activation of E1,4 E2, and E3 enzymes (1), where the E3 enzyme (ubiquitin ligase) has the important function of substrate recognition and transfer of ubiquitin onto that substrate (in Hect E3 ligases) or facilitating transfer of ubiquitin from the E2 onto the substrate (in RING E3 ligases). Nedd4 (neuronal precursor cell expressed developmentally down-regulated 4; or Nedd4-1; Ref. 5) is an E3 ubiquitin ligase from the Hect family that contains a C2 domain, three or four WW domains, and a Hect domain, much like its close relative Nedd4-2, as well as other Nedd4 family members (e.g. Smurf1, Smurf2, Itch, WWP1, WWP2, HECW1, and HECW2) (6, 7). The WW domains of Nedd4 proteins usually bind their substrates by recognizing a short sequence, the PY motif ((L/P)PXY) (8–10).
Nedd4-2 was shown to regulate endocytosis of the epithelial Na+ channel by ubiquitination (11–15), an effect impaired in Liddle syndrome patients in whom the PY motif of the epithelial Na+ channel (binding site for Nedd4-2) is mutated. Other ion channels are also negatively regulated by Nedd4-2 (16–20). Although Nedd4-1 is more widely expressed than Nedd4-2, less is known about its function. Nedd4-1 was reported to bind to and regulate stability or vesicular sorting of several proteins, including CNrasGEF (21); LAPTM5 (22); the viral proteins HTLV-1 Gag (23), Marburg/Ebola (24–26), and MLV Gag (27, 28); AP-1 (27); APOBEC3G (29); Grb10 (30, 31); and c-Cbl (32). Also, the Drosophila homolog of Nedd4-1, dNedd4, was recently shown to regulate neuromuscular synaptogenesis in flies by binding to commissureless and promoting its endocytosis (33). Nedd4-1 was also shown to indirectly regulate the function of the endocytic protein Eps15 (34). The proposed role of Nedd4-1 in regulating PTEN stability and nuclear translocation (35, 36) was recently challenged (37).
Despite the numerous reports published on Nedd4-1 over the past decade, its in vivo function(s) in mammals are less clear (see “Discussion”). Here we describe the knock-out of Nedd4-1 in mice. We show that these mice exhibit severe cardiac and some vascular defects and die at mid-gestation. We also show that thrombospondin-1 (Tsp-1), a known inhibitor of angiogenesis, is an in vivo target of Nedd4-1 and that its expression is increased in the Nedd4-1 knock-out animals and mouse embryonic fibroblasts (MEFs) relative to wild type (WT). Moreover, treatment of heterozygote pregnant mothers with (low dose) aspirin, an inhibitor of Tsp-1, leads to reduction of Tsp-1 levels and rescue of the embryonic lethality.
EXPERIMENTAL PROCEDURES
Mice
All of the described experiments conform to the institutional regulatory standards.
Nedd4-1−/−,trap
The Nedd4-1 trapped embryonic stem cell clone (XB786) was obtained from BayGenomics. The formation of the fusion transcript of Nedd4-1 (the first six exons) and the β-gal present in the trapping vector was confirmed by reverse transcription-PCR. The trapping vector (pGT0pfs) is inserted in an intron in the C2 region ∼7000 bp 3′ of exon 6 (see Fig. 1A) and includes En2-SA-βgeo-IRES-hPLAP-SV40pA genes/sequences. Genotyping was performed by PCR of genomic DNA, with one set of primers flanking the insertion region and a second set within the β-gal gene. The PCR amplification of the WT and knock-out regions yields 447- and 680-bp fragments, respectively (see Fig. 1B). WT and Nedd4-1+/−,trap mice were generated on a 129/Sv: C57B6 background. Genotype of embryos was determined by PCR (as above) using DNA from the yolk sac or embryonic tissue.
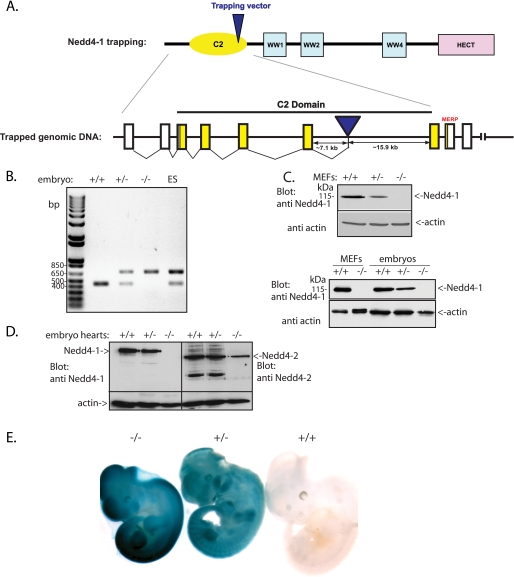
FIGURE 1.
Generation of the Nedd4-1 knock-out (Nedd4-1−/−,trap) mice. A, schematic representation of Nedd4-1, highlighting the insertion locus of the trapping cassette (in an intron within the C2 domain region). B, PCR analysis performed on ES cells and embryonic tissues to genotype the wild type (+/+), Nedd4-1+/−,trap, and Nedd4-1−/−,trap mice. C, immunoblot analysis for Nedd4-1 of MEF cells and embryos from the wild type, Nedd4-1+/−,trap, and Nedd4- 1−/−,trap mice, demonstrating the loss of Nedd4-1 in the Nedd4-1−/−,trap mice and MEFs. D, immunoblot analysis for Nedd4-1 and Nedd4-2, demonstrating the loss of Nedd4-1, but not Nedd4-2, in the hearts of Nedd4-1−/−,trap mice. E, LacZ staining revealing the expression of knock-out cassette (which contains the β-gal gene) in the Nedd4-1+/−,trap and Nedd4-1−/−,trap E11.5 embryos (blue).
Constructs and Antibodies
FLAG-tagged Tsp-1 cDNA was cloned into the pCDNA3 vector (Invitrogen). Mutation in Tsp-1 in the PY motif sequence was generated by site-directed mutagenesis using PCR and verified by sequencing. Monoclonal antibodies against Tsp-1 were purchased from Novous Biological (clone A4.1) or Labvision (antibody Ab-4, clone A6.1). Polyclonal anti-Nedd4-Hect domain antibodies (specific for Nedd4-1) were described earlier (38), and monoclonal anti Nedd4-1 antibody was from BD Biosciences (clone 15). Anti-Nedd4-2 antibodies were generated against a region unique to Nedd4-2, as described (14). Antibodies to annexin II (A2, clone H-50), annexin VII (A7, clone A-1), and Grb10 (K-20) were purchased from Santa Cruz, the antibody to PECAM-1 (CD31, clone 13.3) was from Pharmingen, and the antibody to β-gal (polyclonal antibody 616) was from Abcam. Aspirin (acetyl salicylic acid) was purchased from Sigma, and MG132 was from Boston Biochem.
MEFs
MEFs were generated from WT, Nedd4-1+/−,trap, and Nedd4-1−/−,trap (E14.5) as described (37).
Co-immunoprecipitations, Ubiquitination, and Pulse-Chase Assays
Co-immunoprecipitation, ubiquitination assays, and pulse-chase analysis were performed as described earlier (37) and detailed in the supplemental material.
ICAT Labeling and Mass Spectrometry
Confluent WT and Nedd4-1−/−,trap MEF cells were harvested and lysed in lysis buffer containing 20 mm Tris, pH 7.8, 1 mm EDTA, and 0.5% Triton. Lysate was centrifuged 10,000 rpm for 10 min, and the concentration of proteins was measured by the Bradford assay.
ICAT Labeling
ICAT labeling experiments were performed on the proteins extracted from the MEFs according to the instructions recommended by the manufacturer (Applied Biosystems). Briefly, 100 μg of proteins from the control WT sample, and the Nedd4-1−/−,trap MEFs were initially reduced (50 mm tris(2-carboxyethyl)phosphine) and labeled with light (13C0) and heavy (13C9) cleavable ICAT reagents (Applied Biosystems Inc., Framingham, MA), respectively. The labeled protein samples were then combined and digested with trypsin at 37 °C overnight into small peptides. Selectively, the cysteine-containing peptides were captured by avidin affinity column (supplied with the ICAT reagent kit; loading buffer contained 20 mm NaH2PO4, 150 mm NaCl, pH 7.2; elution buffer contained 30% CH3CN, 0.4% trifluoroacetic acid) following cation exchange purification (loading buffer contained 10 mm NaH2PO4, 25% CH3CN, pH 3; elution buffer contained 10 mm NaH2PO4, 25% CH3CN, 350 mm NaCl, pH 3). The biotin moiety was cleaved by the cleaving reagents with a mixture of trifluoroacetic acid-triethyl saline solution at 37 °C for 2 h, and the sample was finally dried in a SpeedVac concentrator.
NanoLC-ESI MS/MS Analysis
ICAT-labeled peptides were analyzed by on-line nanoLC MS/MS using a Waters CapLC system coupled to a QStar XL quadrupole time-of-flight instrument (Applied Biosystems) with a Nanospray® II source designed by MDS Sciex (Concord, Canada). The sample was dissolved in 0.1% formic acid, and 6.4 μl was loaded into a high pressure liquid chromatography column using the microliter pickup injection. The peptides were trapped by a reversed phase PepMap C18 precolumn (LC Packings, 300-μm inner diameter × 5-mm length) and subsequently separated on a C18 PepMap100 analytical column (75-μm inner diameter × 15-cm length; particle size, 3 μm; pore size, 100 Å). The composition of mobile phases consisted of solvent A, 0.1% (v/v) formic acid; solvent B, 98% (v/v) acetonitrile containing 0.1% formic acid. NanoLC separation was achieved by a 100-min gradient from 5 to 90% of solvent B mixed with solvent A at a flow rate of 300 nl/min. Information-dependent acquisition was employed to obtain MS and MS/MS data using a 1-s survey scan followed by three consecutive 2-s product ion scans of 2+ and 3+ parent ions at a mass range of m/z 350–2000.
Data Processing
The acquired liquid chromatography MS/MS data were searched against the NCBI mouse proteome data base, and the expressed proteins were automatically quantified and identified using ProICAT software 1.1, proteins with confidence scores higher than 95% were considered. Protein identification was also subjected to the NCBI mouse data base search by Mascot, in which the mass tolerance allowed for searches between expected and observed masses was set to 0.1 Da for both MS peaks and MS/MS fragments. The identified sequence of the cysteine-containing peptides and the heavy:light ratio of the cleavable ICAT-labeled counterparts were manually validated in each case.
Histology, X-Gal Staining, and Hematoxilin:Eosin (H&E) Staining
For X-gal staining, uteri from pregnant Nedd4- 1+/−trap mothers (that were crossed with Nedd4-1+/−trap males) were isolated in ice-cold phosphate-buffered saline (PBS) at E10.5, E13.5, or E14.5, and the embryos were processed for histological analysis. The embryos or organs of adult mice were fixed in PBS containing 3.7% formaldehyde overnight. After fixation and three 15-min washes with X-gal wash buffer containing 2 mm MgCl2 and 0.02% Nonidet P-40 in 0.1 m phosphate buffer, embryos or tissues were incubated in 30% sucrose in PBS at 4 °C, embedded in Tissue Tek OCT compound (Sakura), frozen on dry ice, and stored at −80 °C. The frozen sections were cut into 10–15-μm-thick slices in a cryostat. The sections were stained overnight in the dark at 37 °C in wash buffer containing 5 mm K+ ferricyanide, 5 mm K+ ferrocyanide, and 1 mg of X-gal (Invitrogen)/ml. The slides were counterstained with nuclear fast red, dehydrated, and mounted in Entellan mounting medium (39). For histology, the tissues were fixed in 4% paraformaldehyde in PBS, pH 7.6, overnight at 4 °C. After dehydration, the embryos were embedded in paraffin, sectioned (5-μm sections), and stained with H&E.
Immunohistochemistry
Uteri were isolated in ice-cold PBS at E13.5. The embryos were fixed in 4% paraformaldehyde overnight, dehydrated, embedded in wax, and sectioned (5-μm sections). For immunohistochemistry, heat-induced epitope retrieval was carried out in 10 mm citrate buffer, pH 6.0. Endogenous peroxidase was quenched by incubating the sections for 40 min in PBS containing 3% H2O2 and 10% methanol. After blocking with 10% normal serum, the following primary antibodies were used: monoclonal anti Tsp-1 (Ab-4, 1:50), anti-Nedd4-1 (1:50), or polyclonal anti-β-gal (1:100), followed by the appropriate fluorescent conjugated secondary antibodies (Cy3-conjugated donkey anti-rabbit IgG or AlexaFluor 488-conjugated goat anti-mouse IgG, each at 1:200, used at 24 °C in a humidified chamber). Confocal microscopy was performed on a Zeiss Axiovert 200 microscope. Negative controls included the omission of the primary antibody.
Immunostaining for Blood Vessels
Whole mount immunohistochemistry was performed on E10.5 embryos. The embryos were fixed in 4% paraformaldehyde at 4 °C (4 h) and after blocking were incubated with PECAM-1 antibody (5 μg/ml) overnight at 4 °C, followed by horseradish peroxidase-conjugated donkey anti-rat IgG (1:2000, 4 °C, overnight). The embryos were then incubated in Tyramid-fluorophore solution (1:50, 1 h), and the reaction was stopped by incubation in 3% H2O2 in TnT buffer (0.1 m Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20). The vasculature of embryos was then analyzed by optical projection tomography (OPT).
Optical Projection Tomography
OPT was performed as previously described (40) on the fluorescently labeled E10.5 embryos described above. Visualization and manipulation of OPT data were performed with Amira software (V.3.1, TGS Inc.). Surface renderings were created using the Amira “Isosurface” module with a threshold chosen just above the noise floor.
Luciferase Assay for Analysis of Tsp-1 Transcription
MEF cells were seeded in 6-well plates and grown to 80% confluency. For each well 1 μg of Tsp-1 promoter in pGL3 (pTSP-LUC-1.1) (41) or vector alone was co-transfected with 10 ng of Renilla luciferase in pRL-SV40 (Promega), which was used as an internal control, and 0–1 μg of c-Myc in pCDNA3, using ESCORT V (Sigma-Aldrich). The cells were harvested 48 h after transfection, and reporter activity was measured using the dual luciferase assay (Promega) according to the manufacturer's instructions. Experimental luciferase activities were normalized for efficiency against the internal control (Renilla luciferase readings). The experiments were performed in duplicate wells at least three times.
Real Time Quantitative PCR Assay
RNA was extracted using TRIzol (Invitrogen), and quantitative reverse transcription-PCR was performed using a Superscript III platinum two-step quantitative reverse transcription-PCR kit (Invitrogen). β-Actin was used as control. Primer sequences for Tsp-1 were: forward, ctgtctgtaaaggttgtgaactc; reverse, acatcaccactctgatataaccg. For β-actin, the forward primer was cttcttgggtatggaatcctg, and the reverse primer was cttgatcttcatggtgctagg. The PCR cycles were: 2 min at 95 °C, followed by 40 cycles with the annealing temperature of 59 °C. A fluorogenic SYBR Green (Invitrogen) and MJ research detection system were used for real time quantification. The results were presented as parameter threshold cycle (CT) values. ΔCT was the difference in the CT values derived from the specific gene being assayed and β-actin, whereas ΔΔCT represented the difference between the paired samples, as calculated by the formula: ΔΔCT = ΔCT of a sample −ΔCT of the reference. The amount of target, normalized to β-actin and relative to a reference, was expressed as 2−ΔΔCT (42).
Aspirin Treatment of Pregnant Mice and MEFs
After breeding and time mating, females received either placebo or low dose aspirin (20 mg/liter) in the drinking water, which was replaced every other day. Considering each animal drinks an average of 3–4 ml water/day, this would be equal to 60–80 μg of aspirin/30 g of weight for a mouse (equivalent to 120–160 mg/day/60 kg of weight for a person) (43). The animals were sacrificed at the indicated times for analysis of the embryos. For aspirin treatment of MEFs, 24 h after plating, confluent MEF cells were treated with 0, 1, 5, or 10 mm of aspirin, for 4, 8, or 12 h, as indicated. They were then lysed as described below, and proteins were separated on SDS-PAGE and immunoblotted with anti Tsp-1, annexin A2, annexin A7, or Grb10 antibodies.
RESULTS
Generation of Nedd4-1 Knock-out Mice and Mouse Embryonic Fibroblasts
The mouse Nedd4-1 gene was disrupted using a gene trap approach. The β-gal and PLAP genes in the trapping cassette (ES trap clone XB786 from BayGenomics, 129 background) were inserted into the mouse Nedd4-1 locus between exons 6 and 7 (Fig. 1A), as verified by PCR. These ES cells were introduced into pseudopregnant C57B6 females. PCR analysis of the ES cells as well as tissue harvested from embryos of heterozygote crosses verified the presence of the knock-out cassette in the heterozygote (Nedd4-1+/−,trap) and knock-out (Nedd4-1−/−,trap) embryos (Fig. 1B). Analysis of newborns revealed the presence of WT and Nedd4- 1+/−,trap mice, but no Nedd4- 1−/−,trap mice were found, suggesting that ablation of Nedd4-1 leads to embryonic lethality. Similar embryonic lethality was also observed in an independently derived Nedd4-1 knock-out mouse (Nedd4- 1−/−,exon9,10).5 Most of the Nedd4- 1+/−,trap mice appeared normal, grew to adulthood, and were fertile (although occasionally we found some heterozygotes with similar abnormalities as the Nedd4- 1−/−,trap embryos; see below). Analysis of the Nedd4-1−/−,trap embryos demonstrated that they survived until mid-gestation (∼E11.5-E13.5 and rarely to E14.5) but not beyond that stage. Immunoblotting for Nedd4-1 demonstrated the complete absence of Nedd4-1 protein in these embryos (Fig. 1C, lower panel), whereas the closely related Nedd4-2 was still present (Fig. 1D). Likewise, MEF lines generated from E14.5 embryos revealed the absence of the Nedd4-1 (but the presence of Nedd4-2) protein in the Nedd4-1−/−,trap MEFs (Fig. 1C, upper panel, and Ref. 37). Expression of β-gal from the trapping cassette was evident in Nedd4-1+/−,trap and Nedd4-1−/−,trap embryos stained for LacZ (Fig. 1E).
Pattern of Expression of Nedd4-1 during Embryonic Development
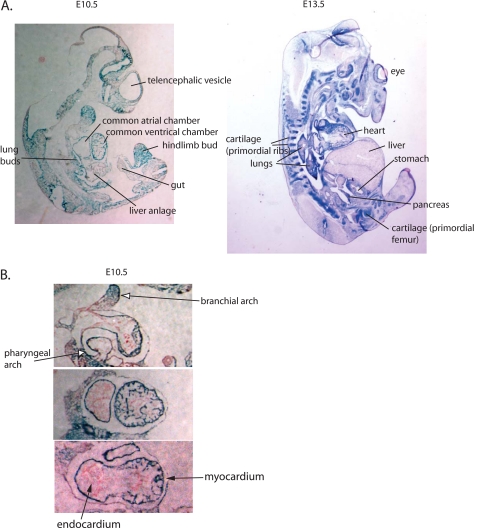
The in vivo expression pattern of Nedd4-1 in mammals during development is unknown. We thus took advantage of the β-gal gene inserted into the knock-out cassette to follow the normal pattern of expression of Nedd4-1 in Nedd4-1+/−,trap mice. As seen in Figs. 1E and 2A, Nedd4-1 expression is quite widespread in several tissues during development, with strong expression in the heart, lungs, brain, somites, as well as kidney (mainly collecting ducts), gastrointestinal tract, pancreas, reproductive organs/urogenital region, mammary glands and ducts, spleen, and vertebra (Fig. 2A and data not shown). In the heart, Nedd4-1 is expressed throughout the myocardium at E10.5 but is excluded from the endocardium. Staining was also apparent in pharyngeal arch and branchial arches adjacent to the heart (Fig. 2B).
FIGURE 2.
Pattern of endogenous expression of Nedd4-1 in embryos and hearts of Nedd4-1+/−,trap mice. A, sagittal sections showing the pattern of expression of Nedd4-1 as revealed from LacZ staining (blue) of E10.5 and E13.5 Nedd4-1+/−,trap embryos. B, cross-section of the heart of E10.5 Nedd4-1+/−,trap embryos, demonstrating Nedd4-1 (LacZ, blue) expression in the myocardium.
Ablation of Nedd4-1 Leads to Cardiac Defects
The embryonic lethality of the Nedd4-1−/−,trap mice observed at mid-gestation prompted us to analyze the heart and placenta in more detail. Although analysis of the placenta did not reveal gross abnormalities,6 there were marked defects in the heart. In particular, an outflow tract defect was observed, revealing a double-outlet right ventricle (DORV) in the knock-out animals (Fig. 3A and supplemental Fig. S1), characterized by the connection of both the pulmonary artery and the aorta to the right ventricle, as opposed to the normal arrangement that connects the aorta to the left ventricle and the pulmonary artery to the right ventricle. The Nedd4-1−/−,trap mice also had endocardial cushion defects resulting in a membranous ventricular septal defect and in more severe cases a common atrioventricular canal defect (Fig. 3, A and B). Similar heart defects were also observed in the Nedd4-1−/−,exon9,10 knock-out mouse (Fig. 3B). Overall, we found the heart defect in 19 of 19 (100%) knock-out embryos we analyzed.
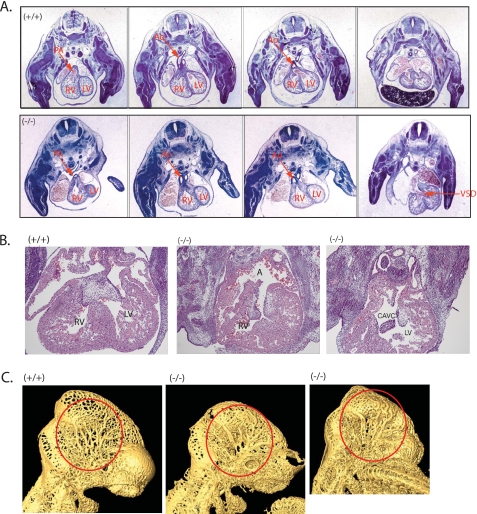
FIGURE 3.
Heart defects and vasculature abnormalities in the Nedd4-1−/−,trap embryos. A, H&E staining of cross-sections from E13.5 WT ((+/+), upper panels) and Nedd4-1−/−,trap ((−/−), lower panels) embryos, revealing DORV and ventricular septal defect in the Nedd4-1−/−,trap embryos. B, heart defects in the Nedd4-1exon9.10 embryos (−/−) relative to WT (+/+), including a common atrioventricular canal defect in some of the knock-out embryos. In A and B, RV, right ventricle; LV, left ventricle; A, atrium; Ao, aorta; PA, pulmonary artery; VSD, ventricular septal defect; CAVC, common atrioventricular canal. More detailed serial sections are shown in supplemental Fig. S1. C, PECAM-1 staining of the vasculature of E10.5 WT (+/+) and Nedd4-1−/−,trap (−/−) embryos followed by OPT analysis. The head region is shown, demonstrating fewer and flatter veins in the cephalic plexus of the knock-out embryos (red circles). A full three-dimensional view of vasculature in the whole embryo is depicted in supplemental Movies S1 and S2.
We also analyzed the vasculature of the Nedd4-1 knock-out mice. Staining of E10.5 WT and Nedd4-1−/−,trap embryos with antibodies to PECAM-1 to decorate the blood vessels followed by optical projection tomography (40) revealed some vasculature abnormalities. In particular, there were fewer veins in the cephalic plexus of some of the knock-out embryos, and they appear thicker and more contorted than in the WT (Fig. 3C and supplemental Movies 1 and 2). In addition to the above pronounced cardiovascular defects, we also noted delayed maturation of the lungs in the knock-out embryos (not shown), where Nedd4-1 is highly expressed (Fig. 2A), and overall smaller size.
Identification of Tsp-1 as an in Vivo Downstream Target of Nedd4-1
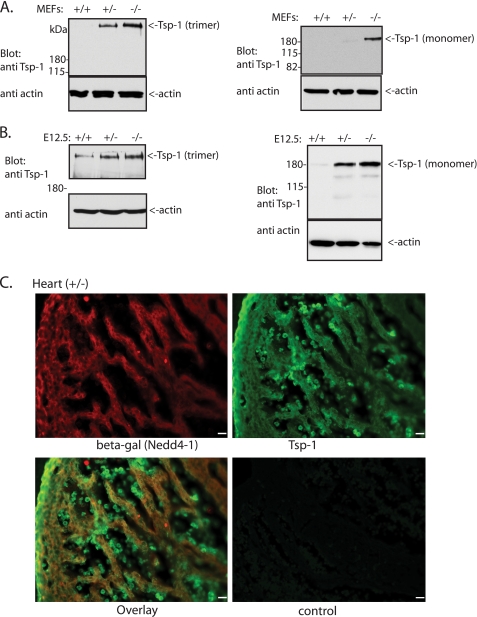
The heart (and vascular) defects we observed could result from one or more biochemical pathways and substrates being affected by the loss of Nedd4-1. Because Nedd4-1 is an E3 ligase, it is likely that at least some of its substrates are stabilized (increased in amounts) in the Nedd4-1−/−,trap cells and animals relative to WT. To identify such putative in vivo substrates, we utilized differential quantitative mass spectrometry analysis (ICAT) to identify proteins that increase in amounts in Nedd4-1−/−,trap cells relative to WT controls. We thus used MEFs generated from Nedd4-1−/−,trap and WT embryos (Fig. 1C and Ref. 37) in a quantitative ICAT assay. A total of 210 proteins were identified by the mass spectrometric analyses on the ICAT-labeled peptides, most of them unaltered in amounts between the WT and knock-out MEFs. As expected, Nedd4-1 itself was absent in the Nedd4-1−/−,trap MEFs (supplemental Fig. S2). Of the putative substrate proteins, the levels of a few (see below) appeared to be more highly elevated in the Nedd4- 1−/−,trap MEFs (consistent in two independent analyses) with the strongest increase (∼8-fold) observed for Tsp-1 (supplemental Fig. S2). Accordingly, we found higher amounts of the Tsp-1 protein in the Nedd4-1−/−,trap MEFs and embryos relative to WT upon immunoblotting with antibodies to Tsp-1 that recognize both the native (trimer) or monomeric forms of the protein (Fig. 4, A and B). Moreover, immunohistochemistry of Nedd4-1−/+,trap embryos revealed a similar expression pattern of Tsp-1 and Nedd4-1 in heart muscle (Fig. 4C).
FIGURE 4.
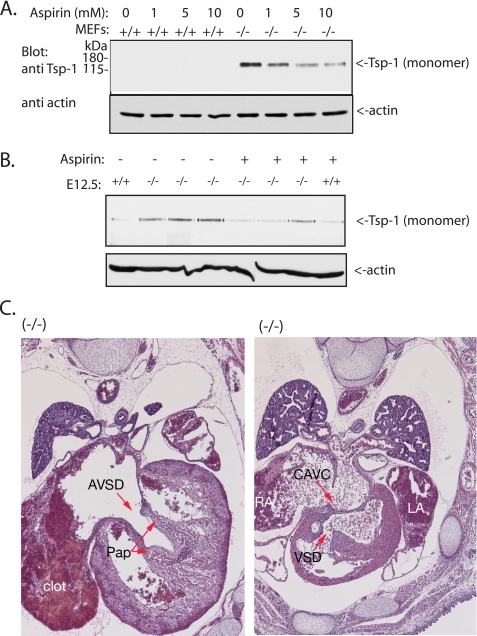
Levels of Tsp-1 are increased in the Nedd4-1+/−,trap MEFs and embryos. A and B, immunoblot analysis of Tsp-1 (either the monomer or trimer (native) forms of the protein) in the WT (+/+), Nedd4-1+/−,trap (+/−), and Nedd4-1−/−,trap (−/−) MEFs (A) or E12.5 embryos (B). For the left panels in A and B, nonreducing conditions and gels were used to observe the native Tsp-1 trimers. Note the increase in Tsp-1 expression in the knock-out embryos and MEFs (in A and B) observed when probing for either the native/trimeric or monomeric Tsp-1. C, similar pattern expression of Nedd4-1 and Tsp-1 in the heart. Embryonic hearts (E13.5, +/−) were immunostained for Tsp-1 and β-gal (Nedd4-1) in parallel, using monoclonal anti Tsp-1 and polyclonal anti β-gal antibodies, respectively. The control panel shows no primary antibodies. Scale bar, 20 μm.
Tsp-1 possesses a PY motif (albeit luminal). We thus tested for interactions between Tsp-1 and Nedd4-1. Our work showed that whereas transfected FLAG-Tsp-1 could co-immunoprecipitate with transfected GFP-Nedd4-1, this interaction was of low stoichiometry, and moreover, a PY motif mutant of Tsp-1 was also able to bind Nedd4-1 (supplemental Fig. S3A), suggesting that the low association between Nedd4-1 and Tsp-1 does not require the PY motif of the latter. We also tested Tsp-1 ubiquitination and stability. We were not able to detect ubiquitination of endogenous Tsp-1 but could detect such ubiquitination following overexpression of Nedd4-1 (but not the catalytically inactive Nedd4-1(CS)) in HEK293T cells. Moreover, pulse-chase analysis performed on the Nedd4- 1−/−,trap MEFs did not reveal any increase in the protein stability of endogenous Tsp-1 relative to its stability in the WT MEFs (supplemental Fig. S1, B–B). In fact, there was a more rapid decline in Tsp-1 stability in the Nedd4-1−/−,trap MEFs, possibly because of activation of the cellular quality control to remove the excess Tsp-1 accumulating in these cells, as was also supported by our observation of cellular toxicity upon overexpression of Tsp-1 (not shown). Collectively, these results suggest that Nedd4-1 likely plays only a minor (if any) role in directly regulating Tsp-1 ubiquitination and stability and that instead it may affect Tsp-1 levels indirectly by other means.
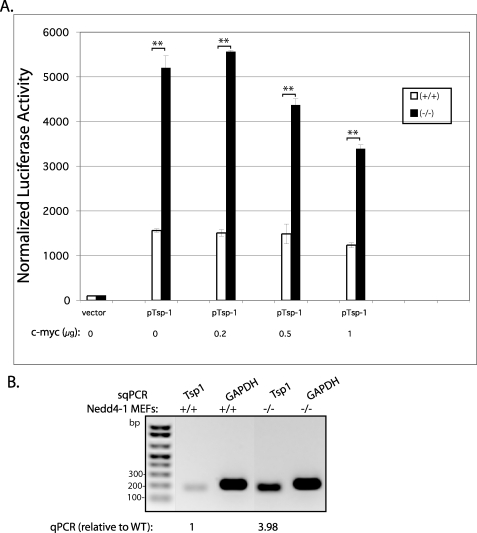
To investigate the latter possibility, we tested whether Nedd4-1 affects Tsp-1 transcription. Thus, we transfected the Tsp-1 promoter (pTSP-LUC 1.1, cloned into the pGL3 enhancer vector) (41) into the Nedd4-1−/−,trap and WT MEFs (along with pRL-SV40) and analyzed its transcription activity by the luciferase assay. As seen in Fig. 5A, Tsp-1 promoter activity was significantly enhanced (3.4-fold) in the knock-out MEFs relative to WT, and moreover, this activity was reduced by co-transfection of increasing amounts of c-Myc cDNA, a known suppressor of Tsp-1 transcription (44). In accord, mRNA levels of Tsp-1, analyzed by semi-quantitative PCR (Fig. 5B) or quantitative PCR, revealed a 3.98-fold increase in the Nedd4- 1−/−,trap relative to WT MEFs (averages of two separate experiments [mean1 = 3.99 and mean2 = 3.97], each performed in triplicate and normalized for actin). These results suggest that Nedd4-1 reduces the mRNA levels of Tsp-1 by inhibiting its transcription.
FIGURE 5.
Elevated transcription from the Tsp-1 promoter in the Nedd4-1−/−,trap MEFs. A, luciferase activity reporting Tsp-1 promoter activity in WT (+/+) or Nedd4-1−/−,trap (−/−) MEFs. The Tsp-1 promoter was expressed from the pGL3 vector in the presence/absence of the indicated amounts of c-Myc cDNA. Shown is a representative experiment, performed in duplicate. **, p ≤ 0.01, unpaired t test. In three separate experiments, each performed in duplicate, there was a 3.3-fold increase in Tsp-1 transcription in the Nedd4-1−/−,trap relative to WT MEFs (p = 3 × 10−9, unpaired t test). B, semi-quantitative PCR (sqPCR) revealing increased mRNA levels in Nedd4-1−/−,trap MEFs relative to WT. Quantification of mRNA levels, analyzed by quantitative PCR, revealed 3.98-fold increase in mRNA of Tsp-1 in the Nedd4-1−/−,trap MEFs relative to WT MEFs.
Aspirin Treatment Decreases Tsp-1 Levels and Partially Rescues the Embryonic Lethality
Tsp-1 is a well known inhibitor of angiogenesis (45–47). Thus, it is possible that the increase in its levels in the heart or developing blood vessels may impair normal heart or vasculature development. To investigate this, we treated the pregnant heterozygote mothers with aspirin, an inhibitor of Tsp-1 (48, 49). As seen in Table 1, treatment with aspirin resulted in a higher percentage of knock-out mice surviving to the late embryonic stage (E17.5–18.5) or even to birth (not shown): 14 of 137 (10%) Nedd4-1−/−,trap embryos in the aspirin-treated animals survived to E17.5–18.5, versus 0 of 118 (0%) of the nontreated embryos (Table 1). Because we expected 25% survival based on Mendelian ratio, the aspirin treatment rescued almost a third (31%) of the expected knock-out embryos. Interestingly, the beneficial effect of aspirin was already apparent at E12.5, where we found ∼13% (13 of 101) Nedd4-1−/−,trap embryos alive after aspirin treatment, relative to ∼8% (11 of 139) in the absence of treatment. Younger embryos (E9.5–10.5) exhibited normal survival rate in the absence or presence of aspirin. Because we observed increased levels of Tsp-1 in the Nedd4-1−/−,trap embryos (and MEFs), we tested whether the rescue of the embryonic lethality by aspirin was correlated with a reduction in the levels of Tsp-1. Our results show that treatment of Nedd4-1−/−,trap MEFs with aspirin for 4, 8 (not shown), or 12 h (Fig. 6A) indeed led to a dose-dependent reduction in levels of Tsp-1. Likewise, we found that the amount of Tsp-1 in E12.5 Nedd4-1−/−,trap embryos was reduced in most aspirin-treated embryos (Fig. 6B) (although, as expected, we observed some variability, possibly correlating with the observation that aspirin treatment did not rescue all the Nedd4- 1−/−,trap embryos).
TABLE 1.
Survival of E17.5–18.5 embryos after aspirin treatment given to pregnant mothers
The control embryos analyzed were from 22 (+/−) mothers, and the aspirin-treated embryos were from 21 (+/−) mothers. There was one dead control (−/−) embryo identified.
| Nedd4-1 | +/+ | +/− | −/− | Total | Total expected |
|---|---|---|---|---|---|
| % | |||||
| Control | 44 | 74 | 0 (0%) | 118 | 157 |
| With aspirin | 54 | 83 | 14 (10%) | 137 | 183 |
FIGURE 6.
Aspirin reduces Tsp-1 levels but does not correct the heart defects. A, WT (+/+) and Nedd4- 1−/−,trap (−/−) MEFs were treated with the indicated amounts of aspirin for 12 h, and the amount of Tsp-1 was analyzed by immunoblotting. The same results were obtained after aspirin treatment for 4 or 8 h (not shown). B, aspirin treatment of pregnant Nedd4-1+/−,trap mothers causes reduction in the levels of Tsp-1 in the embryos. Pregnant mothers were treated with aspirin in the drinking water, embryos were harvested at E12.5, and their tissues were analyzed by immunoblotting for levels of Tsp-1. C, the heart defects in the Nedd4- 1−/−,trap embryos persist after aspirin treatment. Aspirin-treated embryos (E18.5) were harvested, and tissue sections were analyzed by H&E for heart integrity. Shown are the persistent defects in the heart despite aspirin treatment. RA, right atrium; LA, left atrium; AVSD, atrioventricular septal defect; CAVC, common atrioventricular canal; Pap, papillary muscle.
In addition to Tsp-1, we also tested for elevated levels of other top hits of the ICAT analysis (annexin A2 and A7), as well as of other proteins previously reported to be substrates for Nedd4-1, in the Nedd4-1−/−,trap MEFs relative to WT. As seen in supplemental Fig. S4, annexin A7 (which possesses a PY motif), annexin A2, and Grb10 all exhibited increased levels in the knock-out MEFs relative to WT MEFs, albeit to various degrees. However, unlike Tsp-1, the level of these proteins (and others we tested) was not decreased in response to aspirin treatment (supplemental Fig. S4 and data not shown), suggesting that they are not likely to be involved in the aspirin-mediated rescue of the Nedd4-1−/−,trap embryos.
To test whether the aspirin-mediated increase in survival of the Nedd4-1−/−,trap embryos was a result of correction of the heart defects, we analyzed the hearts of these aspirin-rescued embryos at E18.5. As shown in Fig. 6C, the rescued embryos still exhibited severe heart defects. These results suggest that the heart defects we observed were not the cause of death of the Nedd4-1−/−,trap embryos and that aspirin treatment prolonged their survival by affecting other systems or organs.
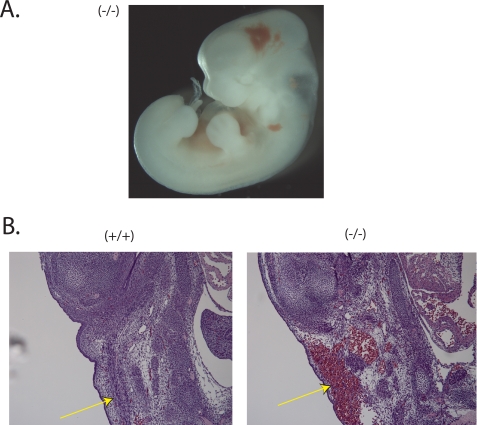
Analysis of several of the (untreated) Nedd4-1−/−,trap embryos harvested at E12.5 reveled hemorrhaging, especially around the brain area (Fig. 7A), as well as in some of the blood vessels (Fig. 7B). Such hemorrhaging is expected to be fatal and could be the cause of death of these embryos. Overall, the prevalence of this bleeding was higher in the untreated embryos relative to aspirin-treated ones.
FIGURE 7.
Bleeding in the Nedd4-1−/−,trap embryos. A, whole Nedd4-1−/−,trap embryo (−/−), E12.5, revealing bleeding, mainly in the head area. B, H&E staining of E13.5 embryos showing blood vessel rupture and bleeding in the (−/−) but not WT (+/+) embryos.
DISCUSSION
In this paper, we show that Nedd4-1 is required for proper heart development and likely vasculature development as well. The heart defect observed, DORV, belongs to the outflow tract abnormalities that are also seen in humans and accounts for 1–3% of all congenital heart defects (50). The type of DORV seen here was of Type II DORV, because it was associated with malformations of the atrioventricular valves and ventricles and was not associated with heterotaxy. The defect did not appear to be related to neural crest migration defects, as septation of the outflow tract was intact. Rather, we suggest that abnormal signaling in the myocardium resulted in defective positioning of the outflow tract relative to the chambers, as well as defective cushion formation. Knock-out of various genes in mice has resulted in DORV (reviewed in Ref. 50), suggesting that DORV represents a final common pathway defect during cardiac outflow tract development. These mouse models point to the involvement of at least several signaling pathways in this aspect of heart development, including the Ras/extracellular signal-regulated kinase (Erk), transforming growth factor β, Wnt, Notch, retinoic acid, and others (reviewed in Ref. 50).
Our work here identified Tsp-1 as a protein exhibiting dramatically increased levels in the Nedd4-1 knock-out MEF cells and embryos. Tsp-1 is a homotrimeric Ca2+-binding protein that is synthesized by several cell types, including vascular endothelial cells, and is a well known inhibitor of angiogenesis. It is also a major constituent of α granules in platelets that is secreted upon platelet activation and is involved in regulating platelet aggregation/thrombosis (51–54). It also regulates the immune response (55). Tsp-1 knock-out mice develop pneumonia because of macrophage infiltration of the lungs (56). A gain of function mutant of Tsp-1 does not exist. We speculate that the vascular and heart defect that we observed in our Nedd4-1 knock-out animals may be at least partially mediated by the highly elevated levels of Tsp-1, which is a known anti-angiogenesis factor.
Although Tsp-1 possesses a PY motif (a binding motif for Nedd4-WW domains), this PY motif is luminal and hence not likely to contribute to binding to Nedd4-1, which is very weak in the first place. Indeed, we found that mutation of this PY motif did not abrogate binding to Nedd4-1, suggesting that it is not required for the interaction. Moreover, pulse-chase analysis of Tsp-1 levels in MEFs revealed lack of stabilization of Tsp-1 in the Nedd4-1−/−,trap MEFs relative to WT, suggesting that Nedd4-1 may not significantly regulate Tsp-1 protein stability. Instead, it appears to suppress the transcription rate of Tsp-1, leading to a reduction in its mRNA levels. Our preliminary chromatin immunoprecipitation analysis suggests that this regulation of transcription may involve the transcription factors Egr1 and Sp1, which exhibit increased binding to the Tsp1 promoter in Nedd4-1−/− MEFs relative to WT. However, these transcription factors are not direct substrates for Nedd4-1. Thus, the exact pathway(s) involved in the Nedd4-1 -mediated regulation of Tsp-1 transcription, as well as heart and vasculature development, are currently unknown. Our recent work identified receptor Tyr kinases (e.g. fibroblast growth factor receptor 1) as targets for Nedd4-1-mediated regulation (57), but it is not clear whether this negative regulation is related to the suppressive effect of Nedd4-1 on Tsp1 transcription. Also, our preliminary work suggests that the levels of c-Myc, a known suppressor of Tsp-1 transcription (44) that is required for proper angiogenesis and vasculogenesis (58), are reduced in Nedd4-1−/−,trap MEFs. Future studies are needed to identify all of the mediator(s) of the Nedd4-1-dependent suppression of Tsp-1 transcription.
Although aspirin treatment of the pregnant mothers did not correct the heart defects, it is possible that the extent of reduction of Tsp-1 levels by aspirin was not sufficient to reverse the phenotype or that other factors are required as well. It is also possible that aspirin treatment led to embryonic rescue by regulating other functions of Tsp-1 (such as thrombosis) or by regulating the function of other proteins. The former possibility is less likely, however, given that aspirin is known to increase bleeding, and we observed greater incidence of bleeding in embryos that were not treated with this drug. Unfortunately, we could not measure platelet activation in the knock-out embryos because of the limited amounts of blood we were able to collect from the young embryos. Future genetic studies are needed to test whether knock-out of Tsp1 could rescue the embryonic defects or lethality caused by a loss of Nedd4-1.
Nedd4-1 is a widely expressed protein and most likely has numerous substrates, as is also suggested by our mass spectrometric analyses. Thus, it is quite possible that the phenotypes we observed in the Nedd4-1 knock-out mice may be mediated, at least in part, by additional Nedd4-1 target proteins. For example, the smaller size of the Nedd4-1−/−,trap embryos and the slower growth of the Nedd4-1−/−,trap MEFs (37) could involve the Nedd4-1 target Grb10, which is a binding partner for Nedd4-1 (30) and is involved in suppression of signaling from the insulin receptor and the insulin-like growth factor-1 receptor (59–61). Indeed, the Grb10 levels were elevated in our knock-out MEFs and mice (supplemental Fig. S2 and data not shown), as also reported by others (62). The rescue of the embryonic lethality by aspirin, however, is not likely to be mediated by most of these targets, because unlike Tsp-1, their levels did not decrease in response to aspirin treatment. Moreover, the aspirin-treated embryos, although viable, were still smaller than their littermates, suggesting that the aspirin-mediated rescue is not acting via regulation of Grb10. In addition to its role in regulating animal growth (62), two recent reports suggested that Nedd4-1 is also involved in regulating T cell activation (63) and, independently, in regulating neuromuscular synaptogenesis (64), similar to our previous observations in flies (33).
Nedd4-1 and Nedd4-2 are closely related family members, yet their in vivo function is clearly not redundant, because their knock-out in mice yields very different phenotypes. Nedd4-2 was demonstrated previously to regulate the epithelial Na+ channel (12, 13) by its ubiquitination and removal from the plasma membrane (14, 15). In accord, a recent knock-out of Nedd4-2 in mice led to salt-induced hypertension (65), which is consistent with an accumulation of the epithelial Na+ channel at the plasma membrane in the distal nephron, exhibiting a phenotype resembling that of Liddle syndrome mice (66) and patients (67). As we described in this paper, knock-out of Nedd4-1 leads to heart, vasculature, and general growth defects, resulting in embryonic lethality. Thus, these very different biological roles of Nedd4-1 and Nedd4-2, despite the close sequence similarity, underscore the importance of determining the in vivo functions of individual E3 ubiquitin ligases, even within closely related family members.
Supplementary Material
Acknowledgments
We thank Drs. J. Rossant, L. Coultas, W. Kahr, C. C. Hui, and K. Koshiba-Takeuchi (The Hospital for Sick Children) for help with setting up some of the assays described in the paper and for very helpful discussions.
This work was supported by Canadian Institute of Health Research Grant MOP 13494 (to D. R.), by the National Cancer Institute of Canada/Canadian Cancer Society Research Institute (to D. R.), by Deutsche Forschungsgemeinschaft Grant SFB271/B11 (to N. B.), and by the European Union (EUSynapse) (to N. B.). This work was also supported in part by the Canadian Foundation for Innovation and the Ontario Research and Development Challenge Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Movies S1 and S2, and Figs. S1–S4.
H. Kawabe and N. Brose, unpublished results.
J. Rossant, F. Fouldakou, and D. Rotin, unpublished results.
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin conjugating enzyme
- E3
- ubiquitin-protein isopeptide ligase
- WT
- wild type
- MEF
- mouse embryonic fibroblast
- Tsp
- thrombospondin
- β-gal
- β-galactosidase
- En
- embryonic day n
- MS/MS
- tandem mass spectrometry
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- H&E
- hematoxilin:eosin
- PBS
- phosphate-buffered saline
- OPT
- optical projection tomography
- DORV
- double-outlet right ventricle
- ICAT
- isotope coded affinity tag.
REFERENCES
- 1.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 2.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 3.Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 [DOI] [PubMed] [Google Scholar]
- 4.Dikic I., Wakatsuki S., Walters K. J. (2009) Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Tomooka Y., Noda M. (1992) Biochem. Biophys. Res. Commun. 185, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 6.Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 7.Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 8.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 9.Kanelis V., Rotin D., Forman-Kay J. D. (2001) Nat. Struct. Biol. 8, 407–412 [DOI] [PubMed] [Google Scholar]
- 10.Kasanov J., Pirozzi G., Uveges A. J., Kay B. K. (2001) Chem. Biol. 8, 231–241 [DOI] [PubMed] [Google Scholar]
- 11.Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abriel H., Loffing J., Rebhun J. F., Pratt J. H., Schild L., Horisberger J. D., Rotin D., Staub O. (1999) J. Clin. Invest. 103, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamynina E., Debonneville C., Bens M., Vandewalle A., Staub O. (2001) FASEB J. 15, 204–214 [DOI] [PubMed] [Google Scholar]
- 14.Lu C., Pribanic S., Debonneville A., Jiang C., Rotin D. (2007) Traffic 8, 1246–1264 [DOI] [PubMed] [Google Scholar]
- 15.Zhou R., Patel S. V., Snyder P. M. (2007) J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 16.van Bemmelen M. X., Rougier J. S., Gavillet B., Apothéloz F., Daidié D., Tateyama M., Rivolta I., Thomas M. A., Kass R. S., Staub O., Abriel H. (2004) Circ. Res. 95, 284–291 [DOI] [PubMed] [Google Scholar]
- 17.Embark H. M., Böhmer C., Palmada M., Rajamanickam J., Wyatt A. W., Wallisch S., Capasso G., Waldegger P., Seyberth H. W., Waldegger S., Lang F. (2004) Kidney Int. 66, 1918–1925 [DOI] [PubMed] [Google Scholar]
- 18.Fotia A. B., Ekberg J., Adams D. J., Cook D. I., Poronnik P., Kumar S. (2004) J. Biol. Chem. 279, 28930–28935 [DOI] [PubMed] [Google Scholar]
- 19.Ekberg J., Schuetz F., Boase N. A., Conroy S. J., Manning J., Kumar S., Poronnik P., Adams D. J. (2007) J. Biol. Chem. 282, 12135–12142 [DOI] [PubMed] [Google Scholar]
- 20.Jespersen T., Membrez M., Nicolas C. S., Pitard B., Staub O., Olesen S. P., Baró I., Abriel H. (2007) Cardiovasc. Res. 74, 64–74 [DOI] [PubMed] [Google Scholar]
- 21.Pham N., Rotin D. (2001) J. Biol. Chem. 276, 46995–47003 [DOI] [PubMed] [Google Scholar]
- 22.Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blot V., Perugi F., Gay B., Prévost M. C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M. C., Pique C. (2004) J. Cell Sci. 117, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 24.Harty R. N., Brown M. E., Wang G., Huibregtse J., Hayes F. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda J., Nakao M., Kawaoka Y., Shida H. (2003) J. Virol. 77, 9987–9992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urata S., Noda T., Kawaoka Y., Morikawa S., Yokosawa H., Yasuda J. (2007) J. Virol. 81, 4895–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camus G., Segura-Morales C., Molle D., Lopez-Vergès S., Begon-Pescia C., Cazevieille C., Schu P., Bertrand E., Berlioz-Torrent C., Basyuk E. (2007) Mol. Biol. Cell 18, 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina G., Pincetic A., Ehrlich L. S., Zhang Y., Tang Y., Leis J., Carter C. A. (2008) Virology 377, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dussart S., Douaisi M., Courcoul M., Bessou G., Vigne R., Decroly E. (2005) J. Mol. Biol. 345, 547–558 [DOI] [PubMed] [Google Scholar]
- 30.Morrione A., Plant P., Valentinis B., Staub O., Kumar S., Rotin D., Baserga R. (1999) J. Biol. Chem. 274, 24094–24099 [DOI] [PubMed] [Google Scholar]
- 31.Monami G., Emiliozzi V., Morrione A. (2008) J. Cell. Physiol. 216, 426–437 [DOI] [PubMed] [Google Scholar]
- 32.Magnifico A., Ettenberg S., Yang C., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. (2003) J. Biol. Chem. 278, 43169–43177 [DOI] [PubMed] [Google Scholar]
- 33.Ing B., Shteiman-Kotler A., Castelli M., Henry P., Pak Y., Stewart B., Boulianne G. L., Rotin D. (2007) Mol. Cell. Biol. 27, 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 35.Trotman L. C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N. P., Carver B. S., Cordon-Cardo C., Erdjument-Bromage H., Tempst P., Chi S. G., Kim H. J., Misteli T., Jiang X., Pandolfi P. P. (2007) Cell 128, 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., Jiang X. (2007) Cell 128, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., Rotin D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staub O., Yeger H., Plant P. J., Kim H., Ernst S. A., Rotin D. (1997) Am. J. Physiol. Cell Physiol. 272, C1871–C1880 [DOI] [PubMed] [Google Scholar]
- 39.Gasca S., Hill D. P., Klingensmith J., Rossant J. (1995) Dev. Genet 17, 141–154 [DOI] [PubMed] [Google Scholar]
- 40.Walls J. R., Coultas L., Rossant J., Henkelman R. M. (2008) PLoS ONE 3, e2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q. W., Liu S., Tian Y., Salwen H. R., Chlenski A., Weinstein J., Cohn S. L. (2003) Cancer Res. 63, 6299–6310 [PubMed] [Google Scholar]
- 42.Mizukami Y., Fujiki K., Duerr E. M., Gala M., Jo W. S., Zhang X., Chung D. C. (2006) J. Biol. Chem. 281, 13957–13963 [DOI] [PubMed] [Google Scholar]
- 43.Cyrus T., Sung S., Zhao L., Funk C. D., Tang S., Praticò D. (2002) Circulation 106, 1282–1287 [DOI] [PubMed] [Google Scholar]
- 44.Watnick R. S., Cheng Y. N., Rangarajan A., Ince T. A., Weinberg R. A. (2003) Cancer Cell 3, 219–231 [DOI] [PubMed] [Google Scholar]
- 45.Good D. J., Polverini P. J., Rastinejad F., Le Beau M. M., Lemons R. S., Frazier W. A., Bouck N. P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6624–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolsma S. S., Volpert O. V., Good D. J., Frazier W. A., Polverini P. J., Bouck N. (1993) J. Cell Biol. 122, 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo N., Krutzsch H. C., Inman J. K., Roberts D. D. (1997) Cancer Res. 57, 1735–1742 [PubMed] [Google Scholar]
- 48.Serebruany V. L., Malinin A. I., Oshrine B. R., Sane D. C., Takserman A., Atar D., Hennekens C. H. (2004) Thromb. Res. 113, 197–204 [DOI] [PubMed] [Google Scholar]
- 49.Coppinger J. A., O'Connor R., Wynne K., Flanagan M., Sullivan M., Maguire P. B., Fitzgerald D. J., Cagney G. (2007) Blood 109, 4786–4792 [DOI] [PubMed] [Google Scholar]
- 50.Obler D., Juraszek A. L., Smoot L. B., Natowicz M. R. (2008) J. Med. Genet 45, 481–497 [DOI] [PubMed] [Google Scholar]
- 51.Xie L., Chesterman C. N., Hogg P. J. (2001) J. Exp. Med. 193, 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pimanda J. E., Ganderton T., Maekawa A., Yap C. L., Lawler J., Kershaw G., Chesterman C. N., Hogg P. J. (2004) J. Biol. Chem. 279, 21439–21448 [DOI] [PubMed] [Google Scholar]
- 53.Bonnefoy A., Daenens K., Feys H. B., De Vos R., Vandervoort P., Vermylen J., Lawler J., Hoylaerts M. F. (2006) Blood 107, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isenberg J. S., Romeo M. J., Yu C., Yu C. K., Nghiem K., Monsale J., Rick M. E., Wink D. A., Frazier W. A., Roberts D. D. (2008) Blood 111, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams J. C., Lawler J. (2004) Int. J. Biochem. Cell Biol. 36, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawler J., Sunday M., Thibert V., Duquette M., George E. L., Rayburn H., Hynes R. O. (1998) J. Clin. Invest. 101, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Persaud A., Alberts P., Amsen E., Xiong X., Wasmuth J., Saadon Z., Fladd C., Parkinson J., Rotin D. (2009) Mol. Syst. Biol. 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baudino T. A., McKay C., Pendeville-Samain H., Nilsson J. A., Maclean K. H., White E. L., Davis A. C., Ihle J. N., Cleveland J. L. (2002) Genes Dev. 16, 2530–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vecchione A., Marchese A., Henry P., Rotin D., Morrione A. (2003) Mol. Cell. Biol. 23, 3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith F. M., Holt L. J., Garfield A. S., Charalambous M., Koumanov F., Perry M., Bazzani R., Sheardown S. A., Hegarty B. D., Lyons R. J., Cooney G. J., Daly R. J., Ward A. (2007) Mol. Cell. Biol. 27, 5871–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Balas B., Christ-Roberts C. Y., Kim R. Y., Ramos F. J., Kikani C. K., Li C., Deng C., Reyna S., Musi N., Dong L. Q., DeFronzo R. A., Liu F. (2007) Mol. Cell. Biol. 27, 6497–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao X. R., Lill N. L., Boase N., Shi P. P., Croucher D. R., Shan H., Qu J., Sweezer E. M., Place T., Kirby P. A., Daly R. J., Kumar S., Yang B. (2008) Sci. Signal 1, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang B., Gay D. L., MacLeod M. K., Cao X., Hala T., Sweezer E. M., Kappler J., Marrack P., Oliver P. M. (2008) Nat. Immunol. 9, 1356–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y., Oppenheim R. W., Sugiura Y., Lin W. (2009) Dev. Biol. 330, 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi P. P., Cao X. R., Sweezer E. M., Kinney T. S., Williams N. R., Husted R. F., Nair R., Weiss R. M., Williamson R. A., Sigmund C. D., Snyder P. M., Staub O., Stokes J. B., Yang B. (2008) Am. J. Physiol. Renal Physiol. 295, F462–F470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pradervand S., Wang Q., Burnier M., Beermann F., Horisberger J. D., Hummler E., Rossier B. C. (1999) J. Am. Soc. Nephrol. 10, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 67.Lifton R. P., Gharavi A. G., Geller D. S. (2001) Cell 104, 545–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.