Abstract
Smad proteins are intracellular molecules that mediate the canonical signaling cascade of TGFβ superfamily growth factors. The TGFβ superfamily comprises two groups of growth factors, BMPs and TGFβs. Both groups can be further divided into several sub-groups based on sequence homologies and functional similarities. Ligands of the TGFβ superfamily bind to cell surface receptors to activate Smad proteins in the cytoplasm; then the activated Smad proteins translocate into the nucleus to activate or repress specific target gene transcription. Both groups of growth factors play important roles in skeletal development and regeneration. However, whether these effects reflect signaling through canonical Smad pathways, or other non-canonical signaling pathways in vivo remains a mystery. Moreover, the mechanisms utilized by Smad proteins to initiate nuclear events and their interactions with cytoplasmic proteins are still under intensive investigation. This review will discuss the most recent progress understanding Smad signaling in the context of skeletal development and regeneration.
1. Introduction
1.1 TGFβ superfamily
BMPs were initially discovered by the fact that demineralized bone matrix can initiate bone formation when transplanted to ectopic sites in rodents [1]. Later, TGFβs were discovered in studies of platelet derived growth factor (PDGF) and epidermal growth factors (EGF/TGFα) [2, 3]. Eventually, other related ligands were identified, leading to the definition of the TGFβ superfamily, consisting of BMPs, TGFβs, and other groups of proteins such as growth and differentiation factors (GDFs), activins, inhibins and Mullerian inhibitory factor (MIF) [4]. Interestingly, although BMPs reserve a bone forming capability in different species, mammalian TGFβs have been found to only induce bone formation with site and tissue specificity in non-human primates [5, 6].
1.2. Smad proteins
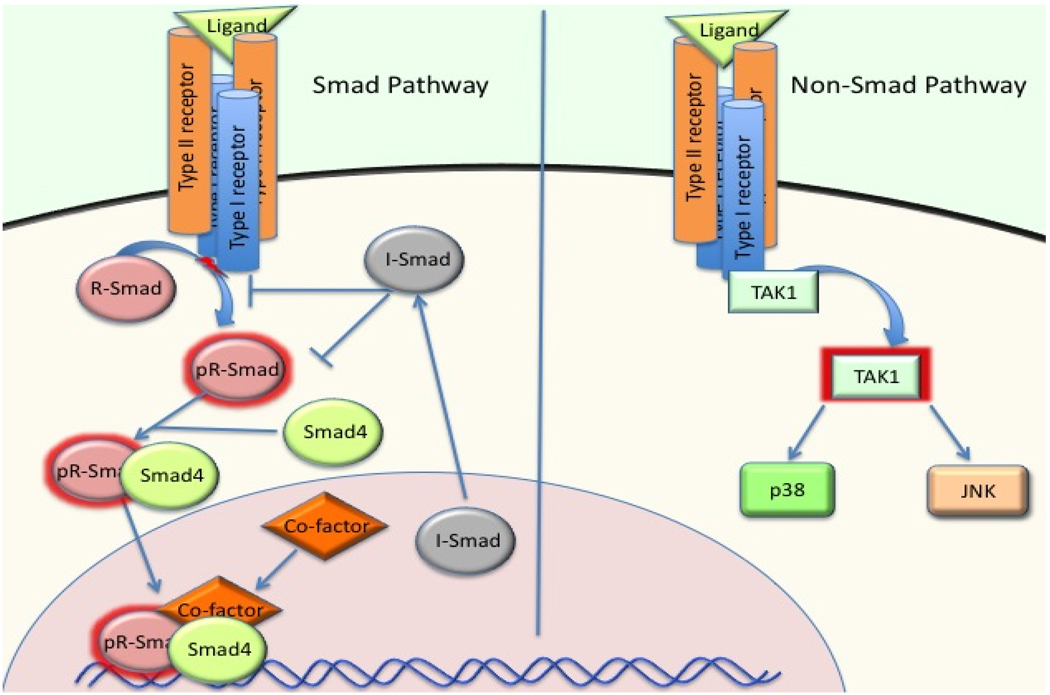
The first description of Smad proteins was the finding of Mothers Against Dpp (MAD) in Drosophila, which modified the phenotype of decapentaplegic (dpp; a BMP ligand) mutants [7]. Later studies identified Sma proteins in C. elegans as closely related to MAD, and both mediated signaling downstream of serine/threonine kinase receptors of TGFβ superfamily proteins [8]. Therefore, homologs of Mad and Sma have been named Smad. So far, 8 mammalian Smad proteins have been isolated, designated Smad1 through Smad8. The Smad proteins are divided into 3 groups according to their functions. The first group is the receptor-regulated Smads (RSmads), which include Smad1, 2, 3, 5 and 8. These Smad proteins bind to membrane bound serine/threonine receptors, and are activated by the kinase activity of the receptors. The second group includes only one member, Smad4. Smad4 acts as a co-factor that binds to the activated R-Smads to form a complex that translocates into the nucleus. Therefore Smad4 has been named Co-Smad. The third group comprises the inhibitory Smads (I-Smads), which includes Smad6 and Smad7. These two Smads exert an inhibitory effect on the signaling cascade by various mechanisms (Figure 1).
Figure 1. The Smad and Non-Smad pathway of BMP/TGFβ signaling.
BMP/TGFβ signaling in vivo is mediated by either the Smad pathway (canonical) or the Non-Smad (non-canonical) pathways. Both pathways could mediate important functions of BMP/TGFβ in skeletogenesis. The non-Smad pathways also regulate Smad proteins by modulating linker region and C-terminal phosphorylation. Moreover, Smad proteins also modulate activities of molecules in Non-Smad pathways. The preference of intracellular pathways by different receptor complexes, the crosstalk between these two pathways, and the exact signaling mechanisms of the Non-Smad pathways in skeletal system remain to be investigated.
Smad proteins also share similar structures. A typical Smad structure includes N-terminal MH1 domain, linker region and a C-terminal MH2 domain. The MH1 domain is highly conserved in all R-Smads and Smad4, but not in I-Smads. The major function of the MH1 domain is to mediate DNA binding of Smad proteins. The linker region is highly variable in different Smads. It is the target of regulation by other intracellular proteins through phosphorylation, ubiquitination, or sumoylation. The MH2 domain is present in all Smads. Activation of R-Smads is through the phosphorylation of a Ser-X-Ser motif in the MH2 domain by activated receptors. The MH2 domain is also responsible for Smad protein interactions with other intracellular proteins and transcriptional activation of target genes [9]. Different receptors in the TGFβ superfamily have different preferences for binding to R-Smad proteins. For example, Smad1, 5 and 8 mediate BMP signaling by interacting with the BMP receptors ALKs 1, 2, 3, and 6, whereas Smad2 and 3 mediate TGFβ and Activin signaling through the TGFβ/activin receptors ALKs 4 and 5. Smad6 is more specific for the inhibition of BMP signaling, whereas Smad7 has inhibitory effects on both BMP and TGFβ signaling (for more detailed reviews, see [9, 10]). The consensus understanding so far is that R-Smads require Smad4 binding before they can translocate into the nucleus, Recent evidence has challenged this dogma, as Smad4 conditional deletion in mice did not cause significant skeletal defects[11]; while, conditional deletion of Smad1/5/8 led to lethality at birth due to severe chondrodysplasia[12].
1.3 Skeletal development and regeneration
Skeletal development in mammals is accomplished via two different mechanisms. In intramembranous bone formation, mesenchymal cells from neural crest and cephalic mesoderm differentiate into osteoblast cells to form the major cranial vault and clavicles. Endochondral bone formation accounts for the development of the majority of the skeleton. Initially, mesenchymal cells from the mesoderm condense at the sites where the bones will form. The condensed mesenchymal cells then proliferate and differentiate into chondrocytes, forming cartilaginous anlagen of the future bones. Later, the chondrocytes undergo terminal differentiation and become replaced by invading osteoblasts to form the mineralized bone tissue (for a more detailed review of endochondral bone formation, see [13]). BMPs are important for condensation, and they are required for the initiation of chondrocyte differentiation by inducing Sox9 expression. BMPs also regulate chondrocyte differentiation at later stages by interacting with other signaling pathways such as Indian Hedgehog (Ihh), Parathyroid Hormone Related Peptide (PTHrP) and Fibroblast Growth Factor (FGF) (see later in this review). In the osteoblasts, BMP signaling is required for the commitment of mesenchymal cells toward the osteoblast lineage. Similarly, TGFβ/Activin signaling is important in chondrogenesis and osteogenesis, working synergistically or antagonistically with BMP signaling, depending on the stage of differentiation.
Skeletal regeneration is a process of new bone formation after trauma or injury. The new bones form at the site of injury, and could involve both intramembranous and endochondral bone formation. Therefore, skeletal regeneration is considered a process recapitulating development. However, there are particular differences between these two processes, and these will be discussed more in this review later.
In the past few decades, studies with gene deletions, targeted gene modification, and overexpression of BMP/TGFβ signaling pathway components have revealed essential roles of BMPs and TGFβs in skeletal development in vivo. Several excellent reviews have addressed this topic. [14–16]). Therefore, in this review, we focus on a less well-understood aspect of BMP/TGFβ biology: the roles of Smad family members. Until recently, it was widely assumed that BMP and TGFβ signaling are mediated predominantly by R-Smad proteins acting in concert with Smad4. However, recent studies have increased awareness of the potential importance of non-Smad mediated BMP and TGFβ signaling [17, 18]. The existence of the non-Smad pathways raises the question of the relative importance of canonical Smads vs. non-canonical (non-Smad) pathways. Moreover, intensive studies have described a sophisticated Smad-dependant network of signaling events that are important in many processes, including normal development, regulation of immune system, tumor initiation, tumor metastasis and others. Conclusions from these studies indicated that the mechanisms of Smad signaling are quite species, tissue, and cell type specific. Therefore, understanding the mechanisms of Smad signaling in the context of skeletal development and regeneration requires additional focused studies that are carried out in vivo under physiological conditions.
2. Role of Smads in Chondrogenesis
Smad proteins are ubiquitously expressed in chondrocytes during the entire process of chondrogenesis [41]. However, due to the different functions of BMP and TGFβ pathways, Smads have overlapping but distinct patterns of activity in different stages of chondrogenesis. Elucidating the roles of Smad proteins in vivo involves several important questions. First, within the two different groups of R-Smads, Smad1/5/8 and Smad2/3, does each of the Smads play distinct roles in signal transduction, or do they have essentially overlapping functions? Second, to what extent is BMP/TGFβ signaling mediated by R-Smads at distinct stages of skeletal development? Third, what is the evidence that Smad4 is required for Smad1/5/8 and/or Smad2/3 signaling? Finally, what role, if any, do the I-Smads play in skeletal development and regeneration? Several mouse models with knockout, conditional knockout, and overexpression of Smad proteins have been generated to study functions of Smads (See table 1). The phenotypes of these mice, if not lethal before skeletogenesis, normally include skeletal defects, confirming the important functions of Smads in this process.
Table 1.
| Gene | Mutation | Promoter | Phenotype | Ref |
|---|---|---|---|---|
| Smad1 | −/− | - | Lethal at E 10.5, defect in extra- embryonic tissues and germ cell formation |
[19] |
| Smad2 | −/− | - | Lethal at E 7.5–12.5, defects in primitive streak formation, A-P axis formation in epiblast, and gastrulation |
[20] [21]; [22]; [23] |
| Smad2 | ES chimera | Lac-z marked Smad2 deficient cells |
Absence of Smad2 deficient cells in definitive endoderm |
[24] |
| Smad3 | −/− | - | Colorectal carcinoma, immuno-function defect, and osteoarthritis later in life |
[25]; [26]; [27]; [28]; [29] |
| Smad4 | −/− | - | Lethal at E 6.5–8.5, defects in gastrulation (mesoderm), anterior truncation of embryos |
[30]; [31] |
| Smad4 | CKO | Col2a1-Cre | Dwarfism | [11] |
| Smad4 | CKO | Osteocalcin-Cre | Reduced osteoblast proliferation and function |
[32] |
| Smad5 | −/− | - | Lethal at E 10.5–11.5, defects in angiogenesis |
[33] |
| Smad6 | −/− | - | Partially lethal, defects in endocardial cushion formation, and aortic ossification and high blood pressure in viable mutants |
[34] |
| Smad6 | Overexpression | Col11a2 | Delayed chondrocyte hypertrophy, dwarfism with osteopenia |
[35] |
| Smad7 | MH2 domain deletion |
- | Partially lethal, defects in endocardial cushion formation |
[36] |
| Smad7 | Exon1 deletion | - | Mutant is smaller, altered B cell response to TGFβ signaling, increased fibrogenesis. |
[37]; [38]; [39] |
| Smad7 | Overexpression |
Prx1-Cre; 11Enh-Cre; 11Prom-Cre |
Decreased chondrocyte proliferation; differentiation |
[40] |
|
Smad1 /5/8 |
Smad1cko;Smad 5cko;Smad8−/− |
Col2a1-Cre | Severe chondrodysplasia with embryonic lethality |
[12] |
Since BMP/TGFβ signaling is important for nearly every aspect of development, it is not surprising that global knockout of an R-Smad generally leads to early embryonic lethality. Smad1 knockout mice die in mid-gestation due to extra-embryonic defects [19]. Smad2 knockout mice die at embryonic day 7.5–12.5 due to defects in primitive streak formation and failure to establish an anterior-posterior axis within the epiblast or defects during gastrulation[20–23]. Moreover, studies of Smad2-deficient chimeric mice revealed that Smad2, but not Smad3, mRNAs were expressed in visceral endoderm, and definitive endoderm formation is Smad2-dependant, indicating a unique function of Smad2 [24]. Global knockout of Smad4 causes early embryonic lethality at day E6.5 to E8.5, due to defects in gastrulation [30, 31], Smad5 knockout mice die between embryonic day 10.5 and 11.5, because of defects in angiogenesis [33]. Interestingly, Smad8 knockout mice do not have apparent defects, suggesting Smad1 and 5 could compensate for most of the functions of Smad8 [12, 42, 43]. Smad3 knockout mice survive birth, but develop colorectal cancer, impaired immunonological functions, and osteoarthritis later in life[25–29]. These studies suggested that Smad1 and Smad5 are both indispensible in BMP signaling, especially in early development, although they might share some overlapping functions. Similarly, Smad2 signaling is indispensible in embryonic development. Smad2 or other mechanisms could compensate for the loss of Smad3 in early development, suggesting the functional overlap of Smad2 and Smad3. However, in later stage of life, Smad3 has several indispensible functions that are different from Smad2. In particularly, Smad3 is required for maintenance of normal immuno-suppressive responses, articular chondrocyte homeostasis, and the tumor suppressive functions of TGFβ.
With the advancement in the genetic technologies, use of Cre-LoxP system allowed conditional deletion of Smad genes in skeletal tissue. Since direct evidence of the functional role of Smad1/5/8 in early chondrogenesis and growth plate chondrocytes is still missing, we have recently generated conditional deletions of Smad1/5/8 in cartilage using Col2a1-Cre [12]. Smad1/5/8 triple deletions yielded early embryonic lethality and closely phenocopied mice lacking the BMP receptors ALK3 (BMPR1A) and ALK6 (BMPR1B) [44]. These mice do not form any endochondral skeleton; condensations form, but with the onset of Col2a1-Cre expression, any further development is blocked. Smad1/5 double mutant mice have very similar phenotypic presentations to Smad1/5/8 triple mutants, suggesting that Smad8 plays a very minor role in chondrogenesis. In contrast, individual loss of Smad1, 5 or 8, and mice carrying only a single allele of Smad5 (Smad1−/−;Smad5+/−;Smad8−/−) in cartilage are viable and form a nearly normal skeleton. This observation demonstrates that Smads1 and 5 exhibit extensive functional overlap. Additional studies demonstrated that the BMP signaling mediated by Smad1/5 is required for the regulation of the Ihh/PTHrP feedback loop and the antagonism between BMP and FGF signaling in the growth plate[12].
An important finding from these studies is that the majority of BMP signaling in endochondral bone formation appears to be mediated by canonical Smad1/5 pathways as opposed to noncanonical pathways. Moreover, it is surprising that Smad4 is not required for skeletogenesis.. As discussed above, the current dogma is that Smad4 is a required co-Smad for canonical BMP and TGFβ signaling, and Smad4 is expressed ubiquitously in all zones of growth plate [41]. However, conditional deletion of Smad4 in cartilage leads to fairly minor defects; Smad4cko mice develop dwarfism post-natally, mainly as a result of a disorganized growth plate. The Smad4- deficient growth plate showed an expanded resting zone, reduced proliferation, accelerated differentiation and increased apoptosis of chondrocytes, as well as ectopic bone collar formation in the perichondrium and loss of responsiveness to TGFβ1 [11]. Given that loss of Smads1 and 5 leads to a total arrest in chondrogenesis at the condensation stage, these data indicate that BMP signaling in skeletogenesis is largely independent of Smad4.
Potential mechanisms by which BMP R-Smads may mediate their effects in skeletal cells have been the topic of numerous in vitro studies. As aforementioned, BMPs induce chondrogenesis by regulating Sox9 expression in mesenchymal cells. However, the molecular mechanism is still unclear. It has been suggested that BMP alone is not sufficient to induce Sox9 expression [45], although later studies indicated that BMP/Smad pathways regulate Sox9 expression through a CCAAT-box in the Sox9 promoter, as well as by chromatin remodeling at the proximal promoter [46, 47]. A number of transcriptional targets of Smads1 and 5 in the growth plate have been described, including Ihh, Col2, Col10, and Runx2 [48–51]. In addition to a role as a transcriptional activator, BMP R-Smads also act as transcriptional repressors through specific recruitment of transcriptional repressor complexes. For example, the Smad1/4 complex is required to recruit a HDAC/Sin3A complex for modulating a transcription repressor, Nkx3.2, which promotes chondrocyte differentiation [52]. At the same time, other nuclear proteins regulate Smad protein functions by regulating Smad stability, DNA binding, and transcriptional activities. For example, most Smads can be degraded by the proteasome through ubiquitination; on the other hand, sumoylation, seems to protect Smads from being ubiquitinated. The capacity of R-Smads to bind DNA is enhanced by the presence of stabilizing co-factors. Some transcriptional co-activators have been shown to interact with the MH2 domain of R-Smads to fully activate target gene transcription [9]. Other protein-protein interactions influence Smad signaling without participating in Smad-DNA complexes. For example, calponin 3, an actin binding protein, has been found to interact directly with Smads 1 and 5 to negatively regulate the BMP–dependent cellular response of human chondrocytes, possibly by sequestering Smads to the cytoskeleton [53]. Jab1, a subunit of the COP9 signalosome [54] interacts directly with Smad5 to attenuate the BMP-signaling response in chondrocytes, possibly by inducing Smad5 degradation. [55].
With respect to the TGFβ/activin R-Smads, even less is known. The extent by which TGFβ/activin signaling in vivo is mediated by Smads2/3 remains an important and unanswered question. It is possible that Smad3 plays an essential role, since the Smad3 null mice phenotype is similar to that of the mice expressing a transgenic dominant negative TGFβ type II receptor (TgfbrII) [26, 56]. However, conditional deletion of the TgfbrII with Col2a1-Cre and Prx1-Cre causes axial skeleton defects, alteration in hypertrophic differentiation in growth plates, and joint fusions in phalanges [57, 58]. These defects are not present in the Smad3 null mice, indicating that either Smad2 is more dominant in mediating TGFβ signaling in skeletal tissue or the non-Smad pathways are major players (Figure 1). Moreover, TGFβ/activin signaling may be more dependent on Smad4 than in BMP signaling, as the cartilage-specific loss of Smad4 resembles in many aspects the phenotype of mice lacking Smad3 [11, 26]. Nevertheless, the full repertoire of effects mediated by TGFβ signaling in cartilage has not yet been defined in vivo, nor have any studies yet addressed potential overlapping functions for Smads2 and 3.
Previous studies have shown that TGFβ may play important functions at early stages of chondrogenesis. An in vitro study showed that Smad3, but not Smad2, forms a complex with Sox9 and CEBP/p300 to activate genes for chondrogenesis [59]. A recent study showed that Smad3 works cooperatively with Sox9 to initiate target gene transcription through chromatin remodeling [60]. However, the fact that Smad3 knockout mice survive birth and only have limited defects in the skeleton argues that the role of Smad3 in early chondrogenesis is not critical, or Smad2 could largely compensate for the loss of Smad3 in early chondrogenesis. Organ culture studies demonstrated that Smad3 is required for TGFβ1-induced chondrocyte proliferation in mice, but share redundant functions with Smad2 in terms of inhibiting hypertrophic differentiation [61]. In the post natal stage of life, Smad3 has been shown to play an essential role in maintaining articular cartilage by preventing articular chondrocytes from undergoing terminal hypertrophic differentiation [28]. In accordance, chondrocytes in Smad3 deficient mice show accelerated differentiation in the growth plate shortly after weaning, resulting in dwarfism. Accelerated differentiation was also observed in articular chondrocytes, such that they escape from quiescence and continue the process of maturation, resulting in the loss of articular cartilage. Later studies carried out with primary chondrocytes isolated from these mice demonstrated increased BMP responsiveness, decreased responsiveness to TGFβ1, and increased apoptosis[29]. Altogether, these observations suggested that for TGFβ signaling, Smad2 could compensate for most of Smad3’s functions in early development. However, Smad3 is required to maintain cartilage homeostasis by mediating the TGFβ signaling that inhibits terminal differentiation of chondrocytes. Whether this is due to elevated levels of expression of Smad3 relative to Smad2 in the chondrocytes, or to a distinctly different activity of Smad3 compared to Smad2, remains to be investigated.
Data from in vitro experiments indicated extensive differences between Smad2 and Smad3 in terms of DNA binding capacity, interactions with other nuclear proteins, and target gene selection. For example, an additional 30 amino acids encoded by exon3 in the MH1 domain of Smad2 prevents its direct binding to DNA, such that a complex of Smad2/4 and other transcription factors is required for DNA binding. However, Smad3 homomers can form DNA binding complexes even without Smad4. A whole list of transcription factors and nuclear proteins that interact with Smad3 has been generated; most of them also interact with Smad2 [62]. Studies with modulating Smad2 and Smad3 levels by siRNA have revealed that Smad2 and Smad3 not only share redundant functions, but also have unique roles. Smad3 appears more important than Smad2 in TGFβ’s function in cell growth arrest (for a detailed review, please see [62] ).
It is becoming increasingly clear that R-Smads also have essential functions in processes other than initiating or repressing transcription directly on DNA. A recent finding linked R-smads to the post-transcriptional processing of microRNAs. Smad1, 3, and 5 interact with primary transcripts of miR-21, in a complex with the RNA helicase p68. This complex is a component of the DROSHA microprocessor complex, which processes primary microRNAs to mature forms. BMP and TGFβ signaling increase the expression of mature miR21 by stimulating the activity of microprocessor in an R-Smad-dependent manner [63]. A noteworthy point is that this process does not need Smad4. Although miR-21 has no known function in skeletal cells, it is reasonable to speculate that Smad signaling has an effect in skeletal tissue by modulating the processing of different miRNAs. For example, miR-141 and miR-200 modulate BMP2-induced pre-osteoblast differentiation through translational repression of the transcription factor Dlx5 [64]. Moreover, a BMP responsive miRNA199a has been shown to target Smad1 and down-regulate its level to negatively regulate BMP2-induced target gene transcription in C3H10T1/2 cells[65].
Taken together, R-Smads 1/5/8 appear to mediate the majority of BMP effects in chondrogenesis, and they may do so through both transcriptional regulation and non-transcriptional regulatory mechanisms. Smads 1 and 5 share a high level of functional redundancy, whereas Smad8 is less important in chondrogenesis. On the other hand, Smad2 and 3 have been shown to have quite unique functions from each other. The non-Smad pathways may significantly contribute to TGFβ signaling in chondrogenesis suggested by the difference between the TgfrIIcko and Smad3−/− phenotypes. However, conditional deletion of either or both of Smads 2 and 3 will reveal additional valuable information.
3. Roles of Smads in Osteogenesis
BMP’s function in osteogenesis is tightly related to runt related factor Runx2 (Cbfa1/AML3). Runx2 is a platform for the assembly of a multi-component regulatory complex that controls activation and repression of genes during cell fate determination and differentiation. In the skeletal system, Runx2 is critical for osteogenic lineage commitment and formation of the skeleton [66, 67]. The interaction of Runx2 with BMP signaling is bidirectional. Runx2 is induced by BMP2 in osteoblast and chondrocyte cultures. On the other hand, Runx2 also induces BMP2 and 4 expressions by binding to a region in their promoters. In the process of osteogenic induction, Runx2 works together with Smads through direct binding in a transcriptional activator complex. Runx2 recruits R-Smads to the complex to initiate BMP responsive gene transcription [68, 69]. The carboxyl terminus of Runx2 interacts with R-Smads via a Smad interacting domain (SMID), which overlaps with the nuclear matrix targeting signal (NMTS) [69]. Furthermore, the specific residues responsible for the interaction of Runx2 with R-Smads have been identified. A triple mutation of amino acids 426–428 (HTY-AAA) in the Runx2 C-terminal domain abolished both interaction with Smads and osteogenic differentiation [70].
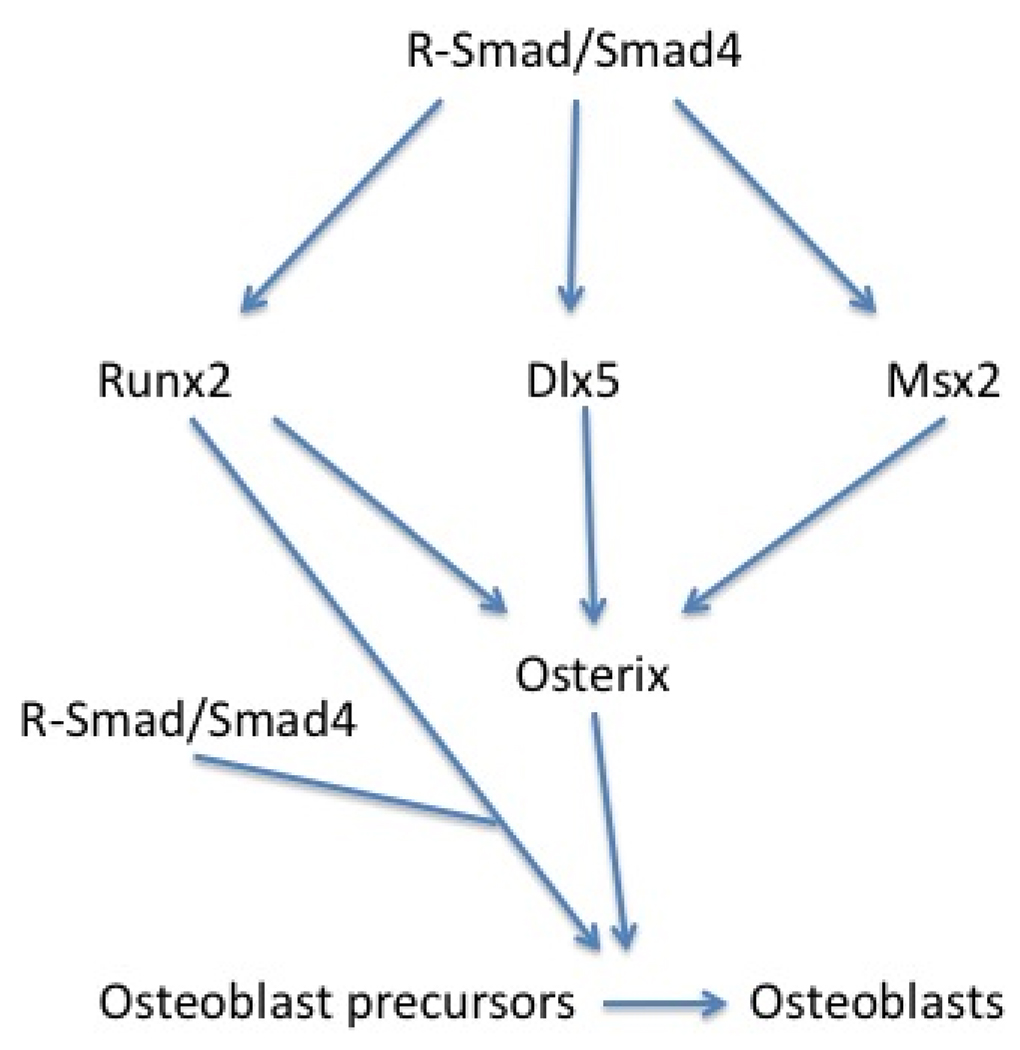
Other mechanisms by which Smad proteins promote osteogenesis have been described. Smad1 interacts with Hoxc8, a transcription inhibitor, and dislodges it from its binding sites to stimulate osteoprotegrin transcription [71]. Osterix, an Sp1 transcription family member, is essential for osteogenesis. It is up regulated by BMP2 during osteoblast differentiation and is considered to work downstream of Runx2 [73]. Recent studies have suggested that in addition to Runx2- mediated induction of Osterix, BMP signaling can also induce Osterix expression through Msx2 [74], which is one of the three members of Msh family of homeobox genes and induced directly by BMP specific R-Smads [75]. Other possible mechanisms of BMP induction of osterix expression could be through Dlx5 (reviewed in [76]). Taken together, BMP R-Smads mediate BMP function in osteogenesis by interacting with Runx2 to activate target gene transcription, in parallel with direct induction of important osteogenic genes like MSX2 and Dlx5, leading to the induction of osterix (Figure 2).
Figure 2.
BMP signaling regulates important osteogenic genes through direct induction of Runx2, Msx2 and possibly Dlx5 in a Smad dependant manner. Then these molecules induce other important osteogenic gene expression, such as osterix. Runx2 work with R-smad in an activator complex to activate BMP responsive genes. Osterix also activate osteogenic gene transcription, promoting the osteoblastic differentiation
On the other hand, TGFβ R-Smads have been shown to work both synergistically and antagonistically with BMP in osteogenesis. For example, Smad3 has been shown to bind to the osteopontin promoter as a sequence specific activator [72]. However, TGFβ activated Smad3 inhibits osteocalcin expression by forming a repressive complex with Runx2 and histone deacetylase (HDAC) at osteocalcin promotor, and this mechanism is cell type and promoter specific [77].
Other transcriptional regulators working upstream, downstream or in parallel with BMP signaling in osteogenesis include Bapx1, Msx1, Dlx6, and Inhibitor of differentiations (Ids) (for a detailed review, see [78]). Although these molecules participate in BMP signaling, direct evidence of interactions with Smad proteins in the context of osteogenesis is still missing. How TGFβ work synergistically or antagonistically with BMPs in this process through different Smad activities requires further investigation.
4. Roles of Smads in Skeletal Tissue Regeneration
Tissue regeneration in bone and cartilage more or less recapitulates the process of development. However, there are noteworthy differences in terms of the cytokines and growth factors involvement. Therefore, it is reasonable to speculate that the process of bone/cartilage tissue regeneration has its own requirements for specific BMP/TGFβ ligands, receptors, and Smads [79]. The bone healing process has been artificially divided to four different stages including 1) Inflammation, 2) cartilage formation and periosteum response, 3) cartilage resorption and primary bone formation, and 4) secondary bone formation and remodeling. Multiple cytokines and growth hormones are involved in the four temporally overlapping stages. These include Interleukins, TNFα, PDGF, VEGF, and BMP/TGFβ. The major challenge in understanding the roles of BMPs and TGFβ in tissue regeneration at the molecular level is to characterize the spatial/temporal activity profile of different BMP/TGFβ ligands, receptors, and Smads, so that appropriate therapeutic strategies can be developed [80]. Fracture healing is the most widely studied process in bone tissue regeneration. Most bones heal by a combination of intramembranous and endochondral ossification. Endochondral bone formation occurs closer to the fracture site, which is mechanically unstable. It occurs external to the periosteum. Intramembranous bone formation occurs at both ends of the callus and internal to the periosteum [81]. During this process, BMP/TGFβ signaling is responsible for recruiting bone-forming cells, initiation of chondrogenesis and osteogenesis, and regulation of bone remodeling. Many studies have been done in animals and humans to elucidate the expression patterns of different ligands, leading to speculation on their functions [79]. A recent study demonstrated an essential role for BMP2 in fracture healing. Mice lacking BMP2 in limb cartilage and bone suffer from spontaneous fractures and an impaired fracture response in which cells are recruited to the site of injury but are unable to commit to chondrogenic or osteogenic fates[82]. For clinical applications, human recombinant BMP2 and BMP7 have been approved for promoting fracture healing. Direct evidence for the specific roles of Smad proteins in this process is very limited. A study in a rat fracture model indicated that in the fracture healing process, Smad1 and 5 expression patterns are similar to those of BMP2 and 7; whereas Smad2 and 3 expression patterns are similar to those of TGFβ[83]. This suggested that, at least partially, BMP/TGFβ signaling in fracture healing is mediated by canonical Smad pathways. However, in addition to mediating the BMP/TGFβ signaling, it is conceivable that Smad proteins have multiple interactions with other signaling molecules to promote chondrogenesis and osteogenesis.
Studies of BMP/TGFβ signaling in cartilage repair or regeneration are mostly within the context of osteoarthritis (OA). TGFβ signaling promotes chondrocyte production of extracellular matrix (ECM), and maintains articular cartilage homeostasis. A number of ECM proteins have been shown to be TGFβ target genes [84]. Its protective role has been confirmed by the fact that Smad3−/− mice develop OA [28]. This also indicated that TGFβ exerts its protective role in articular cartilage at least partially through Smad3, rather than Smad2. Moreover, TGFβ has been shown to promote cartilage repair and to alleviate OA in animals. However, a major complication of applying TGFβ for OA treatment is that TGFβ application also induces the unwanted effects of tissue fibrosis and osteophyte formation. Recent studies have explored the possibility of applying TGFβ I-Smads locally in soft tissue to avoid this problem, but whether this is practicable is still under evaluation [85, 86].
5. Role of I-Smads
I-Smads (Smad6 and 7) are key factors in intracellular regulation of BMP and TGFβ signaling. The protein-binding MH2 domain of I-Smads is structurally similar to that of R-Smads, but lacks the C-terminal Ser-X-Ser motif that is phosphorylated by the activated type I receptor. I-Smads act as competitive inhibitors of R-Smad phosphorylation by forming stable associations with activated type I receptors [87–89]. Smad6 can also inhibit R-Smad signaling in a phosphorylation-independent manner by interacting with receptor-activated Smad1, thus forming an inactive Smad1–Smad6 complex [90]. In addition to their role as competitive inhibitors, I-Smads can inhibit BMP and TGFβ signaling by recruiting E3 ubiquitin ligases to type I receptors, R-Smads or Smad4, leading to their ubiquitination and degradation [91–94]. I-Smads, however, are not immune to E3 ligases. Smad7 can be targeted by the RING-domain E3 ligase, Arkadia [95], resulting in amplified TGFβ signaling. Interestingly, the expression of I-Smads is directly induced by BMP and TGFβ signaling, thus forming a negative feedback loop that limits the intensity and duration of BMP and TGFβ signaling. In addition, the complex interaction between I-Smads and E3 ligases may fine-tune BMP and TGFβ signaling.
I-Smads are strongly expressed in the prehypertrophic and hypertrophic zones of the growth plate [41], suggesting a role for I-Smads in regulating chondrocyte maturation. Indeed, gain-and loss-of-function studies have shown that I-Smads regulate chondrocyte maturation in vitro [96, 97]. In vivo analyses revealed that I-Smads play multiple roles in development. Mice with a global deletion of Smad6 or Smad7 exhibit defects in the cardiovascular system [34, 36]. Moreover, mice with a hypomorphic allele of Smad7 have altered immune responses [37]. Gain of function studies has recently been conducted to determine role of I-Smads in endochondral bone formation. Cartilage-specific overexpression of Smad6 results in delayed chondrocyte hypertrophy leading to dwarfism [35]. Overexpression of Smad7 at various stages of endochondral bone formation in mice results in inhibition of mesenchymal cell condensation and chondrocyte proliferation, as well as delayed chondrocyte maturation [40]. Because of the high levels of I-Smad expression in these transgenic mice, the results may only highlight the pathological role of I-Smads in endochondral bone formation. Furthermore, it is not known whether Smad7 is normally expressed in condensing mesenchymal cells or proliferating chondrocytes. Hence, the physiological role of I-Smads is still unknown.
6. Smads and other signaling pathways
BMP/TGFβ signaling interacts with other signaling pathways to form a complicated network regulating cellular growth, differentiation, migration and apoptosis. These interactions and their end effects are usually species, tissue, and temporal-spatial specific. They also occur at different regulatory levels by engaging ligands, Smads, target genes, and intracellular proteins. In the context of skeletal development, BMP/TGFβ signaling predominantly interacts with Wnt, Indian Hedgehog (Ihh), PTHrP, FGF, PI3K/Akt, and MAPK signaling pathways through Smad-dependant interactions.
6.1 Smad interaction with Hedgehog and PTHrP pathway
Ihh signaling is important in growth plate chondrocytes. By interacting with PTHrP, Ihh promotes proliferation and inhibits differentiation of growth plate chondrocytes, thus regulating bone growth (reviewed in [98]). As mentioned above, Ihh is a direct target gene of BMP [48], and Ihh also promotes BMP expression levels. In vitro studies demonstrated direct association of Smad1 with truncated Gli3 protein[99], indicating direct roles of Smad proteins in the interaction of these two signaling pathways. In vivo data from Smad1/5cko mice showed that Ihh and PTHrP receptor (PPR) mRNA levels were significantly reduced, indicating that BMP regulation of Ihh and PPR is direct and Smad1/5 dependent [12]. Little is known about interaction between TGFβ and Hedgehog signaling. A mouse metatarsal culture study suggested that the signaling relay from Ihh to PTHrP in the growth plate is mediated by TGFβ2 in the perichondrium [100]. Another study has shown that Smad3-dependent TGFβ signaling up-regulates Gli2 expression in fibroblasts, keratinocytes, and several cancer cell lines. Mice with TGFβ1 overexpression showed Smad3- dependent increased expression of Gli1 and Gli2 in the skin. [101]. However, whether these are true in the skeletal system remains to be investigated.
6.2 Smad interaction with FGF pathway
FGF signaling is essential in both endochondral and intramembranous bone formation. Disruption of FGF signaling is the cause of several human craniosynostosis and chondrodysplasia syndromes. FGF controls chondrocyte and osteoblast proliferation and differentiation through Jak/Stat and MEK1 pathways (reviewed in [102]. Previous studies have shown the antagonistic functions of FGF and BMP signaling in chondrocytes, however, the particular mechanisms underlying this antagonism remain largely unknown. Mouse model studies involving deletion of BMPR1A (ALK3) and BMPR1B (ALK6) in chondrocytes have confirmed this antagonism by showing the up-regulation of Stat1, Stat5, ERK1/2 in mutant growth plates, as well as increased levels of FGF receptor I [103]. In terms of Smad involvement, studies in other systems have shown that the linker region of Smad1 can be phosphorylated by MEK1 pathways, thereby inhibiting BMP signaling [104, 105]. However, our recent study in a limb culture system argues this is not true for growth plate chondrocytes in vivo. FGF signaling did not affect linker phosphorylation of Smad. Instead, linker phosphorylation was induced by BMP treatment. Interestingly, the C-terminal phosphorylation of Smads is reduced in cartilage after FGF treatment, suggesting that FGF signaling antagonizes BMP by an indirect mechanisms, possibly by inducing de-phosphorylation of Smads or regulating BMP ligand and receptor expression [12].
6.3 Smad interaction with Wnt signaling
Wnt is another key signaling molecule in skeletal cells, regulating proliferation, differentiation, migration and apoptosis. The interactions between BMP/TGFβ and Wnt pathways are profound and bidirectional. They occur at the level of ligands, cytoplasmic signaling intermediates, and transcriptional targets. First of all, Wnt and BMP/TGFβ pathways regulate ligand expression reciprocally. For example, in chicken embryos, Wnt-8c induces Nodal (a member of TGFβ superfamily) expression in a β-catenin-dependent manner [106], and BMP2 down-regulates Wnt7a and β-catenin in a p38 dependent manner, leading to enhanced chondrogenesis in mesenchymal cells [107]. An interaction also happens through connective tissue growth factor (CTGF). Wnt and BMP co-regulate CTGF expression in mesenchymal stem cells, and its induction inhibits osteoblastic differentiation[108]. CTGF is also co-regulated by Wnt and TGFβ in Xenopus embryos. In this scenario, CTGF interacts directly with BMP4 and TGFβ through its cystine rich (CR) domain. This direct binding prevents BMP4 from binding to its receptor, but enhances TGFβ1-receptor binding [109]. (For a more detailed review, see [110]).
Smad proteins play an important role in the cross talk between BMP/TGFβ and Wnt signaling. The first evidence of direct Smad interaction with Wnt signaling components was in Xenopus embryos. It was shown that Smad4, β catenin and Lef1 form a complex to activate expression of the Wnt target gene twin (Xtwn) [111]. Interestingly, this process does not necessarily require active TGFβ/Activin signaling, implying an independent function of Smad4 [112]. Another study showed a direct interaction of Smad2, 3 and 4 with Lef1/TCF in mammalian cells. But these mechanisms of transcriptional regulation cannot be generalized since not all target genes of Wnt are regulated this way, and BMP signaling does not affect expression of these genes [111–113]. However, there are other genes that are regulated by a similar mechanism, including Msx1, Msx2 and Id2, which are relevant to skeletal system development, although the evidences comes from studies of neural development and human carcinoma [114–116]. In reciprocity, Wnt signaling can regulate BMP/TGFβ signaling by regulating GSK3-β activity. Wnt signaling deactivates GSK3-β and stabilizes Smad1, by preventing the ability of GSK3-β to phosphorylate the Smad linker region [117]. Linker phosphorylation of Smad1/5 by GSK3-β facilitates degradation, and prevents R-Smad interaction with nuclear pores [118]. Axin interacts with Smad3 to facilitate TGFβ signaling [119]. However, a recent study reported that Axin promotes Smad3 degradation to inhibit TGFβ signaling [120]. In a skeletal context, TGFβ1 has been shown to increase β-catenin nuclear translocation and to exert effects similar to those of Wnts on human bone marrow derived mesenchymal stem cells (MSC) (stimulating proliferation while inhibiting differentiation of these cells toward osteoblasts) or adipocytes. This process is Smad3-dependent and involves direct interaction between Smad3 and β-catenin. Using siRNA to knock down Smad3 abolished this effect, suggesting that Smad2 is unable to compensate for Smad3 for this specific function [121]. BMP signaling has been shown to have an opposite effect on Wnt signaling. Smad1 interacts with Dvl-1 at the linker region and this interaction accounts for the inhibitory effect of BMP2 on Wnt signaling in mouse mesenchymal stem cells (MSCs) [122].
6.4 Smad interaction with PI3K/Akt pathway
Phosphatidylinositol 3 kinase converts phosphatidylinositol-4, 5-bisphosphate (PIP2) to phosphatidylinositol-3, 4,5-triphosphate (PIP3). PIP3 then regulates downstream effectors such as Akt, a serine/threonine kinase. This pathway usually promotes cell survival, growth, and migration. The PI3K/Akt pathway is negatively regulated by phosphatase and tensin homolog deleted on chromosome 10 (PTEN), which is a phosphatase that dephosphorylate PIP3 to PIP2 thus deactivating PIP3-dependent pathways. Just like TGFβ-Wnt interactions, interactions between PI3K/Akt and BMP/TGFβ have been discovered in different cell types. PI3K/Akt pathways antagonize the pro-apoptotic effects and cell cycle arrest induced by TGFβ signaling. The mechanism of the pro-apoptotic effect of TGFβ has been shown in different cell types, and is mostly Smad3-dependent [123]. Consistently, the mechanism of PI3K/Akt –mediated restriction of TGFβ signaling appears to be through Smad3. Different studies have suggested mechanisms involving direct or indirect interaction of Akt with Smad3, and enhanced or attenuated TGFβ signaling regulated by PI3K/Akt (for a more detailed review, see [110]). On the other hand, PI3K/Akt signaling is also regulated by BMP/TGFβ in the context of cell migration, epithelial-mesenchymal transition, cell survival, and cell growth; but again, these interactions are all likely cell type dependent [110]. In a more skeletal context, studies carried out in mesenchymal precursor 2T3 cells showed dominant-negative PI3K and dominant- negative Akt have been shown to inhibit Smad5-dependent target gene transcription, as well as nuclear translocation of BMP-specific R-Smads after ligand stimulation [124].
7.4 Smad interaction with MAPK pathway
MAPK pathways are evolutionarily conserved and regulate a variety of cellular events. There are three distinct MAPK pathways, Erk1/2, JNK1/2/3, and p38/MAPKs. The interaction of MAPK pathways with BMP/TGFβ and Smads generates a complicated network, involving transcriptional regulation of Smads, as well as phosphorylation of Smad linker regions. Some of these interactions have been discussed in the context of Smad/FGF interactions. Again, although the interactions have been intensively studied, most of the evidence is derived from in vitro studies. For example, Smad1/5 and Smad2/3 could all be phosphorylated by MAPKs at the linker region, and this phosphorylation had different effects on each individual Smad (reviewed in [110]). So far, in vivo data from the skeletal system are limited.
However, both BMP and TGFβ/activins have been shown to activate certain MAPK pathways, such as TAK1/p38 pathway. These pathways mediate the effects of non-Smad signaling, or non-canonical signaling of TGFβ superfamily proteins [17, 18], although the mechanisms of how BMP/TGFβ activate the MAPK signaling pathways are still under intensive investigation. The most recent study showed that, in glomerular mesangial cells, TGFβ1 activate TAK1 through TAB1-mediated auto-phosphorylation of TAK1, without the requirement of the kinase activity of type I receptor [125]. As mentioned in the beginning of this review, such non-canonical signaling may be more important than suggested by our current understanding. The fact that MAPK pathways also regulate Smad protein activities raised the possibility of cross-talk between these two branches of BMP/TGFβ signaling in skeletal genesis. In deed, in addition to linker region phosphorylation, studies on TAK1 conditional knock out mice in chondrocytes suggested TAK1 also regulate C-terminal phosphorylation of Smad1. Moreover, TAK1 deficient chondrocytes showed reduced activity of Smad1/5/8, and decreased expression level of multiple BMP target genes, suggesting extensive regulation of BMP signaling by TAK1 through acting on both Smad and non-Smad proteins [126].Further investigation is needed to reveal this emerging complex picture of BMP/TGFβ signaling crosstalk.
Perspectives
Smad proteins have been long considered the major intracellular signaling transduction molecules for TFGβ superfamily members. Since TGFβ superfamily signaling has a key role in skeletal development and regeneration, it is then very important to understand the role of Smad proteins in this context. However, fewer studies have been focused on Smad family members than on the corresponding ligands and receptors. With the emerging picture of a complicated network of intracellular molecules involved in TGFβ superfamily signaling, it is more important to unveil the functions of Smad proteins. A major challenge here is that the functions of Smads and their interactions with other signaling molecules seem to be cell type, tissue and species specific. This emphasizes the importance of in vivo studies in animal models. Currently, mouse models are the most accessible and convenient for studying Smad functions in skeletal development. Recent studies have already pointed out the importance of Smad1/5 in skeletal formation; however, roles of TGFβ specific Smads, co-Smad, and inhibitory Smads have not been fully addressed. For the investigation of these topics, conditional knockouts of Smad proteins in different compartments or stages of skeletal development will yield valuable information. At the same time, interactions with other signaling pathways or intracellular molecules could be studied in these animal models. Such studies will reveal more information about the major regulators in skeletal development and regeneration, identify new participants/targets in these processes and lead to new strategies for disease treatment and tissue engineering.
Acknowledgements
Due to the space limit, the authors apologize to the researchers whose important work was not included in this review. The authors want to thank Drs. Paul Benya, Deborah Krakow, Susan Krum, Rosa Serra and Courtney J. Haycraft for helpful suggestions and critical reviewing of the manuscript. Buer Song is supported by Jane Wyman Trust Award from Arthritis Foundation, Southern California Chapter. The studies were funded by grant 5R01AR044528-12 from National Institute of Arthritis and Musculoskeletal and Skin Diseases to Karen M. Lyons.
Biographies

Buer Song
Buer Song received his Bachelor of Medicine and Master of Medicine degree from Peking University Health Science Center in 1999 and 2001 respectively. Then he went to University of Alabama at Birmingham. In 2006, he received a Ph.D degree in Pathology under the guidance of Dr. Rosa Serra. During his graduate study he focused on the research of bone and cartilage development, characterized the function of primary cilia in growth plate chondrocytes. He is currently performing a post doctoral training under the guidance of Dr. Karen M. Lyons at University of California, Los Angeles.

Kristine Estrada
Kristine Estrada is a PhD student in Molecular, Cell and Developmental Biology at the University of California, Los Angeles (UCLA). She obtained her BSc degree in Chemical Engineering at UCLA in 2002 and her MSc in Biomedical Engineering at the University of Wisconsin, Madison in 2005. She joined the laboratory of Dr. Karen Lyons in 2008 and has been working since on identifying the role of inhibitory Smads 6 and 7 on bone development.

Professor Karen M. Lyons is a world-renowned expert in the research of developmental biology. Her research interest has been focused on BMP/TGFβ signaling in bone and cartilage. She received a Ph.D. degree in Medical Genetics from the University of Wisconsin, Madison in 1988. She then completed two postdoctoral fellowships in Developmental Biology. The first fellowship was with Dr. Brigid Hogan at Vanderbilt University in Nashville, Tennessee, where she cloned one of the first BMP genes to be discovered, BMP6. The second fellowship was with Dr. Liz Robertson at Harvard University in Cambridge, Massachusetts, where she generated mice lacking BMPs 6 and 7. In 1995, She accepted a faculty position at the University of California in Los Angeles, where she is currently a full professor in the Department of Orthopaedic Surgery and has a joint appointment in Molecular, Cell and Developmental Biology. At UCLA, Karen generated mice lacking the BMP receptor type IB, and demonstrated that this receptor is essential for chondrogenesis. More recently, she has shown that the majority of BMP signaling in skeletal cells occurs through canonical pathways.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Urist MR, et al. Inductive substrates for bone formation. Clin Orthop Relat Res. 1968;59:59–96. [PubMed] [Google Scholar]
- 2.Anzano MA, et al. Sarcoma growth factor from conditioned medium of virally transformed cells is composed of both type alpha and type beta transforming growth factors. Proc Natl Acad Sci U S A. 1983;80(20):6264–6268. doi: 10.1073/pnas.80.20.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts AB, et al. Transforming growth factors from neoplastic and nonneoplastic tissues. Fed Proc. 1983;42(9):2621–2626. [PubMed] [Google Scholar]
- 4.Burt DW, Law AS. Evolution of the transforming growth factor-beta superfamily. Prog Growth Factor Res. 1994;5(1):99–118. doi: 10.1016/0955-2235(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 5.Ripamonti U, et al. Induction of endochondral bone formation by recombinant human transforming growth factor-beta2 in the baboon (Papio ursinus) Growth Factors. 2000;17(4):269–285. doi: 10.3109/08977190009028971. [DOI] [PubMed] [Google Scholar]
- 6.Ripamonti U, et al. The induction of endochondral bone formation by transforming growth factor-beta(3): experimental studies in the non-human primate Papio ursinus. J Cell Mol Med. 2008;12(3):1029–1048. doi: 10.1111/j.1582-4934.2008.00126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekelsky JJ, et al. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139(3):1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage C, et al. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci U S A. 1996;93(2):790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 10.Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Smad4 is required for the normal organization of the cartilage growth plate. Dev Biol. 2005;284(2):311–322. doi: 10.1016/j.ydbio.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 12.Retting KN, et al. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136(7):1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provot S, Schipani E. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 2005;328(3):658–665. doi: 10.1016/j.bbrc.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 14.Kitisin K, et al. Tgf-Beta signaling in development. Sci STKE. 2007;2007(399):cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- 15.Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357(1):1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao GQ. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35(1):43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 17.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118(Pt 16):3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 18.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102(3):593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128(18):3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 20.Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393(6687):786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 21.Waldrip WR, et al. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92(6):797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 22.Weinstein M, et al. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc Natl Acad Sci U S A. 1998;95(16):9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyer J, et al. Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc Natl Acad Sci U S A. 1999;96(22):12595–12600. doi: 10.1073/pnas.96.22.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremblay KD, et al. Formation of the definitive endoderm in mouse is a Smad2- dependent process. Development. 2000;127(14):3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, et al. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94(6):703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 26.Datto MB, et al. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol. 1999;19(4):2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18(5):1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, et al. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153(1):35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li TF, et al. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J Bone Miner Res. 2006;21(1):4–16. doi: 10.1359/JBMR.050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirard C, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12(1):107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, et al. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc Natl Acad Sci U S A. 1998;95(7):3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X, et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci. 2007;120(Pt 13):2162–2170. doi: 10.1242/jcs.03466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, et al. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development. 1999;126(8):1571–1580. doi: 10.1242/dev.126.8.1571. [DOI] [PubMed] [Google Scholar]
- 34.Galvin KM, et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nat Genet. 2000;24(2):171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 35.Horiki M, et al. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J Cell Biol. 2004;165(3):433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q, et al. Smad7 is required for the development and function of the heart. J Biol Chem. 2009;284(1):292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li R, et al. Deletion of exon I of SMAD7 in mice results in altered B cell responses. J Immunol. 2006;176(11):6777–6784. doi: 10.4049/jimmunol.176.11.6777. [DOI] [PubMed] [Google Scholar]
- 38.Hamzavi J, et al. Disruption of the Smad7 gene enhances CCI4-dependent liver damage and fibrogenesis in mice. J Cell Mol Med. 2008;12(5B):2130–2144. doi: 10.1111/j.1582-4934.2008.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung AC, et al. Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol Dial Transplant. 2009;24(5):1443–1454. doi: 10.1093/ndt/gfn699. [DOI] [PubMed] [Google Scholar]
- 40.Iwai T, et al. Smad7 Inhibits chondrocyte differentiation at multiple steps during endochondral bone formation and down-regulates p38 MAPK pathways. J Biol Chem. 2008;283(40):27154–27164. doi: 10.1074/jbc.M801175200. [DOI] [PubMed] [Google Scholar]
- 41.Sakou T, et al. Localization of Smads, the TGF-beta family intracellular signaling components during endochondral ossification. J Bone Miner Res. 1999;14(7):1145–1152. doi: 10.1359/jbmr.1999.14.7.1145. [DOI] [PubMed] [Google Scholar]
- 42.Orvis GD, et al. Functional redundancy of TGF-beta family type I receptors and receptor-Smads in mediating anti-Mullerian hormone-induced Mullerian duct regression in the mouse. Biol Reprod. 2008;78(6):994–1001. doi: 10.1095/biolreprod.107.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pangas SA, et al. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol. 2008;28(1):248–257. doi: 10.1128/MCB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon BS, et al. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102(14):5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon BS, Lyons KM. Multiple functions of BMPs in chondrogenesis. J Cell Biochem. 2004;93(1):93–103. doi: 10.1002/jcb.20211. [DOI] [PubMed] [Google Scholar]
- 46.Pan Q, et al. Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol. 2008;217(1):228–241. doi: 10.1002/jcp.21496. [DOI] [PubMed] [Google Scholar]
- 47.Pan Q, et al. Bone morphogenetic protein-2 induces chromatin remodeling and modification at the proximal promoter of Sox9 gene. Biochem Biophys Res Commun. 2009;379(2):356–361. doi: 10.1016/j.bbrc.2008.12.062. [DOI] [PubMed] [Google Scholar]
- 48.Seki K, Hata A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. J Biol Chem. 2004;279(18):18544–18549. doi: 10.1074/jbc.M311592200. [DOI] [PubMed] [Google Scholar]
- 49.Hatakeyama Y, et al. Smad signaling in mesenchymal and chondroprogenitor cells. J Bone Joint Surg Am. 2003;85-A Suppl 3:13–18. doi: 10.2106/00004623-200300003-00004. [DOI] [PubMed] [Google Scholar]
- 50.Drissi MH, et al. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem. 2003;90(6):1287–1298. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
- 51.Lee KS, et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol Cell Biol. 2003;23(23):8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haag J, Aigner T. Identification of calponin 3 as a novel Smad-binding modulator of BMP signaling expressed in cartilage. Exp Cell Res. 2007;313(16):3386–3394. doi: 10.1016/j.yexcr.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Bech-Otschir D, Seeger M, Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci. 2002;115(Pt 3):467–473. doi: 10.1242/jcs.115.3.467. [DOI] [PubMed] [Google Scholar]
- 55.Haag J, Aigner T. Jun activation domain-binding protein 1 binds Smad5 and inhibits bone morphogenetic protein signaling. Arthritis Rheum. 2006;54(12):3878–3884. doi: 10.1002/art.22261. [DOI] [PubMed] [Google Scholar]
- 56.Serra R, et al. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J Cell Biol. 1997;139(2):541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baffi MO, et al. Conditional deletion of the TGF-beta type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Dev Biol. 2004;276(1):124–142. doi: 10.1016/j.ydbio.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 58.Seo HS, Serra R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev Biol. 2007;310(2):304–316. doi: 10.1016/j.ydbio.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furumatsu T, et al. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J Biol Chem. 2005;280(9):8343–8350. doi: 10.1074/jbc.M413913200. [DOI] [PubMed] [Google Scholar]
- 60.Furumatsu T, Ozaki T, Asahara H. Smad3 activates the Sox9-dependent transcription on chromatin. Int J Biochem Cell Biol. 2009;41(5):1198–1204. doi: 10.1016/j.biocel.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez J, Serra R. Unique and redundant roles of Smad3 in TGF-beta-mediated regulation of long bone development in organ culture. Dev Dyn. 2004;230(4):685–699. doi: 10.1002/dvdy.20100. [DOI] [PubMed] [Google Scholar]
- 62.Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101(1):9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 63.Davis BN, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse Pre-osteoblast differentiation by targeting distalless homeobox 5. J Biol Chem. 2009 doi: 10.1074/jbc.M109.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin EA, et al. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284(17):11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 67.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 68.Zhang YW, et al. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97(19):10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Afzal F, et al. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204(1):63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 70.Javed A, et al. Specific residues of RUNX2 are obligatory for formation of BMP2- induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs. 2009;189(1–4):133–137. doi: 10.1159/000151719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wan M, et al. Transcriptional mechanisms of bone morphogenetic protein-induced osteoprotegrin gene expression. J Biol Chem. 2001;276(13):10119–10125. doi: 10.1074/jbc.M006918200. [DOI] [PubMed] [Google Scholar]
- 72.Shi X, et al. Hoxa-9 represses transforming growth factor-beta-induced osteopontin gene transcription. J Biol Chem. 2001;276(1):850–855. doi: 10.1074/jbc.M005955200. [DOI] [PubMed] [Google Scholar]
- 73.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 74.Matsubara T, et al. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283(43):29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brugger SM, et al. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development. 2004;131(20):5153–5165. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- 76.Ryoo HM, Lee MH, Kim YJ. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene. 2006;366(1):51–57. doi: 10.1016/j.gene.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 77.Kang JS, et al. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24(14):2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Shi W, Cao X. Multiplicity of BMP signaling in skeletal development. Ann N Y Acad Sci. 2007;1116:29–49. doi: 10.1196/annals.1402.053. [DOI] [PubMed] [Google Scholar]
- 79.Ai-Aql ZS, et al. Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J Dent Res. 2008;87(2):107–118. doi: 10.1177/154405910808700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ripamonti U, Herbst NN, Ramoshebi LN. Bone morphogenetic proteins in craniofacial and periodontal tissue engineering: experimental studies in the non-human primate Papio ursinus. Cytokine Growth Factor Rev. 2005;16(3):357–368. doi: 10.1016/j.cytogfr.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 81.Dimitriou R, Tsiridis E, Giannoudis PV. Current concepts of molecular aspects of bone healing. Injury. 2005;36(12):1392–1404. doi: 10.1016/j.injury.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 82.Tsuji K, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38(12):1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- 83.Yu Y, et al. TGF-beta, BMPS, and their signal transducing mediators, Smads, in rat fracture healing. J Biomed Mater Res. 2002;60(3):392–397. doi: 10.1002/jbm.1289. [DOI] [PubMed] [Google Scholar]
- 84.Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem. 2001;276(20):17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- 85.Schiller M, Javelaud D, Mauviel A. TGF-beta-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J Dermatol Sci. 2004;35(2):83–92. doi: 10.1016/j.jdermsci.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 86.Blom AB, van der Kraan PM, van den Berg WB. Cytokine targeting in osteoarthritis. Curr Drug Targets. 2007;8(2):283–292. doi: 10.2174/138945007779940179. [DOI] [PubMed] [Google Scholar]
- 87.Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 88.Imamura T, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389(6651):622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 89.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 90.Hata A, et al. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12(2):186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6(6):1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 92.Murakami G, et al. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14(7):2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki C, et al. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 2002;277(42):39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 94.Moren A, et al. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280(23):22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 95.Koinuma D, et al. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003;22(24):6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X, et al. Smad6 is induced by BMP-2 and modulates chondrocyte differentiation. J Orthop Res. 2003;21(5):908–913. doi: 10.1016/S0736-0266(03)00008-1. [DOI] [PubMed] [Google Scholar]
- 97.Valcourt U, et al. Functions of transforming growth factor-beta family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J Biol Chem. 2002;277(37):33545–33558. doi: 10.1074/jbc.M202086200. [DOI] [PubMed] [Google Scholar]
- 98.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 99.Liu F, Massague J, Ruiz i Altaba A. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat Genet. 1998;20(4):325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- 100.Alvarez J, et al. TGFbeta2 mediates the effects of hedgehog on hypertrophic differentiation and PTHrP expression. Development. 2002;129(8):1913–1924. doi: 10.1242/dev.129.8.1913. [DOI] [PubMed] [Google Scholar]
- 101.Dennler S, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67(14):6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 102.Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16(2):205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoon BS, et al. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133(23):4667–4678. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 104.Kretzschmar M, et al. A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes Dev. 1999;13(7):804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pera EM, et al. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17(24):3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez-Esteban C, et al. Wnt signaling and PKA control Nodal expression and leftright determination in the chick embryo. Development. 2001;128(16):3189–3195. doi: 10.1242/dev.128.16.3189. [DOI] [PubMed] [Google Scholar]
- 107.Jin EJ, et al. BMP-2-enhanced chondrogenesis involves p38 MAPK-mediated down-regulation of Wnt-7a pathway. Mol Cells. 2006;22(3):353–359. [PubMed] [Google Scholar]
- 108.Luo Q, et al. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279(53):55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 109.Abreu JG, et al. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4(8):599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97(15):8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nishita M, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature. 2000;403(6771):781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 113.Laurent MN, Cho KW. Bone morphogenetic protein antagonism of Spemann's organizer is independent of Wnt signaling. Dev Biol. 1999;206(2):157–162. doi: 10.1006/dbio.1998.9143. [DOI] [PubMed] [Google Scholar]
- 114.Theil T, et al. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129(13):3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- 115.Hussein SM, Duff EK, Sirard C. Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J Biol Chem. 2003;278(49):48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- 116.Willert J, et al. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fuentealba LC, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131(5):980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sapkota G, et al. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25(3):441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 119.Furuhashi M, et al. Axin facilitates Smad3 activation in the transforming growth factor beta signaling pathway. Mol Cell Biol. 2001;21(15):5132–5141. doi: 10.1128/MCB.21.15.5132-5141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo X, et al. Axin and GSK3- control Smad3 protein stability and modulate TGF-signaling. Genes Dev. 2008;22(1):106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jian H, et al. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;20(6):666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Z, et al. A dishevelled-1/Smad1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J Biol Chem. 2006;281(25):17156–17163. doi: 10.1074/jbc.M513812200. [DOI] [PubMed] [Google Scholar]
- 123.Ramesh S, Wildey GM, Howe PH. Transforming growth factor beta (TGFbeta)- induced apoptosis: the rise & fall of Bim. Cell Cycle. 2009;8(1):11–17. doi: 10.4161/cc.8.1.7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ghosh-Choudhury N, et al. Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J Biol Chem. 2002;277(36):33361–33368. doi: 10.1074/jbc.M205053200. [DOI] [PubMed] [Google Scholar]
- 125.Kim SI, et al. TGF-{beta}1 activates TAK1 via TAB1-mediated autophosphorylation, independent of TGF-{beta} receptor kinase activity in mesangial cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shim JH, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28(14):2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]