Summary
Voltage-dependent K+ channels play key roles in shaping electrical signaling in both excitable as well as non-excitable cells. These channels open and close in response to the voltage changes across the cell membrane. Many studies have been carried out in order to understand the voltage sensing mechanism. Our laboratory recently determined the atomic structures of a mammalian voltage-dependent K+ channel Kv1.2 and a mutant of Kv1.2 named the ‘paddle-chimera’ channel, in which the voltage sensor paddle was transferred from Kv2.1 to Kv1.2. These two structures provide atomic descriptions of voltage-dependent channels with unprecedented clarity. Until now the functional integrity of these two channels biosynthesized in yeast cells have not been assessed. Here we report the electrophysiological and pharmacological properties of Kv1.2 and the paddle chimera channels in planar lipid bilayers. We demonstrate that Pichia yeast produce ‘normally functioning’ mammalian voltage-dependent K+ channels with qualitatively similar features to the Shaker K+ channel in the absence of the N-terminal inactivation gate, and that the paddle chimera mutant channel functions as well as Kv1.2. We find, however, that in several respects the Kv1.2 channel exhibits functional properties that are distinct from Kv1.2 channels reported in the literature.
Keywords: potassium channels, voltage-dependent gating, voltage sensor paddle, inactivation, planar lipid bilayer
Introduction
For many decades voltage-dependent ion channels have been the subjects of careful study and analysis1. The impetus for their study stems from their central importance to electrical signaling in biology and from the desire to understand how can a protein ‘sense’ the membrane voltage. Our laboratory recently determined the atomic structures of the eukaryotic voltage-dependent K+ (Kv) channel Kv1.2 and a mutant of Kv1.2 named the ‘paddle-chimera’ channel, in which the helix-turn-helix element termed the voltage sensor paddle has been mutated to correspond to that from Kv2.12; 3; 4. Definition of the chimera boundaries was based on the work of Swartz and colleagues5. The structure of the paddle chimera channel was determined at 2.4 Å resolution in a membrane like environment formed by co-crystallization of the ion channel protein, detergent, and lipid molecules. These structures provide atomic descriptions of voltage-dependent channels with unprecedented clarity (Figure 1), but we have yet to assess their functional properties. Since the channels were biosynthesized in yeast cells instead of in their native neuronal environment, and then purified in mixtures of lipid and detergent, we have set out to assess their functional integrity. Electrophysiological and pharmacological properties of Kv1.2 and paddle chimera channels reconstituted into lipid vesicles of defined lipid composition are described here. Both channels are associated with β2 subunits, which are also present in the crystal structures6.
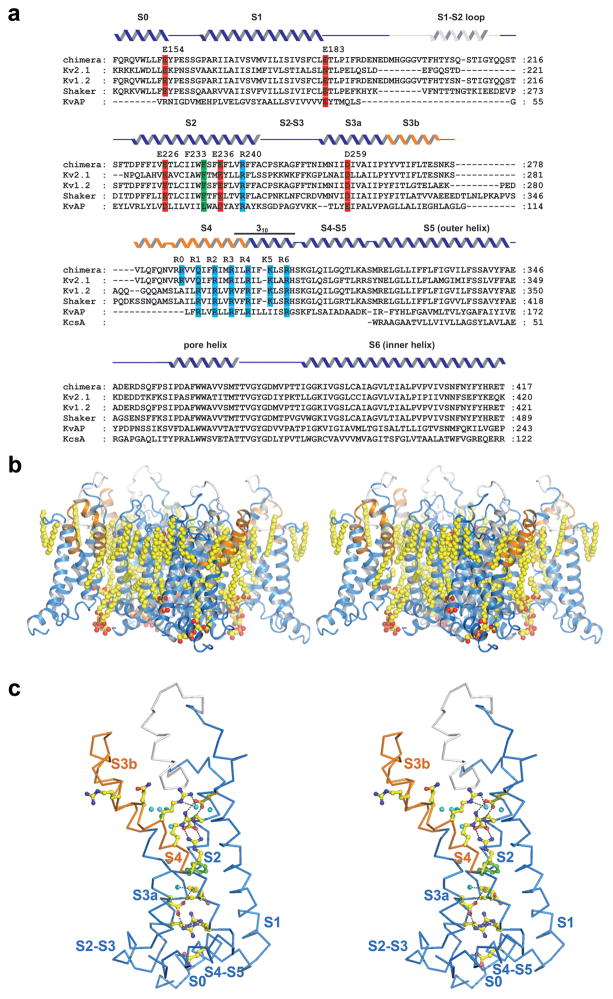
Figure 1.
a, Sequence alignment of the paddle chimera channel, rat Kv2.1 (GI: 24418849), rat Kv1.2 (GI: 1235594), Shaker Kv (GI: 13432103), KvAP (GI: 5104624), and KcsA (GI: 61226909) transmembrane regions. Secondary structure elements are indicated above the sequences: grey, the S1–S2 loop; orange, the S3b-S4 region of Kv2.1 inserted into the Kv1.2 channel (blue). Selected conserved residues are highlighted red (negatively charged), blue (positively charged), and green (phenylalanine 233). b, Side view of the paddle chimera channel transmembrane region (ribbon representation, colored as in a) in stereo, showing the bilayer-like structure formed by the lipid molecules (sphere representation, colored according to atom type: yellow, carbon; red, oxygen; orange, phosphorous, PDB accession code, 2R9R). c, Stereo representation of a voltage sensor and the S4–S5 linker helix of the paddle chimera channel viewed from the pore (α-carbon trace, colored as in a). Side chains of residues highlighted in a are shown as ball and stick and colored according to atom type: yellow, carbon; red, oxygen; blue, nitrogen; green, phenylalanine; cyan, water. Panel b and c are made with PyMOL34.
Results
Voltage-dependent activation
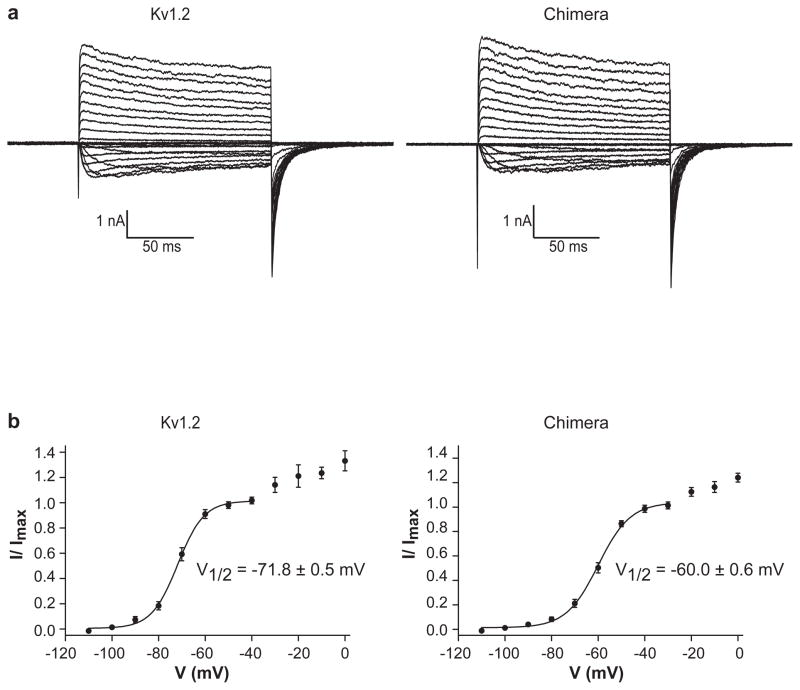
Channels were reconstituted into lipid vesicles composed of 3:1 (w:w) POPE:POPG and fused into planar bilayers of the same lipid composition in decane7. Membranes were held at −110 mV, stepped to more positive voltages in 10 mV increments to open channels, and then back to −110 mV to again close the channels (Figure 2a). The solution on both sides of the membrane contained 150 mM KCl. The direction and magnitude of K+ currents during the depolarizing steps depend on both the channel open probability and the electrochemical driving force for K+, which produces inward (downward) current at negative voltages and outward (upward) current at positive voltages under these ionic conditions. Immediately following the depolarizing step, when the membrane voltage is returned to −110 mV but the channels have not had sufficient time to close, the electrochemical driving force is the same for every trace, but the open probability is not the same because it was determined by the preceding depolarization voltage. The magnitude of inward current at −110 mV, normalized by the maximum value, is graphed as a function of the preceding depolarization voltage and fit to a two-state Boltzmann equation (Figure 2b). Kv1.2 and paddle chimera channels turn on steeply as a function of voltage followed by a shallower increase. The midpoint of the voltage for the steep phase is −72 mV for Kv1.2 and −60 mV for the paddle chimera. The more gradual current increase at more positive depolarization voltages is typical of many Kv channels, such as the Shaker K+ channel and KvAP. These data show that both channels function as strongly voltage-dependent K+ channels.
Figure 2.
Channel activation. a, Currents are shown of Kv1.2 (left) and the paddle chimera channel (right) with CTX-insensitive leak and capacitance currents subtracted. Voltage pulses: holding potential (h.p.) −110 mV, depolarizing steps: −110 mV to +110 mV, ΔV = 10 mV, tail currents measured following a step back to −110 mV. b, The voltage activation curve. Fraction of the maximal current (I/Imax, mean ± s.e.m.) is plotted as a function of the depolarization voltage and fitted with the Boltzmann equation (see methods, n = 9 for Kv1.2 and n = 15 for paddle chimera). Apparent valence Z is 4.6 for Kv1.2 and 3.8 for paddle chimera (calculated at 25 °C)
Inactivation
Many Kv channels undergo inactivation, a phenomenon that refers to spontaneous closure during prolonged depolarization8. There are two types of inactivation processes described so far: N-type and C-type9; 10; 11. The N-type inactivation is generally fast (within a few milliseconds) and involves the cytoplasmic N-terminus of either the alpha subunit or the auxiliary β subunit (β1 subunit), which occludes the pore by entering the intracellular entryway9; 10; 12. The C-type inactivation is generally a slower process and its mechanism is still not well understood.
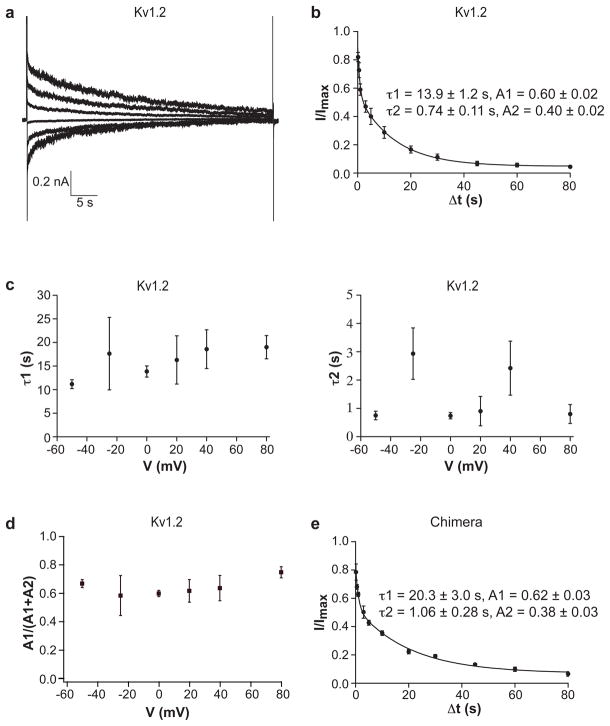
Both Kv1.2 and the paddle chimera channel exhibit inactivation during long depolarizing steps (Figure 3a, the paddle chimera data not shown). The slow rate of inactivation required very long depolarizing steps, which tended to compromise the stability of the planar lipid membranes. In order to follow the time course of inactivation with improved signal relative to background noise, we used a distinct protocol from that of Figure 3a, in which the membrane was held at −110 mV, stepped briefly to +110 mV to record the current level prior to inactivation, then to the test voltage at which inactivation occurred, and finally back to +110 mV to record the fraction of current remaining. Inactivation determined by this pulse protocol, shown in Figure 3b for the Kv1.2 channel, follows a double exponential time course. Variation of the test voltage had little systematic effect on the fast or slow time constants for inactivation (Figure 3c) or the ratio of fast and slow time constants (Figure 3d). The paddle chimera channel also follows a double exponential time course with slightly longer time constants (slower rates) when compared to Kv1.2 (Figure 3e). The slow inactivation rates of Kv1.2 and paddle chimera channels are consistent with the C-type inactivation.
Figure 3.
Channel inactivation. a, A representative trace of Kv1.2 recorded with voltage pulses: h.p. −110 mV, 45 s pulse at voltages ranging from −50 mV to +75 mV in 25 mV steps, stepped back to −110 mV. b, Fraction of non-inactivated currents of Kv1.2 is graphed as a function of inactivation time (inactivation voltage 0 mV). The smooth curve is a fit to a double exponential decay (n = 4). c, The inactivation time constants of Kv1.2 (left, major component τ1; right, minor component τ2) are graphed as a function of the inactivation voltage. d, Fraction of the amplitude of the slow component of Kv1.2 is graphed as a function of the inactivation voltage. e, Fraction of non-inactivated currents of the paddle chimera channel is graphed as a function of inactivation time (inactivation voltage 0 mV). The smooth curve is a fit to a double exponential decay (n = 3).
The time course of recovery from inactivation was monitored by first inactivating the channels with a long depolarizing step to 0 mV, then stepping to −110 mV for variable time intervals, followed by a depolarizing step to measure recovered channels (Figure 4). Kv1.2 and the paddle chimera channels both follow a single exponential recovery time course. The rate of recovery is approximately three times faster for the paddle chimera channel.
Figure 4.
Channel recovery from inactivation. Fraction of recovery as a function of the recovery time (top, Kv1.2 n=3; bottom, paddle chimera n =3) follows a single exponential time course. Data within the first 5 s as well as the time constant obtained from the fitting of all the data are shown as inset.
When a voltage-dependent channel is held at a membrane voltage at which a fraction of the channels open, then inactivation can also occur. The degree of ‘steady-state’ inactivation can be assessed during a depolarizing pulse from different holding voltages. Figure 5a shows that as the holding voltage is made more positive the magnitude of current elicited during a depolarizing step decreases. Figure 5b quantifies this behavior for Kv1.2 and the paddle chimera channels. The midpoint voltage for steady-state inactivation occurs at −91 mV and −83 mV.
Figure 5.
Steady-state inactivation. a, Representative traces of Kv1.2 (left) and the paddle chimera channel (right) recorded with voltage pulses: 30 s at holding voltages ranging from −50 mV to −130 mV (Kv1.2), −50 mV to −150 mV (paddle chimera) in −10 mV steps, stepped to +110 mV and back to −90 mV. 60 s duration prepulses gave similar result to 30 s prepulses. b, Fraction of activatable currents measured in a is graphed as a function of holding voltage (left, Kv1.2 n = 11; right, paddle chimera n = 8). The smooth curves are fits to a two-state Boltzmann equation (see methods). Apparent valence Z is 4.4 for Kv1.2 and 3.3 for paddle chimera (calculated at 25 °C).
Sensitivity to CTX and VsTx1
Kv channels are targets for natural toxins of venomous animals. Scorpions produce protein toxins that plug the extracellular pore entryway13. Spiders produce voltage sensor toxins that interfere with voltage-dependent gating by binding to the voltage sensor14; 15. Charybdotoxin (CTX) is the archetypal pore-blocking scorpion toxin16; 17; 18. It binds to the pore with a stoichiometry of one toxin molecule per channel tetramer. Kv1.2 and the paddle chimera channels are inhibited by CTX with inhibition constants of 25 nM and 19 nM, respectively (Figure 6). The very similar affinities are consistent with the fact that Kv1.2 and paddle chimera differ only in the paddle region of the voltage sensors, which means their pores have identical amino acid sequences.
Figure 6.
CTX inhibition. Fraction of unblocked current (I/Imax, mean ± s.e.m, n= 3 for Kv1.2 and n= 5 for paddle chimera) is graphed as a function of log(CTX concentration) and fitted with equation I/Imax = (1 + [CTX]/Kd)−1. Voltage pulses: h.p. −110 mV, depolarized to +110 mV followed by a step back to −110 mV.
VsTx1 was first described as an inhibitor of the archaebacterial Kv channel KvAP7; 19. Mutational analysis has shown that VsTx1 targets the lipid-exposed face of the voltage sensor paddle in a chimera in which the voltage sensor paddle from KvAP was transferred to Kv2.15. Therefore VsTx1, like hanatoxin (HaTx) from the same tarantula species, interferes with gating by binding to the voltage sensor paddle. The stoichiometry for inhibition is one toxin per subunit, which corresponds to four toxin molecules per channel at saturating concentrations of toxin (one toxin molecule on each voltage sensor)15.
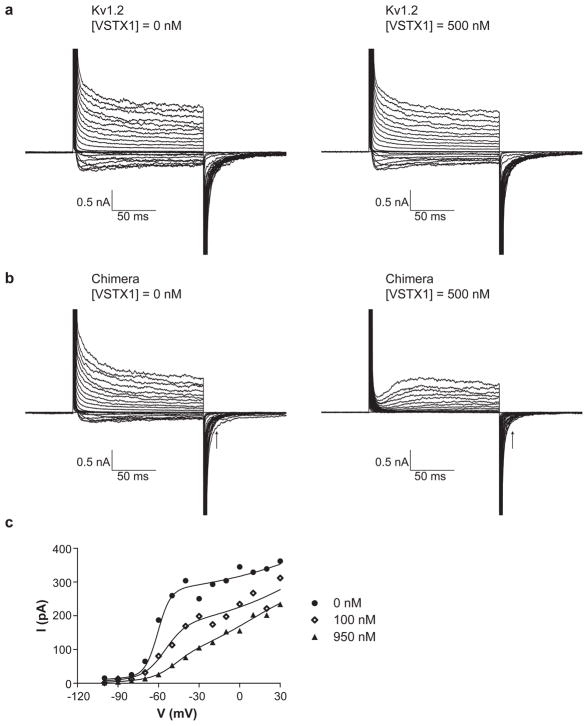
For some Kv channels voltage sensor toxins result in disappearance of current, whereas for other Kv channels the voltage activation curve is shifted and other kinetic aspects of the gating are affected but the current does not disappear7; 15. Figure 7 shows current traces for Kv1.2 and paddle chimera channels in the absence and presence of VsTx1. It is evident that Kv1.2 is relatively insensitive to VsTx1, whereas the paddle chimera channel is sensitive. The paddle chimera channel is not strongly inhibited (i.e. the current level does not disappear), but the rate of voltage-dependent activation is dramatically slowed and the activation curve is shifted toward more positive voltages (Figure 7c). That VsTx1 modifies gating of the paddle chimera channel in these experiments is unexpected since VsTx1 was discovered as an inhibitor of the KvAP channel and has been found to be ineffective against Kv2.1 channels, at least when they are expressed in Xenopus oocytes 5; 7. The effect of VsTx1 on the paddle chimera channel in planar lipid bilayers is qualitatively similar to the effect that hanatoxin has on Kv2.1 channels expressed in Xenopus oocytes15. The only amino acid compositional difference (other than the four cysteine to serine mutations which have essentially no effect on the function of Kv1.2 in lipid bilayers, see methods) between Kv1.2 and paddle chimera occurs within the voltage sensor paddle. Therefore, the differential sensitivities of Kv1.2 and paddle chimera are consistent with the voltage sensor paddle being the target of VsTx1.
Figure 7.
Effect of VsTx1. a, Current traces of Kv1.2 in the absence and presence of VsTx1 (500 nM). b, Current traces of the paddle chimera channel in the absence and presence of VsTx1 (500 nM). Pulse protocols are the same as in Figure 2a. c, Tail currents in the absence and presence of 100 nM and 950 nM VsTx1 are plotted as a function of the depolarization voltage. The smooth curves are fits to a sum of two two-state Boltzmann equations (0 nM and 100 nM VsTx1) and three two-state Boltzmann equations (950 nM VsTx1). Voltage pulses: h.p. −100 mV, depolarizing steps: −100 mV to +100 mV, ΔV = 10 mV, tail currents measured at 15 ms (indicated by the arrow) following a step back to −100 mV.
Discussion
The crystal structures of Kv1.2 and the paddle chimera channels are the only two structures of eukaryotic Kv channels determined so far and they have provided significant insights into the underlying molecular mechanism of voltage-dependent gating2; 3; 4. Moreover, the paddle chimera crystal structure, determined only recently, was crystallized with lipid molecules that form a visible bilayer structure around the channel, creating a native-like environment (Figure 1b). What has been missing so far is a functional characterization of these channels in order to address the following questions: do yeast produce ‘normally functioning’ Kv channels encoded by genes of mammalian origin, is the paddle chimera mutant a properly functioning Kv channel, and what do the functional properties tell us about the conformational state of the channel in the crystal structures?
These data show that both Kv1.2 and the paddle chimera channel produced in Pichia yeast, purified and reconstituted into lipid vesicles of POPE and POPG, produce functional voltage-dependent channels in planar bilayers of the same lipid composition. Both channels exhibit the characteristic features of neuronal delayed rectifier K+ channels: voltage-dependent activation upon depolarization followed by slow inactivation, presumably C-type. The channels are qualitatively similar to Shaker Kv channels following removal of the N-terminal inactivation gate10. The main differences between Kv1.2, paddle chimera and Shaker are quantitative (i.e. they exhibit different values of the midpoint activation voltage, absolute rates of activation and inactivation, and sensitivity to pharmacological reagents). We conclude that Pichia yeast produce ‘normally functioning’ mammalian Kv channels and that the paddle chimera mutant functions as well as Kv1.2. The similarity of Kv1.2 and paddle chimera to the Shaker channel in terms of both amino acid sequence and function suggest that their atomic structures represent valid models with which functional data on the Shaker channel can be interpreted.
What do the electrophysiological properties tell us about the conformation of Kv1.2 and the paddle chimera in the crystal structures? The channels were crystallized in the absence of a transmembrane voltage. The electrophysiological data in Figure 2 inform us that at zero mV we should expect the voltage sensors to be in their open (meaning ‘up’ or ‘depolarized’) conformation. Based on the electrophysiological data in Figure 3 we should also expect the pore’s activation gate (the inner helix bundle) to be open, but that the inactivation gate (see below) should be closed since at zero mV the channel conducts transiently and then inactivates over a period of many seconds4; 20; 21.
How does the atomic structure compare with the above expectations? The conformation of the voltage sensor in the crystal structures is consistent with the open state, or a conformation very similar to the open state, because the arginine residues that carry most of the gating charge are near the extracellular surface of the channel2; 3; 4; 22; 23. The pore’s activation gate (inner helix bundle) is also open: the inner helix bundle is sufficiently wide to allow passage of a fully hydrated K+ ion4; 20; 21. Therefore, regarding the voltage sensor and the pore’s activation gate, there seems to be good agreement between electrophysiological states of the channel at zero mV and protein conformations observed in the crystal structure.
With respect to inactivation the agreement between electrophysiological and structural data is not so clear. Many studies support the hypothesis that C-type inactivation involves a conformational change within the selectivity filter 24; 25. From the crystal structures of KcsA in complex with Fab fragments and the paddle chimera channel we can conclude that the selectivity filter main chain structures are essentially identical in conformation within a few tenths of an angstrom4; 26. Added to this comparison, Perozo and colleagues have provided electrophysiological evidence that the selectivity filter of the KcsA complex with Fab fragments is conductive27. Thus, we should conclude that the paddle chimera crystal structure does not reflect an inactivated conformation, even though the electrophysiological data inform us that at zero mV the channel should inactivate. We suggest three possible explanations for this apparent conflict. First, the conditions of the crystal may be sufficiently different than those of the electrophysiological experiment so that the inactivated conformation is not favored at zero mV. Second, conformational changes associated with inactivation may be so subtle as to be not apparent in the crystal structure (i.e. the non-inactivated and inactivated states look almost identical). Third, C-type inactivation does not have the same structural basis in all Kv channels: our expectations based on studies of other Kv channels may not apply to the paddle chimera and Kv1.2 channels. Regarding the mechanisms of C-type inactivation we would conclude that there are many uncertainties and we do not understand the process.
Because Kv1.2 and paddle chimera differ mainly in the amino acid sequence of the voltage sensor paddle (see methods), it is interesting to ask how do these differences affect function? On first inspection, given that the voltage sensor paddles differ at 27 positions along the sequence, what seems most surprising is how similar the channels are to each other in gating properties. Many of the amino acid differences occur within the extracellular loop between S3b and S4, but still, many occur within the structured helical regions: in S3b the sequence is PYYVTIFLTE for paddle chimera and PYFITLGTEL for Kv1.2 and in S4 the sequence is VVQIFRIMRILR for paddle chimera and ILRVIRLVRVFR for Kv1.2. The hydrophobic versus polar nature of a given amino acid position tends to be conserved, especially for amino acids within the membrane. However, there is enough chemical variation to expect large functional differences especially when one considers that many rather conservative point mutations in Kv channels have large functional effects. Why might the effects on gating be relatively small? The crystal structures of Kv1.2 and paddle chimera offer a possible answer. The voltage sensor paddle has the interesting structural property of being tilted away from other helices in the channel (Figure 1b, c). Consequently, the only direct contacts between the voltage sensor paddle and the S1–S2 half of the voltage sensor are through the third and fourth arginine residues, which form ionized hydrogen bonds with acidic amino acids forming what we call the external negative cluster4. In other words the structure implies, at least for the open conformation, that a complementary protein-protein interaction surface does not exist between the voltage sensor paddle and other regions of the channel. If this is also true for the closed conformation then it would help explain why Kv1.2 and paddle chimera have very similar functional properties. We note in contrast to the variation of amino acid sequence of the voltage sensor paddle, there is a high degree of sequence identity when comparing Kv1.2 and Kv2.1 in the region of S4 ‘below’ the tilted paddle, where contacts with other protein surfaces are extensive (sequence IFKLSRH) (Figure 1).
The ‘normally functioning’ Kv1.2 channels in this study are actually quite dramatically different in many respects from Kv1.2 channels reported in the literature, expressed in Xenopus oocytes28. For example, the activation midpoint of Kv1.2 is −72 mV in the present study and −3.5 mV in oocytes28. The rate of activation in oocytes is slow compared to the present study to the extent that the currents are unrecognizable as being carried by the same channels. Perhaps what is most surprising is the demonstration here that the paddle chimera is sensitive to VsTx1. VsTx1 was discovered as an inhibitor of KvAP7; 19. This toxin does not inhibit Kv2.1 (in Xenopus oocytes), and yet it does inhibit the paddle chimera, which contains the Kv2.1 voltage sensor paddle5. Therefore Kv1.2 and paddle chimera produced in Pichia yeast and reconstituted into planar lipid bilayers produce functional voltage-dependent K+ channels, but many of their properties do not match our expectation based on past studies in Xenopus oocytes. There are several possible reasons for this. In the present study the Kv channels were associated with a β subunit and in the oocyte studies they were not. However, it is unlikely that the β subunit accounts for the differences because, despite great efforts to discover ways in which β subunits influence Kv channels, so far they have only been shown to influence N-type inactivation rates and expression or current levels29; 30; 31. Another possibility is that channel proteins produced in yeast are not identical to those produced in oocytes, perhaps due to post-translational modifications. Finally, there might be something different about the cellular system compared to the bilayer, either in the cytoplasm or in the membrane, which influences the function of the channel. Further studies will be needed to understand the differences.
Methods
Protein preparation
Kv1.2 and the paddle chimera channels were purified as previously described2; 4, with minor modifications. In brief, the two channels were co-expressed with rat β2-core gene in Pichia pastoris and extracted with DDM (n-dodecyl-β-D-maltopyranoside, Anatrace), and purified by the cobalt affinity column. The elutions were then concentrated and further purified on a Superdex-200 gel filtration column. The gel filtration buffer contained 20 mM Tris-HCl pH 7.5, 150 mM KCl, 6 mM DM (n-decyl-β-D-maltopyranoside, Anatrace), 1 mM EDTA (ethylenediaminetetraacetic acid), 2 mM TCEP (Tris[2-carboxyethyl] phosphine), 10 mM DTT (dithiothreitol) and 0.1 mg/ml lipid −3:1:1 (mass ratio) POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine):POPE (1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine):POPG (1-palmitoyl-2-oleoyl-glycero-3-phospho-1-glycerol, Anatrace).
The two channels in these studies contain two amino acid substitutions (L15H and N207Q). In addition, four amino acid substitutions were made to reduce sensitivity to oxidation (C31S, C32S, C435S and C482S) in the paddle chimera channel. These four mutations do not affect the function of Kv1.2 in the POPE:POPG lipid bilayers (data not shown).
Channel reconstitution
Purified channels were reconstituted into OM (octyl-β-D-maltopyranoside, Anatrace) solubilized 3:1 (w:w) POPE:POPG lipid vesicles as described4. Detergents were removed by dialyzing against reconstitution buffer consisting of 10 mM HEPES pH 7.5, 450 mM KCl with 2 mM freshly added DTT at 4°C for 4–6 days. The reconstituted channels were flash-frozen in liquid nitrogen and stored at −80°C. PAGE analysis of reconstituted channels confirms the presence of both α and β subunits (data not shown) in the membranes.
Electrophysiology
Bilayer experiments were performed as previously described7; 32. In brief, planar lipid bilayers of 20 mg/ml 3:1 (w:w) POPE:POPG in decane were formed by painting over a 300-μM hole in a polystyrene partition separating two chambers. Vesicles were added to the chamber (cis) containing 4 ml of 150 mM KCl and 10 mM HEPES pH 7.5. The other chamber (trans) contained 3 ml of 15 mM KCl and 10 mM HEPES pH 7.5. Before measurements, 135 mM KCl was added to the trans chamber to equalize the K+ concentration. All the measurements were performed using the voltage-clamp method in whole-cell mode. An axopatch 200B amplifier, a Digidata 1322A analogue to digital converter, and Axoclamp software (Axon patch) were used to control membrane voltage and record current. Analogue data were filtered at 1 kHz (lowpass Bessel filter) and digitized at 10 kHz for Figures 2, 5, 6, and 7, and 1–10 kHz for Figures 3 and 4 (due to the long pulses and instrument limitation). CTX was used to block channels in one direction while channels from the other direction were being recorded. CTX-insensitive leak and capacitance currents were subtracted from Figure 2a. All electrophysiological recordings were performed at room temperature. All statistical fits and figure plotting were done using GraphPad Prism version 5.01 software33.
The voltage-dependent activation and steady-state inactivation data were fitted with the Boltzmann equation:
where I/Imax is the fraction of the maximal current, V is the depolarization voltage to open the channels, V1/2 is the voltage at which the channels have reached 50% of their maximal current, F is the Faraday constant, R is the gas constant, T is the absolute temperature, and Z is the apparent valence of voltage dependence.
The CTX inhibition data were fitted with the equation:
where I/Imax is the fraction of unblocked current, [CTX] is the concentration of CTX, Kd is the CTX concentration at which 50% of the maximal current is blocked.
Acknowledgments
We thank Alice L. MacKinnon for providing CTX and VsTx1, Stephen B. Long for help and advice on biochemistry and reconstitution of the channels, Joel Butterwick and Anirban Banerjee for comments on the manuscript. R.M. is an investigator in the Howard Hughes Medical Institute. This work was supported by NIH grant (GM43949) to R.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates, Inc; Sunderland, MA: 2001. [Google Scholar]
- 2.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 3.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–8. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 4.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–82. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 5.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–5. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K + channel beta subunit. cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 7.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–5. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 8.Connor JA, Stevens CF. Inward and delayed outward membrane currents in isolated neural somata under voltage clamp. J Physiol. 1971;213:1–19. doi: 10.1113/jphysiol.1971.sp009364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–71. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 10.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–8. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 11.Hoshi T, Zagotta WN, Aldrich RW. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–56. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- 12.Heinemann S, Rettig J, Scott V, Parcej DN, Lorra C, Dolly J, Pongs O. The inactivation behaviour of voltage-gated K-channels may be determined by association of alpha- and beta-subunits. J Physiol Paris. 1994;88:173–80. doi: 10.1016/0928-4257(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 13.Garcia ML, Gao Y, McManus OB, Kaczorowski GJ. Potassium channels: from scorpion venoms to high-resolution structure. Toxicon. 2001;39:739–48. doi: 10.1016/s0041-0101(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 14.Swartz KJ, MacKinnon R. Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron. 1997;18:675–82. doi: 10.1016/s0896-6273(00)80307-4. [DOI] [PubMed] [Google Scholar]
- 15.Swartz KJ, MacKinnon R. Hanatoxin modifies the gating of a voltage-dependent K+ channel through multiple binding sites. Neuron. 1997;18:665–73. doi: 10.1016/s0896-6273(00)80306-2. [DOI] [PubMed] [Google Scholar]
- 16.Miller C, Moczydlowski E, Latorre R, Phillips M. Charybdotoxin, a protein inhibitor of single Ca2+-activated K+ channels from mammalian skeletal muscle. Nature. 1985;313:316–8. doi: 10.1038/313316a0. [DOI] [PubMed] [Google Scholar]
- 17.MacKinnon R, Miller C. Mechanism of charybdotoxin block of the high-conductance, Ca2+-activated K+ channel. J Gen Physiol. 1988;91:335–49. doi: 10.1085/jgp.91.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia ML, Knaus HG, Munujos P, Slaughter RS, Kaczorowski GJ. Charybdotoxin and its effects on potassium channels. Am J Physiol. 1995;269:C1–10. doi: 10.1152/ajpcell.1995.269.1.C1. [DOI] [PubMed] [Google Scholar]
- 19.Ruta V, MacKinnon R. Localization of the voltage-sensor toxin receptor on KvAP. Biochemistry. 2004;43:10071–9. doi: 10.1021/bi049463y. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;417:523–6. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 21.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–77. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 23.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–67. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 24.Yellen G, Sodickson D, Chen TY, Jurman ME. An engineered cysteine in the external mouth of a K+ channel allows inactivation to be modulated by metal binding. Biophys J. 1994;66:1068–75. doi: 10.1016/S0006-3495(94)80888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Jurman ME, Yellen G. Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron. 1996;16:859–67. doi: 10.1016/s0896-6273(00)80106-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–8. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 27.Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, Perozo E. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–8. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 28.Koopmann R, Scholle A, Ludwig J, Leicher T, Zimmer T, Pongs O, Benndorf K. Role of the S2 and S3 segment in determining the activation kinetics in Kv2.1 channels. J Membr Biol. 2001;182:49–59. doi: 10.1007/s00232-001-0029-x. [DOI] [PubMed] [Google Scholar]
- 29.Shi G, Nakahira K, Hammond S, Rhodes KJ, Schechter LE, Trimmer JS. Beta subunits promote K + channel surface expression through effects early in biosynthesis. Neuron. 1996;16:843–852. doi: 10.1016/s0896-6273(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 30.Lazaroff MA, Taylor AD, Ribera AB. In vivo analysis of Kvbeta2 function in Xenopus embryonic myocytes. J Physiol. 2002;541:673–83. doi: 10.1113/jphysiol.2002.016568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional Coupling between the Kv1.1 Channel and Aldoketoreductase Kv{beta}1. J Biol Chem. 2008;283:8634–42. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C. Ion Channel Reconstitution. Plenum Press; New York: 1986. [Google Scholar]
- 33.Software G. GraphPad Prism version 5.01 for Windows. San Diego, California, USA: http://www.graphpad.com. [Google Scholar]
- 34.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. http://www.pymol.org. [Google Scholar]