Abstract
The Notch signaling pathway plays an essential role in the regulation of cell specification by controlling differentiation, proliferation, and apoptosis. Numb is an intrinsic regulator of the Notch pathway and exists in four alternative splice variants that differ in the length of their phosphotyrosine-binding domain (PTB) and proline-rich region domains. The physiological relevance of the existence of the Numb splice variants and their exact regulation are still poorly understood. We previously reported that Numb switches from isoforms containing the insertion in PTB to isoforms lacking this insertion in neuronal cells subjected to trophic factor withdrawal (TFW). The functional relevance of the TFW-induced switch in Numb isoforms is not known. Here we provide evidence that the TFW-induced switch in Numb isoforms regulates Notch signaling strength and Notch target gene expression. PC12 cells stably overexpressing Numb isoforms lacking the PTB insertion exhibited higher basal Notch activity and Notch-dependent transcription of the transient receptor potential channel 6 (TRPC6) when compared with those overexpressing Numb isoforms with the PTB insertion. The differential regulation of TRPC6 expression is correlated with perturbed calcium signaling and increased neuronal vulnerability to TFW-induced death. Pharmacological inhibition of the Notch pathway or knockdown of TRPC6 function ameliorates the adverse effects caused by the TFW-induced switch in Numb isoforms. Taken together, our results indicate that Notch and Numb interaction may influence the sensitivity of neuronal cells to injurious stimuli by modulating calcium-dependent apoptotic signaling cascades.
Keywords: Apoptosis, Calcium/Calcineurin, Cell/Trafficking, Diseases/Neurodegeneration, Diseases/Neurological, Receptors/Endocytosis, Signal Transduction/Calcium, Transcription/Regulation
Introduction
The Notch pathway is required for the embryonic and post-natal development of many organs by regulating cell proliferation, differentiation, and apoptosis through direct cell to cell contact (1). Notch consists of a family of single-pass transmembrane receptors (Notch1–4), which can be activated by interaction with membrane-tethered ligands of the Delta/Serrate/Lag2 family expressed on neighboring cells (2). Upon activation, Notch is cleaved by γ-secretase (3), releasing from the membrane the Notch intracellular domain (NICD)3 that translocates to the nucleus. NICD associates with CSL (an acronym for the mammalian CBF-1, Drosophila Suppressor of Hairless and Caenorhabditis elegans Lag-1 transcription factors (4)) to control the expression of target genes, most notably the basic helix-loop-helix transcription factors belonging to the Hes (hairy and enhancer of split) (5) and Herp (Hes-related protein) family (6). These transcription factors in turn regulate the ability of the cells to respond to environmental clues. In the absence of Notch signaling, CSL represses transcription of Notch target genes, and following activation by Notch, CSL is converted into a transcriptional activator and activates transcription of the same genes (7). Disruption of the Notch gene in mice results in severe developmental defects and embryonic lethality, supporting a major role for Notch in regulating cell fate (8).
Alterations in Notch signaling are associated with tumorigenesis suggesting that dysfunction of intracellular Notch prevents differentiation, ultimately guiding undifferentiated cells toward malignant transformation (9, 10). One key regulator of Notch signaling is Numb, an evolutionarily conserved adapter protein identified by its ability to control cell fate in the nervous system of Drosophila (11). Loss of Numb expression has been shown to promote tumorigenesis (12) suggesting that the function of Numb is important for limiting signaling through the Notch pathway. Numb is lost in >50% of human breast carcinoma samples, and its level is inversely correlated with grade and proliferation rate (12). Numb contains two protein-protein interaction domains, a phosphotyrosine-binding (PTB) domain and a proline-rich region (PRR) (13). Whereas only one form of Numb has been identified in Drosophila, mammals produce four Numb isoforms that differ in the length of the PTB (lacking or containing an 11-amino acid insert) and PRR (lacking or containing a 48-amino acid insert) domains (13, 14).
Emerging evidence supports the notion that the Numb isoforms are functionally significant and have different biologic effects (13–16). In the mouse P19 embryonic carcinoma cell line, it was shown that expression of Numb isoforms with short PRR promote differentiation but not proliferation, whereas expression of Numb isoforms with long PRR promote proliferation but not differentiation (15). Expression of Numb isoforms that lack the insertion in the PTB (short PTB or SPTB) but not those with the insertion (long PTB or LPTB) increases neuronal vulnerability to apoptotic insults through a mechanism that involves perturbation of calcium (Ca2+) homeostasis (16).
The intracellular messenger Ca2+ controls a vast array of cellular functions ranging from short term responses, such as motility and secretion, to longer term control of gene expression, cell division, and various metabolic reactions (17). Another major role for Ca2+ is in cell death in physiological settings or during injury or diseases (18–21, 49). The mechanisms that control cellular Ca2+ dynamics are complex, with ion channels and pumps in the plasma membrane and endoplasmic reticulum (ER) playing major roles in regulating both rapid and long term changes in the cytoplasmic free Ca2+ concentration ([Ca2+]i). Ca2+ entry is controlled by voltage-gated and ligand-gated channels, store-operated channels (SOCs), and receptor-operated channels (ROCs). The latter are classified according to the initial stimulus for channel activation. Whereas SOCs are recruited in response to the release of Ca2+ from intracellular stores (22, 23), ROCs are activated by G-protein-coupled receptors (24). Although the exact molecular configuration of SOCs and ROCs has not been conclusively identified, much attention has been focused on the role of canonical transient receptor potential channel (TRPC) subfamily in this process (25). The TRPC subfamily of proteins consists of seven mammalian TRPC isoforms numbered from 1 to 7 and is expressed in a variety of tissues, with surprisingly high levels of expression seen in the brain and heart (26). Functional studies showed that they form voltage-independent cationic channels with distinct gating and biophysical properties when heterogeneously expressed in various cell lines (26). Among the TRPC members, TRPC6 has been linked to diverse cellular functions, and its aberrant expression has been associated with various disease states such as cardiac hypertrophy, pulmonary hypertension, and glomerulosclerosis (27–29).
In the present study we found that the switch of Numb isoforms increases neuronal vulnerability to TFW-induced death by a mechanism that involves Notch-dependent transcription of TRPC6. Our results may have important implications for the development of therapeutic modalities to counteract perturbations of Ca2+ signaling in various disease states.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies used for immunodetection of Notch and NICD were anti- Notch1 (G-20, Santa Cruz Biotechnology) and Val-177 anti-NICD (Upstate), respectively. The polyclonal antibodies to Numb were obtained from Santa Cruz (sc-15590) and Upstate (anti-pan-Numb). The antibodies to TRPC1–7 were kindly provided by Dr. Schilling (Case Western University). Additional antibodies included: anti-Rab5 (Santa Cruz Biotechnology) and actin (Sigma). Immunofluorescence-conjugated secondary antibodies (Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 594-conjugated rabbit anti-goat IgG) were obtained from Vector Laboratories (Burlingame, CA). All commercial antibodies were used within the concentration ranges recommended by the manufacturer. Additional reagents included: Lipofectamine 2000, TRIzol, and Opti-MEM I-reduced medium (Invitrogen); DAPT (N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester) and β-Secretase Inhibitor II (Calbiochem); oleoyl-2-acetyl-sn-glycerol (OAG), carbachol, cyclopiazonic acid, and thapsigargin (Sigma); SKF96365 and nifedipine (Tocris); ECL-Plus kit (Amersham Biosciences); propidium iodide, Hoechst, and Fura-2AM (Molecular Probes); and a γ-secretase fluorometric (EDANS/DABCYL) assay kit (R&D Systems).
Plasmid Constructs, Transfection, and Generation of Stably Transfected Cells
The pcDNA3.1 constructs for all four human Numb isoforms have been described previously (30). Pheochromocytoma 12 (PC12) cells were transfected with the expression vector pcDNA3.1 with or without cDNAs for each of the human Numb isoforms. Stably expressing clones were selected with G418 and maintained as previously described (30, 31). Cultures were maintained at 37 °C (5% CO2 atmosphere) in RPMI medium supplemented with 10% heat-inactivated horse serum, 5% heat-inactivated fetal bovine serum, antibiotic, and 0.5 mg/ml G418.
Experimental Treatments
Trophic factor withdrawal (TFW) was accomplished by washing cultures four times (2 ml per wash) with Locke's buffer (154 mm NaCl, 5.6 mm KCl, 2.3 mm CaCl2, 1.0 mm MgCl2, 3.6 mm NaHCO3, 5 mm glucose, and 5 mm HEPES, pH 7.2) with subsequent incubation in 1 ml of Locke's buffer (30, 31). DAPT and FK506 were dissolved in DMSO and used to inhibit γ-secretase and calcineurin activity, respectively. Control cultures were treated with equivalent volumes of DMSO.
Quantification of Cell Death
Cell viability was assessed using the fluorescent DNA binding dye Hoechst 33342 as described previously (32). Briefly, after treatment, cells were stained for 15 min at 37 °C with 2 μm propidium iodide and 5 μg/ml Hoechst dye in PBS. Cells were washed twice with PBS and fixed with 4% paraformaldehyde for 1 h at room temperature. Coverslips were mounted at microscope slides using DPX mounting medium for histology (Fluka) and visualized under a fluorescence microscope (Zeiss). Fluorescent images of three random fields per coverslip were acquired, and morphologically distinct apoptotic cells with condensed nuclei were counted. The data from three independent experiments were statistically examined by paired t test and are expressed as means ± S.E. Cells were counted, and the percent cell death was assessed using a ratio of propidium iodide-stained cells to total number of cells.
siRNA Constructs and Transfection
Stably transfected PC12 cells were cultured as described above to 50–70% confluency. TRPC3 and TRPC6 were knocked down using Stealth siRNA (Invitrogen): TRPC3, UCAUCUUCCUGGGUCUGCUUUGUGUU and TRPC6, AACAUUCCAAAGUCAAGCAUAUUCC. A Stealth RNAi Negative duplex with limited homology to any known sequences in human, mouse, and rat genomes was used as control (Invitrogen). Notch siRNA consisting of a pool of three to five target-specific sequences was obtained from Santa Cruz Biotechnology. Cells were transfected with 100 nm siRNA using Lipofectamine 2000 in Opti-MEM I-reduced medium for 24–48 h and then subjected to the experimental treatments described above.
Ca2+ Imaging
Real-time fluorescence measurements of intracellular Ca2+ concentration ([Ca2+]i) were performed as described previously (16). Briefly, cells were incubated for 10 min in the presence of 2.5 μm of the acetoxymethylester form of fura-2 (Molecular Probes) at 37 °C, washed three times with serum-free RPMI. Coverslips containing cells were mounted in a closed, heated chamber on the stage of an inverted microscope (Nikon TSE 100 Eclipse). Cells were superfused with Krebs Ringer bicarbonate (KRB) solution containing (in mm): 118 NaCl, 4.7 KCl, 0.57 MgSO4, 1.18 KH2PO4, 25 NaHCO3, 2.5 CaCl2, and 10 glucose. Cells were perfused for 15 min prior to beginning experiments to remove extracellular dye. Ratiometric measurement of fura-2 fluorescence was performed using a collimated light beam from a xenon arc lamp filtered at 340 and 380 nm and focused onto Cells via a 20× fluorescence objective (Super Fluor 20, Nikon). The fluorescence was collected from 30–50 cells simultaneously monitored. Only one field was used per dish. Protocols were executed, and data were collected on-line with InCyte software (Intracellular Imaging Inc.). Background fluorescence signals were collected at the same rate for the same wavelengths and were subtracted from the corresponding fluorescence images. In some experiments Ca2+ was omitted from the KRB buffer without (Ca2+-deficient buffer) or with 100 μm EGTA (Ca2+-free buffer). SOCE was induced by stimulation with thapsigargin (TG, 1 μm) as described previously (33, 34). Measurements were taken until [Ca2+]i reached resting level. Cells were then superfused with 2 mm Ca2+-containing KRB. Measurements were continued for 2–4 min after Ca2+ peak was recorded. In most instances, fluorescence ratios (fluorescence at 510 nm with excitation at 340 nm divided by that with excitation at 380) were reported.
Total RNA Isolation and Real-time PCR
Total RNA was extracted using the TRIzol reagent according to the manufacturer's recommendation, and 1 μg of total extracted RNA was used for synthesis of first-strand cDNA using SuperScriptTM III RNase H reverse transcriptase and 1 μg of oligo(dT). Successively, 2 μl of each cDNA was PCR-amplified on the ABI Prism 7700 sequence detection system by using SYBR Green PCR core reagents and 300 nm primers in a 25-μl reaction volume. PCR primers for the TRPC homologs were designed based on published sequences in GenBankTM (Table 1). The integrity of the RT-PCR products was confirmed by melting curve analysis. Melting curves for all TRPCs showed one specific peak. We used 18 S mRNA as an endogenous control to normalize variations in RNA extraction, the degree of RNA degradation, and variability in RT efficiency. We quantified the results with the comparative Ct method (35).
TABLE 1.
Oligonucleotide sequences of TRPC primers used for real-time RT-PCR
| Target | GenBankTM accession no. | Orientation | Sequence (5′–3′) | Predicted size | Nucleotide location |
|---|---|---|---|---|---|
| bp | |||||
| TRPC1 | AF061266 | Forward | CGACACCTTCCACTCGTTCA | 64 | 1716–1780 |
| Reverse | GCGCTAAGGAGAAGATGTACCAGA | ||||
| TRPC2 | NM017011 | Forward | GTTCCAGTTTCTCTTCTGGACCAT | 190 | 1804–1994 |
| Reverse | CAGCATCGTCCTCGATCTTCT | ||||
| TRPC3 | AB022331 | Forward | CATGCAGTGCAAAGACTTCGTAG | 74 | 531–605 |
| Reverse | TTCAGAATGGCTTCCACCTCTT | ||||
| TRPC4 | NM_080396 | Forward | CTCTGCAGATATCTCTGGGAAGAAT | 67 | 1514–1581 |
| Reverse | CACGAGGCAGTATATGAATAAGAACTTT | ||||
| TRPC5 | AY064411 | Forward | TGAAACCCTTCAGTCACTCTTCTG | 102 | 2038–2140 |
| Reverse | AGTAGCTCCCACAAACTCCGTG | ||||
| TRPC6 | NM_053559 | Forward | AGTGTACAGAATGCAGCCAGAAAC | 79 | 758–837 |
| Reverse | CAGCCCTTTGTAGGCATTGATC | ||||
| TRPC7 | XM_225159 | Forward | CCCTTTAACCTGGTGCCGA | 113 | 2665–2778 |
SDS-PAGE and Western Blotting
Cells were harvested and lysed in buffer (100 mm Tris, pH 6.8, 1% SDS, 10 mm EDTA, 5 mm EGTA, 2 mm phenylmethylsulfonyl fluoride, 25 μg/ml leupeptin, 5 μg/ml pepstatin). Protein quantification, electrophoretic separation, and transfer to nitrocellulose membranes were performed as described previously (33). Membranes were incubated in blocking solution (5% milk in Tween Tris-buffered saline (TTBS)) overnight at 4 °C followed by a 1-h incubation in primary antibody diluted in blocking solution at room temperature. Membranes were then incubated for 1 h in secondary antibody conjugated to horseradish peroxidase (Vector Laboratories), and bands were visualized using a chemiluminescence detection kit (ECL, Amersham Biosciences). The primary antibodies included rabbit polyclonal antibodies against TRPC3 and TRPC6. Membranes were stripped and re-probed with the actin antibody to verify and normalize protein loading (50 μg of total protein, unless stated otherwise).
For immunodetection of blots, enhanced chemiluminescence was applied. The intensity of the signals obtained was evaluated by densitometry and semiquantified using National Institutes of Health Image software based on the ratio between the intensity of the protein of interest divided by the intensity of actin in each experiment. Each experiment presented was repeated at least three times using different cell lysates.
Immunofluorescence Labeling
Following experimental treatments, cultured cells were fixed for 20 min with 4% paraformaldehyde in PBS. Cells were then incubated for 5 min in a solution of 0.2% Triton X-100 in PBS and for 1 h in blocking solution (0.02% Triton X-100, 5% normal horse or goat serum in PBS). After nonspecific binding was blocked, coverslips were incubated with a polyclonal antibody to TRPC6 or a polyclonal Numb antibody (1:500) in combination with a monoclonal antibody to the organelle marker Rab5 (early endosomes). All antibodies were diluted in blocking solution. After three washes, coverslips were incubated with fluorescein isothiocyanate-conjugated anti-rabbit and Cy3-conjugated anti-mouse secondary antibodies and then mounted. Immunofluorescence staining was examined by confocal microscopy (Zeiss) using the ×63 oil immersion objective lens, and images were processed by using the imaging software.
γ-Secretase Activity Assay
The activity of γ-secretase in cell extracts was determined using a commercially available secretase kit according to the manufacturer's protocol (R&D Systems). The method is based on the γ-secretase-dependent cleavage of a specific peptide conjugated to the fluorescent reporter molecules EDANS and DABCYL, which results in the release of a fluorescent signal that can be detected using a fluorescence microplate reader (excitation at 355 nm/emission at 510 nm). The level of γ-secretase enzymatic activity is proportional to the fluorometric reaction (36).
Calcineurin Activity Assay
Cellular calcineurin phosphatase activity was measured using a commercial non-radioactive Calcineurin Cellular Activity Assay Kit, according to the manufacturer's instruction (Biomol). The RII phosphopeptide (DLDVPIPGRFDRRVpSVAAE) derived from the RII subunit of cAMP-dependent kinase, was used as the substrate for calcineurin. PC12 cells were lysed in ice-cold lysis buffer and centrifuged at 100,000 × g for 45 min. After removal of free phosphate from the supernatant in the Desalting Column Resin, the lysates were incubated with RII phosphopeptide (1.64 mg/ml) in assay buffer containing 100 mm NaCl, 50 mm Tris-HCl (pH 7.5), 6 mm MgCl2, 0.5 mm dithiothreitol, 0.025% Nonidet P-40 with 0.5 mm CaCl2 or with 10 mm EGTA. Purified recombinant calcineurin was used as the positive control. After 30 min at 30 °C, reactions were terminated by adding 100 μl of Biomol GREENTM reagent, and absorbance was measured at A620 nm using a 96-well plate reader (Bio-Rad). A standard curved was plotted as A620 nm versus nanomoles of PO4 (used as the standard). Calcineurin activity was determined by subtracting the EGTA-insensitive activity from total activity.
Statistical Analysis
Comparison between two groups was performed using Student's t test, whereas multiple comparisons between more than two groups was analyzed by one-way ANOVA and post hoc tests by least significant difference. Data evaluated for the effects of two variables was analyzed using two-way ANOVA. Results are presented as means ± S.E. For all analyses, statistical significance is defined as a p value of ≤0.05.
RESULTS
TFW Activates the Notch Signaling Pathway
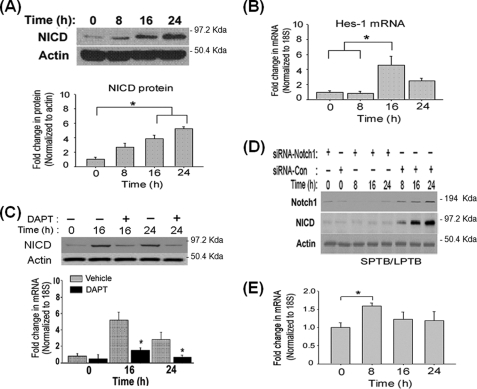
To determine whether the Notch pathway is activated in response to TFW, we measured the level of NICD, the activated form of Notch (1), by immunoblotting with the antibody Val-177, specific for NICD. In untreated cultures of PC12 cells, the NICD protein level was low but was rapidly up-regulated after TFW (Fig. 1A). Consistent with activation of the Notch pathway, the nuclear accumulation of NICD (supplemental Fig. S1) and the expression of the Notch downstream gene Hes1 (Fig. 1B) were detected after TFW. To further confirm the involvement of the Notch pathway in TFW-induced death, we employed the small molecule γ-secretase inhibitor DAPT to pharmacologically inhibit the proteolytic processing of Notch to NICD (36). Notch pathway inhibition was confirmed by immunoblotting and immunostaining using an antibody to NICD. Pre-treatment of PC12 cells with DAPT strongly reduced the amount of NICD, although it was not completely abolished (Fig. 1C). Immunolabeling indicated that pretreatment with DAPT greatly abrogated TFW-induced nuclear accumulation of NICD (data not shown). No staining was observed when the primary antibody was omitted or when the antibody was blocked (data not shown). DAPT also substantially inhibited Hes1 mRNA expression after TFW (Fig. 1C), which confirms the efficacy of DAPT in inhibiting the TFW-induced activation of the Notch pathway. To establish that Notch1 is responsible for TFW-induced Notch signaling, we inhibited Notch1 expression by RNA interference. The effectiveness of silencing the expression of endogenous Notch1 was determined by immunoblotting (Fig. 1D). Inhibition of Notch1 substantially inhibited TFW-induced accumulation of NICD (Fig. 1D), which confirms the findings obtained with pharmacological inhibition of the Notch pathway (Fig. 1C).
FIGURE 1.
TFW activates the Notch signaling pathway by inducing a switch in Numb isoforms. A, representative immunoblot showing the time course of NICD protein level in cultures of PC12 cells subjected to TFW. For each lane, 40 μg of total protein extract was applied. Equal protein load was confirmed by reprobing the blots for actin. The histogram shows the density of the NICD protein band normalized to actin. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cells. B, time course of steady-state levels of Hes1 transcript in PC12 cells after TFW. The level of Hes-1 transcript was determined in triplicate by real-time PCR and was normalized to the expression of housekeeping gene 18S. Values are the means ± S.E. of determinations made of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cultures. C, representative immunoblot showing the time course of TFW-induced NICD protein in cultures that were treated with DAPT (20 μm) or vehicle (DMSO). The histogram shows the density of the NICD protein band normalized to actin. Values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cells at each time point. D, representative immunoblot showing the time course of Notch1 and NICD in cultures that were transfected with a siRNA targeting Notch1 (siRNA-Notch1) or non-silencing control siRNA (siRNA-Con). Each siRNA (100 nm) was added to the cultures 24 h prior to TFW. At the indicated time points following TFW, total protein lysates were analyzed by immunoblotting using an antibody to either the N-terminal region of Notch1 or NICD. Equal protein loading was confirmed by reprobing the blots with an antibody to actin. Shown is the extracellular domain of Notch1 (∼180 kDa). E, time course of steady-state levels of SPTB- and LPTB-Numb transcripts in cultures of PC12 cells before and after TFW. Levels of each Numb transcripts were determined by real-time PCR, normalized to the housekeeping gene 18S, and expressed in -fold change relative to the non-treated cultures. Values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cultures.
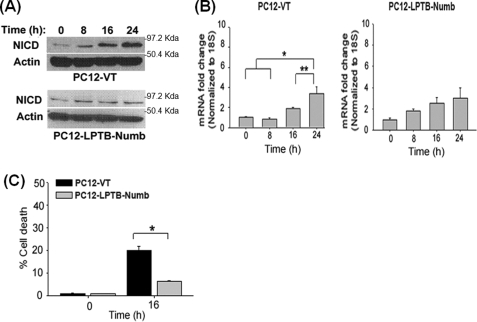
The TFW-induced Switch in Numb Isoform Is Reversible and Precedes Transcriptional Activation of Notch-regulated Gene Targets
We previously reported that TFW induces the switch in Numb from isoforms containing the PTB insertion to isoforms lacking this insertion in PC12 cells (30, 31). To determine whether the Numb isoform switch preceded activation of the Notch pathway, we quantitatively assessed transcripts levels of the Numb proteins that differ in the PTB domain in PC12 cells by RT-PCR using primer sets described previously (31). Time-course study showed that the SPTB-Numb transcripts increased rapidly and peaked at 8 h (Fig. 1E), suggesting that the TFW-induced switch in Numb isoforms is an early event that precedes the time point during which NICD accumulates in the nucleus (Fig. 1B) or before the appearance of the Hes1 transcript (Fig. 1C). The level of SPTB-Numb transcripts returned to near basal level in cultures that had been subjected to TFW for up to 6 h and then resupplied trophic support for an additional 2 h (supplemental Fig. S2) suggesting that TFW-induced switch in Numb isoforms is reversible.
Numb-mediated Control of the Notch Signaling Pathway Is PTB Domain-specific
The functions of the Numb proteins are defined by their interaction with other cellular proteins via the PTB domain (37, 38). Although inhibition of Notch signaling by Numb is critical for many cell fate decisions (39, 40), it is not clear whether all Numb isoforms demonstrate similar influence on modulating the Notch signaling events.
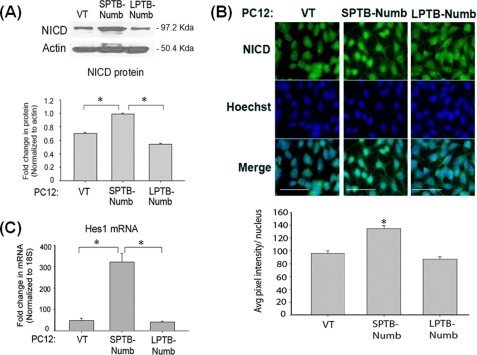
To determine whether the Numb proteins modulate activation of the Notch pathway in an isoform-specific manner, we measured protein level of NICD in PC12 clones stably overexpressing human Numb isoforms with long PTB and short PRR (LPTB/SPRR) and short PTB and short PRR (SPTB/SPRR) (30, 31). PC12 clones overexpressing SPTB-Numb (PC12-SPTB-Numb) exhibited higher basal levels of NICD compared with PC12-LPTB-Numb or control vector-transfected cells (PC12-VT) (Fig. 2A). By contrast, basal levels of NICD in PC12-LPTB-Numb were lower but did not reach statistical significance when compared with PC12-VT (Fig. 2A). NICD was detected in the nuclei of PC12-SPTB-Numb but not PC12-VT and PC12-LPTB-Numb (Fig. 2B). Hes-1 mRNA levels were also highest in PC12-SPTB-Numb (Fig. 2C) confirming that steady-state Notch activity is elevated in PC12-SPTB-Numb. Collectively, the data indicate that the Numb proteins that differ in the length of the PTB domain affect the processing of Notch in a distinct and contrasting manner.
FIGURE 2.
Numb modulates the Notch signaling pathway in an isoform-specific manner. A, representative immunoblot of the NICD protein in the PC12 clones stably overexpressing the Numb isoforms that differ in the PTB domain. Equal protein load was confirmed by reprobing the blots for actin. The histogram shows the density of the NICD protein band normalized to actin. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cells at each time point. B, representative micrographs of NICD immunoreactivity in the nuclei of the indicated clones of PC12 cells. Scale bars represent 15 μm. The histogram shows the average NICD immunoreactivity per nucleus. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with PC12-VT cells. C, steady-state level of Hes-1 transcript in the indicated PC12 clones. The level of Hes-1 transcript was determined in triplicate by real-time PCR and normalized to the expression of the housekeeping gene 18S. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests).
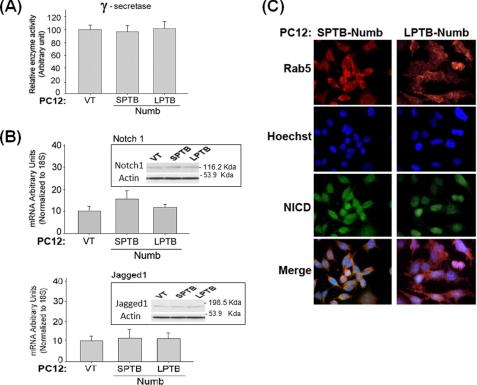
PTB Domain-specific Control of the Notch Pathway Is Independent of γ-Secretase Activity or the Expression of Notch Pathway Genes
Next, we determined the underlying mechanism whereby the Numb proteins differentially impact signaling through the Notch pathway. To exclude the possibility that the Numb isoforms may alter the activity of the Notch processing enzyme γ-secretase, we quantified γ-secretase activity in lysates prepared from each stable PC12 clone (31). The activity of γ-secretase was not affected by the presence of the different Numb isoforms (Fig. 3A). To determine whether the Numb isoforms may alter the expression of Notch pathway components, we measured transcripts levels of Notch1 and its ligand Jagged1 (41) in PC12 clones by qRT-PCR. Levels of Notch1 and Jagged1 transcripts were not markedly affected by expression of the Numb isoforms (Fig. 3B). These data indicate that Numb-mediated control of Notch signaling strength was not attributable to changes in the expression of components of the Notch pathway. Because the clones expressing the SPTB-Numb and LPTB-Numb are phenotypically similar with respect to Notch signaling, we performed forthcoming experiments with two representative clones of Numb that differ only in the PTB domain: SPTB/SPRR and LPTB/SPRR.
FIGURE 3.
Numb alters the endocytic trafficking of Notch in an isoform-specific manner. A, γ-secretase activity in the PC12 clones stably overexpressing the indicated Numb isoforms that differ in the PTB domain. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests). B, steady-state level of Notch1 and Jagged1 transcripts in the indicated PC12 clones. The level of these transcripts was determined in triplicate by real-time PCR and normalized to the expression of the housekeeping gene 18S. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with vector-transfected (VT) clones. The corresponding proteins are shown in the inset. C, representative confocal micrographs showing the localization of Notch1 to Rab5a-labeled endocytic compartments in the indicated PC12 clones. Following fixation and permeabilization, rabbit anti-Rab5a and goat anti-rabbit CY3-conjugated secondary antibody were used to label the endocytic compartments (red signal), and goat anti-Notch1 (raised to the N-terminal extracellular domain) and goat anti-goat-fluorescein isothiocyanate-conjugated secondary antibody to detect Notch1 protein (green signal). The bottom images show the merged images of Notch and Rab5a, depicting co-localization of Notch1 with early endosomes. The images are representative of those obtained from at least two stably transfected clonal cell lines.
Numb Regulates Post-endocytic Sorting and Activation of Notch in a Manner Dependent on the PTB Domain
Numb is an adapter protein that interacts with components of the endocytic machinery (42) and plays roles in the endocytic trafficking of several transmembrane proteins, including the neuronal cell adhesion molecule L1 (43), E-cadherin (44), and amyloid precursor protein (APP) (31). Given that the Numb proteins differentially impact the trafficking and processing fate of APP (31) and that Notch is a membrane protein that undergoes similar proteolytic processing as APP (45, 46), we next tested the hypothesis whether the Numb proteins may also regulate the trafficking of the Notch receptor within the endocytic pathway. Rab5A is a small GTPase localized on early endosomes and controls endosome fusion along the endocytic pathway. Double immunofluorescence labeling showed that the Notch protein co-localized with Rab5-labeled early endocytic compartments in PC12-SPTB-Numb (Fig. 3C). By contrast, PC12-VT and PC12-LPTB-Numb exhibited little or undetectable co-labeling of Notch with the Rab5 protein marker (Fig. 3C). These data are consistent with the notion that the differential activation of Notch signaling by the Numb proteins is mediated at the level of post-endosomal sorting of the Notch receptor (48).
Notch Signaling Modulates TRPC6 Expression in a Manner Dependent on the PTB Domain of Numb
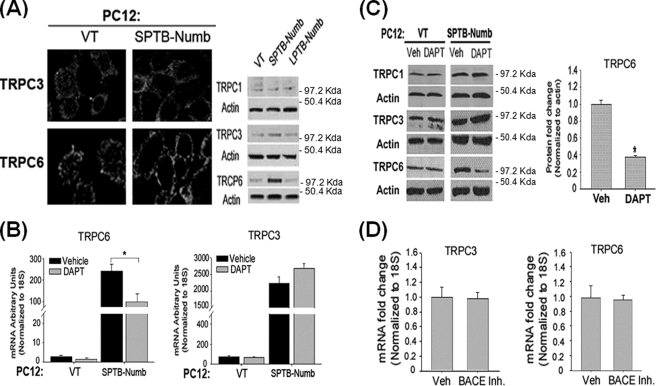
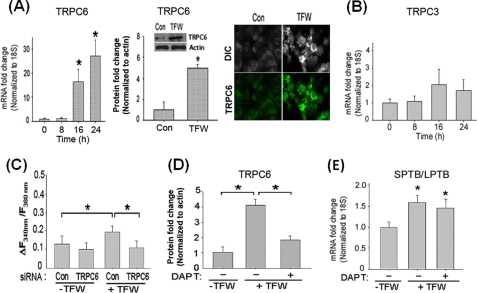
We previously reported that Numb modulates intracellular Ca2+ homeostasis in a manner dependent on the PTB domain (16, 30). Because destabilization of intracellular Ca2+ homeostasis was a critical component of the cell death-promoting effect of the SPTB-Numb proteins (16, 30), we next determined whether the PTB domain-specific effect of the Numb proteins on Ca2+ homeostasis may be mediated through the differential activation of the Notch pathway. First, we determined whether Notch signaling may regulate the expression of Ca2+ regulatory protein(s) that are important for Ca2+ signaling during differentiation, proliferation, migration, and cell death. We previously showed that PC12-SPTB-Numb exhibited a dramatically greater Ca2+ response to carbachol and bradykinin (16, 30), two agonists that not only induce Ca2+ release in PC12 cells but also stimulate Ca2+ entry across the plasma membrane through SOCs and ROCs. Because members of the TRPC family have been postulated as candidates for forming both SOCs and ROCs, we measured the expression of TRPC transcripts in PC12-Numb clones by qRT-PCR using primer sets listed in Table 1. PC12 cells express most of the TRPC (TRPC1–7) members except for TRPC2 and TRPC7 (50). Levels of TRPC3 and TRPC6 transcripts were markedly increased in PC12-SPTB-Numb when compared with PC12-VT and PC12-LPTB-Numb (supplemental Fig. S3). Although TRPC1 is the most abundant transcript, no significant change in the level of the corresponding protein was detected among the PC12 clones. By contrast, TRPC4, TRPC5, and TRPC7 proteins are expressed at very low or undetectable level in PC12 cells (data not shown (50)).
To examine the corresponding changes in TRPC3 and TRPC6 protein levels, we performed immunostaining and immunoblotting (Fig. 4A). Whereas TRPC1 protein was not markedly different among the PC12 clones, levels of TRPC3 and TRPC6 protein were significantly elevated only in PC12-SPTB-Numb cells by ∼2- and 10-folds, respectively (Fig. 4A).
FIGURE 4.
Numb modulates subtype-specific TRPC expression in an isoform-dependent manner via the Notch signaling pathway. A, levels of TRPC1, TRPC3, and TRPC6 protein in the indicated PC12 clones were assessed by immunoblotting (left panels) and immunostaining (right panels). Cultures were fixed and stained with antibodies to TRPC3 and TRPC6. B and C, effects of DAPT (20 μm) on the levels of TRPC6 and TRPC3 transcripts (B) and proteins (C). The amount of each transcript was determined in triplicate by real-time PCR and normalized to the expression of the housekeeping gene 18S. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with vehicle (Veh)-treated cultures. Protein amount was determined by immunoblotting using antibodies to TRPC3 and TRPC6. Equal protein loading was confirmed by reprobing the blots with an antibody to actin. The histogram shows the density of TRPC6 protein band normalized to actin. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with Veh-treated cultures. D, effects of β-secretase inhibitor II (BACE Inh., 10 μm) on the expression of TRPC6 and TRPC3. The level of each transcript was quantitated by real-time RT-PCR and normalized to the expression of the housekeeping gene 18S. The values are the means ± S.E. of three independent experiments.
Pharmacological inhibition of the Notch pathway in PC12-SPTB-Numb by using DAPT resulted in markedly lower levels of TRCP6 transcript (Fig. 4B) and protein (Fig. 4C) but had little impact on TRPC3 expression. TRPC1 protein level was not altered in the presence of DAPT (Fig. 4C). Treatment with a β-secretase inhibitor, which is known to inhibit the processing of the amyloid precursor protein but not the Notch receptor, had no measurable effect on either TRPC6 or TRPC3 transcript level in PC12-SPTB-Numb (Fig. 4D). The latter indicates that the Numb-isoform-specific effects on TRPC6 expression are not caused by altered APP processing (31). Collectively, these data suggest that the PTB domain-specific regulation of the Notch pathway is correlated with subtype-specific expression of TRPC.
Notch-mediated Expression of TRPC6 Augments ROCE
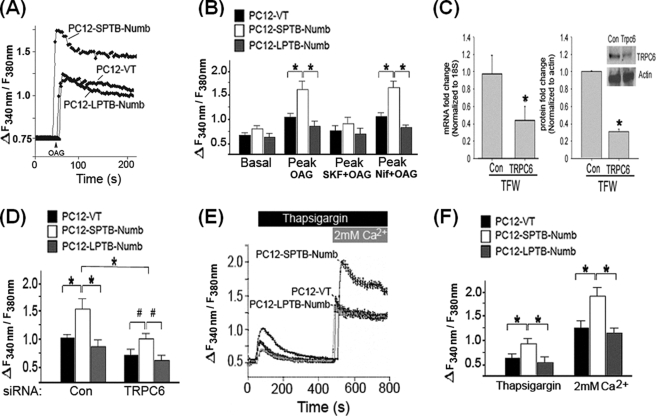
To determine whether Notch-induced expression of TRPC6 differentially impacts Ca2+ entry in PC12 cells, we applied OAG (100 μm), the membrane permeant analogue of diacylglycerol, which induces Ca2+ entry through receptor-operated TRPC3, -6, and -7 (51, 52). Unlike carbachol, OAG activates SOCs without causing ER Ca2+ release that may in turn activate SOCE. PC12-SPTB-Numb exhibited slightly higher resting [Ca2+]i level compared with PC12-LPTB-Numb, but this difference did not reach statistical significance (Fig. 5, A and B). The magnitude of the OAG-evoked Ca2+ transients were significantly greater in PC12-SPTB-Numb when compared with PC12-LPTB-Numb and PC12-VT (Fig. 5, A and B) suggesting that the expressed TPRC proteins assembled into functional ROCs. The OAG-evoked Ca2+ transients were not observed when the cells were bathed in a Ca2+-free medium (data not shown). Furthermore, the OAG-induced Ca2+ responses were unaffected by the L-type voltage-dependent Ca2+ channel blocker nifedipine (Fig. 5B), but they were strongly inhibited by SKF-96365 (Fig. 5B), a commonly used blocker of TRPCs (53).
FIGURE 5.
Numb modulates intracellular calcium homeostasis in an isoform-specific manner. A, representative recordings showing the mean [Ca2+]i before and after the addition of OAG in the indicated PC12 clones. Cells were loaded with fura-2, and [Ca2+]i was recorded as described under “Experimental Procedures.” The arrow indicates time of addition of OAG (100 μm). B, effect of SKF96365 (50 μm) and nifedipine (5 μm) on basal [Ca2+]i and the peak [Ca2+]i after exposure to OAG. Ca2+ measurements are shown as F340/F380 ratios obtained from four separate cultures (30–50 cells assessed/culture); *, p < 0.01 compared with PC12-SPTB-Numb. C, siRNA-mediated knockdown of TRPC6 reduces transcript and protein levels. PC12-SPTB-Numb were transfected with siRNA (100 nm) targeting TRPC6 (siRNA-TRPC6) or non-silencing control siRNA (siRNA-Con) for 24 h. The level of TRPC6 transcript was determined in triplicate by real-time PCR and normalized to the expression of housekeeping gene 18S. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with cultures transfected with siRNA-Con. D, effect of TRPC6 knockdown on the OAG-induced peak of [Ca2+]i. The indicated PC12 clones were transfected with either siRNA-TRPC6 or siRNA-Con (100 nm) for 24 h and the OAG-induced increase in [Ca2+]i was recorded as described under “Experimental Procedures.” Ca2+ measurements are shown as F340/F380 ratios obtained from four separate cultures (30–50 cells assessed/culture); *, p < 0.01; #, p < 0.05 compared with PC12-SPTB-Numb. E, representative recordings showing the mean [Ca2+]i after the sequential addition of 1 μm thapsigargin and 2 mm Ca2+ in the perfusate containing 5 μm nifedipine. The indicated PC12 clones were incubated in Ca2+ free KRB buffer and the change in [Ca2+]i upon the addition of thapsigargin and Ca2+ was recorded as described under “Experimental Procedures.” F, the peak increase of [Ca2+]i after exposure to thapsigargin and re-addition of Ca2+ in the indicated clones of PC12 cells. Ca2+ measurements are shown as F340/F380 ratios obtained from four separate cultures (30–50 cells assessed/culture); *, p < 0.01; #, p < 0.05 compared with PC12-SPTB-Numb.
To verify that TRPC6 was mainly responsible for the OAG-induced Ca2+ transients in PC12-SPTB-Numb, we selectively knocked down TRPC6 expression by RNA interference. qRT-PCR analysis was utilized to assess the efficiency of suppression of TRPC6 expression and to evaluate whether the siRNA construct had any cross-reactivity in terms of suppressing non-targeted TRPC homologs. Transfection of PC12-SPTB-Numb with siRNA-TRPC6 but not siRNA-Con substantially suppressed levels of TRPC6 transcript (Fig. 5C). Knockdown of TRPC6 did not alter the expression of all other TRPC mRNAs (data not shown). The efficiency of suppression of TRPC6 protein was confirmed by immunostaining (supplemental Fig. S4) and immunoblotting (Fig. 5C) and was in the range between 60 and 70% (Fig. 5C). Although TRPC6 knockdown in PC12-SPTB-Numb failed to abrogate OAG-stimulated Ca2+ entry, the OAG-dependent increase of [Ca2+]i was substantially reduced in the presence of siRNA-TRPC6 (Fig. 5D). Hence, despite TRPC6 knockdown, the OAG-stimulated receptor-operated Ca2+ entry (ROCE) activity remained higher in PC12-SPTB-Numb compared with PC12-LPTB-Numb and PC12-VT cells (Fig. 5D), and this may be attributed to the differential expression of TRPC3 (Fig. 4A), which is known to assemble into homomeric ROCs (51, 52). The OAG-stimulated ROCE was slightly but not significantly decreased in PC12-VT and PC12-LPTB-Numb (Fig. 5D), and this may be attributable to siRNA-TRPC6-mediated reduction of basal TRPC6 protein level. Consistent with the notion that Notch signaling modulates TRPC6-mediated ROCE, we found that pretreatment with DAPT at a dose that inhibited TRPC6 expression (Fig. 4C) markedly inhibited OAG- evoked Ca2+ transients in PC12-SPTB-Numb (supplemental Fig. S5A).
Notch-mediated Expression of TRPC6 Augments SOCE
Although TRPC6 overexpression studies uniformly support the formation of ROCs (51, 52), several recent studies suggest a role for TRPC6 in forming native SOCs (54, 55). To test whether the Notch-mediated up-regulation of TRPC6 also enhances SOCE activity, we used the Ca2+ add-back assay previously validated to activate store-operated Ca2+ entry (SOCE) (33). Cultures of PC12 cells were perfused with the irreversible sarco(endo)plasmic reticulum pump inhibitor TG in Ca2+-free medium containing nifedipine to prevent Ca2+ entry through L-type voltage-dependent Ca2+ channels and EGTA to chelate any residual Ca2+ (33). In the absence of external Ca2+, TG-dependent inhibition of Ca2+ sequestration elevated [Ca2+]i due to passive release of Ca2+ from the ER. The TG-induced elevation of [Ca2+]i was significantly higher in PC12-SPTB-Numb (Fig. 5, E and F) suggesting that expression of the Numb proteins differentially modulates refilling of the ER Ca2+ store. Following the return of Ca2+ to basal level, Ca2+ was re-added into the medium to a final concentration of 2.0 mm, and SOCE was evaluated by the increase in [Ca2+]i, reflecting Ca2+ entry into the cytoplasm (Fig. 5E). The initial peak of the increase in [Ca2+]i (increase in fluorescence ratio/min) observed upon the admission of external Ca2+ was significantly greater in PC12-SPTB-Numb compared with PC12-VT and PC12-LPTB-Numb (Fig. 5F). Because the activation of SOCs depends on the filling state of the stores, we next determined whether TG was equally depleting inositol 1,4,5-trisphosphate-sensitive stores in the clones examined. Addition of carbachol following TG treatment had no additional effect on Ca2+ release in all clones suggesting that TG had completely emptied intracellular stores (data not shown). To verify that the response to Ca2+ re-addition reflected Ca2+ influx, we checked whether Mn2+ could quench the fura-2 fluorescence excited at 360 nm (F360) in PC12-SPTB-Numb after perfusion with Ca2+-free KRB buffer containing nifedipine. SOCE was evaluated by measuring the rate at which the fluorescence intensity at 360 nm (F360) was quenched by Mn2+, a Ca2+ surrogate that enters the cells, and reduces fluorescence upon binding to the dye (34). Rates of Mn2+ inflow (decrease in fluorescence units/min) following addition of cyclopiazonic acid, a reversible sarco(endo)plasmic reticulum pump inhibitor, were significantly reduced in the presence of siRNA-TRPC6, suggesting that Mn2+ entry through SOCs was dependent on TRPC6 (supplemental Fig. S6). Treatment with DAPT at the dose that markedly inhibited TRPC6 expression (Fig. 4C) resulted in a significant inhibition of SOCE in PC12-SPTB-Numb cells (supplemental Fig. S5B), suggesting that Notch modulation of TRPC6 expression could also impact SOCE activity. Because TRPC3 expression is not affected by DAPT (Fig. 4D), the residual SOCE activity detected PC12-SPTB-Numb cells following DAPT treatment (supplemental Fig. S5B) could be attributed to TRPC3, because this TRPC is capable of combining with TRPC1 and TRPC7 to form native SOCs (56).
Stress-induced Expression of TRPC6 Is Sensitive to Notch Pathway Modulation
Having shown that overexpression of the Numb proteins in PC12 cells differentially modulates cellular Ca2+ homeostasis via Notch-dependent induction of TRPC6 expression, we next examined whether the stress-induced switch of Numb isoforms (Fig. 1) was sufficient to up-regulate TRPC6 expression in a manner dependent on activation of the Notch pathway. Increased Notch receptor localization to early endosomes was detected prior to accumulation of NICD in the nucleus of naïve PC12 cells subjected to TFW (supplemental Fig. S7). Real-time PCR, immunoblotting, and immunolabeling experiments confirmed the TFW-induced increase of TRPC6 mRNA and protein in PC12 (Fig. 6A). Both TRPC6 transcript and protein were significantly increased after TFW. By contrast, expression of TRPC3 was not substantially altered after TFW (Fig. 6B). The Notch-induced TRPC6 expression in PC12 cells following TFW was associated with an increase in steady-state [Ca2+]i (Fig. 6C). Treatment with siRNA-TRPC6 but not siRNA-Con markedly attenuated TFW-induced elevation of [Ca2+]i in PC12 cells 16 h after TFW (Fig. 6C).
FIGURE 6.
TFW-induced activation of the Notch pathway is sufficient to increase TRPC6-mediated Ca2+ entry. A, time course of TRPC6 transcript and protein levels in cultures of PC12 cells subjected to TFW. The amount of transcript was determined in triplicate by real-time PCR and normalized to the expression of housekeeping gene 18S (left panel). Protein amount was determined by immunoblotting (center panel) and immunostaining (right panel). The increase of TRPC6 protein is shown in the inset. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cultures. B, time course of TRPC3 transcript in cultures of PC12 cells subjected to TFW was determined as described in A. C, cultures of PC12 cells were incubated with siRNA-TRPC6 or siRNA-Con (100 nm) for 24 h prior to TFW. Cells were then loaded with fura-2, and steady-state [Ca2+]i was recorded as described under “Experimental Procedures.” Ca2+ measurements are shown as F340/F380 ratios obtained from four separate cultures (30–50 cells assessed/culture); *, p < 0.05 compared with untreated and TFW-treated cultures incubated with siRNA-TRPC6. D, cultures of PC12 cells were incubated with 20 μm DAPT (+) or vehicle (−) for 24 h prior to TFW. At 12 h following TFW, levels of TRPC6 and actin in cell lysates were determined by immunoblotting. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with untreated cultures and TFW-treated cultures incubated with DAPT. E, cultures of PC12 cells were incubated with 20 μm DAPT (+) or vehicle (−) for 24 h prior to TFW. At 12 h following TFW, levels of the SPTB- and LPTB-Numb transcripts were determined in triplicate by real-time PCR, normalized to the expression of housekeeping gene 18S, and expressed in -fold change relative to the non-treated cultures. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests).
To establish that TFW-induced TRPC6 expression requires activation of the Notch pathway, PC12 cells were treated with DAPT for 2 h prior to TFW. We found that incubation of DAPT at concentrations that significantly blocked TFW-induced Notch activation (Fig. 1C) robustly reduced both transcript and protein level of TRPC6 in PC12 cells after TFW (Fig. 6D). DAPT did not prevent the TFW-induced switch in Numb isoforms (Fig. 6E) further confirming the notion that this switch is an early event (Fig. 2) that precedes Notch activation and, hence, is independent of Notch activation. Treatment of PC12 cells with siRNA-Notch1 (supplemental Fig. S8A) and DAPT (supplemental Fig. S8B) replicated the results obtained with siRNA-TRPC6 (Fig. 6C) further confirming that TFW-induced Notch signaling mediates Ca2+ entry through TRPC6.
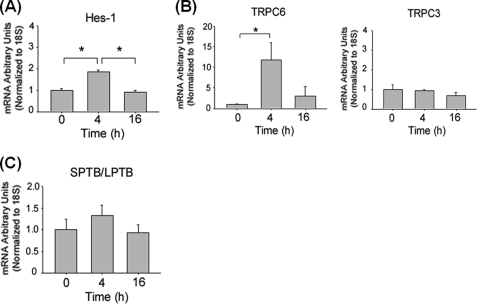
Next, we treated PC12 cells with the Jagged1 peptide ligand to activate Notch signaling (36). The Jagged1 peptide increased the level of Hes-1 transcript confirming activation of the Notch pathway (Fig. 7A). Concomitantly, the Jagged1 peptide increased TRPC6 but not TRCP3 transcripts in PC12 cells (Fig. 7B). Similar to DAPT (Fig. 6E), treatment with the Jagged1 peptide alone did not induce a switch of Numb isoforms (Fig. 7C).
FIGURE 7.
Direct activation of the Notch pathway increases TRPC6 expression. A and B, time course of Hes1, TRPC6, and TRPC3 transcripts in cultures of PC12 cells that were treated with Jagged1 or a scramble peptide (15 μm). The amount of each transcript was determined in triplicate by real-time RT-PCR and normalized to the expression of housekeeping gene 18S. The values are the means ± S.E. of three independent experiments. *, p < 0.01 (ANOVA with Scheffe post-hoc tests) compared with untreated cultures. C, time course of the SPTB- and LPTB-Numb transcripts in cultures of PC12 cells that were treated with either Jagged1 or scramble peptide (15 μm). Transcript levels were determined in triplicate by real-time PCR. The amount of each transcript was normalized to the expression of housekeeping gene 18S and expressed in -fold change relative the scramble peptide-treated cultures.
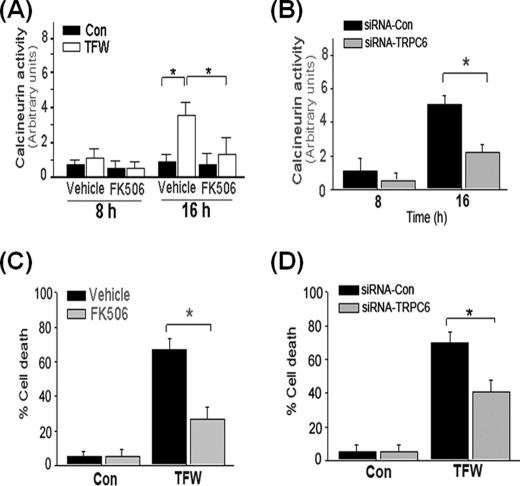
Expression of TRPC6 Increases Cellular Vulnerability to TFW via Activation of Calcineurin
Next we investigated the functional implications of Notch-induced TRPC6 expression. Previous reports suggest a cross-talk of the Notch pathway with calcineurin (57), also known as protein phosphatase 2B (58). Calcineurin is activated by sustained increases in [Ca2+]i and, therefore, serves to couple intracellular Ca2+ to the dephosphorylation of selected substrates such as nuclear factor of activated T cells (58, 59). Calcineurin and FKBP12, the 12-kDa FK506-binding protein, have been shown to form a functional multiprotein complex with TRPC6 (60). To determine whether TFW-induced TRPC6 expression is linked to the activation of the calcineurin pathway, we measured calcineurin activity in PC12 cells. TFW markedly increased calcineurin activity (Fig. 8A), which was reversed by knocking down TRPC6 (Fig. 8B). Prior studies indicated that a high level of calcineurin activity may render neuronal cells susceptible to serum deprivation-induced apoptosis, which was prevented by Bcl-2 overexpression or incubation with cyclosporin A or FK506 (61). FK506 is an immunosuppressant that directly binds to FKBP12 and inhibits formation of complexes with TRPC6 (60). Treatment with FK506 significantly ameliorated TFW-induced activation of calcineurin (Fig. 8A) and protected PC12 cells from TFW-induced death (Fig. 8C). Similarly to FK506, TRPC6 knockdown markedly decreased TFW-induced cell death (Fig. 8D) further establishing that TRPC6 up-regulation is a required step for the Ca2+ dependence of calcineurin activation.
FIGURE 8.
TFW-induced cell death is dependent on TRPC6-mediated activation of calcineurin. A, time course of calcineurin activity in cultures of PC12 cells that were treated with FK506 (1 μm) 1 h prior to TFW. At the indicated time points following TFW, calcineurin activity in cell lysates was measured by a colorimetric assay using the RII phosphopeptide as substrate. Values are shown as arbitrary units and are the means ± S.E. of three independent experiments performed in duplicate. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with the indicated cultures. B, time course of calcineurin activity in cultures of PC12 cells that were treated with siRNA-Con and siRNA-TRPC6 (100 nm) for 18 h prior to TFW. The values are the means ± S.E. of three independent experiments performed in duplicate. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with the indicated cultures. C, cultures of PC12 cells were exposed to the calcineurin inhibitor FK506 (1 μm) 1 h prior to TFW. Cell viability was determined 24 h after TFW by Hoechst staining. Forty fields compromising at least 30–50 cells were counted. Results shown are the means ± S.E. of three independent experiments performed in duplicate. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with vehicle-treated cultures. D, cultures of PC12 cells were exposed to siRNA-Con and siRNA-TRPC6 (100 nm) for 18 h prior to TFW. Cell viability was determined 24 h after TFW by Hoechst staining. The values are the means ± S.E. of three independent experiments performed in duplicate. *, p < 0.01; **, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with the indicated cultures.
Expression of LPTB-Numb Decreases Neuronal Vulnerability to TFW
Because LPTB-Numb exerted opposite action compared with SPTB-Numb, we next determined the role of LPTB-Numb in TFW-induced Notch activation and cell death. Immunoblotting showed that TFW-induced Notch activation is significantly attenuated in PC12-LPTB-Numb compared with PC12-VT (Figs. 9, A and B). PC12-LPTB-Numb were also more resistant to TFW-induced death when compared with PC12-VT (Fig. 9C).
FIGURE 9.
Expression of LPTB-Numb ameliorates TFW-induced Notch activation and cell death. A, representative immunoblots showing the time course of NICD protein level in PC12-VT and PC12-LPTB-Numb clones after TFW. B, the histogram shows the density of the NICD protein band normalized to actin. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with cultures at the indicated time points. C, time course of the TFW-induced cell death in PC12-VT and PC12-LPTB-Numb clones. Cell viability was determined 16 h after TFW by Hoechst staining. Forty fields compromising at least 60 cells were counted. The values are the means ± S.E. of three independent experiments. *, p < 0.05 (ANOVA with Scheffe post-hoc tests) compared with PC12-VT.
DISCUSSION
Beyond their role in stem cell maintenance, Notch and Numb are also essential in the formation of neuronal cell shape and synaptic connection in the developing brain (62, 63). Increased Notch activity was associated with decreased neurite outgrowth, and this effect was reversed by expression of Numb (62). Although both Notch and Numb remain to be expressed in adult tissues, including the brain (64), little is yet known about their interaction and functional roles.
Deficiency of the Notch pathway resulted in specific neurocognitive deficits related to long term memory formation in Drosophila melanogaster and mice (65, 66). A link between Notch and memory deficits is suggested by data showing that the presenilin protein required for proteolytic cleavage of NICD from Notch is mutated in familial Alzheimer disease (45, 46). Recent studies also implicate aberrant Notch signaling in stroke (36), cancers (9, 10), and diabetic nephropathy (67). Increased Notch activation is associated with dendritic atrophy in prion-infected brains of mice (68). Hence, the Notch signaling pathway is tightly regulated both temporally and spatially.
Several HECT-domain E3 ubiquitin-ligases regulate the number and availability of Notch receptors at the cell surface by ubiquitinylating and targeting Notch for degradation (47). The potency of Notch signaling also depends on intrinsic regulators that positively or negatively influence the strength and duration of the Notch signal. Numb is a critical negative regulator of Notch, and Numb deficiency affects the same developmental steps that are physiologically regulated by the Notch pathway. Numb is an endocytic adapter involved in the internalization and recycling of cell surface proteins (42) suggesting that Numb likely influences Notch signaling activity through vesicular trafficking. In the D. melanogaster nervous system, Numb triggers the endocytosis of Sanpodo, a positive regulator of Notch, targeting it to the late endosomes (69).
The role of Numb in regulating Notch activity in vertebrates is less clear. Recent studies showed that endosomal entry is required for Notch activation (70) and that post-endosomal sorting of the Notch receptor plays a role in signal modulation (71). We previously reported that the Numb proteins regulate the post-endosomal sorting of APP (31). This trafficking function of Numb appears to be important in the regulation of APP proteolytic processing and the generation of amyloid β-peptide (Aβ), the principle component of senile plaques in Alzheimer disease. Numb proteins that differ in the PTB domain have opposite effects on the endosomal sorting and processing fate of APP (31). Expression of SPTB-Numb isoforms results in the retention of APP and C-terminal APP fragments (APP-CTFs) within the early endosomes, which in turn favors the production of Aβ locally by γ-secretase. In contrast, expression of LPTB-Numb isoforms reduces the dwelling time of APP in the early endosomes and increases lysosome-mediated turnover of APP, resulting in a substantial reduction in the cellular content of APP and APP-CTFs (31). Collectively, our data suggest that Numb influences post-endocytic sorting and processing fates of APP in a manner dependent on the PTB-domain. Although the detailed molecular mechanism of Numb-mediated post-endocytic trafficking is not known, the insertion in the PTB domain of Numb may induce structural elements that are sufficient to mediate APP recycling and to strongly inhibit post-endocytic degradation.
In the present study we found that the Numb proteins also have disparate consequences for Notch activation by regulating intracellular trafficking of the Notch receptor. By evaluating Notch localization in the various endocytic compartments, we uncovered that, similarly to APP (31), the degree of Notch co-localization with the Rab5-labeled early endosomes is greater in PC12 clones expressing SPTB-Numb compared with those clones expressing LPTB-Numb (Fig. 3C). Notch pathway activity was also robust in clones expressing SPTB-Numb as evident by the elevated steady-state levels of NICD protein (Fig. 2A) and Hes-1 transcript (Fig. 2B), suggesting that Notch retention in the early endosomes promotes γ-secretase cleavage. These findings are consistent with a previous report showing that γ-secretase cleavage of Notch is reduced in D. melanogaster mutants that impair endosomal entry but is enhanced in mutants that increase endosomal retention (70).
Although basal Notch signaling activity in PC12-LPTB-Numb was slightly but not significantly lower compared with PC12-VT (Fig. 2A), the TFW-induced Notch activation was markedly attenuated in PC12-LPTB-Numb compared with PC12-VT (Fig. 9A). It remains to be determined how the LPTB-Numb isoforms suppress TFW-induced Notch activation. LPTB-Numb isoforms may promote the trafficking of the Notch receptor to the degradative pathway because Notch has been shown to be degraded in the lysosomes (71, 72). It is also worth noting that the PTB insertion is required for the recruitment of the HECT-type E3 ligase Itch and for targeting Notch for Itch-dependent ubiquitination and proteasomal degradation (47, 71, 73). Collectively, these data suggest that the Numb proteins differentially influence the endocytic activation of Notch without altering the expression of the components of the Notch pathway.
Several lines of evidence imply that the Numb isoforms have distinct cellular functions (13, 14). Ectopic expression of Numb isoforms without the insertion in the PRR promotes differentiation, whereas those isoforms with the insertion direct the proliferation of neural crest stem cells (13). The Numb proteins also differentially modulate the responses of PC12 cells to the neurotrophic factor nerve growth factor, namely, stimulation of neurite outgrowth and cell survival dependence (31).
Changes in intracellular Ca2+ are known to be involved in key processes during development such as neuronal cell proliferation, differentiation, migration, and cell death (19–21). Because of the interplay between Numb and Notch in various cellular processes important for development, it is tempting to speculate that Notch and Numb modulate these diverse processes, in part, by modifying Ca2+-sensitive signaling pathways. We previously reported that the Numb proteins alter intracellular Ca2+ homeostasis in a manner dependent on the PTB domain and that destabilization of intracellular Ca2+ homeostasis is a critical component of the cell death-promoting effect of SPTB-Numb (16). It is not known how the Numb proteins impact the Ca2+ homeostatic mechanisms that influence neuronal life/death cell fate decisions. In this study, we examined whether alterations in cellular Ca2+ homeostasis may be the direct consequence of the differential modulation of the Notch pathway by the Numb proteins. We found that activation of the Notch pathway is required for the expression of TRPC6 and that the Numb proteins differentially modulate TRPC6 expression by altering the signaling strength of the Notch pathway (Figs. 3 and 4, A–C).
The Notch-induced TRPC6 expression in PC12-SPTB-Numb clones was accompanied by heightened OAG-induced SKF96365-sensitive Ca2+ transients suggesting that the expressed TRPC6 functionally augmented Ca2+ entry (Fig. 5, A and B). Increased TRPC6 expression in PC12-SPTB-Numb clones also augmented SOCE in response to TG-induced store depletion (Fig. 5, E and F). Knockdown of TRPC6 markedly reduced both OAG- and TG-stimulated Ca2+ entry indicating that TRPC6 was mainly responsible for enhanced Ca2+ entry through ROCs (Fig. 5, A and B) and SOCs (Fig. 5, E and F). Pretreatment with DAPT also markedly inhibited the OAG- and TG-stimulated Ca2+ entry (supplemental Fig. S5) consistent with the notion that Notch signaling mediates TRPC6-dependent Ca2+ entry.
The secretase activities involved in Notch processing are also involved in APP processing. As reported previously, the Numb proteins differentially modulate APP processing leading to the accumulation of APP derivatives in PC12-SPTB-Numb but not PC12-LPTB-Numb (31). Both Aβ and APP-CTFs can alter cellular Ca2+ homeostasis (74). The γ-secretase-dependent proteolytic release of the intracellular domain of APP and its subsequent translocation to the nucleus suggest that it plays a signaling role analogous to NICD (75). A functional role for the intracellular domain of APP in regulating phosphoinositide-mediated Ca2+ signaling has been reported (76). To exclude the possibility that the effects of the Numb proteins on cellular Ca2+ homeostasis were mediated by either intracellular domain of APP or Aβ, we treated PC12 cells with a β-secretase inhibitor that blocks the production of these APP derivatives (31). Treatment of PC12-SPTB-Numb with β-secretase inhibitor had no impact on TRPC6 expression in PC12-SPTB-Numb (Fig. 4D) thus excluding a direct role of these derivatives of APP proteolysis in the observed TRPC6-dependent changes in cellular Ca2+ signaling and homeostasis.
Despite their similarity in amino acid sequence, members of the TRPC subfamily have different modes of activation (25, 26). There is still considerable controversy regarding which TRPC proteins form native SOCs or ROCs. Although TRPC6 overexpression studies uniformly support the formation of ROCs, several recent studies suggest a role for TRPC6 in forming native SOCs (54–56). Considering that TRPCs probably function as tetramers, overexpression may result in a predominance of homotetrameric structures, whereas endogenous expression may reflect heteromers between TRPC channel subtypes (25). TRPC6 may form a functional heteromeric complex with TRPC3, which has been implicated as a ROC and SOC (50).
Our data are consistent with the notion that Notch-dependent up-regulation of endogenous TRPC6 can enhance both ROC and SOC function (Fig. 5). While endogenous TRPC6 can elicit a SOCE event in certain cell types under specific conditions, it is also likely that Notch may regulate the expression of additional components required to generate a properly regulated and store-operated TRPC6. We cannot rule out the possibility that increased TRPC6-mediated Ca2+ entry could be in part attributed to changes in the physicochemical characteristics of this channel. These interesting possibilities warrant future investigations.
Both SOCs and ROCs are present in non-excitable and excitable cells such as cardiac and skeletal muscle cells (77) and neurons (78). SOCE may be critical for neuronal migration and neurite outgrowth during cortical development (79) and for synaptic plasticity in the adult brain (80). Each of these processes has been shown to be impacted by altered Numb (31, 43, 44) or Notch function (63, 66), although it is yet not clear how the Numb and Notch interaction influences the downstream signaling mechanisms that regulate these processes.
Calcineurin is a highly conserved serine-threonine protein phosphatase that is present in all tissues in mammals, with notably high levels in brain where it may account for 1% of the total protein (58). Activation of the calcineurin signaling pathway is important in axonal growth and guidance during brain development and in the regulation of synaptic plasticity (81). We found that Notch activation during TFW resulted in the up-regulation of TRCP6 that is mainly responsible to the activation of calcineurin as knockdown of TRPC6 markedly suppressed TFW-induced calcineurin activity (Fig. 8, A and B).
Aberrant activation of calcineurin has been linked to various pathological conditions, including Alzheimer disease and Huntington disease, cardiomyopathy, and stroke (82–84). Deregulated calcineurin activity is implicated in Aβ-mediated inhibition of long term potentiation (77) and can predispose vulnerable neurons to N-methyl-d-aspartic acid-mediated cell death (76). We showed that pharmacological inhibition of calcineurin or knockdown of TRPC6 expression significantly attenuated TFW-induced death (Fig. 8, C and D), although the underlying mechanism remained to be determined. Previous studies demonstrated that aberrant calcineurin activation can prime apoptosis by transcriptional-dependent and -independent mechanisms (85, 86).
Collectively, these findings suggest that the effects of Numb and Notch proteins on cell fate decisions are likely mediated by TRPC6-mediated enhancement of SOC and ROC entries leading to the activation of Ca2+-dependent signaling pathways (Fig. 10). Whether the Notch pathway is responsible for TRPC6 up-regulation in several disease states such as diabetic nephropathy (66), focal segmental glomerulosclerosis (29), and fatal cardiomyopathy (27, 28) require future investigations.
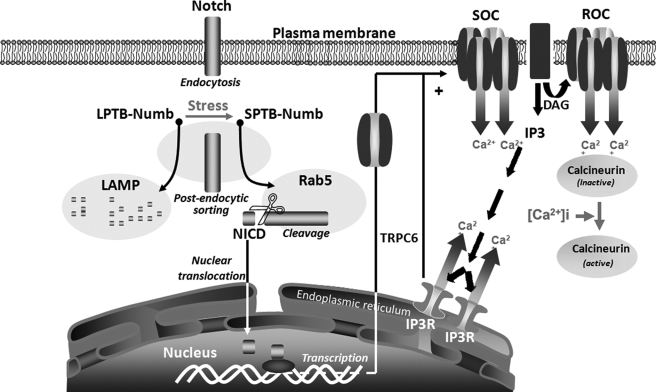
FIGURE 10.
A schematic diagram summarizing the isoform-specific effects of the Numb proteins on the activation of the Notch pathway and the regulation of cellular Ca2+-regulating systems in PC12 cells under stress conditions. In addition to activation of the Notch pathway, trophic factor withdrawal-induced stress promotes an early switch in the expression of the alternatively spliced Numb isoforms that differentially modulate Notch signaling strength. Whereas the LPTB-Numb isoforms reduce the dwelling time of the internalized Notch in Rab5-labeled early endosomes, the SPTB-Numb proteins increase the retention of Notch and its processing to NICD in these endocytic compartments. Consequently, the stress-induced accumulation of Numb-SPTB promotes NICD-dependent transcription of TRPC6, which assembles into native ROCs and SOCs on the plasma membrane. Increased Ca2+ entry through these channels leads to the subsequent activation of calcineurin in cells expressing the SPTB-Numb isoforms.
Supplementary Material
Acknowledgments
We are grateful to Dr. Schilling for providing TRPC antibodies and members of the Chan laboratory for helpful discussions and comments on the manuscript.
This work was supported by start-up funds from the University of Central Florida.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- NICD
- Notch intracellular domain
- APP
- amyloid precursor protein
- APP-CTF
- APP C-terminal fragment
- Aβ
- amyloid β-peptide
- ER
- endoplasmic reticulum
- KRB
- Kreb's Ringer bicarbonate
- PC12
- pheochromocytoma12
- OAG
- 1-oleoyl-2-acetyl-sn-glycerol
- PTB
- phosphotyrosine binding
- PRR
- proline-rich region
- ROC
- receptor-operated channel
- siRNA
- small interfering RNA
- SOC
- store-operated channel
- TFW
- trophic factor withdrawal
- TG
- thapsigargin
- TRPC
- transient receptor potential channel
- DAPT
- N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester
- PBS
- phosphate-buffered saline
- ANOVA
- analysis of variance
- RT
- reverse transcription
- qRT
- quantitative RT
- SPRR
- short PRR
- E3
- ubiquitin-protein isopeptide ligase.
REFERENCES
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Lai E. C. (2002) EMBO Rep. 3, 840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 4.Furukawa T., Maruyama S., Kawaichi M., Honjo T. (1992) Cell 69, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 5.Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999) EMBO J. 18, 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iso T., Kedes L., Hamamori Y. (2003) J. Cell. Physiol. 194, 237–255 [DOI] [PubMed] [Google Scholar]
- 7.Schweisguth F. (2004) Curr. Biol. 14, R129–R138 [PubMed] [Google Scholar]
- 8.Conlon R. A., Reaume A. G., Rossant J. (1995) Development 121, 1533–1545 [DOI] [PubMed] [Google Scholar]
- 9.Stylianou S., Clarke R. B., Brennan K. (2006) Cancer Res. 66, 1517–1525 [DOI] [PubMed] [Google Scholar]
- 10.Miele L., Golde T., Osborne B. (2006) Curr. Mol. Med. 6, 905–918 [DOI] [PubMed] [Google Scholar]
- 11.Rhyu M. S., Jan L. Y., Jan Y. N. (1994) Cell 76, 477–491 [DOI] [PubMed] [Google Scholar]
- 12.Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., Di Fiore P. P. (2004) J. Cell Biol. 167, 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verdi J. M., Schmandt R., Bashirullah A., Jacob S., Salvino R., Craig C. G., Program A. E., Lipshitz H. D., McGlade C. J. (1996) Curr. Biol. 6, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 14.Dho S. E., French M. B., Woods S. A., McGlade C. J. (1999) J. Biol. Chem. 274, 33097–33104 [DOI] [PubMed] [Google Scholar]
- 15.Verdi J. M., Bashirullah A., Goldhawk D. E., Kubu C. J., Jamali M., Meakin S. O., Lipshitz H. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan S. L., Pedersen W. A., Zhu H., Mattson M. P. (2002) Neuromolecular Med. 1, 55–67 [DOI] [PubMed] [Google Scholar]
- 17.Mattson M. P., Cheng B., Davis D., Bryant K., Lieberburg I., Rydel R. E. (1992) J. Neurosci. 12, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattson M. P., Chan S. L. (2001) J. Mol. Neurosci. 17, 205–224 [DOI] [PubMed] [Google Scholar]
- 19.Munaron L., Antoniotti S., Lovisolo D. (2004) J. Cell Mol. Med. 8, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattson M. P., LaFerla F. M., Chan S. L., Leissring M. A., Shepel P. N., Geiger J. D. (2000) Trends Neurosci. 23, 222–229 [DOI] [PubMed] [Google Scholar]
- 21.Berridge M. J., Bootman M. D., Roderick H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 22.Putney J. W., Jr. (1986) Cell Calcium 7, 1–12 [DOI] [PubMed] [Google Scholar]
- 23.Birnbaumer L., Zhu X., Jiang M., Boulay G., Peyton M., Vannier B., Brown D., Platano D., Sadeghi H., Stefani E., Birnbaumer M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15195–15202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulay G., Zhu X., Peyton M., Jiang M., Hurst R., Stefani E., Birnbaumer L. (1997) J. Biol. Chem. 272, 29672–29680 [DOI] [PubMed] [Google Scholar]
- 25.Venkatachalam K., Montell C. (2007) Annu. Rev. Biochem. 76, 387–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clapham D. E., Runnels L. W., Strübing C. (2001) Nat. Rev. Neurosci. 2, 387–396 [DOI] [PubMed] [Google Scholar]
- 27.Kuwahara K., Wang Y., McAnally J., Richardson J. A., Bassel-Duby R., Hill J. A., Olson E. N. (2006) J. Clin. Invest. 116, 3114–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue R., Jensen L. J., Shi J., Morita H., Nishida M., Honda A., Ito Y. (2006) Circ. Res. 99, 119–131 [DOI] [PubMed] [Google Scholar]
- 29.Möller C. C., Wei C., Altintas M. M., Li J., Greka A., Ohse T., Pippin J. W., Rastaldi M. P., Wawersik S., Schiavi S., Henger A., Kretzler M., Shankland S. J., Reiser J. (2007) J. Am. Soc. Nephrol. 18, 29–36 [DOI] [PubMed] [Google Scholar]
- 30.Pedersen W. A., Chan S. L., Zhu H., Abdur-Rahman L. A., Verdi J. M., Mattson M. P. (2002) J. Neurochem. 82, 976–986 [DOI] [PubMed] [Google Scholar]
- 31.Kyriazis G. A., Wei Z., Vandermey M., Jo D. G., Xin O., Mattson M. P., Chan S. L. (2008) J. Biol. Chem. 283, 25492–25502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan S. L., Fu W., Zhang P., Cheng A., Lee J., Kokame K., Mattson M. P. (2004) J. Biol. Chem. 279, 28733–28743 [DOI] [PubMed] [Google Scholar]
- 33.Chan S. L., Liu D., Kyriazis G. A., Bagsiyao P., Ouyang X., Mattson M. P. (2006) J. Biol. Chem. 281, 37391–37403 [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Shimoda L. A., Sylvester J. T. (2004) Am. J. Physiol. Lung Cell Mol. Physiol. 286, L848–858 [DOI] [PubMed] [Google Scholar]
- 35.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 36.Arumugam T. V., Chan S. L., Jo D. G., Yilmaz G., Tang S. C., Cheng A., Gleichmann M., Okun E., Dixit V. D., Chigurupati S., Mughal M. R., Ouyang X., Miele L., Magnus T., Poosala S., Granger D. N., Mattson M. P. (2006) Nat. Med. 12, 621–623 [DOI] [PubMed] [Google Scholar]
- 37.Li S. C., Zwahlen C., Vincent S. J., McGlade C. J., Kay L. E., Pawson T., Forman-Kay J. D. (1998) Nat. Struct. Biol. 5, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 38.Zwahlen C., Li S. C., Kay L. E., Pawson T., Forman-Kay J. D. (2000) EMBO J. 19, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo M., Jan L. Y., Jan Y. N. (1996) Neuron 17, 27–41 [DOI] [PubMed] [Google Scholar]
- 40.Spana E. P., Doe C.Q. (1996) Neuron 17, 21–26 [DOI] [PubMed] [Google Scholar]
- 41.Lindsell C. E., Shawber C. J., Boulter J., Weinmaster G. (1995) Cell 80, 909–917 [DOI] [PubMed] [Google Scholar]
- 42.Santolini E., Puri C., Salcini A. E., Gagliani M. C., Pelicci P. G., Tacchetti C., Di Fiore P. P. (2000) J. Cell Biol. 151, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura T., Kaibuchi K. (2007) Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 44.Wang Z., Sandford S., Wu C., Li S. S. (2009) EMBO J. 28, 2360–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopan R., Goate A. (2000) Genes Dev. 14, 2799–2806 [DOI] [PubMed] [Google Scholar]
- 46.Selkoe D., Kopan R. (2003) Annu. Rev. Neurosci. 26, 565–597 [DOI] [PubMed] [Google Scholar]
- 47.McGill M. A., McGlade C. J. (2003) J. Biol. Chem. 278, 23196–23203 [DOI] [PubMed] [Google Scholar]
- 48.Jafar-Nejad H., Norga K., Bellen H. (2002) Dev. Cell 3, 155–156 [DOI] [PubMed] [Google Scholar]
- 49.Guo Q., Sopher B. L., Furukawa K., Pham D. G., Robinson N., Martin G. M., Mattson M. P. (1997) J. Neurosci. 17, 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tesfai Y., Brereton. H. M., Barritt G. J. (2001) Biochem. J. 358, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofmann T., Obukhov A. G., Schaefer M., Harteneck C., Gudermann T., Schultz G. (1999) Nature 397, 259–263 [DOI] [PubMed] [Google Scholar]
- 52.Dietrich A., Kalwa H., Rost B. R., Gudermann T. (2005) Pflugers Arch. 451, 72–80 [DOI] [PubMed] [Google Scholar]
- 53.Merritt J. E., Armstrong W. P., Benham C. D., Hallam T. J., Jacob R., Jaxa-Chamiec A., Leigh B. K., McCarthy S. A., Moores K. E., Rink T. J. (1990) Biochem. J. 271, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Erxleben C., Yildirim E., Abramowitz J., Armstrong D. L., Birnbaumer L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4682–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bréchard S., Melchior C., Plançon S., Schenten V., Tschirhart E. J. (2008) Cell Calcium 44, 492–506 [DOI] [PubMed] [Google Scholar]
- 56.Zagranichnaya T. K., Wu X., Villereal M. L. (2005) J. Biol. Chem. 280, 29559–29569 [DOI] [PubMed] [Google Scholar]
- 57.Mammucari C., Tommasi di Vignano A., Sharov A. A., Neilson J., Havrda M. C., Roop D. R., Botchkarev V. A., Crabtree G. R., Dotto G. P. (2005) Dev Cell. 8, 665–676 [DOI] [PubMed] [Google Scholar]
- 58.Rusnak F., Mertz P. (2000) Physiol. Rev. 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 59.Hogan P. G., Chen L., Nardone J., Rao A. (2003) Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 60.Kim J. Y., Saffen D. (2005) J. Biol. Chem. 280, 32035–32047 [DOI] [PubMed] [Google Scholar]
- 61.Snyder S. H., Sabatini D. M., Lai M. M., Steiner J. P., Hamilton G. S., Suzdak P. D. (1998) Trends Pharmacol. Sci. 19, 21–26 [DOI] [PubMed] [Google Scholar]
- 62.Sestan N., Artavanis-Tsakonas S., Rakic P. (1999) Science 286, 741–746 [DOI] [PubMed] [Google Scholar]
- 63.Redmond L., Oh S. R., Hicks C., Weinmaster G., Ghosh A. (2000) Nat. Neurosci. 3, 30–40 [DOI] [PubMed] [Google Scholar]
- 64.Roncarati R., Sestan N., Scheinfeld M. H., Berechid B. E., Lopez P. A., Meucci O., McGlade J. C., Rakic P., D'Adamio L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7102–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costa R. M., Honjo T., Silva A. J. (2003) Curr. Biol. 13, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 66.Wang Y., Chan S. L., Miele L., Yao P. J., Mackes J., Ingram D. K., Mattson M. P., Furukawa K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9458–9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niranjan T., Bielesz B., Gruenwald A., Ponda M. P., Kopp J. B., Thomas D. B., Susztak K. (2008) Nat. Med. 14, 290–298 [DOI] [PubMed] [Google Scholar]
- 68.Ishikura N., Clever J. L., Bouzamondo-Bernstein E., Samayoa E., Prusiner S. B., Huang E. J., DeArmond S. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hutterer A., Knoblich J. A. (2005) EMBO Rep. 6, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008) J. Cell Biol. 180, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGill M. A., Dho S. E., Weinmaster G., McGlade C. J. (2009) J. Biol. Chem. 284, 26427–26438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jehn B. M., Dittert I., Beyer S., von der Mark K., Bielke W. (2002) J. Biol. Chem. 277, 8033–8040 [DOI] [PubMed] [Google Scholar]
- 73.Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M. A., Alimandi M., Giannini G., Maroder M., Screpanti I., Gulino A. (2006) Nat. Cell Biol. 8, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 74.Kim H. S., Park C. H., Cha S. H., Lee J. H., Lee S., Kim Y., Rah J. C., Jeong S. J., Suh Y. H. (2000) FASEB J. 14, 1508–1517 [DOI] [PubMed] [Google Scholar]
- 75.Cao X., Südhof T. C. (2001) Science 293, 115–120 [DOI] [PubMed] [Google Scholar]
- 76.Leissring M. A., Murphy M. P., Mead T. R., Akbari Y., Sugarman M. C., Jannatipour M., Anliker B., Müller U., Saftig P., De Strooper B., Wolfe M. S., Golde T. E., LaFerla F. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4697–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uehara A., Yasukochi M., Imanaga I., Nishi M., Takeshima H. (2002) Cell Calcium 31, 89–96 [DOI] [PubMed] [Google Scholar]
- 78.Tu P., Kunert-Keil C., Lucke S., Brinkmeier H., Bouron A. (2009) J. Neurochem. 108, 126–138 [DOI] [PubMed] [Google Scholar]
- 79.Bouron A., Altafaj X., Boisseau S., De Waard M. (2005) Brain Res. Dev. Brain Res. 159, 64–71 [DOI] [PubMed] [Google Scholar]
- 80.Baba A., Yasui T., Fujisawa S., Yamada R. X., Yamada M. K., Nishiyama N., Matsuki N., Ikegaya Y. (2003) J. Neurosci. 23, 7737–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winder D. G., Sweatt J. D. (2001) Nat. Rev. Neurosci. 2, 461–474 [DOI] [PubMed] [Google Scholar]
- 82.Xifró X., García-Martínez J. M., Del Toro D., Alberch J., Pérez-Navarro E. (2008) J. Neurochem. 105, 1596–1612 [DOI] [PubMed] [Google Scholar]
- 83.Dineley K. T., Hogan D., Zhang W. R., Taglialatela G. (2007) Neurobiol. Learn Mem. 88, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharkey J., Butcher S. P. (1994) Nature 371, 336–339 [DOI] [PubMed] [Google Scholar]
- 85.Latinis K. M., Norian L. A., Eliason S. L., Koretzky G. A. (1997) J. Biol. Chem. 272, 31427–31434 [DOI] [PubMed] [Google Scholar]
- 86.Wang H. G., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., Shibasaki F., McKeon F., Bobo T., Franke T. F., Reed J. C. (1999) Science 284, 339–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.