Abstract
Plastid genomes of the grasses (Poaceae) are unusual in their organization and rates of sequence evolution. There has been a recent surge in the availability of grass plastid genome sequences, but a comprehensive comparative analysis of genome evolution has not been performed that includes any related families in the Poales. We report on the plastid genome of Typha latifolia, the first non-grass Poales sequenced to date, and we present comparisons of genome organization and sequence evolution within Poales. Our results confirm that grass plastid genomes exhibit acceleration in both genomic rearrangements and nucleotide substitutions. Poaceae have multiple structural rearrangements, including three inversions, three genes losses (accD, ycf1, ycf2), intron losses in two genes (clpP, rpoC1), and expansion of the inverted repeat (IR) into both large and small single-copy regions. These rearrangements are restricted to the Poaceae, and IR expansion into the small single-copy region correlates with the phylogeny of the family. Comparisons of 73 protein-coding genes for 47 angiosperms including nine Poaceae genera confirm that the branch leading to Poaceae has significantly accelerated rates of change relative to other monocots and angiosperms. Furthermore, rates of sequence evolution within grasses are lower, indicating a deceleration during diversification of the family. Overall there is a strong correlation between accelerated rates of genomic rearrangements and nucleotide substitutions in Poaceae, a phenomenon that has been noted recently throughout angiosperms. The cause of the correlation is unknown, but faulty DNA repair has been suggested in other systems including bacterial and animal mitochondrial genomes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-009-9317-3) contains supplementary material, which is available to authorized users.
Keywords: Plastid genomics, Molecular evolution, Poales, Poaceae, Grass genomes, Typha latifolia
Introduction
The monocot order Poales comprises 16 families and approximately 18,000 species (sensu APG II 2003), and relationships among families are generally well-resolved and supported (Chase 2004; Graham et al. 2006). The largest family within the order, Poaceae (the grasses), has been the focus of many biological studies due to its ecological, economical, and evolutionary importance. Poaceae include species that are the primary source of nutrition for humans and grazing animals, e.g., wheat (Triticum aestivum), maize (Zea mays), rice (Oryza sativa), rye (Lolium perenne), oats (Avena sativa), sorghum (Sorghum bicolor), and barley (Hordeum vulgare). Grasses have also received much attention as sources of biofuels (Carpita and McCann 2008; Rubin 2008). Furthermore, there is considerable interest in using plastid genetic engineering in the grasses for crop improvement and for producing biopharmaceuticals and vaccines (Verma and Daniell 2007).
With its relatively small nuclear genome size and a high degree of gene synteny with other major cereal grasses, rice is commonly used as the model monocot plant system. The first draft of the rice nuclear genome (Indica group) was made available just two years after the completion of the Arabidopsis genome (Yu et al. 2002). Sequencing of six additional grass nuclear genomes is in progress, and these include Brachypodium, maize, rice, foxtail millet (Setaria italica), sorghum, switchgrass (Panicum virgatum), and wheat (Garvin et al. 2008; Rubin 2008). In addition, plastid genome sequences are currently available for 13 grass genera; Brassicaceae is the only family that is as densely sampled (13 total). Unlike the highly conserved sequences of Brassicaceae, Poaceae plastid genomes have experienced several evolutionary phenomena, including accelerated rates of sequence evolution, gene and intron loss, and genomic rearrangements. For these reasons, Poaceae provide an excellent system to examine plastid genome evolution.
The plastid genomes of land plants are generally highly conserved in terms of gene content, order, and organization (Bock 2007; Palmer 1991; Raubeson and Jansen 2005). The genome is circular with a quadripartite structure composed of two copies of a large inverted repeat (IR) separated by large and small single-copy regions (LSC and SSC, respectively). These genomes usually range in size from 100 to 200 kb and contain 100–130 different genes. The majority of the genes, approximately 80, encode proteins involved in photosynthesis and gene expression and the remaining code for tRNAs and rRNAs. While rates of nucleotide substitutions are low in plastid genomes relative to nuclear genomes, a few lineages have experienced rate acceleration. Plastid genomes in the flowering plant family Geraniaceae exhibit extreme rate heterogeneity, and ribosomal protein, RNA polymerase, and ATPase genes were shown to evolve more rapidly than photosynthetic genes (Guisinger et al. 2008). Aside from this recent example, the first and best documented example of rate heterogeneity among photosynthetic angiosperm plastid genomes occurs for grass lineages (Gaut et al. 1993). Notably, the long-branch leading to the grasses has been shown to impede phylogenetic inference (Soltis and Soltis 2004; Stefanovic et al. 2004), although Leebens-Mack et al. (2005) improved relationship resolution among angiosperms using increased taxon sampling. This and other studies emphasize the importance of taxon sampling in phylogenetic and comparative genomic studies in order to accurately infer molecular evolutionary relationships, rates, and patterns (reviewed in Heath et al. 2008).
Previous studies used methods that do not detect the extent of rate acceleration and genome evolution in grasses; relative rate tests limited the number of taxa to three that were examined (Gaut et al. 1993; Muse and Gaut 1994) or non-Poales sequences were used as outgroups (Bortiri et al. 2008; Chang et al. 2006; Matsuoka et al. 2002). More comprehensive analyses using additional Poales plastid genome sequences are needed to better understand the patterns and causes of genome evolution in this group. There are three major goals in the current study. First, we present the complete plastid genome sequence of Typha latifolia L. (Typhaceae), the first non-grass Poales sequenced to date. Second, we characterize Poales genome organization and evolution using nine fully annotated grass plastid genomes. Third, we examine rates and patterns of sequence evolution within and between grasses relative to other monocot and angiosperm plastid genomes. Jansen et al. (2007) described a positive correlation between genomic changes (gene/intron loss and gene order changes) and lineage-specific branch length. In the current study, we use a genome-wide approach to test the degree and nature of rate acceleration, and we specifically examine genomic rearrangements and substitution patterns for the branch leading to the grasses.
Materials and Methods
DNA Source, Plastid Isolation, Genome Amplification, and Sequencing
Leaf material of T. latifolia was field collected in Arizona (R.C. Haberle 188, Arizona, Yavapai Co., TEX). Plastids were isolated from 21 g of fresh leaves using the sucrose-gradient method (Palmer 1986), as modified by Jansen et al. (2005). They were then lysed and the entire plastid genome was amplified by rolling circle amplification (RCA), using the REPLI-g™ whole genome amplification kit (Qiagen Inc., Valencia, CA, USA) following the methods outlined in Jansen et al. (2005). The RCA product was then digested with the restriction enzymes EcoRI and BstBI, and the resulting fragments were separated in a 1% agarose gel to determine the quality of plastid DNA. The RCA product was sheared by serial passage through a narrow aperture using a Hydroshear device (Gene Machines, San Carlos, CA, USA), and the resulting fragments were enzymatically repaired to blunt ends, gel purified, and ligated into pUC18 plasmids. The clones were introduced into Escherichia coli by electroporation, plated onto nutrient agar with antibiotic selection, and grown overnight. Colonies were randomly selected and robotically processed through RCA of plasmid clones, sequencing reactions using BigDye chemistry (Applied Biosystems, Foster City, CA, USA), reaction cleanup using solid-phase reversible immobilization, and sequencing using an ABI 3730 XL automated DNA sequencer were performed. Detailed protocols are available at http://www.jgi.doe.gov/sequencing/index.html.
Genome Assembly and Annotation
Sequences from randomly chosen clones were processed using PHRED and assembled based on overlapping sequence into a draft genome sequence using PHRAP (Ewing and Green 1998; Ewing et al. 1998). Quality of the sequence and assembly was verified using Consed (Gordon et al. 1998). In most regions of the genome, we had 6–12-fold coverage, but there were a few areas with gaps or low depth of coverage. PCR and sequencing at the University of Texas at Austin were used to bridge gaps and fill in areas of low coverage in the genome. Additional sequences were added until a completely contiguous consensus was created representing the entire plastid genome with a minimum of 2X coverage and a consensus quality score of Q40 or greater. The genome was annotated using DOGMA (Dual Organellar GenoMe Annotator, http://dogma.ccbb.utexas.edu; Wyman et al. 2004).
Comparisons of Gene Content and Gene Order
Gene content comparisons were performed using Multipipmaker (Schwartz et al. 2003). Comparisons involved 10 Poales genomes, including Typha latifolia (current study) and nine grasses: Agrostis stolonifera (NC_008591), Brachypodium distachyon (NC_011032), Hordeum vulgare subsp. vulgare (NC_008590), Lolium perenne (NC_009950), Oryza sativa (NC_001320), Saccharum officinarum (NC_006084), Sorghum bicolor (NC_008602), Triticum aestivum (NC_002762), and Zea mays (NC_001666). T. latifolia was used as the reference genome by including an exon file in the analysis. Gene orders were examined by pair-wise comparisons between all 10 genomes using PipMaker (Elnitski et al. 2002).
Genome and Gene Sampling
Phylogenetic and evolutionary rate comparisons were performed for a total of 47 taxa (Supplementary Table S1), including nine grasses, one other member of the Poales (T. latifolia, current study), and representatives from all major angiosperm clades. The plastid genome of O. sativa Indica group is not completely annotated and was not included in our analyses. In addition, the genomes of Festuca (NC_011713), Coix (NC_013273), and two recently published bamboos (Wu et al. 2009) were not included, because they were not publicly available at the time our comparisons were performed. Our analyses included seven non-Poales monocot genome sequences. Nonetheless, we chose not to include Phalaenopsis, because we wanted to include as many protein-coding genes in our analyses as possible, and all 11 ndh-genes have been lost from this genome (Chang et al. 2006). Protein-coding sequences for 73 genes were used with several exclusions (Supplementary Table S2).
Phylogenetic Analyses
Amino acid sequences were aligned using Multiple Sequence Web viewer and Alignment Tool (MSWAT, http://mswat.ccbb.utexas.edu) and manually adjusted, and the amino acid alignment was used to constrain the nucleotide alignment. Maximum parsimony (MP) and maximum likelihood (ML) analyses were performed using PAUP* version 4.10b10 (Swofford 2003) and GARLI version 0.942 (Zwickl 2006), respectively. MP analyses were performed with 100 random addition replicates and TBR branch swapping with the Multrees option. Non-parametric bootstrap analyses were performed for 100 replicates with 1 random addition replicate and TBR branch swapping with the Multrees option. Four independent ML analyses were performed using GARLI under the default settings, and bootstrap values were generated for 100 replicates and the default settings. Likelihood scores were obtained from PAUP*, because it is better at optimizing branch lengths on the final topology (Zwickl 2006).
Evolutionary Rate Estimation
The program codeml implemented in the software package PAML (Yang 2007) was used to estimate dN, dS, and dN/dS. The ML tree generated above was used as a constraint tree, but branch lengths were generated in PAML. Control files were used with the following settings: CodonFreq = 2 (codon frequency model F3x4), NSsites = 0 (no variation among sites for ω), cleandata = 1 (exclude gapped regions), fix_kappa = 0 (kappa to be estimated), and fix_omega = 0 (omega to be estimated). Using the method of Yang (1998), values of dN, dS, and dN/dS were generated. The null model (H0), where dN/dS was averaged and fixed across all taxa, was compared to two alternative models (H1 and H2). The H1 model allowed for two values of dN/dS across the tree: (1) dN/dS for the lineage leading to the grasses and (2) dN/dS for all other taxa. The H2 model allowed for three values of dN/dS across the tree; (1) dN/dS for the lineage leading to the grasses, (2) dN/dS for all other monocot branches, and (3) dN/dS for all other angiosperms. Likelihood ratio tests were used to test the fit of alternative models and model improvement, and correction for multiple comparisons used the Holm’s method (i.e., sequential Bonferroni correction; Holm 1979) and the false discovery rate method (Benjamini and Hochberg 1995). Gene groups were categorized according to gene function or subunits that form a functional complex; values were, respectively, combined for atp-, ndh-, pet-, psa-, psb-, rpl-, rps-, and rpo-genes, according to previous studies (Chang et al. 2006; Guisinger et al. 2008; Matsuoka et al. 2002). Statistical analyses were conducted using the R software package (http://www.r-project.org), and correction for multiple comparisons was employed using Holm’s method (1979).
Results
Size, Gene Content, and Organization of the Typha Plastid Genome
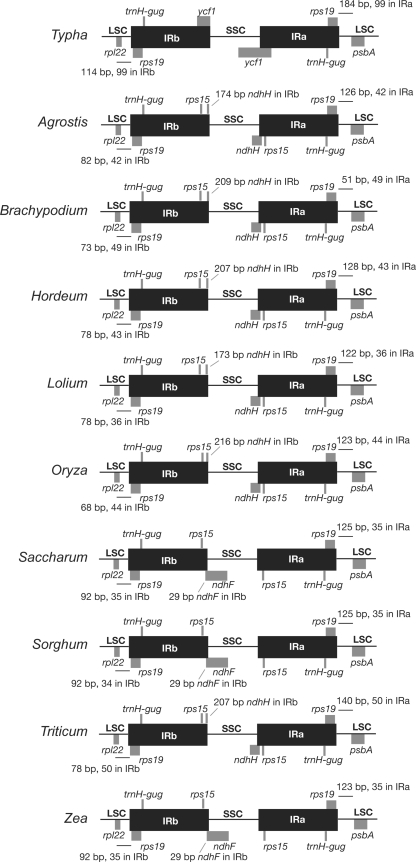
The complete T. latifolia plastid genome is 161,572 base pairs (bp) in length (Supplementary Fig. S1, Table 1, GenBank accession number GU195652). Each IR is 26,390 bp, and the two IR copies are separated by a LSC region of 89,140 bp and a SSC region of 19,652 bp. There are 131 predicted coding regions, 113 of which are different, and 18 that are duplicated in the IR. The coding regions include 79 protein-coding genes, 30 tRNAs, and 4 rRNAs. The T. latifolia plastid genome has 57.1% coding sequence and a 33.8% GC content. Eighteen genes contain introns, including 12 protein-coding genes and 6 tRNAs. The IRs on the LSC boundaries include the duplication of trnH-gug and rps19 and extend 99 bp into the intergenic spacer regions between rps19 and psbA on the IRa/LSC boundary and rps19 and rpl22 on the IRb/LSC boundary (Fig. 1, Supplementary Fig. S1).
Table 1.
Comparison of major features of Typha and nine grass plastid genomes
| Typha | Agrostis | Brachypodium | Hordeum | Lolium | Oryza | Saccharum | Sorghum | Triticum | Zea | |
|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | 161,572 | 136,584 | 135,199 | 136,462 | 135,282 | 134,525 | 141,182 | 140,754 | 134,545 | 140,384 |
| LSC length (bp) | 89,140 | 80,546 | 79,447 | 80,600 | 79,972 | 80,592 | 83,048 | 82,688 | 80,348 | 82,352 |
| SSC length (bp) | 19,652 | 12,740 | 12,668 | 12,704 | 12,428 | 12,335 | 12,544 | 12,502 | 12,791 | 12,536 |
| IR length (bp) | 26,390 | 21,649 | 21,542 | 21,579 | 21,441 | 20,799 | 22,795 | 22,782 | 20,703 | 22,748 |
| Total number of genesa | 131 | 128d | 128e | 128d | 128 | 128 | 128 | 128d | 128 | 128 |
| Number of genes duplicated in IRb | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 |
| Number of genes | ||||||||||
| With introns | 18 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| % GC content | 33.8 | 37.4 | 37.5 | 37.3 | 37.2 | 37.9 | 37.4 | 37.4 | 37.2 | 37.4 |
| % Coding3 | 57.1 | 53.6 | 55.5 | 56.7 | 55.1 | 55.6 | 54.0 | 52.1% | 55.5 | 54.6 |
aOnly includes named genes and ycfs, not orfs; both ycf15 and ycf68 were not included based on comparisons reported in Raubeson et al. (2007)
bDoes not include rps12, which is split between the LSC and IR
cIncludes protein-coding genes, tRNAs, and rRNAs
dAnnotations for Agrostis stolonifera (NC_008591), Hordeum vulgare subsp. vulgare (NC_008590), Sorghum bicolor (NC_008602) in Saski et al. (2007) incorrectly included two extra tRNAs (trnM-cau between trnG-ucc and trnT-ggu; trnfM-cau between trnR-ucu and rps14). These have been deleted in the calculation of number of genes
eBortiri et al. (2008) reported 136 putatively functional genes; the difference in the number reported here is due to annotation errors in the Brachypodium distachyon (NC_011032) genome sequence. These have been deleted in the calculation of number of genes
Fig. 1.
Extent of the inverted repeat (IR) in 10 Poales plastid genomes. Selected genes or portions of genes are indicated by gray boxes above or below the genome. Gene and IR lengths are not to scale (see Table 1 for Poales IR lengths)
Comparisons of Genome Organization of Poales
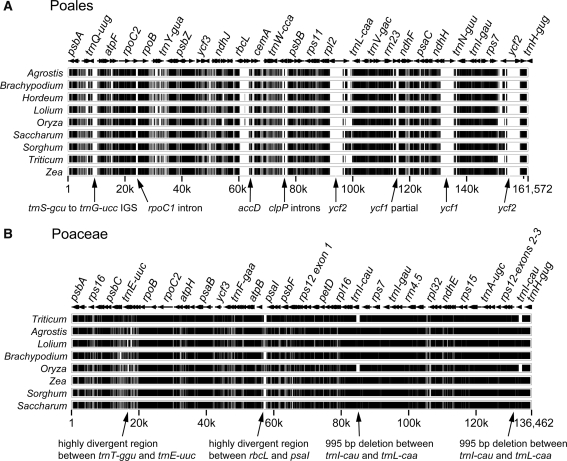
The complete plastid genome sequences for nine genera of grasses (Asano et al. 2004; Bortiri et al. 2008; Hiratsuka et al. 1989; Maier et al. 1995; Ogihara et al. 2000; Saski et al. 2005) and T. latifolia (current study) enable a comparison of genome organization for two families of Poales (Poaceae and Typhaceae, Table 1). Gene and intron content among all 10 genomes are highly conserved with five differences between the grasses and T. latifolia. Relative to the early diverging Poales T. latifolia, all grasses have lost introns in clpP and rpoC1, as well as the three genes accD, ycf1, and ycf2 (Fig. 2a). In the case of gene losses, there has been a progressive degradation of the gene sequences, because differing lengths of residual sequence remain in several taxa (see arrows in Fig. 2a; Table 2; Supplementary Fig. S2). This is especially evident for ycf2, where the first 200 bp of the gene is present and conserved in all of the grasses, and additional remnants of the last 2,300 bp remain in various grasses. The length of residual ycf2 sequence varies from 698 bp in Oryza to 2,089 bp in Zea. The remnant sequences are conserved with pair-wise divergence ranging from 15.0 to 19.1% relative to T. latifolia. In the case of ycf1, the first 250 bp and the final 700 bp are present and conserved across the grasses. Unlike ycf2, the length of residual ycf1 sequence is conserved, ranging from 837 to 867 bp but with higher levels of sequence divergence (23.4–25.6%). The pattern for accD is quite different, because there are few if any small remnants of this gene in grass plastid genomes, and those that do remain are more divergent (Table 2).
Fig. 2.
Multipip analyses (Schwartz et al. 2003) showing overall sequence similarity of plastid genomes based on complete genome alignment. Levels of sequence similarity are indicated by black (75–100%), gray (50–75%), and white (<50%). a Comparison of 10 members of Poales, using Typha latifolia as the reference genome. Arrows indicate gene/intron losses and deletions; partial duplication of ycf1 is due to IR expansion. b Comparison of nine Poaceae genomes using Hordeum vulgare as the reference genome. Arrows indicate deletions; 995 bp deletion is present twice because it is in the IR
Table 2.
Variation in accD, ycf1, and ycf2 in Poales (length in bp/percent divergence relative to Typha)
| Taxon | accD | ycf1 | ycf2 |
|---|---|---|---|
| Typha | 1509 bp | 5508 bp | 6882 bp |
| Agrostis | 0 bp | 851 bp/24.3% | 1380 bp/15.0% |
| Brachypodium | 0 bp | 851 bp//24.1% | 1412 bp/16.4% |
| Hordeum | 134 bp/30% | 855 bp/24.0% | 1413 bp/16.3% |
| Lolium | 132 bp/32.5% | 845 bp/23.4% | 1314 bp/16.5% |
| Oryza | 253 bp/29.2% | 845 bp/24.3% | 698 bp/17.5% |
| Saccharum | 0 bp | 863 bp/25.0% | 2063 bp/18.3% |
| Sorghum | 0 bp | 863 bp/25.1% | 2061 bp/18.0% |
| Triticum | 0 bp | 837 bp/24.1% | 704 bp/17.6% |
| Zea | 0 bp | 867 bp/25.6% | 2089 bp/19.1% |
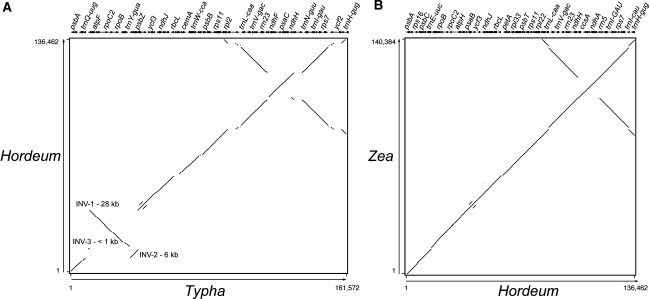
Gene order between the grasses and T. latifolia differs as a result of three inversions of 28, 6, and <1 kb (Fig. 3a). These inversions have been known for 20 years from both gene mapping and genome sequencing (Doyle et al. 1992; Hiratsuka et al. 1989; Howe et al. 1988; Katayama and Ogihara 1993), and the two larger inversions overlap, making it possible to determine that the 28 kb inversion occurred prior to the 6 kb inversion (Doyle et al. 1992; Hiratsuka et al. 1989). Within the grasses, gene order is identical in all sequenced plastid genomes (Fig. 3b).
Fig. 3.
Percent identity plot (Elnitski et al. 2002). a Typha latifolia compared to Hordeum vulgare. Numbers along the x-axis indicate the coordinates for Typha and along the y-axis for Hordeum. INV inversion. b Hordeum vulgare compared to Zea mays. Numbers along the x-axis indicate the coordinates for Hordeum and along the y-axis for Zea
Alignment of the complete plastid genomes indicates that there is a high level of sequence divergence between T. latifolia and the nine grasses (Fig. 2a). Most of the divergent regions (shown in gray or white in Fig. 2a) represent the intron and gene losses, and intergenic regions are the least conserved, containing a few highly divergent regions with large indels (shown in gray or white Fig. 2b). Sequence conservation among the nine grasses is much higher, and again intergenic spacer regions are the most divergent and contain a few large indels (shown in gray or white in Fig. 2b).
The IR in plastid genomes has four boundaries, IRb/LSC, IRb/SSC, IRa/LSC, and IRa/SSC, and there is variation in the extent of duplication of sequences at each of these boundaries in the Poales. All members of the order have expanded the IRb/LSC and IRa/LSC to add both trnH-gug and rps19 to the IR (Fig. 1). However, the extent of IR expansion into the intergenic spacer regions between rps19 to psbA and rps19 to rpl22 varies from 34 to 99 bp among members of the Poales (Fig. 1). At the IR/SSC boundary, the grasses have expanded the IR to duplicate rps15, but expansion beyond rps15 varies within the family. In six of the nine genera, IRa has expanded to duplicate 173–209 bp of ndhH, whereas in the three genera Saccharum, Sorghum, and Zea, IRb has expanded to duplicate 29 bp of ndhF (Fig. 1).
Phylogenetic Relationships
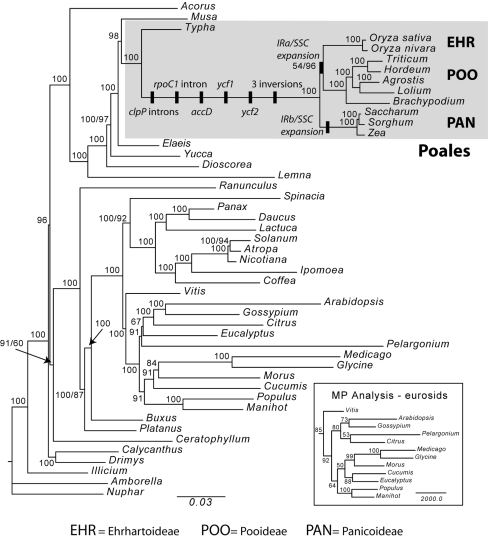
Phylogenetic analyses were performed on an aligned data matrix that included 47 taxa of angiosperms and 73 protein-coding genes. The total length of the aligned data set was 57,603 nucleotides and the Nexus file is available at http://www.biosci.utexas.edu/IB/faculty/jansen/lab/research/data_files/JME-Poales.nex.htm. The MP analysis generated one most parsimonious tree with a length of 104,284, a consistency index (excluding uninformative characters) of 0.36, and a retention index of 0.62. The ML analysis resulted in a tree with –lnL = 568622.59691. The ML and MP trees were largely congruent with each other and with recent phylogenetic analyses based on complete plastid genomes (Jansen et al. 2007; Moore et al. 2007). The only topological differences occurred in the eurosid clade, and these were relatively minor (see inset in Fig. 4). There was strong bootstrap support in all but one node. Further description of results will be limited to monocots and especially Poales. There is strong support for the monophyly of monocots (100% bootstrap values in both ML and MP trees). Acorus is the earliest diverging lineage followed by Lemna, Dioscorea, Yucca, Elaeis, Musa, and finally the Poales, represented by T. latifolia (Typhaceae) and the nine genera of grasses (Poaceae). All of these monocot nodes have bootstrap values >95% and most are 100% in both ML and MP trees. Within grasses all nodes except for one have strong bootstrap support of 100% in both ML and MP trees. The nine genera represent 3 of the 12 recognized subfamilies (sensu GPWG 2001) of grasses (Ehrhartoideae, Panicoideae, and Pooideae), and the monophyly of each is strongly supported. The Ehrhartoideae are sister to the Pooideae, although support for this relationship is weak (54%) in the ML tree.
Fig. 4.
ML tree of 47 taxa for 73 protein-coding genes (−lnL = 568622.59691). MP analysis was generally congruent, but topological differences are shown in the inset. Bootstrap values are shown at nodes for ML/MP; and only one statistic is reported where values are the same except in the eurosid clade where ML values are shown on the full tree and MP values are on the inset. The Poales clade is shaded and genomic changes within Poales are indicated by black bars. Subfamilies sampled are shown (EHR Ehrhartoideae, POO Pooideae, PAN Panicoideae)
The distribution of 10 plastid structural rearrangements in the Poaceae is plotted on the ML tree (Fig. 4). Eight of these rearrangements (intron losses from two genes, three gene losses, and three inversions) occur on the branch leading to the grasses. The other two changes involve small expansions of the IR/SSC boundaries. The first IR expansion on the IRb/SSC boundary has duplicated 29 bp of ndhF, and this change is restricted to members of the subfamily Panicoideae. The second IR expansion occurs in the IRb/SSC boundary and has resulted in duplication of 173–209 bp of ndhH. This structural change provides further support for the sister relationship between the subfamilies Ehrhartoideae and Pooideae. The ycf2 gene has retained various sized remnant fragments ranging from 698 to 2,089 bp (Table 2). The distribution of the sizes of these remnants is congruent with the phylogenetic tree; the largest fragments are in the early diverging Panicoideae lineage, and smaller fragments are present in the subfamilies Ehrhartoideae and Pooideae (Fig. 4).
Evolutionary Rate Comparisons
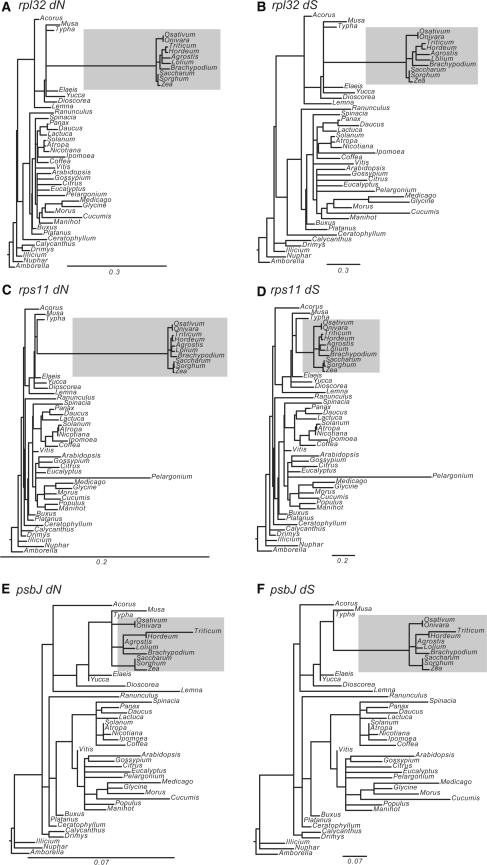
Values of dN/dS, dN, and dS were compared within grasses and between grasses and other angiosperm plastid genomes. Individual gene trees indicate a rapid acceleration of nucleotide substitutions for the branch leading to the grasses (Fig. 5a–c, f). Wilcoxon rank sum tests were used to determine that over all gene types values of dN and dS (both P < 0.0001) are significantly different for the branch leading to the grasses relative to all other branches (Table 3). However, values of dN/dS over all gene types are not significantly different (P = 0.0984). The phylogenetic trees in Fig. 5a–f illustrate the degree of substitution variability for the branch leading to the grasses relative to other angiosperm branches. Both dN and dS are highly accelerated in the gene rpl32 (a, b), dN is high for the gene rps11 (c, d), and dS is relatively high for the gene psbJ (e, f).
Fig. 5.
Sample trees from codeml analyses showing rate acceleration (dN or dS) for three plastid genes. a, b Large subunit ribosomal protein L32. c, d Small subunit ribosomal protein S11. e, f Photosystem II protein J. The Poaceae clade is shaded
Table 3.
Branch comparisons of dN/dS, dN, and dS for gene groups
| Gene groups | Branch comparisons | dN/dS | dN | dS |
|---|---|---|---|---|
| All 73 genes | Branch leading to Poaceae vs. all other branches | 0.0984 | <0.0001** | <0.0001** |
| Branch leading to Poaceae vs. internal Poaceae | 0.0872 | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. non-Poaceae monocots | 0.0904 | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. other angiosperms | 0.1103 | <0.0001** | <0.0001** | |
| Internal Poaceae vs. non-Poaceae monocots | na | <0.0001** | <0.0001** | |
| Internal Poaceae vs. other angiosperms | 0.6577 | <0.0001** | <0.0001** | |
| Non-Poaceae monocots vs. other angiosperms | 0.7123 | 0.8426 | 0.6613 | |
| Photosynthetic apparatus (psa-, psb-, and pet-genes) | Branch leading to Poaceae vs. all other branches | 0.0001** | 0.2746 | <0.0001** |
| Branch leading to Poaceae vs. internal Poaceae | 0.0001** | 0.0053* | <0.0001** | |
| Branch leading to Poaceae vs. non-Poaceae monocots | 0.0001** | 0.6835 | <0.0001** | |
| Branch leading to Poaceae vs. other angiosperms | 0.0001** | 0.4589 | <0.0001** | |
| Internal Poaceae vs. non-Poaceae monocots | na | <0.0001** | <0.0001** | |
| Internal Poaceae vs. other angiosperms | 0.0116 | <0.0001** | <0.0001** | |
| Non-Poaceae monocots vs. other angiosperms | 0.0223 | 0.2702 | 0.9622 | |
| Genes expression (rpl-, rps, and rpo-genes) | Branch leading to Poaceae vs. all other branches | <0.0001** | <0.0001** | <0.0001** |
| Branch leading to Poaceae vs. internal Poaceae | <0.0001** | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. non-Poaceae monocots | <0.0001** | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. other angiosperms | <0.0001** | <0.0001** | <0.0001** | |
| Internal Poaceae vs. non-Poaceae monocots | na | <0.0001** | <0.0001** | |
| Internal Poaceae vs. other angiosperms | <0.0001** | <0.0001** | <0.0001** | |
| Non-Poaceae monocots vs. other angiosperms | <0.0001** | 0.1513 | 0.5967 | |
| Photosynthetic metabolism (atp- and ndh-genes) | Branch leading to Poaceae vs. all other branches | 0.0123 | <0.0001** | <0.0001** |
| Branch leading to Poaceae vs. internal Poaceae | 0.0411 | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. non-Poaceae monocots | 0.0427 | <0.0001** | <0.0001** | |
| Branch leading to Poaceae vs. other angiosperms | 0.0065 | <0.0001** | <0.0001** | |
| Internal Poaceae vs. non-Poaceae monocots | na | <0.0001** | <0.0001** | |
| Internal Poaceae vs. other angiosperms | 0.1186 | <0.0001** | <0.0001** | |
| Non-Poaceae monocots vs. other angiosperms | 0.1574 | 0.7511 | 0.8547 |
P-values were generated using Wilcoxon rank sums tests and asterisks show significant values after correction for multiple comparisons using Holm’s method, i.e., sequential Bonferroni correction (** α = 0.01, * α = 0.05). The value ‘na’ is due to model parameters in PAML analyses
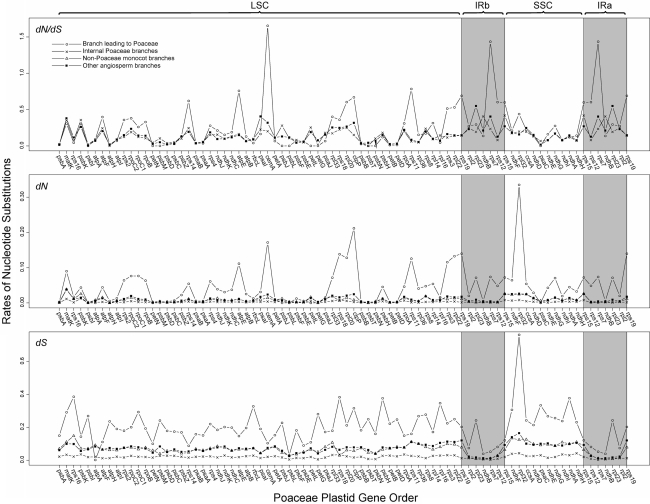
To better understand rates and patterns of nucleotide substitutions, average dN/dS, dN, and dS values per gene were plotted across the length of the plastid genome using the grass gene order (Fig. 6). Values for the branch leading to Poaceae, internal Poaceae branches, non-Poaceae monocot branches, and other angiosperm branches were compared. In general, substitution rates for the branch leading to grasses (shown as circles) are high relative to rates for all other branches. Although dN/dS and dN are highly variable across the genome, dS is broadly accelerated relative to values from other branches. For both dN and dS, values for the internal Poaceae branches (shown as “x”s) are lower across the genome relative to other branches. Aside from the branch leading to Poaceae, rates of sequence evolution in the IR region are low, a phenomenon previously described by Wolfe et al. (1987). Notably, for the branch leading to the grasses, the genes cemA and rps7 exhibit dN/dS ratios greater than 1 (1.66 and 1.44, respectively); however, the raw values of dN and dS are not out of line. Likelihood ratio tests were used to test the fit of a null model to two alternative models (Supplementary Table S3). Significant improvement in likelihood scores was found for a number of genes, notably ATPase, ribosomal protein, and RNA polymerase genes.
Fig. 6.
Average dN/dS, dN, and dS values per gene plotted across the length of the plastid genome using the grass gene order. Values for the branch leading to Poaceae (circles), internal Poaceae branches (“x”s), non-Poaceae monocot branches (triangles), and other angiosperm branches (squares) were compared. For values of dN/dS, black squares show both non-Poaceae monocot and other angiosperm branches due to PAML model parameters. Note that the scales are different for dN/dS, dN, and dS plots
Modest rate heterogeneity among gene types in angiosperm plastid genomes has been previously described (Gaut et al. 1993; Logacheva et al. 2007), and we used Wilcoxon rank sum tests to compare rates among branches for all genes and for genes encoding subunits of the photosynthetic apparatus, genes involved in gene expression, and genes involved in metabolism (Table 3). For the ratio dN/dS, the branch leading to Poaceae is elevated for subunits of the photosynthetic apparatus relative to all other branches, internal Poaceae, non-Poaceae monocot, and other angiosperm branches (all P = 0.0001). Values of dN/dS for genes involved in gene expression are significantly different in all comparisons; the result “na” is due to model parameters in PAML analyses. For subunits of the photosynthetic apparatus, dN is significantly different for the internal Poaceae branches relative to non-Poaceae monocots and other angiosperm branches (P < 0.0001). Values of dN are highly variable for genes involved in gene expression and metabolism (Table 3), and P-values are less than 0.0001 in all but two comparisons. A similar trend was found for values of dS for all gene groups; values of dS are significantly different (P < 0.0001) in all but four comparisons (Table 3). For all gene types, rates of sequence evolution are significantly similar in the non-Poaceae monocots and other angiosperm branches, and this likely reflects the relative extent of rate homogeneity among the majority of angiosperm plastid genomes.
The degree of rate acceleration for the branch leading to Poaceae for individual genes and gene groups was estimated (Tables 4, 5 6). These results show that the branch leading to Poaceae is not significantly elevated for values of dN/dS (Table 4). However, values of dN for the branch leading to Poaceae are highly accelerated relative to internal Poaceae branches (P < 0.001) and moderately accelerated relative to the non-Poaceae monocot branches (P < 0.0056) (Table 5). The greatest degree of dN increase was found for the individual genes clpP and cemA, but ATPase (atp) and ribosomal protein genes (both large and small subunit; rpl and rps, respectively) are high relative to photosynthetic genes (psa, psb, and pet) (Table 5). In terms of values of dS, the branch leading to Poaceae is significantly accelerated relative to all other branches, and the degree of increase is consistent among genes and gene types (Table 6). For values of both dN and dS, the internal Poaceae branches indicate a strong degree of sequence conservation, and the branch leading to Poaceae is evolving on average 10- to 20-fold faster (P < 0.001) than internal Poaceae branches.
Table 4.
The degree of rate acceleration for the ratio of nonsynonymous to synonymous substitutions (dN/dS) on the branch leading to Poaceae relative to other branches in the phylogeny
| Gene name/gene group | Leading to Poaceae | Internal Poaceae | Increase dN/dS | Other monocots | Increase dN/dS | Others angiosperms | Increase dN/dS |
|---|---|---|---|---|---|---|---|
| atp | 0.2678 | 0.1077 | 2.49 | 0.1077 | 2.49 | 0.0985 | 2.72 |
| ccsA | 0.2234 | 0.1945 | 1.15 | 0.1945 | 1.15 | 0.2481 | 0.90 |
| cemA | 1.6600 | 0.2016 | 8.23 | 0.2016 | 8.23 | 0.3203 | 5.18 |
| clpP | 0.6711 | 0.1513 | 4.44 | 0.1513 | 4.44 | 0.3187 | 2.11 |
| matK | 0.3096 | 0.3502 | 0.88 | 0.3502 | 0.88 | 0.3820 | 0.81 |
| ndh | 0.1652 | 0.1483 | 1.11 | 0.1483 | 1.11 | 0.1281 | 1.29 |
| pet | 0.0149 | 0.0790 | 0.19 | 0.0790 | 0.19 | 0.0780 | 0.19 |
| psa | 0.0607 | 0.1137 | 0.53 | 0.1137 | 0.53 | 0.1336 | 0.45 |
| psb | 0.0532 | 0.1025 | 0.52 | 0.1025 | 0.52 | 0.0776 | 0.69 |
| rbcL | 0.0391 | 0.0963 | 0.41 | 0.0963 | 0.41 | 0.0889 | 0.44 |
| rpl | 0.3400 | 0.1655 | 2.05 | 0.1655 | 2.05 | 0.2034 | 1.67 |
| rps | 0.5426 | 0.1766 | 3.07 | 0.1766 | 3.07 | 0.1923 | 2.82 |
| rpo | 0.3207 | 0.1553 | 2.07 | 0.1553 | 2.07 | 0.1853 | 1.73 |
| Average | 0.3591 | 0.1571 | 2.0879 | 0.1571 | 2.0879 | 0.1888 | 1.6155 |
| P-value | P = 0.2642 | P = 0.2642 | P = 0.5114 |
Asterisks show P-values that are significant after correction for multiple comparisons using Holm’s method, i.e., sequential Bonferroni correction (** α = 0.01, * α = 0.05)
Table 5.
The degree of rate acceleration for nonsynonymous substitutions (dN) on the branch leading to Poaceae relative to other branches in the phylogeny
| Gene name/gene group | Leading to Poaceae | Internal Poaceae | Increase dN | Other monocots | Increase dN | Others angiosperms | Increase dN |
|---|---|---|---|---|---|---|---|
| atp | 0.0424 | 0.0018 | 23.02 | 0.0073 | 5.83 | 0.0069 | 6.16 |
| ccsA | 0.0540 | 0.0049 | 11.06 | 0.0244 | 2.21 | 0.0249 | 2.17 |
| cemA | 0.1715 | 0.0040 | 42.70 | 0.0141 | 12.19 | 0.0238 | 7.21 |
| clpP | 0.2117 | 0.0025 | 85.63 | 0.0085 | 25.05 | 0.0197 | 10.76 |
| matK | 0.0900 | 0.0114 | 7.88 | 0.0386 | 2.33 | 0.0378 | 2.38 |
| ndh | 0.0378 | 0.0030 | 12.49 | 0.0109 | 3.47 | 0.1281 | 0.30 |
| pet | 0.0026 | 0.0016 | 1.64 | 0.0054 | 0.48 | 0.0053 | 0.48 |
| psa | 0.0107 | 0.0019 | 5.54 | 0.0073 | 1.47 | 0.0068 | 1.58 |
| psb | 0.0099 | 0.0022 | 4.40 | 0.0055 | 1.81 | 0.0043 | 2.30 |
| rbcL | 0.0128 | 0.0026 | 4.84 | 0.0069 | 1.86 | 0.0064 | 2.00 |
| rpl | 0.0962 | 0.0040 | 24.30 | 0.0108 | 8.92 | 0.0130 | 7.42 |
| rps | 0.0784 | 0.0032 | 24.43 | 0.0106 | 7.39 | 0.0120 | 6.56 |
| rpo | 0.0656 | 0.0041 | 15.96 | 0.0116 | 5.68 | 0.0146 | 4.51 |
| Average | 0.0680 | 0.0036 | 20.2983 | 0.0124 | 6.0537 | 0.0233 | 4.1396 |
| P-value | P < 0.001** | P = 0.0056* | P = 0.0378 |
Asterisks show P-values that are significant after correction for multiple comparisons using Holm’s method, i.e., sequential Bonferroni correction (** α = 0.01, * α = 0.05)
Table 6.
The degree of rate acceleration for synonymous substitutions (dS) on the branch leading to Poaceae relative to other branches in the phylogeny
| Gene name/gene group | Leading to Poaceae | Internal Poaceae | Increase dS | Other monocots | Increase dS | Other angiosperms | Increase dS |
|---|---|---|---|---|---|---|---|
| atp | 0.1868 | 0.0179 | 10.44 | 0.0679 | 2.75 | 0.0718 | 2.60 |
| ccsA | 0.2416 | 0.0251 | 9.63 | 0.1255 | 1.93 | 0.1003 | 2.41 |
| cemA | 0.1033 | 0.0199 | 5.18 | 0.0698 | 1.48 | 0.0743 | 1.39 |
| clpP | 0.3155 | 0.0164 | 19.23 | 0.0559 | 5.64 | 0.0617 | 5.11 |
| matK | 0.2907 | 0.0327 | 8.90 | 0.1101 | 2.64 | 0.0990 | 2.94 |
| ndh | 0.2280 | 0.0244 | 9.34 | 0.0872 | 2.62 | 0.0858 | 2.66 |
| pet | 0.1847 | 0.0190 | 9.74 | 0.0676 | 2.73 | 0.0653 | 2.83 |
| psa | 0.2004 | 0.0199 | 10.06 | 0.0686 | 2.92 | 0.0649 | 3.09 |
| psb | 0.1875 | 0.0188 | 9.99 | 0.0527 | 3.56 | 0.0571 | 3.28 |
| rbcL | 0.3270 | 0.0276 | 11.85 | 0.0716 | 4.57 | 0.0720 | 4.54 |
| rpl | 0.2773 | 0.0246 | 11.27 | 0.0715 | 3.88 | 0.0820 | 3.38 |
| rps | 0.1652 | 0.0195 | 8.49 | 0.0690 | 2.39 | 0.0693 | 2.38 |
| rpo | 0.2085 | 0.0250 | 8.33 | 0.0740 | 2.82 | 0.0778 | 2.68 |
| Average | 0.2243 | 0.0224 | 10.1888 | 0.0763 | 3.0708 | 0.0755 | 3.0230 |
| P-value | P < 0.001** | P < 0.001** | P < 0.001 ** |
Asterisks show P-values that are significant after correction for multiple comparisons using Holm’s method, i.e., sequential Bonferroni correction (** α = 0.01, * α = 0.05)
Discussion
Plastid Genome Organization and Evolution
Our survey of nine Poaceae plastid genomes and the sequence of T. latifolia (Typhaceae) shows that genome organization and rates of sequence evolution are unusual in the Poaceae. Our analyses included the earliest diverging Poales lineage (Typhaceae), but the closest relatives of the Poaceae have not been sequenced. Doyle et al. (1992) surveyed for the distribution of inversions among Poales, and showed that changes were not confined to the Poaceae. Likewise, gene and intron losses may have a broader distribution among Poales, and more data are needed to fully characterize Poales genome evolution. Nonetheless, Poaceae plastid genomes have experienced genomic change relative to T. latifolia and most other angiosperms. Based on our data, gene content in Poales plastid genomes is identical (Table 1) except for the loss of three genes (accD, ycf1, and ycf2) in the Poaceae. The Festuca arundinacea plastid genome sequence on GenBank apparently also lacks intact copies of the genes psbF, rps14, rps18, and ycf4, but these surprising gene losses should be confirmed. Although other differences can be found among the annotations of the published grass genomes, all are due to annotation errors, both for protein-coding genes and tRNA genes. The recently published Brachypodium genome reported 136 genes (Bortiri et al. 2008), but this included ycf68 that has been shown to be non-functional (Raubeson et al. 2007). Three other recently published Poaceae plastid genomes (Saski et al. 2007) also incorrectly identified 32 tRNAs instead of the 30 found in the other sequenced genomes.
Organization and evolution of Poaceae plastid genomes have been examined extensively in early studies using restriction site and gene mapping approaches (Bowman and Dyer 1986; Howe 1985; Howe et al. 1988; Katayama and Ogihara 1993; Prombona and Subramanian 1989; Quigley and Weil 1985; Shimada and Sugiura 1989) and later based on complete genome sequences (Asano et al. 2004; Bortiri et al. 2008; Hiratsuka et al. 1989; Maier et al. 1995; Ogihara et al. 2000; Saski et al. 2007; Wu et al. 2009). These comparisons identified a number of unusual features, including the presence of three inversions in the LSC, the loss of introns from two genes (clpP and rpoC1), the loss of three genes (accD, ycf1, and ycf2), and expansions of the IR/SC boundaries to duplicate trnH-gug and rps19 on the IR/LSC boundary and rps15 on the IR/SSC boundary. Furthermore, the phylogenetic distribution of some of these rearrangements has been examined by combining genome sequencing and PCR-based surveys (Downie et al. 1996; Doyle et al. 1992; Wang et al. 2008). Our comprehensive comparisons of the complete plastid genomes of nine Poaceae and T. latifolia confirm the presence of all of these genomic rearrangements. We will briefly review the conclusions from the above studies, and then highlight the novel aspects resulting from our comprehensive comparisons of sequences of nine published genomes and the related T. latifolia genome.
Among monocots most of the rearrangements identified in Poaceae plastid genomes appear to be restricted to this family, including the loss of introns from two genes, three gene losses, and the smallest of the three genome inversions. In most cases, losses of these same genes and introns have occurred independently elsewhere in angiosperms (Jansen et al. 2007), including four losses of accD, one loss of ycf1, two losses of both clpP introns, and numerous rpoC1 intron losses. In a survey of inversions for 12 of 16 families of Poales, the first and largest 28 kb inversion is shared by the two closely related families Joinvilleaceae and Restionaceae, and the second 6 kb inversion is present in the Joinvilleaceae, supporting its placement as sister to Poaceae (Doyle et al. 1992). Expansions and contractions of the IR have been documented throughout angiosperm plastid genomes (Goulding et al. 1996; Wang et al. 2008). The expansion of the IR/LSC boundary in grasses to duplicate trnH-gug is characteristic of all monocots and some early diverging eudicots, but further expansion to include a complete duplication of rps19 is restricted to a more derived clade of monocots including Asparagales, Commelinales, Zingiberales, Arecales, and Poales (Wang et al. 2008). Our comparisons of nine plastid genomes of Poaceae confirm the expansion of the IR at the IR/LSC boundary resulting in the duplication of both trnH-gug and rps19 and demonstrate that the endpoint of the IR is highly conserved with only 35–99 bp duplicated beyond rps19 (Fig. 1).
Expansion of the IR into the SSC region is much less common, and only a few angiosperm families exhibit this phenomenon, including Campanulaceae (Cosner et al. 1997; Haberle et al. 2008; Knox and Palmer 1999), Geraniaceae (Chumley et al. 2006; Palmer et al. 1987), and Polygonaceae (Aii et al. 1997; Logacheva et al. 2008). Earlier investigations of one or two plastid genomes of Poaceae (Hiratsuka et al. 1989; Maier et al. 1995; Prombona and Subramanian 1989) identified expansion of the IR into the SSC region resulting in the duplication of rps15. In the comparison of rice and maize, variation in the extent of expansion of the IR/SSC boundary beyond rps15 was examined (Maier et al. 1995). In maize, there was an additional expansion of IRb to duplicate 29 bp of ndhF, whereas in rice IRa expanded to duplicate 216 bp of ndhH. Our comparison of the IR/SSC boundaries among the nine sequenced Poaceae plastid genomes (Fig. 1) demonstrates that the pattern of expansion is congruent with phylogenetic relationships, with the IRb expansion restricted to the Panicoideae, and the IRa expansion shared by the subfamilies Ehrhartoideae and Pooideae. This structural feature supports the sister relationships between the latter two tribes, which is congruent with the tree based on nucleotide sequences (Fig. 4).
Based on comparisons of rice and maize plastid genome sequences, Maier et al. (1995) suggested that accD has been completely lost but that ycf2 represents different stages of gene deletion. This conclusion was supported by the fact that neither species had any residual sequence left for accD but that ycf2 has different sized residual fragments in rice and maize. Our comparisons of the sequences of all three missing genes confirms that accD has been almost completely lost from all grasses, and three genera, Hordeum, Lolium, and Oryza, have small, highly divergent remnant sequences (Table 2). The residual sequence in this gene does not correlate with phylogenetic relationships (Fig. 4). The situation in ycf2 is much more interesting based on the more extensive sampling reported here. For this gene, there are three distinct size classes of remnant sequences (698–700, 1,314–1,413, and 2,061–2,089 bp), and these sizes correlate with phylogenetic relationships among grasses. The largest remnant occurs in the earliest diverging panicoid clade, suggesting that there has been a progressive degeneration of ycf2 within grasses. The loss of ycf1 shows yet another pattern in which all nine Poaceae maintain a similar sized remnant sequence of the gene. In the case of both ycf1 and ycf2, the larger amount of residual sequence for these genes could be attributed to their presence in the IR, which is known to be more highly conserved than single-copy regions (Wolfe et al. 1987). A similar argument was made for the high level of sequence conservation of ycf15 and ycf68 (Raubeson et al. 2007). We do not know the extent of gene and intron loss on Poales lineages leading to the Poaceae, and data gathered through future sequencing projects will certainly shed light on rates and mechanisms of gene and intron loss in plastid genomes.
Rates and Patterns of Sequence Evolution in Grass Plastid Genomes
In the current study, we characterize rates and patterns of sequence evolution for angiosperm plastid genomes, and we specifically test the degree and nature of rate acceleration for the branch leading to Poaceae. Our results are consistent with early models of plastid genome evolution; rates of both dN and dS vary across lineages, rates of dS are relatively homogeneous across loci, and rates of dN vary across loci (Muse and Gaut 1994). However, the degree of rate heterogeneity for the branch leading to Poaceae is highly unusual. Aside from a recent study demonstrating extreme rate heterogeneity in Geraniaceae genome sequences (Guisinger et al. 2008), accelerated rates of nucleotide substitutions are typically not found in photosynthetic angiosperm plastid genomes. Results from the current study indicate a high degree of positive or relaxed selection on the branch leading to Poaceae, and the genes cemA and rps7 exhibit dN/dS ratios greater than 1 (1.66 and 1.44, respectively). This ratio is often used as a measure of selective pressures with dN/dS = 1, >1, and <1 indicating neutral mutation rates, positive selection, and purifying selection, respectively (Yang 1998). Additional analyses are needed to determine amino acid sites that may be under positive selection, and it should be noted that the models used in our analyses do not allow for heterogeneous dN/dS ratios among sites (Yang and Nielsen 2002; Yang et al. 2000). The results from likelihood ratio tests (Supplemental Table S3) indicate that a number of genes are accumulating nonsynonymous mutations at a significantly high rate, suggesting that either positive or relaxed selection at nucleotide sites is acting on these genes. The majority of these are ATPase, ribosomal protein, and RNA polymerase genes.
In addition to better characterizing rates of sequence evolution for the branch leading to the grasses, our results are generally consistent with other findings regarding grass plastid genome evolution. The individual genes clpP, cemA, and rpl32 seem to be evolving rapidly, photosynthetic genes are evolving slower than ribosomal protein genes and appear to be under stronger purifying selection, dN varies across loci, dS is uniform across loci, and substitution rates are accelerated for the grasses relative to other angiosperms (Chang et al. 2006; Matsuoka et al. 2002; Muse and Gaut 1997). Although Chang et al. (2006) found that values of dS were not significantly different for the grasses relative to one other monocot (the orchid Phalaenopsis), we show that dS is significantly different between grass and monocot branches. We chose to exclude the genome sequence of Phalaenopsis (Chang et al. 2006) from our analyses, because all 11 ndh-genes have been lost. Furthermore, we were able to include plastid genome sequences from seven other monocots than the previously mentioned study. In our analyses, the branch leading to the Poaceae and internal Poaceae branches are compared separately, and only the branch leading to the Poaceae exhibits a significant amount of rate acceleration. Moreover, internal Poaceae branches are evolving at a slower rate than other branches in the phylogeny (Figs. 5, 6). It appears that after a rapid burst in sequence evolution, rates decelerated in grass plastid genomes, and this deceleration occurred subsequent to grass diversification. Using increased taxon sampling and methods that detect the degree and nature of rate change on specific branches in a phylogeny, we are able to better characterize grass plastid genome sequence evolution.
Factors affecting rates of sequence evolution in plastid genomes have been extensively examined. Speciation rates (Barraclough et al. 1996; Bousquet et al. 1992), generation time (Chang et al. 2006; Smith and Donoghue 2008), substitution and codon bias (Morton 2003; Morton and Clegg 1995), gene function (Matsuoka et al. 2002), gene copy number (genes duplicated in the IR evolve slower than single-copy genes (Wolfe et al. 1987)), and genome copy number (Khakhlova and Bock 2006) have been shown to influence substitution rates. However, a recent phylogenetic analysis of 81 plastid genes from 64 seed plants described a positive correlation between genomic rearrangements and lineage-specific rate acceleration (Jansen et al. 2007). Furthermore, the highly rearranged plastid genomes of the plant family Geraniaceae exhibit the greatest degree of rate acceleration among photosynthetic angiosperms (Guisinger et al. 2008). As shown in Fig. 4, there are eight major structural changes on the branch leading to grasses, including the loss of introns in two genes, three gene losses, and three inversions. We hypothesize that rates of sequence evolution may be correlated to genomic changes in grass plastid genomes. It should be noted that after the divergence of the grasses no major genomic changes occurred aside from minor expansions of the IR region, a very common process that accounts for size variation in plastid genomes throughout angiosperms (Aii et al. 1997; Goulding et al. 1996; Plunkett and Downie 2000) including the monocots (Wang et al. 2008).
A correlation between genomic changes and rates of sequence evolution has been previously described for bacterial (Belda et al. 2005) and animal mitochondrial genomes (Shao et al. 2003; Xu et al. 2006). Mechanisms have been proposed to explain this correlation, and it is possible that similar mechanisms are responsible for the unusual evolution of grass plastid genomes. One mechanism involves homologs to the eubacterial gene recA. In E. coli, this gene is responsible for DNA repair during homologous recombination and strand exchange (Lin et al. 2006). Homologs of recA are found in plant and algal nuclear genomes (Lin et al. 2006), and gene products are localized to plastids and mitochondria in Arabidopsis (Cao et al. 1997; Cerutti et al. 1992; Khazi et al. 2003). It is possible that genomic changes and accelerated rates of sequence evolution are the result of mutations in plastid-targeted rec-genes, although their presence and function in plastid genomes has not been thoroughly tested.
Conclusion
In agreement with earlier studies using large data sets of morphological, anatomical, and single plastid gene sequence characters (Barker et al. 1995; Clark et al. 1995; Kellogg and Watson 1993), our study suggests that the grasses have experienced rapid molecular diversification relative to other monocots and to early diverging members of the Poales, i.e., T. latifolia. This point was made well by Chase (2004), who noted that there is a pattern of small, “insignificant” sets of taxa sister to Poaceae. Graham et al. (2006) performed a phylogenetic analysis of 17 plastid protein-coding genes and included taxa from Poaceae, Typhaceae, and eight additional Poales families. Branch lengths for most members of the Poales were long except for the three earliest diverging families, including Typhaceae. These data would suggest that genomic changes and accelerated rates of sequence evolution may not be limited to the Poaceae only, and that a positive correlation between these two phenomena can be shown for lineages leading to the Poaceae. We emphasize that additional Poales genome sequences are needed to fully understand the evolution of Poales and Poaceae plastid genomes. Nonetheless, we show the extent to which plastid genomes within the Poaceae are experiencing rapid rates of genomic change and sequence evolution. We also show that the rates of plastid genome evolution for internal Poaceae branches have decelerated. Whatever the cause of rapid change in the branch leading to the grasses, subsequent deceleration indicates that the factors responsible may no longer be driving genome evolution in this family.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank C. Blazier, D. Hansen, and R. Steele for reading an earlier version of the manuscript, R. Haberle for providing material of Typha latifolia, and C. Zhenqiu for access to the alignment tool MSWAT.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aii J, Kishima Y, Mikami T, Adachi T. Expansion of the IR in the chloroplast genomes of buckwheat species is due to incorporation of an SSC sequence that could be mediated by an inversion. Curr Genet. 1997;31:276–279. doi: 10.1007/s002940050206. [DOI] [PubMed] [Google Scholar]
- APG II An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x. [DOI] [Google Scholar]
- Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res. 2004;11:93–99. doi: 10.1093/dnares/11.2.93. [DOI] [PubMed] [Google Scholar]
- Barker NP, Linder HP, Harley EH. Polyphyly of Arundinoideae (Poaceae): evidence from rbcL sequence data. Syst Bot. 1995;20:423–435. doi: 10.2307/2419802. [DOI] [Google Scholar]
- Barraclough TG, Harvey PH, Nee S. Rate of rbcL gene sequence evolution and species diversification in flowering plants (angiosperms) Proc R Soc Lond B Biol Sci. 1996;263:589–591. doi: 10.1098/rspb.1996.0088. [DOI] [Google Scholar]
- Belda E, Moya A, Silva FJ. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol Biol Evol. 2005;22:1456–1467. doi: 10.1093/molbev/msi134. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and molecular biology of plastids. Berlin/Heidelberg: Springer; 2007. pp. 1610–2096. [Google Scholar]
- Bortiri E, Coleman-Derr D, Lazo GR, Anderson OD, Gu YQ. The complete chloroplast genome sequence of Brachypodium distachyon: sequence comparison and phylogenetic analysis of eight grass plastomes. BMC Res Notes. 2008;1:61. doi: 10.1186/1756-0500-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet J, Strauss SH, Doerksen AH, Price RA. Extensive variation in evolutionary rate of rbcL gene-sequences among seed plants. Proc Natl Acad Sci USA. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman CM, Dyer T. The location and possible evolutionary significance of small dispersed repeats in wheat ctDNA. Curr Genet. 1986;10:931–941. doi: 10.1007/BF00398291. [DOI] [Google Scholar]
- Cao J, Combs C, Jagendorf AT. The chloroplast-located homolog of bacterial DNA recombinase. Plant Cell Physiol. 1997;38:1319–1325. doi: 10.1093/oxfordjournals.pcp.a029124. [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC. Maize and sorghum: Genetic resources for bioenergy grasses. Trends Plant Sci. 2008;13:415–420. doi: 10.1016/j.tplants.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Osman M, Grandoni P, Jagendorf AT. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA. 1992;89:8068–8072. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, Cheng CH, Lin CY, Liu SM, Chaw SM. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chase MW. Monocot relationships: an overview. Am J Bot. 2004;91:1645–1655. doi: 10.3732/ajb.91.10.1645. [DOI] [PubMed] [Google Scholar]
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. The complete chloroplast genome sequence of Pelargonium X hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Clark LG, Zhang W, Wendel JF. A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Syst Bot. 1995;20:436–460. doi: 10.2307/2419803. [DOI] [Google Scholar]
- Cosner ME, Jansen RK, Palmer JD, Downie SR. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet. 1997;31:419–429. doi: 10.1007/s002940050225. [DOI] [PubMed] [Google Scholar]
- Downie SR, Llanas E, Katz-Downie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA’s. Syst Bot. 1996;21:135–151. doi: 10.2307/2419744. [DOI] [Google Scholar]
- Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae) Proc Natl Acad Sci USA. 1992;89:7722–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Riemer C, Petrykowska H, Florea L, Schwartz S, Miller W, Hardison R. PipTools: a computational toolkit to annotate and analyze pairwise comparisons of genomic sequences. Genomics. 2002;80:681–690. doi: 10.1006/geno.2002.7018. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Garvin DF, Gu Y-Q, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 2008;48:S-69–S-84. doi: 10.2135/cropsci2007.06.0332tpg. [DOI] [Google Scholar]
- Gaut BS, Muse SV, Clegg MT. Relative rates of nucleotide substitution in the chloroplast genome. Mol Phylogenet Evol. 1993;2:89–96. doi: 10.1006/mpev.1993.1009. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- GPWG Phylogeny and subfamilial classification of the grasses. Ann Mo Bot Gard. 2001;88:373–457. doi: 10.2307/3298585. [DOI] [Google Scholar]
- Graham SW, Zgurski JM, McPherson MA, Cherniawsky DM, Saarela JM, Horne ESC, Smith SY, Wong WA, O’Brien HE, Biron VL, Pires JC, Olmstead RG, Chase MW, Rai HS. Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. Aliso. 2006;22:3–20. [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci USA. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Heath TA, Zwickl DJ, Kim J, Hillis DM. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst Biol. 2008;57:160–166. doi: 10.1080/10635150701884640. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, Li YQ, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequential rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Howe CJ. The endpoints of an inversion in wheat chloroplast DNA are associated with short repeated sequences containing homology to att-lambda. Curr Genet. 1985;10:139–145. doi: 10.1007/BF00636479. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barker RF, Bowman CM, Dyer TA. Common features of three inversions in wheat chloroplast DNA. Curr Genet. 1988;13:343–349. doi: 10.1007/BF00424430. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui L. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Ogihara Y. Structural alterations of the chloroplast genome found in grasses are not common in monocots. Curr Genet. 1993;23:160–165. doi: 10.1007/BF00352016. [DOI] [PubMed] [Google Scholar]
- Kellogg EA, Watson L. Phylogenetic studies of a large data set. I. Bambusoideae, Andropogonodae, and Pooideae (Gramineae) Bot Rev. 1993;59:273–343. doi: 10.1007/BF02857419. [DOI] [Google Scholar]
- Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- Khazi FR, Edmondson AC, Nielsen BL. An Arabidopsis homologue of bacterial RecA that complements an E. colirecA deletion is targeted to plant mitochondria. Mol Genet Genomics. 2003;269:454–463. doi: 10.1007/s00438-003-0859-6. [DOI] [PubMed] [Google Scholar]
- Knox EB, Palmer JD. The chloroplast genome arrangement of Lobelia thuliniana (Lobeliaceae): expansion of the inverted repeat in an ancestor of the Campanulales. Plant Syst Evol. 1999;214:49–64. doi: 10.1007/BF00985731. [DOI] [Google Scholar]
- Leebens-Mack J, Raubeson LA, Cui L, Kuehl JV, Fourcade MH, Chumley TW, Boore JL, Jansen RK, dePamphilis CW. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Penin AA, Samigullin TH, Vallejo-Roman CM, Antonov AS. Phylogeny of flowering plants by the chloroplast genome sequences: in search of a “lucky gene”. Biochem (Mosc) 2007;72:1324–1330. doi: 10.1134/S0006297907120061. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale—a wild ancestor of cultivated buckwheat. BMC Plant Biol. 2008;8:59. doi: 10.1186/1471-2229-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Kossel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K. Whole chloroplast genome comparison of rice, maize, and wheat: Implications for chloroplast gene diversification and phylogeny of cereals. Mol Biol Evol. 2002;19:2084–2091. doi: 10.1093/oxfordjournals.molbev.a004033. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton BR. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J Mol Evol. 2003;56:616–629. doi: 10.1007/s00239-002-2430-1. [DOI] [PubMed] [Google Scholar]
- Morton BR, Clegg MT. Neighboring base composition is strongly correlated with base substitution bias in a region of the chloroplast genome. J Mol Evol. 1995;41:597–603. doi: 10.1007/BF00175818. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. Comparing patterns of nucleotide substitution rates among chloroplast loci using the relative ratio test. Genet. 1997;146:393–399. doi: 10.1093/genetics/146.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, Takumi S, Ikeo K, Gojobori T, Murai R, Murai K, Matsuoka Y, Ohnishi Y, Tajiri H, Tsunewaki K. Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep. 2000;18:243–253. doi: 10.1007/BF02823995. [DOI] [Google Scholar]
- Palmer JD. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. doi: 10.1016/0076-6879(86)18072-4. [DOI] [Google Scholar]
- Palmer JD. Plastid chromosomes: structure and evolution. In: Hermann RG, editor. The molecular biology of plastids. Cell culture and somatic cell genetics of plants. Vienna: Springer; 1991. pp. 5–53. [Google Scholar]
- Palmer JD, Nugent JM, Herbon LA. Unusual structure of geranium chloroplast DNA: a triple-sized repeat, extensive gene duplications, multiple inversions, and new repeat families. Proc Natl Acad Sci USA. 1987;84:769–773. doi: 10.1073/pnas.84.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst Bot. 2000;25:648–667. doi: 10.2307/2666726. [DOI] [Google Scholar]
- Prombona A, Subramanian AR. A new rearrangement of angiosperm chloroplast DNA in rye (Secale cereale) involving translocation and duplication of the ribosomal rps15 gene. J Biol Chem. 1989;264:19060–19065. [PubMed] [Google Scholar]
- Quigley F, Weil JH. Organization and sequence of five tRNA genes and of an unidentified reading frame in the wheat chloroplast genome: Evidence for gene rearrangements during the evolution of chloroplast genomes. Curr Genet. 1985;9:495–503. doi: 10.1007/BF00434054. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Cambridge, MA: CABI Publishing; 2005. pp. 45–68. [Google Scholar]
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Program NCS, Green ED, Hardison RC, Miller W. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- Shimada H, Sugiura M. Pseudogenes and short repeated sequences in the rice chloroplast genome. Curr Genet. 1989;16:293–301. doi: 10.1007/BF00422116. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Amborella not a “basal angiosperm”? Not so fast. Am J Bot. 2004;91:997–1001. doi: 10.3732/ajb.91.6.997. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Rice DW, Palmer JD. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evol Biol. 2004;4:35. doi: 10.1186/1471-2148-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*: phylogenetic analysis using parsimony (*and other methods). version 4. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Kan D, Lee S, Daniell H, Lee Y, Lin C, Lin N, Lin C. Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol. 2009;29:847–856. doi: 10.1093/treephys/tpp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 2006;63:375–392. doi: 10.1007/s00239-005-0246-5. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genet. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ (2006) GARLI: genetic algorithm for rapid likelihood inference, version 0.951. GARLI: genetic algorithm for rapid likelihood inference, version 0.951. Available at http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.