Summary
The rise of systems biology implied a growing demand for highly sensitive techniques for the fast and consistent detection and quantification of target sets of proteins across multiple samples. This is only partly achieved by classical mass spectrometry or affinity-based methods. We applied a targeted proteomics approach based on selected reaction monitoring (SRM) to detect and quantify proteins expressed to a concentration below 50 copies/cell in total S. cerevisiae digests. The detection range can be extended to single-digit copies/cell and to proteins which were undetected by classical methods. We illustrate the power of the technique by the consistent and fast measurement of a network of proteins spanning the entire abundance range over a growth time-course of S. cerevisiae transiting through a series of metabolic phases. We therefore demonstrate the potential of SRM-based proteomics to provide assays for the measurement of any set of proteins of interest in yeast at high-throughput and quantitative accuracy.
Keywords: targeted proteomics, S. cerevisiae, selected / multiple reaction monitoring, MRM/SRM, dynamic range
Introduction
The ability to reliably identify and accurately quantify any protein or set of proteins of interest in a proteome is an essential task in life science research. This has been attempted by two general experimental approaches. The first is based on the generation of affinity reagents exemplified by highly specific antibodies and the development of an array of methods to deploy them for detecting and quantifying specific proteins in complex samples. The second is mass spectrometry (MS)-based quantitative proteomics that attempts to identify and quantify all proteins contained in a sample.
Multiple versions of affinity reagent based methods (e.g. the broadly used Western blot or ELISA approaches) have been implemented. They differ in the type of affinity reagent and detection system used (Uhlen, 2008). The methods with the highest sensitivity have the potential to detect, in principle, low abundant proteins, with zeptomole detection limits already demonstrated (Pawlak et al., 2002). However, the development of sets of reagents of suitable specificity and affinity to support the conclusive detection and quantification of target protein(s) remains challenging, expensive and arduous, and coordinated efforts to develop validated affinity reagents are just getting underway (Taussig et al., 2007; Uhlen and Hober, 2008). The methods based on affinity reagents are therefore limited by slow assay development and, usually, also by the inability to significantly multiplex detection of proteins in the same sample.
Similarly to affinity based methods, a wide range of MS-based proteomic methods have been developed. The most successful of these, in terms of number of proteins identified, use a shotgun strategy in which a subset of peptides present in a tryptic digest of a proteome is selected in an intensity-dependent manner for collision-induced dissociation by a tandem mass spectrometer (Aebersold and Mann, 2003). The resulting fragment ion (MS/MS) spectra are then assigned to sequences in a peptide database and the corresponding peptides and proteins are thus identified. This method provides accurate quantitative data if suitable stable isotope-labeled references are available and included in the analysis (Desiderio and Kai, 1983; Gerber et al., 2003). However, these methods are non-targeted, i.e. in each measurement they stochastically sample a fraction of the proteome that is usually biased towards the higher end of the abundance scale (Domon and Broder, 2004; Picotti et al., 2007). Each repeat analysis required for comparing a proteome at different states, will sample only a subset of the proteins it contains, and not necessarily the same subset in each repeat, thus precluding the generation of complete and consistent datasets (Wolf-Yadlin et al., 2007). More extensive, although still incomplete and stochastic proteome coverage can be achieved in large proteome mapping experiments, whereby the proteome is extensively fractionated by multiple approaches and the content of each fraction is sequenced to saturation (Chen et al., 2006; de Godoy et al., 2008; de Godoy et al., 2006). Such studies carry a significant experimental and computational overhead, and are therefore time/labor consuming and can be performed only in highly specialized laboratories. In addition they mostly retain the bias against low abundance proteins, albeit to a reduced degree. This makes such approaches impractical in cases that require the consistent quantification of sets of proteins of all abundances across a variety of different samples and replicates.
To alleviate these limitations, we demonstrate in this study the potential of selected reaction monitoring (SRM)-based targeted mass spectrometry (Anderson and Hunter, 2006; Lange et al., 2008) to provide specific assays for the detection and quantification of proteins over the whole range of cellular concentrations in S. cerevisiae. We deploy the approach to quantitatively monitor the dynamics of a protein network containing proteins spanning a broad range of abundances, across numerous samples and replicates, at high speed and quantitative accuracy.
Results
Detection of low abundance proteins in a total yeast cell lysate by SRM
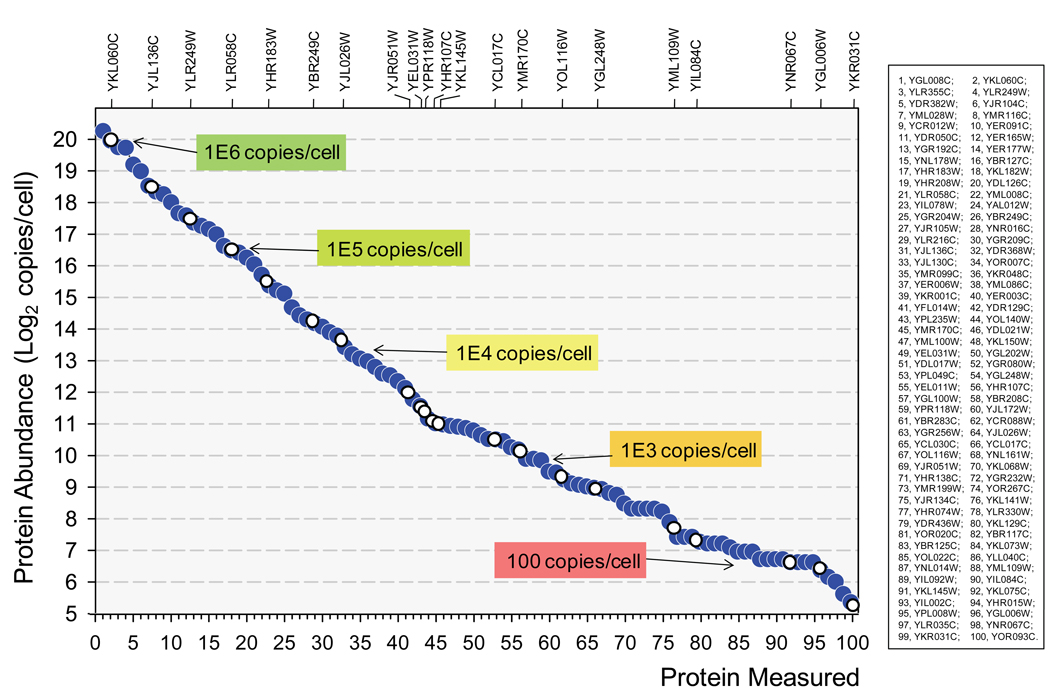
We challenged the dynamic range of SRM-based targeted proteomics by applying it to the detection and quantification of yeast proteins distributed across the whole range of cellular abundance, with the purpose of determining to what concentration (in copies per cell) yeast proteins can be detected in a tryptic digest of a total cell lysate. We selected protein targets based on the list of absolute protein abundances generated by orthogonal methods (i.e. by quantitative Western blotting against a tandem affinity purification (TAP) tag engineered into S. cerevisiae genes (Ghaemmaghami et al., 2003)). We selected a set of 100 target proteins evenly distributed across all levels of cellular abundance, from 1.3E6 to 41 copies per cell (Fig. 1) in S. cerevisiae. These proteins were grouped into the copy number classes indicated in Table 1 (Groups 1–14). Each class minimally contained five proteins. The classes with proteins of lower abundance contained a higher number of proteins to increase the significance of low copy number-protein detection. For each protein, five proteotypic peptides (unique peptides preferentially detectable by MS, PTPs (Brunner et al., 2007; Kuster et al., 2005)) were selected. PTPs were derived by screening a large yeast proteomic data repository, PeptideAtlas (>36,000 unique peptides observed in an array of shotgun proteomic experiments (Deutsch et al., 2008), for the most frequently observed peptides for each target protein. For proteins for which fewer than five PTPs could be extracted from PeptideAtlas, additional peptides with favorable MS properties were derived by bioinformatic prediction using the tool PeptideSieve (Mallick et al., 2007). For each PTP an SRM assay consisting of three optimal and validated precursor ion to fragment ion transitions was developed on a triple quadrupole mass spectrometer (see Experimental Procedures). For low abundant proteins critical MS parameters were specifically optimized to maximize the sensitivity of the corresponding assay. For each protein the SRM assays associated with the two best responding PTPs were used in final measurements. The resulting assays were then applied to detect and/or quantify the target proteins in unfractionated trypsinized extracts of S. cerevisiae cells, grown under the conditions described by Ghaemmaghami et al. (2003). Data were acquired in time-scheduled SRM mode to maximize throughput and sensitivity (Stahl-Zeng et al., 2007). The results indicate that proteins spanning a range of literature abundance values from over a million down to 41 copies per cell could be unambiguously detected. Approximately 10% of the targeted proteins could not be detected. A list of these proteins and a rationale for the inability to detect them is presented in Tab. S1. Both the cellular abundances (1.3E6 to 41 copies/cell) of the set of proteins detected in an unfractionated yeast digest and the associated SRM signal intensities covered a range of ∼4.5 orders of magnitude. The linear correlation between the abundance of proteins and the SRM signal intensity of the respective most intense PTPs (log scale) is shown in Fig S3. These results demonstrate that SRM-based proteomics has the power to reliably detect proteins expressed in the whole range of documented cellular abundance, down to a concentration < 50 protein copies/cell, in S. cerevisiae total cell digests, without the need of sample fractionation or enrichment.
Figure 1. Cellular concentrations of the set of measured proteins.
Protein abundances are derived from Ghaemmaghami et al. (2003). Proteins detected by SRM assays are sorted by abundance to show the even distribution across the whole range of concentration (blue solid circles). Proteins for which the absolute abundance was measured using isotopically-labelled standards are indicated on top of the graph (open circles).
Table 1.
Summary of proteins detected and quantified in each group of cellular abundance.
| Abundance Range (copies/cell) |
Group | Proteins measured |
Protein namesa | Absolute Quantificationb |
|---|---|---|---|---|
| 524288–1255722 | 1 | 5 | YGL008C, YKL060C, YLR355C, YLR249W, YDR382W | YKL060C |
| 262144–524288 | 2 | 5 | YJR104C, YML028W, YMR116C, YCR012W, YER091C | YJL136C |
| 131072–262144 | 3 | 5 | YDR050C, YER165W, YGR192C, YER177W, YNL178W | YLR249W |
| 65536–131072 | 4 | 5 | YBR127C, YHR183W, YKL182W, YHR208W, YDL126C | |
| 32768–65536 | 5 | 5 | YLR058C, YML008C, YIL078W, YAL012W, YGR204W | YHR183W, YLR058C, |
| 16384–32768 | 6 | 5 | YBR249C, YJR105W, YNR016C, YLR216C, YGR209C | YBR249C |
| 8192–16384 | 7 | 5 | YJL136C, YDR368W, YJL130C, YOR007C, YMR099C | YJL026W |
| 4096–8192 | 8 | 5 | YKR048C, YER006W, YML086C, YKR001C, YER003C | |
| 2048–4096 | 9 | 5 | YFL014W, YDR129C, YPL235W, YOL140W, YMR170C | YEL031W, YHR107C, YPR118W, YJR051W |
| 1024–2048 | 10 | 10 | YDL021W, YML100W, YKL150W, YEL031W, YGL202W, YDL017W, YGR080W, YPL049C, YGL248W, YEL011W | YMR170C, YCL017C |
| 512–1024 | 11 | 10 | YHR107C, YGL100W, YBR208C, YPR118W, YJL172W, YBR283C, YCR088W, YGR256W, YJL026W, YCL030C | YOL116W |
| 256–512 | 12 | 10 | YCL017C, YOL116W, YNL161W, YJR051W, YKL068W, YHR138C, YGR232W, YMR199W, YOR267C, YJR134C | YGL248W |
| 128–256 | 13 | 10 | YKL141W, YHR074W, YLR330W, YDR436W, YKL129C, YOR020C, YBR117C, YBR125C, YKL073W, YOL022C YLL040C, YNL014W, YML109W, YIL092W, YIL084C, | YIL084C, YML109W |
| <128 | 14 | 15 | YKL145W, YKL075C, YIL002C, YHR015W.YPL008W, YGL006W, YKR031C, YLR035C, YNR067C, YOR093C | YGL006W, YNR067C, YKR031C |
| no expression detecteda | 15 | 15 | YDR381W, YNL208W, YHR020W, YNL160W, YEL024W, YJL008C, YJL111W, YFL037W, YDR023W, YJR123W, YLR340W, YJR077C, YDR321W, YCL018W, YER055C | |
| below QOD (<50 copies/cell)a | 16 | 6 | YBR006W, YCL043C, YDR150W, YOR120W, YJL167W, YBR006W | |
| WB band not quantifiablea | 17 | 6 | YHR029C, YNL055C, YJL080C, YDL140C, YIR006C, YGR284C | |
| never observed in publicly accessible proteomics datasetsc | 18 | 10 | YDL017W, YOL116W, YBR117C, YIL092W, YKL075C, YIL002C, YHR015W, YPL026C, YLR035C, YOR093C | |
Abundances and categorization according to Ghaemmaghami et al. (2003); WB, Western blot
Abundances are as derived from SRM measurements.
As derived from the absence of the protein in PeptideAtlas.
Absolute quantification of proteins in a total cell lysate
To confirm that the abundance range of the detected proteins truly reflected the literature values described by Ghaemmaghami et al. (2003), we used stable isotope labeled reference peptides to absolutely quantify 21 selected proteins distributed across all levels of cellular abundances (Tab. 2). In most cases the measured absolute protein abundances closely matched the published values. For 17 of these proteins the two measured abundance values deviated by less than a factor of three, with the closest values deviating by less than 10%. Four proteins deviated more than fivefold from the published values. The protein with the lowest abundance was measured at ∼40 copies per cell and the highest at over 1E6 copies/cell. These results confirm the quality of the reference list (Ghaemmaghami et al., 2003) and demonstrate that proteins expressed to a concentration of < 50 copies per cell could be detected and quantified by SRM in total yeast lysates.
Table 2.
Absolute quantification of proteins spanning the whole range of abundances using heavy peptide standards.
| Protein | Measured Abundancea (copies/cell) |
Literature Valueb (copies/cell) |
StDev of Measured Abundance |
|---|---|---|---|
| YKL060C | 996503 | 1018216 | 51818 |
| YLR249W | 189235 | 870578 | 30864 |
| YHR183W | 48926 | 101441 | 7354 |
| YLR058C | 98940 | 67559 | 15860 |
| YBR249C | 19459 | 26272 | 5083 |
| YJL136C | 370314 | 15300 | 41142 |
| YMR170C | 1114 | 2072 | 217 |
| YEL031W | 3125 | 1873 | 472 |
| YGL248W | 501 | 1404 | 68 |
| YHR107C | 2317 | 1169 | 144 |
| YPR118W | 2631 | 922 | 477 |
| YJL026W | 13871 | 538 | 1144 |
| YCL017C | 1508 | 504 | 229 |
| YOL116W | 614 | 491 | 154 |
| YJR051W | 3912 | 432 | 911 |
| YIL084C | 162 | 105 | 31 |
| YML109W | 215 | 105 | 39 |
| YKL145W | 2114 | 105 | 234 |
| YGL006W | 90 | 99 | 19 |
| YNR067C | 103 | 64 | 14 |
| YKR031C | 39 | 49 | 9 |
data are expressed as mean values. StDev, standard deviation
as derived from Ghaemmaghami et al. (2003)
Increase of sensitivity and dynamic range by a simple sample fractionation step
We next asked whether the addition of a single sample fractionation step would further increase the detection sensitivity of the method, due to reduction of sample complexity, to single digit copy per cell numbers. We therefore fractionated peptides in a tryptic yeast digest by isoelectric focusing (Malmstrom et al., 2006) using an off-gel electrofocusing (OGE) system, with 24 fractions collected, spanning a 3–10 pI range. Peptides in each fraction were analyzed via scheduled SRM, using the previously validated SRM assays. Overall 260 peptides were monitored across the 24 OGE fractions and for each peptide the fraction associated with the highest SRM signal and the corresponding signal intensity were derived. Fig. 3 shows for every peptide the maximum signal intensity gain achieved by OGE fractionation compared to the signal obtained for the same peptide in the unfractionated peptide mixture. Overall the average signal gain obtained by peptide fractionation was ∼10 fold. The highest signal gain was realized in the fractions corresponding to low-pI peptides (fractions 1–5, pI < 5), with a maximum in the first fraction (average ∼25 fold increase and up to ∼80 fold) (see Supplemental Data for a discussion of the underlying reasons). The signal gain showed no correlation with the protein abundance or the retention time of the peptide (data not shown). These results show that pI-based enrichment of the targeted PTPs by OGE realized a maximal signal gain of more than 50 fold and an average gain of about tenfold compared to the signal recorded from an unfractionated sample. This demonstrates that proteins expressed at a single digit number of copies per cell can be detected by SRM coupled to a simple, fast and predictable sample fractionation step.
Figure 3. SRM analysis of a biological protein network.
(A) Schematic representation of a core protein network (glycolysis/gluconeogenesis/TCA cycle/glyoxylate cycle) in the central carbon metabolism of S. cerevisiae. Proteins are colored according to their absolute abundances as measured by Ghaemmaghami et al. (2003). (B) Distribution of the cellular abundances of the 45 proteins composing the network. For each protein a SRM assay was developed and applied to detect the protein in an unfractionated yeast digest, using scheduled SRM, in a single MS analysis. (C) LC-SRM chromatogram comprising the whole set of SRM assays for proteins in the network. Each peak represents an SRM assay that detects a tryptic peptide of one of the target proteins in the total yeast digest.
Detection of previously undetected S. cerevisiae proteins
Given the analytical depth achieved by SRM-based approach we asked whether the method has the potential to detect proteins that have been undetectable by other techniques. We assembled a set of proteins (Tab. 1) that were not detected before, either by the affinity-based technique (Ghaemmaghami et al., 2003) (Tab. 1, groups 15–17), or by in-depth shotgun proteomics (Tab. 1, group 18), as determined by their absence in the largest publicly accessible proteomic database PeptideAtlas. To target proteins that had not been detected by the affinity based method we followed the approach described above. To target proteins never observed in proteomics experiments we used unpurified synthetic peptides to generate SRM assays for peptides from each of the targeted proteins. For each protein five PTPs likely to be observed by MS were predicted with PeptideSieve and synthesized on a microscale using the SPOT-synthesis (Hilpert et al., 2007; Wenschuh et al., 2000). The peptides were used as a reference for deriving the final optimal coordinates of the SRM assays and for validating the assays. The assays developed were then applied to detect the proteins in a total yeast cell lysate, by scheduled SRM. Overall, of the 45 targeted proteins 37 could be unambiguously detected in the unfractionated yeast digest (Table 1 and see also Tab S1). The observed SRM signal intensities covered a range of ∼2 orders of magnitude. The highest signals were related to proteins that did not express in a tagged format (Ghaemmaghami et al., 2003). The lowest signal intensities were from proteins not previously detected by proteomics techniques, or from those below the detection/quantification limits in the reference method (Ghaemmaghami et al., 2003) (Tab. 1). These results demonstrate that SRM-based proteomics is capable to measure proteins which have been undetectable by other methods and therefore to detect and quantify previously unknown segments of the yeast proteome. Cumulatively, these results demonstrate the potential of SRM-based proteomics to map out the whole MS-observable yeast proteome and provide assays for the detection/ quantification of proteins expressed at a concentration above single digit copies/cell in S. cerevisiae.
Application to a biological protein network
In order to demonstrate the power of the technique when applied to the analysis of a biological system, we targeted a network of 45 proteins in the central carbon metabolism of S. cerevisiae, Fig. 4A. This pathway is an ideal example to demonstrate the dynamic range of the technique since it contains proteins which range from extremely high concentrations (ALF/YKL060C, 1.0E6 copies/cell, third most abundant protein in S. cerevisiae, based on Ghaemmaghami et al., 2003) to very low (ADH4/YGL256W, < 128 copies/cell) and also contains proteins whose abundance could not be measured by the TAP-tagging approach (e.g. LSC2/YGR244C), Fig. 4A, B (Ghaemmaghami et al., 2003). For each protein in the set we developed SRM assays as described above and applied them to measure the proteins in an unfractionated yeast digest, using scheduled SRM, Fig. 4C. The protein network composed of proteins at all abundance levels could be measured using a single 30-minute chromatographic gradient, corresponding to < 1 hour total MS analysis time. In most cases proteins could be measured via two peptides/protein, each peptide was monitored via three SRM transitions and each SRM-chromatographic peak contained at least eight data points which ensured reliable quantification of the eluting peptide. The SRM assays used in this study are available in Table S5 and Table S6 or via the MRMAtlas interface (www.mrmatlas.org, (Picotti et al., 2008)).
Figure 4. Time-course analysis of the central carbon metabolism protein network along the dynamic growth profile of S. cerevisiae in a glucose-rich medium.
(A) Growth profile , as followed by monitoring the optical density at 600 nm (OD600) of the culture, and the extracellular glucose and ethanol concentrations. (B) Different growth phases occurring during S. cerevisiae growth in a glucose-rich medium. The growth profile is plotted using a log10 scale, to better highlight the transition regions. Protein sampling time-points are represented as open squares in panels A and B (T1, 6.5 hrs; T2, 7.6 hrs; T3, 8.7 hrs; T4, 9.6 hrs; T5, 10.6 hrs; T6, 11.7 hrs; T7, 19.8 hrs; T8, 25.0 hrs; T9, 33.7 hrs; T10, 43.2 hrs). (C) Measured abundance profiles for all the proteins in the protein network under study, along the growth curve. Mean abundance changes out of three biological replicates with respect to time point 1 (6.5 hrs) are plotted using a log10 scale. Profiles of the three clusters deriving from the clustering analysis are shown in color. A schematic representation of the system under study highlights proteins belonging to each cluster in the corresponding color. (D–F) Abundance profiles for each protein belonging to cluster 1 (D), 2 (E) and 3 (F). Abundance changes relative to time point 1 are represented as mean (three biological replicates) +/− standard deviation.
We next applied the validated SRM transitions for the whole protein network described above to generate complete quantitative profiles for each protein over a time-course of the dynamic growth of S. cerevisiae, covering a series of different growth phases and a metabolic shift. We sampled the cultures at ten time points in biological triplicate while they transited from exponential growth in a glucose-rich medium, through the diauxic shift and consequent fermentative growth on ethanol, to the entrance in the stationary phase (Fig. 5A and B). The temporal boundaries of the growth phases were established by monitoring the cell density in the medium as a measure of the growth rate, the consumption of extracellular glucose and the accumulation of ethanol in the medium (Fig. 5A and B). The total 30 samples were subjected to scheduled SRM analysis and the resulting data were compiled to derive the quantitative profile, normalized to the first time point (6.5 hrs), for each protein over the dynamic growth profile. Average values are shown out of the three biological replicates in Fig 5C. Proteins were grouped by unsupervised clustering on the basis of their expression profiles into three clusters. Cluster 1 (Fig. 5C and D) predominantly contained enzymes in the glyoxylate cycle and enzymes responsible for the catalysis of the backward or shortcut reactions required to revert flux directions upon the shift (DeRisi et al., 1997). These proteins showed a marked induction (up to 380 fold, average cluster 210 fold) upon the diauxic shift and their levels remained constant or showed only slight reduction during the respiratory growth and at the entrance of the stationary phase. Cluster 2 (Fig. 5C and E) mostly contained enzymes involved in the tricarboxylic acid (TCA) cycle, that were coordinately induced upon the diauxic shift by up to 22 fold (cluster average induction 8 fold) and then showed a slow decrease in abundance, during the slow respiratory growth and the beginning of the stationary phase. Cluster 3 (Fig. 5C and F) predominantly contained glycolytic enzymes which did not show pronounced abundance changes throughout the whole series of different metabolic phases. The post-diauxic induction was statistically significant (p-value <0.05 and fold change >2, see Tab. S4) for all proteins in clusters 1 and 2, except for pyk2 (p=0.063). The coefficient of variation (CV%) of the measurements was on average ∼15% and the data required in total < 2 days of MS time. These data demonstrate the capacity of SRM-based proteomics to comprehensively and reproducibly measure sets of biologically related proteins spanning the whole range of abundances in a single MS run, at high-throughput and quantitative accuracy.
Figure 5. Comparison between targeted proteomics and gene expression data.
(A) Correlation between a protein and the corresponding transcript abundance fold change. Each point represents a given protein at a given time point. Four regions are highlighted: 1) protein and transcript abundance are both increasing; 2) both decreasing; 3) protein abundance increases while transcript abundance decreases, and 4) protein abundance decreases and transcript abundance increases. (B–H) Overlay of a protein and the corresponding transcript abundance profiles. Representative examples are selected out of the 4 regions highlighted in panel A. Protein abundance changes are reported as mean (three biological replicates) +/−standard deviation. Transcript abundance changes are from DeRisi et al, 1997, after the realignment described in the Experimental Procedures.
We next correlated the protein profiles generated in this study with corresponding transcript profiles to detect potential post-transcriptional regulation during the metabolic shift. We compared the protein abundance profiles to a reference microarray dataset for the diauxic shift in S. cerevisiae (DeRisi et al., 1997). The two datasets were acquired under closely similar experimental conditions and the time-course profiles were realigned, normalized and compared in the growth region associated to the shift (timepoints 1 to 6). We detected four general correlation patterns between protein and transcript abundance changes (Fig. 6A). The first pattern (Fig. 6A, region 1) showed cases where protein and mRNA abundances were both increasing. In the second pattern (Fig 6a, region 2) both types of molecules decreased abundances. The third pattern showed cases where protein abundance increased and mRNA abundance decreased and the fourth pattern showed cases where protein abundance decreased and mRNA abundance increased. Most of the measurements populated group 1 or 2, indicating that predominantly the same direction of regulation is observed for a protein and its corresponding transcript. Representative examples for each pattern are shown in Fig. 6B–H, as overlaid time-course profiles for a protein and the corresponding transcript. The availability of full protein and transcript time-course data also allowed us to compare the magnitude and time dependency of the two datasets. In several cases we observed a striking synchronism and closely similar abundance fold changes (e.g. sdh3 and cit1 from group 1 or eno2 from group 2). In other cases we observed a delayed response at the protein level (e.g. fum1, group 1). Further, in several cases at the decaying tail of an induction curve the transcript and protein profiles diverged, the protein levels persisting while the transcripts decayed (e.g. hxk2, group 2). Finally for some proteins the direction of the abundance change was fully unanticipated by the gene expression data (e.g. lsc1, group 3; ald6, group 4). The time-course proteomic dataset and comparisons of the transcripts and protein profiles for each gene are available in the (Supplemental Data Fig. S4 and Supplemental Data Tab. S3).
Discussion
A common requirement in molecular and cellular biology research is the ability to detect and quantify target proteins in biological samples, a need that became even more apparent with the rise of systems biology. For the generation of mathematical models of protein networks (e.g. metabolic or signaling networks), in the context of systems biology research, it is crucial to measure all the elements that constitute the network under a set of different perturbing conditions. Frequently, such sets of proteins cover a wide range of physico-chemical properties and cellular abundances that complicate their detection and quantification. Comprehensive proteomic measurements to face such challenge still suffer substantial limitations. This is optimally illustrated with the relatively simple eukaryotic organism S. cerevisiae, the species with the best characterized proteome to date. In spite of the considerable efforts worldwide, applying a range of experimental approaches, about 20% of the predicted yeast proteome has never been detected and, more generally, to date no proteome has been fully mapped yet. A main reason for this is the difficulty in detecting low abundant proteins. However low-copy-number proteins (< 1000 copies per cell) are extremely attractive targets in systems biology research, since they often play a crucial role, e.g. as signal transducers, isoenzymes or regulators of cellular processes. In the literature there are anecdotal reports claiming the detection of low abundant proteins. However, these results were generally achieved in targeted studies in which a specific protein was studied, e.g. after affinity enrichment (Ghaemmaghami et al., 2003) or by large-scale proteomic studies whereby with intensive efforts the proteome was extensively fractionated via multiple separation/enrichment steps prior to shotgun MS, resulting in identification of thousands of proteins (Chen et al., 2006; de Godoy et al., 2008; de Godoy et al., 2006). Although powerful, the latter studies require weeks of data acquisition and instrumental analysis per each single sample and are thus not practical when multiple samples and replicates have to be consistently analyzed, as is the case in dosage series or time-course experiments required for systems biology.
Here we demonstrate that SRM-based proteomics has the power to detect and quantify yeast proteins expressed in the whole range of cellular abundance, down to less than 50 protein molecules per cell. Proteins could be detected in total yeast cell digests, without the need for sample fractionation or enrichment, making the use of the technique fast and practical. The technique is also highly multiplexed, supporting the detection and quantification of more than one hundred different proteins, deliberately chosen and spanning all levels of abundance, in a single analysis. This allows to comprehensively monitor entire protein networks in a 1-hour MS run and thus to analyze in a reasonable time the effects on the system under study of different perturbing conditions and replicates. This satisfies in an ideal way the growing demand of systems biology for consistent, complete, and quantitative data sets from cells in differentially perturbed states. We illustrate the performance of targeted proteomics and the utility of the data by analyzing the dynamics of the proteins in the central carbon metabolism of S. cerevisiae over a complete growth time-course including a series of growth phases and a metabolic shift. This is, to date, the most complete quantitative dataset describing the responses of each protein in the network to the series of events occurring during S. cerevisiae growth, at high temporal resolution. The whole SRM analysis required < 2 days, and 30 samples were analyzed at the speed of 1 sample/hour, resulting in total 1,350 protein abundance measurements. Previous attempts to capture by proteomics the events occurring upon glucose consumption during S. cerevisiae growth resulted in low coverage of the system under study ((Futcher et al., 1999) and (Haurie et al., 2004), 13 and 15 out of the 45 proteins composing the system detected, respectively, with a clear bias towards the most abundant components). Likely, the application of advanced shotgun proteomic methods involving extensive sample fractionation (de Godoy et al., 2008) would increase the coverage achieved by these studies, albeit at a significant cost in time, material and labor.
Due to to the lack of comprehensive, quantitative proteomic datasets, thus far the closest descripiton of enzyme abundance changes upon glucose exhaustion in S. cerevisiase was derived from microarray studies (Brauer et al., 2005; DeRisi et al., 1997; Gasch et al., 2000; Radonjic et al., 2005)). The strong induction observed in our study of all TCA and glyoxylate cycle enzymes, of fba1 and adh2 agrees with what previously extrapolated from transcriptomics data sets (although with different timing and regulation extent) and biochemical analyses. Thus, our data confirm the current view of a metabolic remodelling that redirects carbon fluxes from fermentative to respiratory pathways upon glucose exhaustion and thus release from glucose repression (Brauer et al., 2005; DeRisi et al., 1997; Polakis and Bartley, 1965). Specifically, this entails 1. the activation of the anaplerotic glyoxylate cycle to regenerate TCA intermediates 2. the reversion of glycolysis with consequent induction of key enzymes such as fbp1, that switch irreversible glycolytic steps and 3. the activation of respiratory enzyme isoforms (e.g adh2) (Brauer et al., 2005; DeRisi et al., 1997; Gasch and Werner-Washburne, 2002). Most of the protein expression differences persist through the post diauxic phase until entrance to the stationary phase (this study, (Radonjic et al., 2005)). In addition, based on transcriptomic analyses it has been assumed that glycolytic enzymes and enzymes that control flow of metabolites to ethanol during fermentation (pdc1–6) undergo a decrease in abundance upon glucose exhaustion (Brauer et al., 2005; DeRisi et al., 1997; Gasch et al., 2000). This is in agreement with the reported lower glycolytic and pdc activity during respiratory growth (Entian and Zimmermann, 1980). Our data show that this extrapolation from transcriptomic data is not correct. Instead the abundance of glycolytic and pdc enzymes is not significantly changing throughout the whole growth profile, even though the corresponding transcripts decrease (see pdc1–6, fba1, pyk1, gpm1, gpm3, pgk1, pgi1, adh1, pfk1, Fig. S3). This suggests that potential post-trascriptional regulation of glycolysis and pdc- activity occurs upon the diauxic shift in S. cerevisiae, in analogy to what recently proposed for the metabolic adaptation of yeast to benzoic acid treatment and oxygen deprivation (Daran-Lapujade et al., 2007). Therefore our dataset confirms previous knowledge but also carries a significant amount of new information, in terms of timing and regulation extent, for the protein network upon the metabolic shift that was not apparent from transcriptomic analyses alone, thus highlighting points of potential post-transcriptional regulation. This shows that accurate proteomic datasets such as the one generated in this study are required to provide a detailed picture of how protein networks adapt to changing conditions. The dataset in particular provides an ideal framework for the improvement of mathematical models of metabolic reprogramming in S. cerevisiae. This overall confirms that the SRM approach provides a simple, economical and fast way to explore the dynamics of cellular pathways, which will find broad applications in systems biology, but also in medical and pharmaceutical research (e.g. drug screening).
The results of the study also show that SRM detects proteins which have not been detectable before either by mass spectrometry or quantitative Western blotting, indicating that the number of proteins expressed in S. cerevisiae cells in log-phase growth is higher than previously reported (Ghaemmaghami et al., 2003). The data also suggest that it will now be possible, for the first time, to map out previously unknown segments of the yeast proteome, an advance that will also have significant implications for genome annotation.
The SRM technology showed a high success rate (∼90%) in detecting proteins expressed in yeast cells. Examples of failed detection include highly modified proteins, cell-wall or membrane proteins or low abundant proteins which lack PTPs with good MS properties (see Table S1). Variations of the technology applied here, e.g. the use of proteases different from trypsin, testing a higher number of PTPs in the case of highly modified proteins or adapting the protein extraction procedure to detect membrane/cell-wall proteins will further increase the success in detecting previously undetected proteins.
The addition of a single-step of peptide fractionation, performed by the well-established and fast technique of off-gel electrofocusing further increased the sensitivity by on average one order of magnitude, thereby allowing the detection of proteins expressed at a single digit copies/cell concentration. The additional sensitivity provided by OGE might be exploited to detect difficult, low abundant proteins which do not contain PTPs with good MS response, or to follow the down-regulation of proteins of low abundance. Targeting PTPs with pIs in the range 3–4.5, which showed the strongest signal gain in OGE fractions, can significantly increase the sensitivity of the SRM assay by up to 80 times (compared to the unfractionated samples).
When developing the SRM assays used in this work, a bottleneck step, in particular for low abundant proteins, was the validation of the SRM transitions that constitute the definitive mass spectrometric assay in the type of mass spectrometer used for the measurements. This is typically achieved by acquiring a full fragment ion spectrum of the targeted peptides. For low intensity peptides high quality fragment ion spectra are difficult to generate from full yeast digests due to the interference of the high level background. In such cases, validation required acquisition of the fragment ion spectra for the peptide in lower complexity sample (e.g. using OGE fractions). To facilitate the process we used unpurified synthetic peptides to develop and validate SRM assays for proteins that proved undetectable by other techniques. The use of such artificial proteomes strongly increased the confidence of the assay validation and increased the throughput in generating SRM assays. It is also particularly advantageous for targeting previously undetected proteins and therefore represents a significant advance in achieving complete proteome coverage by MS.
The final coordinates of SRM assays become universally useful and exportable. To this purpose, we developed a centralized web-based resource (Picotti et al., 2008) to store and allow the fast diffusion and usage of the SRM assays across different laboratories. The resource currently contains SRM assays for >1,500 yeast proteins, including complete cellular pathways, such as the one shown in Fig. 4.
In conclusion this study demonstrates the potential of SRM-based proteomics to map out the whole observable yeast proteome by using assays to detect and quantify virtually any protein expressed at a concentration above single digit copies/cell. It shows that quantitative assays for complete protein networks of interest can be developed and deployed to monitoring the dynamics of any network under study, across a high number of samples and replicates, at unprecedented speed, and with high quantitative accuracy. The described development of highly specific assays is generally applicable to any protein and proteomics project. These advances open a new avenue in the quantitative and qualitative analysis of proteins in the context of systems biology research and make the fast quantitative analysis of any protein in a proteome a concrete possibility.
Experimental Procedures
Detailed protocols are available in the Supplemental Experimental Procedures.
Sample preparation
Yeast cells were grown in two biological replicates to log-phase at conditions closely matching those of Ghaemmaghami et al. (2003). Before lysis aliquots of the cell suspension were subjected to cell counting in a Neubauer chamber and averaged results were expressed as cells/ml. Pelleted cells were disrupted by glass bead beating, proteins were precipitated by cold acetone, reduced with 12 mM dithiotreitol and alkylated with 40 mM iodoacetamide and digested with sequencing grade trypsin (Promega). Peptide samples were cleaned by Sep-Pak tC18 cartridges (Waters, Milford, MA, USA). The peptide mixtures were either directly destined to MS analysis or firstly separated by off-gel electrofocusing (OGE) using a pH 3–10 IPG strip (Amersham Biosciences, Otelfingen, Switzerland), and a 3100 OFFGEL Fractionator (Agilent Technologies) with collection in 24 wells. Peptides collected in each well were cleaned as previously described and all peptide samples were evaporated to dryness and resolubilized in 0.1% formic acid for MS analysis.
In the time-course experiments yeast cells were grown in yeast extract peptone dextrose (YEPD, 20 g/L glucose) medium in triplicate. Cells sampled at each timepoint (from inoculation: 6.5, 7.6, 8.7, 9.6, 19.6, 11.7, 19.8, 25.0, 33.7 and 43.2 hrs) were subjected to protein extraction and digestion. The optical density (OD) at 600 nm of the yeast cultures was measured regularly. Aliquots of the culture broth were analyzed with an HPLC system (Agilent HP1100), equipped with a refractive index detector and an UV/Vis-detector (DAD) to determine the extracellular concentration of glucose and ethanol, using calibration curves constructed with external standards.
Design of SRM assays
A set of S. cerevisiae proteins was selected containing proteins evenly distributed across the full range of cellular concentrations (Ghaemmaghami et al., 2003). Proteins which could not be detected by previous proteomic (Deutsch et al., 2008) or affinity-based (Ghaemmaghami et al., 2003) techniques were added to the list. For the network analysis a set of 45 proteins composing the core of carbon metabolism in S. cerevisiae was assembled. For each protein 3–5 PTPs were selected based on previous evidence (www.peptideatlas.org (Deutsch et al., 2008),) or by bioinformatic prediction using PeptideSieve (Mallick et al., 2007). For each peptide 3–8 transitions for each of the two main charge states were calculated, corresponding to y-series fragment ions. The transitions were used to detect the peptides in whole yeast protein digests or in OGE peptide fractions by SRM and to trigger acquisition of a MS/MS spectrum for each peptide. For proteins not observed in PeptideAtlas the five predicted PTPs were synthesized in an unpurified format, via the SPOT-synthesis (JPT Peptide Technology, Berlin, Germany) and used as a reference to develop the corresponding SRM assays.
Mass spectrometry analysis
MS analyses were performed on a hybrid triple quadrupole/ion trap mass spectrometer (4000QTrap, ABI/MDS-Sciex, Toronto). Chromatographic separations of peptides were performed on a Tempo nano LC system (Applied Biosystems) coupled to a 16 cm fused silica emitter, 75 µm diameter, packed with a Magic C18 AQ 5 µm resin (Michrom BioResources, Auburn, CA, USA). Peptides (up to 3.5 micrograms of total protein digest) were separated with a linear gradient from 5 to 30% acetonitrile in 30 or 60 minutes, at a flow rate of 300 nl/min. In the SRM assays validation phase the mass spectrometer was operated in MRM mode, triggering acquisition of a full MS/MS spectrum upon detection of an SRM trace. Each SRM assay was validated by acquiring a full MS/MS spectrum for the peptide. For detailed information on the MS operating conditions see Supplemental Experimental Procedures. Upon validation, the SRM assays were used to detect and/or quantify the proteins in total cell lysates and in each of the 24 OGE fractions, using scheduled SRM mode (retention time window, 180 s; target scan time, 3.5 sec). Blank runs were performed regularly, in which the same set of transitions was monitored as in the following (sample) run. Blank runs were perfomed until no signal was detected for all transition traces, in particular prior any measurement of low abundant proteins. Where synthetic peptides were available, validation of peptide identities in yeast samples was based on the analogy of chromatographic and fragmentation properties to those of the standard. For low abundant proteins the relative intensities of SRM traces were confirmed to match those of the corresponding fragment ions in the MS/MS spectrum of the peptide.
Database search and extraction of optimal SRM transitions
MS/MS spectra were assigned to peptide sequences by a target-decoy sequence database searching strategy using the tool Sequest. The search results were validated and assigned probabilities using the PeptideProphet program (Deutsch et al., 2008), with decoy-assisted semiparametric model and filtered as in (Picotti et al., 2008). For each peptide, the three fragment ions resulting in the highest signals were extracted from the triple quadrupole MS2 spectra (Picotti et al., 2008). The selected transitions were reanalyzed in scheduled SRM mode and the two peptides resulting in the maximal intensities were selected as final SRM assay for each protein. To accept validation of a set of SRM traces we checked that the retention time at which the MS2 spectrum was acquired matched that of the SRM peaks for the target peptide and we confirmed “coelution” of all SRM traces for the peptide. The SRM assay dataset was uploaded to the MRMAtlas (www.mrmatlas.org) (Picotti et al., 2008) and is available in Table S5 and Table S6.
Quantitative analysis
For absolute quantification, isotopically labeled synthetic versions of the selected PTPs (see Supplemental Experimental Procedures) were purchased from Thermo Scientific (Ulm, Germany). The synthetic peptides were used for collision energy and declustering potential optimization. A known amount of heavy peptide peptides was added prior to trypsinization to the protein mixtures. For relative quantification in the time course analyses each protein sample was mixed prior to trypsinization to an equal amount of yeast proteins extracted from 15N-completely labeled yeast cells, used as an internal reference (see Supplemental Experimental Procedures). The SRM assays were used to measure the peptides in the heavy and light versions using scheduled SRM.
Peak height for the transitions associated to the heavy and light peptides were quantified using the software MultiQuant v. 1.1 Beta (Applied Biosystems). Each transition for a given peptide was treated as an independent abundance measurement. Absolute quantification was obtained from the ratio between the light and heavy SRM peak height, multiplied by the known amount of the standard. Results were related to the number of cells processed and expressed in protein copies/cell as mean out of the different transitions, peptides and the two biological replicates, +/− the standard deviation. The potential contamination of the heavy peptide preparations with the corresponding unlabelled peptides was tested by injecting the heavy peptides alone and monitoring the transitions for both the heavy and light peptide forms. At the concentration used for quantitative measurements no signal was detectable in the ‘light’ transitions.
For relative quantification of each protein across the growth time-course the ratio between the light and heavy SRM peaks height was calculated and normalized to that obtained at the growth timepoint 1 (6.5. hrs). Results were expressed as mean out of the different transitions/peptide, peptides/protein and the three replicate cultures, +/− the standard deviation. Outlier transitions (e.g. shouldered transition traces, or noisy transitions with S/N < 3) were not considered in the calculations.
Time-course data analysis. Protein time-course profiles were compared to transcript data (DeRisi et al. 1997). The datasets were realigned using the transition midpoints and the glucose consumption curves and overlaid (Fig. 6, B–H). A comparison of the protein and transcript fold changes was performed in the growth time frame covered by both datasets, normalizing transcript fold changes to the first common sampling point. When sampling was performed at different time intervals, transcript fold changes at matching points was linearly extrapolated from the two closest measured time points.
The log10 transformed profiles of the mean protein fold changes were subjected to K-means clustering (Macqueen, 1967) (4 initial clusters). A Student’s paired t-test was performed to determine statistically significant changes in protein abundances upon the metabolic shift. The threshold for statistical significance was p < 0.05 and an abundance change > 2 fold (Table S4).
Supplementary Material
Supplemental Data include Supplemental Experimental Procedures, Supplemental Discussion, four figures, and six tables and can be found with this article online at http:
Figure 2. Analysis of the SRM signal gain obtained by peptide fractionation.
Peptide fractionation was achieved by off-gel electrofocusing of a total yeast digest in 24 wells, using 3–10 pI strips. For each peptide the signal gain compared to the unfractionated peptide mixture resulting from the highest SRM transition and the highest-concentration fraction is reported using a logarithmic scale (left axis). Vertical bars show the mean signal gain in each fraction. The peptide pI associated to each fraction is also reported (right axis) as mean +/−standard deviation, as derived from a large-scale shotgun proteomic experiments in which more than 55,000 yeast peptides were identified across the 24 OGE fractions (data not shown).
Acknowledgements
We acknowledge Holger Wenschuh (JPT Peptide Technologies), Marko Jovanovic, Vinzenz Lange, Nichole King, and Matthias Heinemann for insightful discussions. We are also grateful to Roeland Costenoble and Uwe Sauer for providing 15N-labeled S. cerevisiae cells. This project has been funded in part by ETH Zurich, the Swiss National Science Foundation, with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract No. N01-HV-28179 and by SystemsX.ch the Swiss initiative for systems biology. B.B. is the recipient of a fellowship from the Boehringer Ingelheim Fonds. PP is the recipient of a Marie Curie Intra-European fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT) Mol Cell Proteomics. 2006;5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Rossell S, van Gulik WM, Luttik MA, de Groot MJ, Slijper M, Heck AJ, Daran JM, de Winde JH, Westerhoff HV, et al. The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc Natl Acad Sci U S A. 2007;104:15753–15758. doi: 10.1073/pnas.0707476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Desiderio DM, Kai M. Preparation of stable isotope-incorporated peptide internal standards for field desorption mass spectrometry quantification of peptides in biologic tissue. Biomed Mass Spectrom. 1983;10:471–479. doi: 10.1002/bms.1200100806. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Broder S. Implications of new proteomics strategies for biology and medicine. J Proteome Res. 2004;3:253–260. doi: 10.1021/pr034082c. [DOI] [PubMed] [Google Scholar]
- Entian KD, Zimmermann FK. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980;177:345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Haurie V, Sagliocco F, Boucherie H. Dissecting regulatory networks by means of two-dimensional gel electrophoresis: application to the study of the diauxic shift in the yeast Saccharomyces cerevisiae. Proteomics. 2004;4:364–373. doi: 10.1002/pmic.200300564. [DOI] [PubMed] [Google Scholar]
- Hilpert K, Winkler DF, Hancock RE. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat Protoc. 2007;2:1333–1349. doi: 10.1038/nprot.2007.160. [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, et al. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics. 2008;7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen J. Some methods for classification and analysis of multivariate observations. Proc 5th Berkeley Symp on Math Statist Probab; Berkley, CA. IEEE; 1967. pp. 281–297. [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- Malmstrom J, Lee H, Nesvizhskii AI, Shteynberg D, Mohanty S, Brunner E, Ye M, Weber G, Eckerskorn C, Aebersold R. Optimized peptide separation and identification for mass spectrometry based proteomics via free-flow electrophoresis. J Proteome Res. 2006;5:2241–2249. doi: 10.1021/pr0600632. [DOI] [PubMed] [Google Scholar]
- Pawlak M, Schick E, Bopp MA, Schneider MJ, Oroszlan P, Ehrat M. Zeptosens' protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics. 2002;2:383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics. 2007;6:1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- Picotti P, Lam H, Campbell D, Deutsch E, Mirzaei H, Ranish J, Domon B, Aebersold R. A database of mass spectrometric assays for the yeast proteome. Nat Methods. 2008;5:1–2. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis ES, Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965;97:284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- Taussig MJ, Stoevesandt O, Borrebaeck CA, Bradbury AR, Cahill D, Cambillau C, de Daruvar A, Dubel S, Eichler J, Frank R, et al. ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. tNat Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- Uhlen M. Affinity as a tool in life science. Biotechniques. 2008;44:649–654. doi: 10.2144/000112803. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Hober S. Generation and validation of affinity reagents on a proteome-wide level. J Mol Recognit. 2008 doi: 10.1002/jmr.891. [DOI] [PubMed] [Google Scholar]
- Wenschuh H, Volkmer-Engert R, Schmidt M, Schulz M, Schneider-Mergener J, Reineke U. Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers. 2000;55:188–206. doi: 10.1002/1097-0282(2000)55:3<188::AID-BIP20>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Supplemental Experimental Procedures, Supplemental Discussion, four figures, and six tables and can be found with this article online at http: