Summary
Sexual reproduction requires the unique cell division called meiosis, in which a diploid cell undergoes a reductional division to generate haploid gametes. A hallmark of meiotic prophase is the formation of pairwise linkages between homologous chromosomes, which later enable them to segregate from each other. In most organisms the pairing of homologous chromosomes is reinforced by synapsis, the polymerization of the synaptonemal complex (SC) between paired chromosome axes. The primary questions addressed here are: 1) how pairing is accomplished and 2) how synapsis is regulated so that it occurs selectively between homologs. We provide evidence that a connection between the chromosomes and the microtubule cytoskeleton via a bridge across the nuclear envelope is critical for both of these mechanisms. Our results indicate the existence of a mechanism that uses dynein to assess homology before licensing SC polymerization. The molecular components of this mechanism are conserved from fungi to mammals.
Introduction
Accurate segregation of homologous chromosomes during meiosis requires that they first form stable pairwise interactions. In most species, these are established through homolog pairing, synapsis (formation of the synaptonemal complex, or SC), and crossover recombination. While these processes are closely coordinated, they can be separated by mutation in different organisms (reviewed by Bhalla and Dernburg, 2008). A key question is how pairing between appropriate partners is normally recognized and selectively reinforced by synapsis. In C. elegans, special regions known as Pairing Centers (PCs) promote both pairing and synapsis of homologous chromosomes (MacQueen et al., 2005). Our previous work has shown that these sites are associated with the nuclear envelope (NE) during the leptotene and zygotene stages of meiotic prophase, during which pairing and synapsis are executed (Phillips and Dernburg, 2006; Phillips et al., 2005). This dynamic phase is marked by a polarized nuclear appearance, defined as an asymmetric distribution of chromosomes within the nuclear volume, with the nucleolus also displaced to one side (MacQueen and Villeneuve, 2001). In premeiotic nuclei and meiotic prophase nuclei that have completed synapsis, the nucleolus occupies a more central position within the nucleus, and the chromosomes are evenly distributed throughout the surrounding space.
This polarized configuration with chromosome sites associated with the NE is reminiscent of the bouquet stage of meiosis in other organisms, in which telomeres associate with the NE and often cluster together. The bouquet appears transiently during meiotic prophase, approximately concomitant with pairing and synapsis initiation, but its function is not well understood (reviewed by Harper et al., 2004; Scherthan, 2001, 2007). Meiotic interactions between telomeres and the NE have been most extensively analyzed in the fission yeast Schizosaccharomyces pombe. During meiotic prophase in this organism, a protein bridge links chromosomes, via their telomeres, to the spindle pole body (SPB) and cytoplasmic microtubules. Clustering of telomeres, together with dynein-driven “ horsetail ” movement of the entire nucleus, plays a major role in aligning chromosomes and facilitating homologous recombination (Chikashige et al., 1994; Miki et al., 2002). A complex of NE proteins, including Sad1p and Kms1p, is required for telomere attachment and clustering. Sad1p is a member of the SUN domain family of inner NE proteins, which anchor diverse partner proteins in the outer NE through interaction with their C-terminal KASH domains (Starr and Fischer, 2005). Sad1p interacts with Kms1p, which spans the outer NE and interacts with cytoplasmic dynein. Mutations in sad1 or kms1 severely reduce crossing-over and impair homolog segregation, while mutations in dynein cause more subtle defects. Since S. pombe lacks transverse SC components, telomere attachment and motion do not play roles in synapsis per se, but they do appear to both facilitate homologous chromosome interactions and restrict non-homologous, or ectopic, interactions (Davis and Smith, 2006; Ding et al., 2004; Niwa et al., 2000).
In C. elegans, a mutation in the SUN domain protein sun-1 disrupts normal homolog synapsis (Penkner et al., 2007), but the source and contribution of force-generating mechanisms in pairing and synapsis have not yet been determined. The generality of interactions between chromosomes and cytoskeletal elements and their significance for meiosis remain important unresolved questions.
Here we show that in C. elegans, a bridge of SUN/KASH proteins is dynamically assembled during meiosis to connect chromosomes to the microtubule cytoskeleton. Our data provide direct evidence that microtubules, acting through this NE bridge, promote homolog pairing. We also demonstrate that dynein and NE components collaborate to license the initiation of synapsis and to restrict SC formation to appropriate, homologous pairs.
Results
Reorganization of the nuclear envelope coincides with initiation of chromosome pairing and synapsis
Cells within the germline of C. elegans are organized in a temporospatial gradient, making it straightforward to visualize both premeiotic proliferation and meiotic progression. During the stages of pairing and SC formation in C. elegans, the Pairing Center (PC) of each chromosome is bound by one of four related zinc finger proteins, named HIM-8, ZIM-1, -2, and -3, which show intimate association with the NE (Phillips and Dernburg, 2006; Phillips et al., 2005). In these early meiotic nuclei, we found that the sites where PCs contact the NE also showed striking enrichment of ZYG-12, a KASH domain protein with homology to S. pombe Kms1 (Figure 1A). This transmembrane protein is distributed evenly around the nuclear surface in all interphase cells, and also associates with centrosomes during mitosis. It is required to link centrosomes to the NE in C. elegans embryos (Malone et al., 2003), and also plays an essential role in nuclear positioning in the C. elegans germline (Zhou et al., 2009).
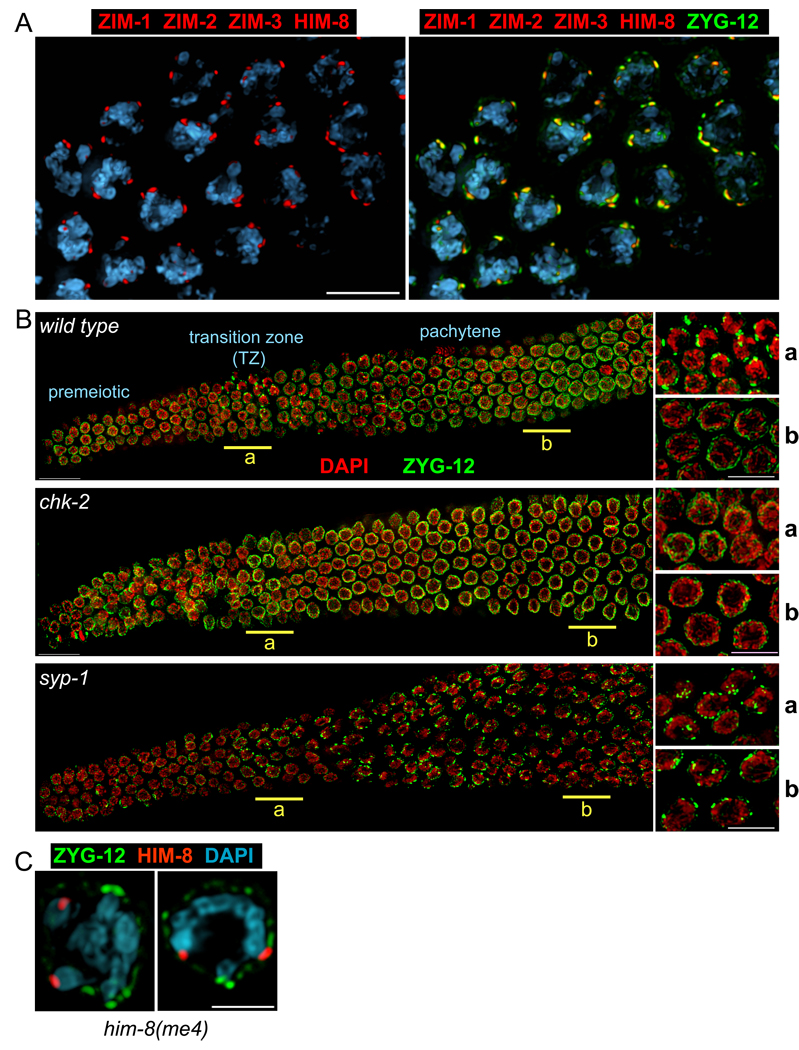
Figure 1. Dynamic association of ZYG-12 with Pairing Centers during early meiosis.
(A) Left: Pairing Centers, marked by ZIM-1, -2, -3 and HIM-8 (red), are dispersed along the NE in transition zone nuclei. Right: the NE protein ZYG-12 (green), here detected with anti-GFP antibodies, coincides with the sites of PC association. (B) Composite images of whole gonads from wild-type, chk-2(me64), and syp-1(me17) animals, each expressing zyg-12::gfp, stained with anti-GFP (green) and DAPI (red) are shown with the distal tip oriented to the left, and meiotic progression advancing from left to right. Higher magnification views from the underlined regions (a/b) are shown to the right. In wild type animals, ZYG-12 patches appear transiently in the transition zone. At pachytene, ZYG-12 is redistributed throughout the NE. Patches are not detected in chk-2 mutants (middle), whereas the region of patches is greatly extended in syp-1 hermaphrodites (bottom) and other mutants that fail to synapse one or more chromosomes (Figure S3). Nuclei with patches also show polarized morphology by DAPI staining. Scale bars, 5µm. (C) The him-8(me4) missense mutation disrupts HIM-8 association with ZYG-12 patches. Cross sections of transition zone nuclei from him-8(me4) zyg-12:gfp animals stained with antibodies against HIM-8 (red) and GFP (green), and DAPI (blue) are shown. Scale bar, 3µm.
Upon completion of meiotic S-phase, concomitant with the appearance of polarized “ transition zone ” nuclear morphology, prominent “ patches ” of ZYG-12 appeared at the NE (Figure 1B, 2, Movie S1). Individual nuclei typically displayed 1–9 large ZYG-12 patches that could be resolved by wide-field deconvolution microscopy (Figure 1). 3D-Structured Illumination Microscopy (3D-SIM; Gustafsson et al., 2008) revealed that many of these patches represent the convergence of multiple chromosomes (Figure 2, Movie S1). Real-time imaging of ZYG-12:GFP with fast-3D microscopy has revealed that ZYG-12 patches are highly dynamic (Movie S2 and Movie S3, Figure S1). Upon completion of synapsis, ZYG-12 redistributed throughout the NE, except for one small focus that remained associated with the paired HIM-8 signal throughout pachytene (Figure S2).
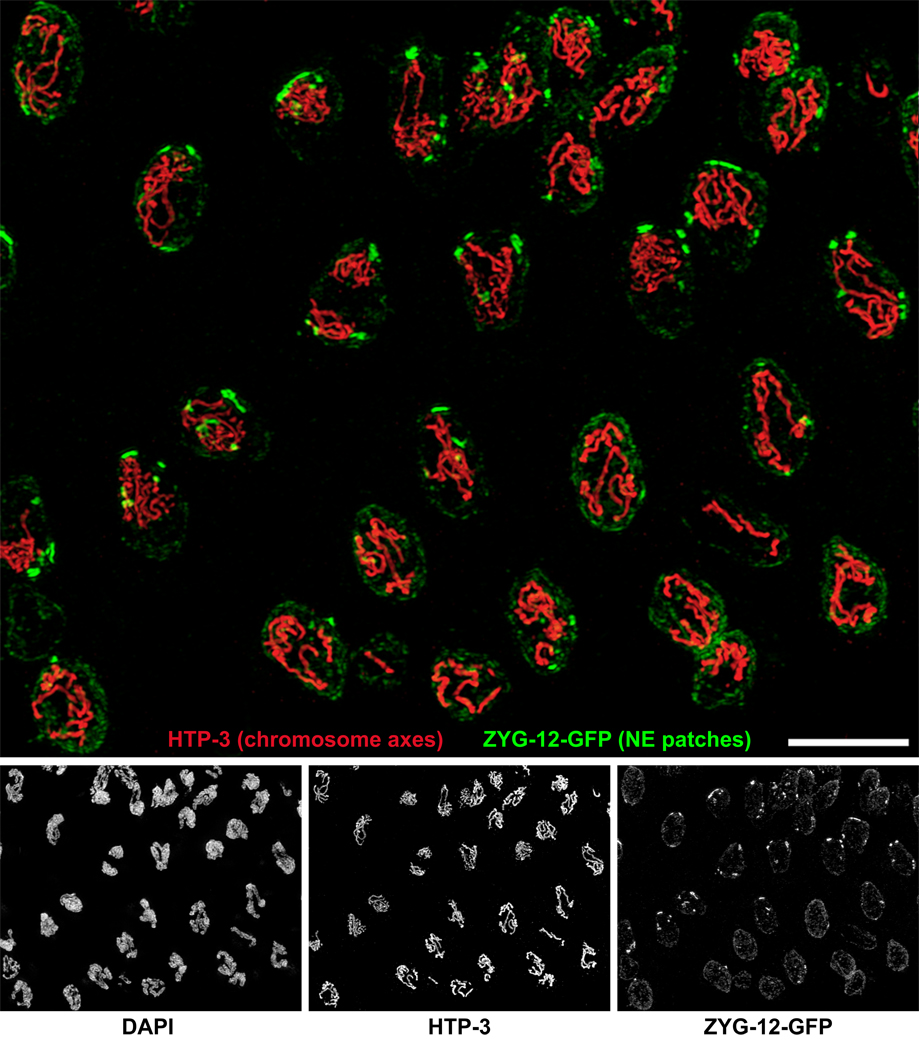
Figure 2. Chromosomes cluster at nuclear envelope patches.
3D Structured Illumination Microscopy (Gustafsson et al., 2008) was used to image transition zone and early pachytene nuclei in wild type (zyg-12:gfp) animals. The images are maximum-intensity projections through a 5-µm sample depth. Separate images are shown below the large merged image. Axial elements were visualized with anti-HTP-3 antibodies (red). ZYG-12-GFP was visualized with anti-GFP (green). The DAPI counterstain is not shown in the merged image. Nuclei in the upper left region are mostly in leptotene/zygotene, based on the presence of ZYG-12 patches, DAPI morphology, and incomplete synapsis, while pachytene nuclei, which show fewer and thicker stretches of axial elements, predominate to the lower right. Transition zone nuclei often contain multiple “mini-bouquets,” in which variable numbers of chromosomes are associated with large NE patches (see also Supplemental Movie 1). In early pachytene nuclei, ZYG-12 is redistributed throughout the NE, but a small focus remains. Scale bar, 5µm.
ZYG-12 is likely localized to the outer NE (Malone et al., 2003; Starr and Fischer, 2005). In C. elegans embryos, the inner nuclear envelope protein SUN-1 is required to anchor ZYG-12 within the NE (Malone et al., 2003). A missense mutation in sun-1 results in pairing defects and non-homologous synapsis during meiosis (Penkner et al., 2007). We found that SUN-1 was closely associated with ZYG-12 patches and showed essentially the same NE dynamics as ZYG-12 in the germline: transient concentration at the sites of PC-NE apposition in transition zone nuclei, with a fairly uniform distribution surrounding premeiotic, pachytene, and later-stage meiotic nuclei (Figure 3 and data not shown). By contrast, other known NE components, including lamin (LMN-1), emerin (EMR-1), LEM-2, and nuclear pore complexes were not concentrated at the PC sites (data not shown; for lamin, see Phillips et al., 2005; Phillips and Dernburg, 2006).
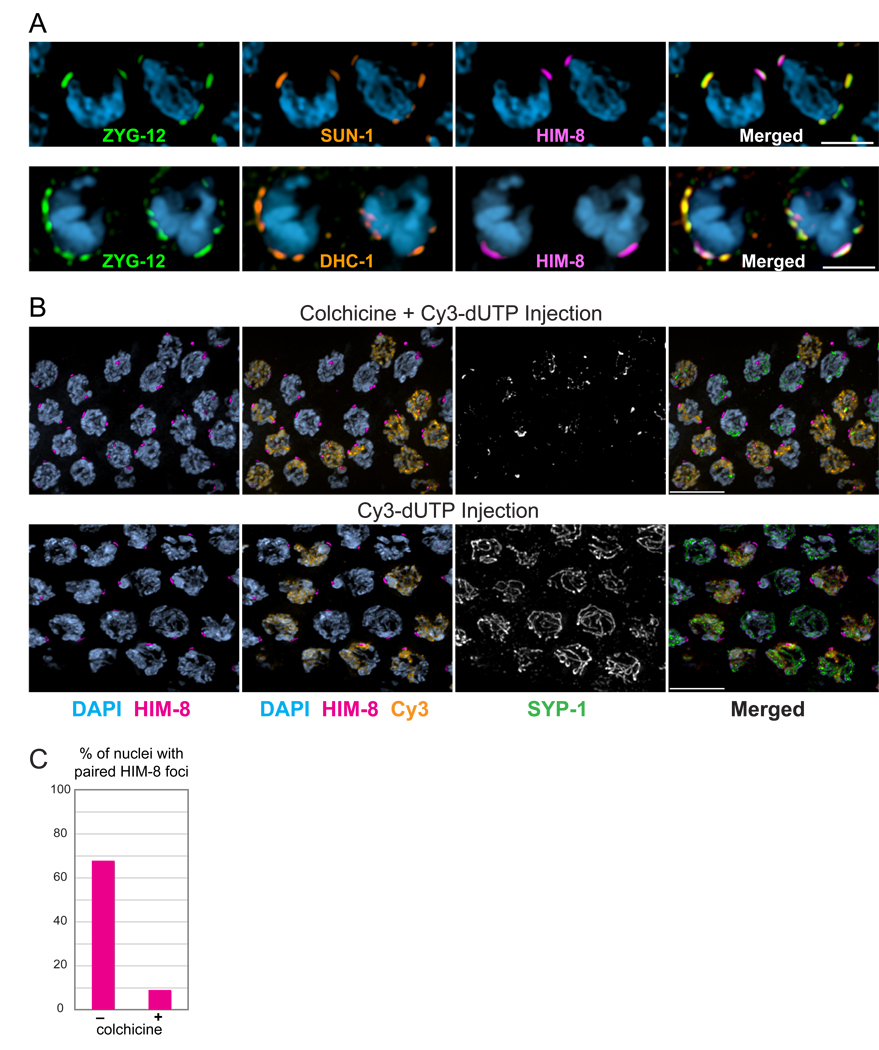
Figure 3. A nuclear envelope bridge connects PCs to dynein and microtubules.
(A) SUN-1 and dynein are associated with NE patches. Two adjacent transition zone nuclei showing immunolocalization of ZYG-12:GFP (green), SUN-1 or DHC-1 (orange), and HIM-8 (magenta), and DAPI staining (blue). Scale bars, 3µm. (B) Disruption of microtubules abrogates homolog pairing. Transition zone nuclei from wild type hermaphrodites injected with Cy3-dUTP and colchicine or Cy3-dUTP only (control). Injected animals were dissected after 6 hours. Pairing of the X chromosome PC was assessed following HIM-8 immunostaining (magenta). Cy3 incorporation is shown in orange, and SYP-1 immunofluorescence in white (green in merged image). Scale bars, 5µm. (C) Quantification of the experiment shown in (B). Pairing was assessed in nuclei positive for both Cy3 and nuclear SYP-1. Colchicine injection significantly reduced HIM-8 pairing compared to the control (p<0.0001).
Previous characterization of a unique allele of him-8 revealed that association of the protein with the X PC and the NE is not sufficient to promote pairing or synapsis (Phillips et al., 2005). him-8(me4) contains a S85F missense mutation, but its C-terminal DNA binding domain (Phillips et al., 2009) is intact, and it retains normal binding to the X chromosome PC. In him-8(me4) mutants, we found that all ZIM proteins were associated with SUN-1/ZYG-12 patches, but that HIM-8 foci were not adjacent to a NE patch, although they localized to the nuclear periphery (Figure 1D, S3B and data not shown). This suggests that HIM-8me4 is defective in an activity that promotes patch formation or association.
For a number of reasons, we favor the idea that the patches are nucleated by association of NE components with the high density of HIM-8 or ZIM binding sites that confer PC function. We have never observed a NE patch that lacked a closely apposed focus of HIM-8 or ZIM proteins, either in wild-type or in mutant animals. Further, when “ artificial PCs ” consisting of high-copy arrays of HIM-8 or ZIM binding sites are introduced, they are consistently associated with large SUN-1/ZYG-12 patches (Phillips et al., 2009). In premeiotic nuclei, two foci of HIM-8 are associated with the X chromosome PCs and the NE, prior to the appearance of NE patches (data not shown). This suggests that HIM-8 requires (a) yet-unknown factor(s) to trigger patch formation in early meiosis.
Formation of SUN-1/ZYG-12 patches coincides temporally with the period of pairing and synapsis initiation. The patches are physically associated with PCs, which mediate synapsis-independent pairing between homologous chromosomes, and probably also act as major sites of synapsis initiation (MacQueen et al., 2005). These observations, along with previous functional evidence (Penkner et al., 2007), suggested important roles for these NE components in meiotic chromosome dynamics. We therefore explored whether the early prophase reorganization of SUN-1 and ZYG-12 might be altered in situations with perturbed pairing or synapsis. We found that appearance of the SUN-1/ZYG-12 patches requires the function of chk-2 (Figure 1B), a serine/threonine kinase required for many early meiotic events, including nuclear polarization, chromosome pairing, timely synapsis, and meiotic recombination (MacQueen and Villeneuve, 2001). Conversely, mutation of the SC transverse filament component syp-1, which eliminates synapsis and results in an extended region of polarized nuclei (MacQueen et al., 2002), also resulted in an extended region of nuclei with SUN-1/ZYG-12 patches (Figure 1B and data not shown). Other mutations that impair synapsis of one or more chromosomes, including mutations in the him-8 or zim genes, resulted in a similar extension of the “patchy” region of the gonad, throughout which the ZIM proteins remain focused and closely apposed to NE patches. Notably, HIM-8 or ZIM proteins associated with synapsed chromosomes remained patch-associated in the presence of a single unsynapsed chromosome (Figure S3 and data not shown), indicating that exit from the patch-associated, polarized configuration occurs on a nucleus-by-nucleus, rather than chromosome-by-chromosome basis. Thus, the appearance of polarized nuclei is correlated with the presence of SUN-1/ZYG-12 patches, and these features are normally restricted to nuclei in which chromosome pairing has initiated but has not been fully stabilized by synapsis.
The nuclear envelope proteins connect meiotic chromosomes with dynein motors during early meiosis
ZYG-12 interacts with cytoplasmic dynein in a yeast two-hybrid assay (Malone et al., 2003). Consistent with this prior data, immunofluorescence revealed that dynein heavy chain (DHC-1), dynein light intermediate chain (DLI-1), and the dynein regulators DNC-1 (dynactin/p150) and LIS-1 were enriched at the patches of SUN-1 and ZYG-12 in the transition zone (Figure 3A, S4).
ZYG-12 is likely to interact with microtubules, based on its association with dynein and its homology to HOOK proteins in other organisms (Malone et al., 2003). The observed colocalization of dynein, ZYG-12, and PCs led us to investigate how microtubules and the dynein motor complex might contribute to meiotic chromosome dynamics. Microtubules and dynein are required for embryonic and larval development, and for germline mitotic divisions. We therefore designed experiments to deplete these proteins in adult worms using RNAi, temperature-sensitive mutations, and/or pharmacological agents under conditions in which mitotic defects did not impair our ability to analyze the consequences for meiosis
Microtubules mediate homolog pairing in early prophase
Homolog pairing can be analyzed using locus-specific markers such as the PC proteins or FISH. SC polymerization can be detected as stretches of transverse filament proteins, such as SYP-1, which coincide with DAPI-staining regions or axial elements. Axial elements lacking transverse filament proteins are defined as unsynapsed (cf. MacQueen et al., 2005). To investigate the role of microtubules in meiotic chromosome behavior, we injected the microtubule-depolymerizing drug colchicine directly into the syncytial gonads of adult worms. Based on the knowledge that meiotic S-phase immediately precedes the onset of transition zone morphology (Figure S5), we co-injected fluorescent deoxynucleotides (Cy3-dUTP) to mark the pool of nuclei that were in S-phase at the time of colchicine introduction, and would potentially be affected by loss of microtubule function as they exited S-phase and initiated pairing. Animals were fixed 6 hours after injection, and pairing and synapsis were assessed in nuclei positive for both Cy3 and nuclear SYP-1, which indicated entry into meiotic prophase. In animals injected with Cy3-dUTP alone, most (Cy3 and SYP-1)-positive nuclei displayed merged HIM-8 foci, and had also undergone extensive synapsis (Figure 3B, C). By contrast, in worms coinjected with colchicine and Cy3-dUTP, most (Cy3 + SYP-1)-positive meiotic nuclei had unpaired HIM-8 foci and showed only aggregates of SYP-1. These data indicate that microtubules are essential for pairing of the X chromosomes within the 6-hour time span of this experiment, and that in the absence of pairing, colchicine-treated nuclei do not undergo synapsis. Disruption of microtubules by colchicine injection also resulted in smaller and more dispersed NE patches (Figure S6) as well as loss of chromosome polarization (data not shown). These data suggest that small patches require functional microtubules to coalesce into normal large patches, and that nuclear polarization depends on microtubules.
Because colchicine targets other than microtubules have been reported, we tested whether other microtubule drugs could produce analogous effects on chromosome and patch dynamics. We observed an identical spectrum of defects in animals incubated for 6 hours in liquid or on plates containing 1–4µM HTI-286 or 20µM cryptophycin (Figure S7 and data not shown). Both are potent inhibitors of microtubule polymerization that bind to tubulin at sites distinct from colchicine (Lo et al., 2004; Smith and Zhang, 1996) and show high efficacy in C. elegans (Zubovych et al., 2006). These observations strongly reinforce the conclusion that microtubules are essential for timely homolog pairing. By contrast, injection of the actin-depolymerizing agent Latrunculin A (2µM) did not result in any defects in homolog pairing, synapsis or chromosome polarization (data not shown). We also did not detect meiotic defects following soaking or injection of the microtubule-stabilizing agent taxol. This suggests that homolog pairing is not highly sensitive to perturbations in microtubule dynamics; however, it is possible that our experiments may not have delivered sufficient concentrations of taxol to induce microtubule stabilization.
Tomographic reconstruction of electron micrographs revealed microtubules associated with the cytoplasmic surface of the NE in transition zone nuclei. (Figures S8, S9). In some images, these were associated with fibrillar structures that appeared to traverse the lumen between the inner and outer nuclear membranes and protrude into the cytoplasm (Figure S9). We speculate that these may correspond to the SUN-1/ZYG-12 patches observed by fluorescence microscopy, although we have not yet obtained immuno-EM data to support this hypothesis. Future studies will investigate microtubule organization and dynamics associated with pairing at the NE.
Dynein is required for SC polymerization
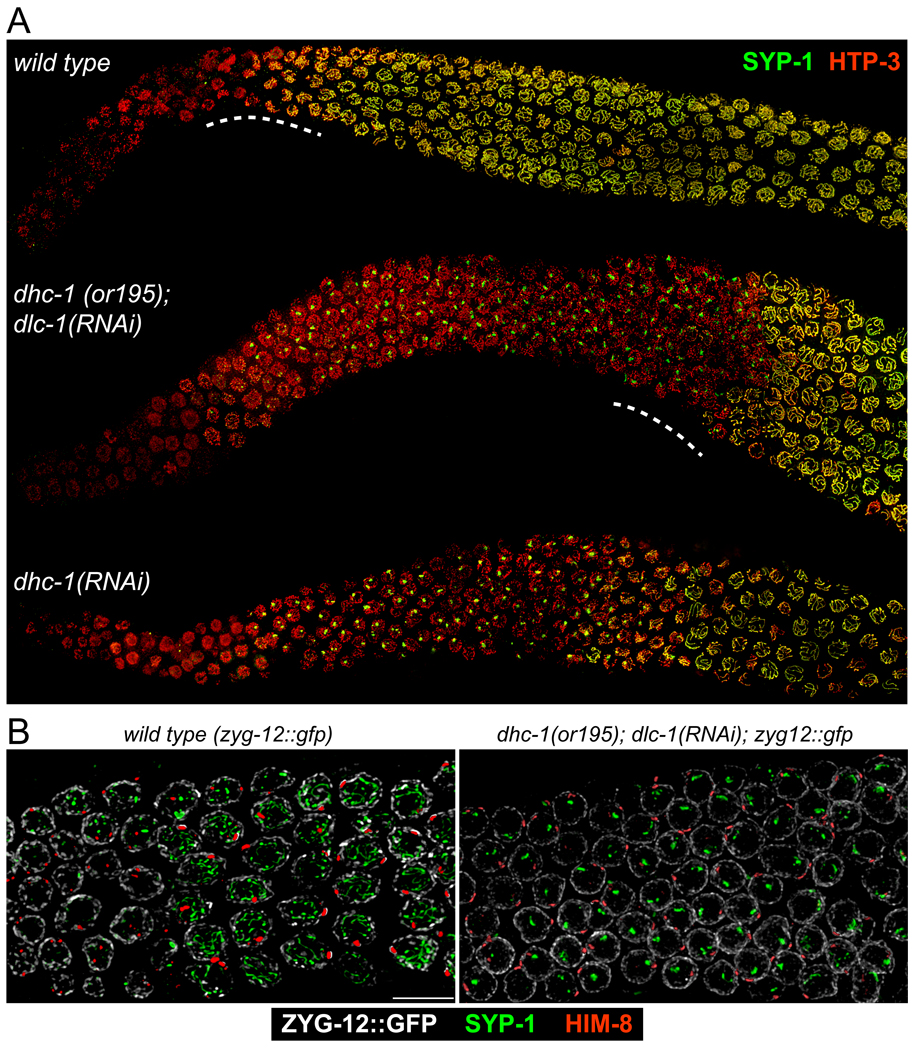
To test the consequences of dynein depletion, we took advantage of an observation that knockdown of the dynein light chain gene dlc-1 partially suppresses mitotic defects in animals carrying the temperature-sensitive dynein heavy chain allele dhc-1(or195) (O'Rourke et al., 2007). dhc-1(or195); dlc-1(RNAi) animals showed only occasional defects in the pre-meiotic region of the germline, and most nuclei entered meiosis on schedule with a normal complement of chromosomes. However, we observed striking meiotic defects in these animals. We first investigated the dynamics of homologous PC association. This analysis was restricted to nuclei containing both HIM-8 and ZIM-2 foci, which correspond to the transition zone (Phillips and Dernburg, 2006). This region was divided into four zones of equal length to evaluate the steady-state level of pairing as a function of meiotic progression (Figure 4A). Homolog pairing was markedly delayed by dynein knockdown, with a stronger effect on Chromosome V than the X chromosome (Figure 4B). Real-time imaging has indicated that patch dynamics are also markedly reduced following dynein inactivation (D.J.W. and P.M.C., unpublished observations).
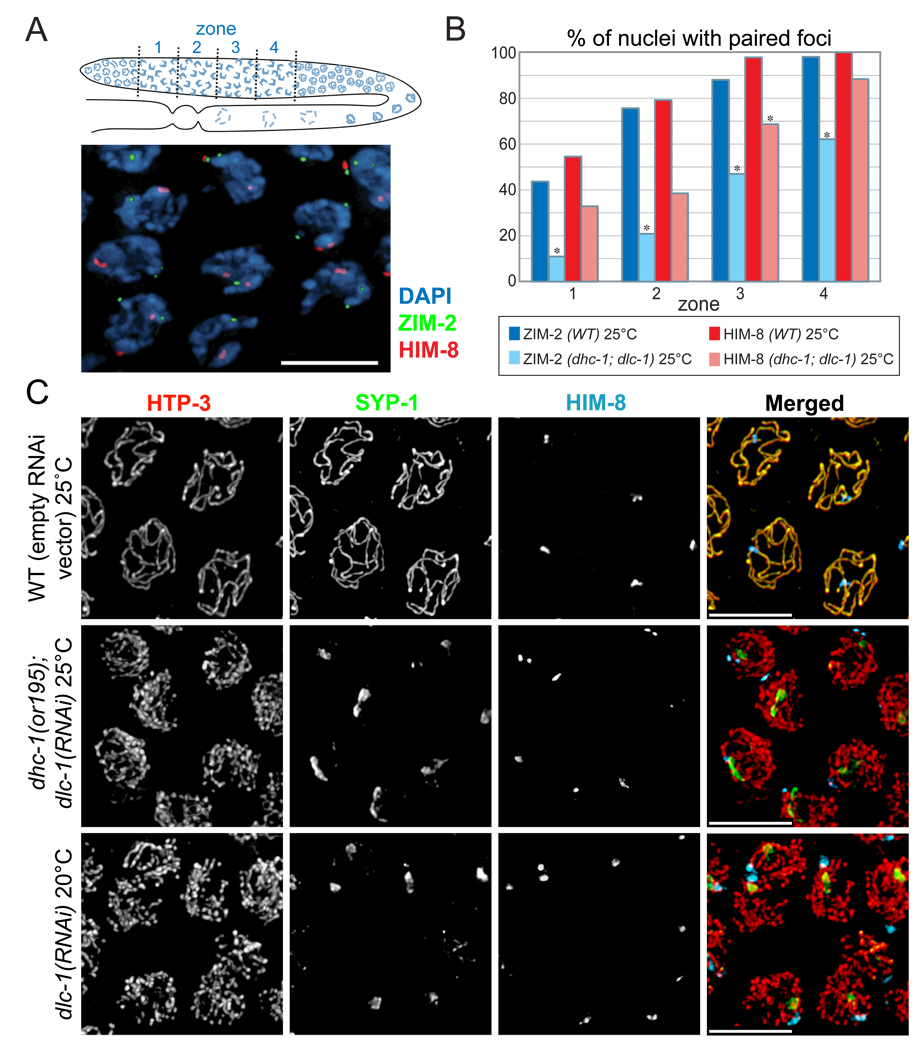
Figure 4. Disruption of the dynein motor complex delays homolog pairing and abrogates synapsis.
(A) Top panel: Diagram of the adult hermaphrodite germline with nuclei depicted in blue. The region in which both HIM-8 and ZIM-2 foci were detected, corresponding to the extended transition zone, was divided into four zones of equal length, designated as zones 1, 2, 3 and 4. Bottom panel: Representative image of transition zone nuclei showing ZIM-2 (green), HIM-8 (red) and DAPI (blue). Scale bar, 5µm. (B) Pairing of HIM-8 and ZIM-2 was scored in control animals and in dhc-1(or195) animals subjected to dlc-1 RNAi for 48 hours. Animals were shifted from 15° C to 25° C 24 hours before fixation. Asterisks indicate values different from wild-type controls at P<0.03. (C) Representative nuclei from the medial germline, which corresponds to pachytene in wild type animals and zone 4 in Figure 4B. Germlines were stained with SYP-1 (green), HTP-3 (red), and HIM-8 (blue) antibodies. In dhc-1(or195); dlc-1(RNAi) animals (48 hours of RNAi and 24 hours at 25° C), and dlc-1(RNAi) animals (55 hours of RNAi at 20° C), SYP-1 failed to polymerize between chromosomes, and instead formed aggregates or polycomplexes. The bottom panels show wild-type animals fed dlc-1 RNAi bacteria for 55 hours at 20° C, then dissected and stained. Abrogated synapsis caused by dlc-1 RNAi at 20°C was less penetrant than dhc-1(or195); dlc-1(RNAi) at 25° C, but ~60% of gonads were affected and showed essentially the same phenotype, indicating that elevated temperature is not required to abrogate synapsis. Control animals (top panels; N2) were fed the same bacteria carrying empty vector (L4440) for 48 hours and shifted to 25° C for the final 24 hrs. Scale bar, 5µm
Strikingly, even once homologous PCs did pair in dynein-depleted animals (e.g., 90% of HIM-8 foci were paired in zone 4 in Figure 4B), the SC failed to polymerize, and central region proteins instead remained localized to prominent polycomplexes (Figure 4C, 5A). By contrast, in wild-type animals, no lag is apparent between PC pairing and synapsis: nuclei with paired HIM-8 signals that lack extensive SYP-1 staining are rarely, if ever, observed (Figure 5B). The accumulation of homologously paired but unsynapsed chromosomes indicates that dynein function is required for homologs that attain pairing at their PCs to initiate synapsis. The loading of axial components of the SC, as visualized by immunofluorescence detection of HTP-1/2, HTP-3 or HIM-3, occurred normally in dynein-depleted animals (Figure 4, 5 and data not shown), indicating that dynein is specifically required for transverse filament loading, rather than axis assembly.
Figure 5. Global view of meiotic phenotypes resulting from loss of dynein function.
(A) Low-magnification images of germlines stained with SYP-1 and HTP-3 antibodies. Normal loading of HTP-3 and SYP-1 was observed in the controls. By contrast, synapsis was severely inhibited in both dhc-1(or195); dlc-1(RNAi) animals. dhc-1 RNAi in wild-type animals was carried out at 25° C for 50 hours. Nuclei in the proximal region (right side of image) of dhc-1(or195); dlc-1(RNAi) and dhc-1(RNAi) animals presumably achieved synapsis prior to reduction of dynein function by RNAi. (B) Nuclei from wild-type (zyg-12::gfp) controls and dhc-1(or195); dlc-1(RNAi) animals corresponding to the regions indicated by the dotted lines in 5A, stained with antibodies against GFP (white), SYP-1(green) and HIM-8 (red). Left panel: In wild-type animals, extensive SYP-1 loading is seen in all nuclei with paired HIM-8 foci, indicating that the SC polymerizes concomitantly with PC pairing. Right panel: Following dynein knockdown, aggregates of SYP-1 persist and polymerization along chromosomes is not detected, despite robust pairing of the X chromosome PC.
Essentially identical effects on pairing and synapsis were observed following dhc-1 RNAi, dlc-1 RNAi, or dnc-1 RNAi in wild-type animals (Figure 4C, 5A and data not shown), but these single-gene knockdowns also resulted in more pronounced mitotic defects in the distal germline. In each case, most nuclei attained homologous pairing of PCs by the middle region of the germline, corresponding to pachytene in wild type, and yet failed to load stretches of SYP-1 between homologs. (Figure 4C, 5). When dynein function was inhibited for longer periods (RNAi feeding for more than 55 hours at 25° C against dlc-1 or dhc-1) mitotic defects accumulated and highly aberrant nuclei, including abundant macro- and micro-nuclei, were distributed with irregular spacing throughout the germline. These defects likely reflect requirements for dynein in accurate mitotic division (O’Rourke et al., 2007) and germline nuclear positioning (Zhou et al., 2009). Some nuclei in the pachytene region also accumulated stretches of SYP-1, although synapsis never appeared to be normal or complete, and large polycomplexes remained (Figure S10). Because of the extensive polyploidy, we could not determine whether SYP-1 loaded between homologous or heterologous chromosomes regions, or perhaps along unpaired chromosomes.
Disruption of the NE bridge complex interferes with meiotic chromosome dynamics
We tested whether NE association of PCs requires SUN-1 or ZYG-12. Homozygous mutant animals derived from heterozygous mothers carrying deletion alleles of sun-1(gk199) or zyg-12(ie14) survive to adulthood (Fridkin et al., 2004; Supplemental Results), likely due to maternal contribution of protein and/or message. Homozygous animals produced by heterozygous mothers have a disorganized mitotic germline region containing fused and polyploid nuclei, fail to execute early stages of meiosis, and are completely sterile. Despite their mitotic and meiotic defects, HIM-8 and ZIM foci were still associated with the NE in zyg-12(ie14) and sun-1(gk199) germlines, suggesting that other membrane-associated proteins likely tether PCs to the NE (Figure S11). Consistent with this, no physical interaction was detected between the N-terminus of SUN-1 (aa1-108) with HIM-8 or ZIM-3 in pairwise yeast two-hybrid assays (Figure S12). However, this could reflect a requirement for meiosis-specific modification of SUN-1 or the zinc finger proteins to promote their interaction, such as the SUN-1 N-terminal phosphorylation described by Penkner et al. (this issue).
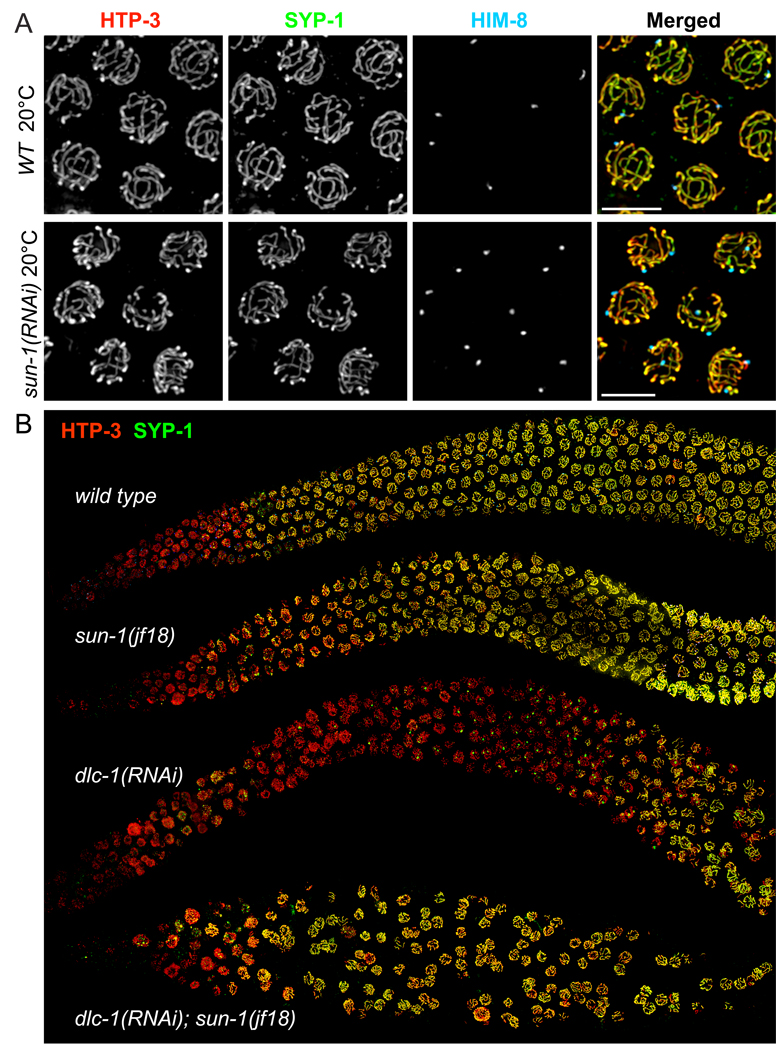
Because of the extensive mitotic errors observed in zyg-12 and sun-1 deletion homozygotes, we primarily analyzed the meiotic roles of these components in adults following RNAi or shifts of temperature-sensitive mutants to restrictive conditions. A missense mutation in sun-1 results in reduced homolog pairing and extensive non-homologous synapsis (Penkner 2007). We recapitulated this phenotype by reducing the amount of SUN-1 protein by sun-1 RNAi (Figure 6A). SUN-1 retained its localization at the NE both in the jf18 mutant and following RNAi of adults. In sun-1(jf18) meiotic nuclei, reduced staining of ZYG-12 was observed (Penkner et al., 2007 and Figure S6) and DHC-1 was undetectable at the NE by immunofluorescence (Figure S6G). Based on our analysis of sun-1(RNAi) and the sun-1(gk199) deletion mutant, sun-1(jf18) behaves as a meiotic hypomorph that does not fully disrupt homolog pairing (Figure S13), but lacks both the ability to inhibit premature synapsis and to sustain the polarized morphology indicative of active chromosome pairing, suggesting that these latter two mechanisms are closely coupled.
Figure 6. The dependence of SC polymerization on dynein function is bypassed by reduction of SUN-1 function.
(A) Reduction of SUN-1 function by RNAi results in promiscuous, non-homologous synapsis, very similar to the sun-1(jf18) phenotype (Penkner et al., 2007). Images show nuclei from the pachytene region stained for SYP-1 (green), HTP-3 (red), and HIM-8 (blue). (B) Low-magnification views of germlines stained for SYP-1 (green) and HTP-3 (red). sun-1(jf18) animals show extensive SC loading in the same region of the gonad as wild-type controls. Similar to dhc-1(RNAi) (shown in 5A), RNAi against dlc-1 at 20° C (55 hours) in wild-type animals abrogated SC polymerization, and also caused mitotic defects. However, in sun-1(jf18) animals, dlc-1 RNAi did not delay loading of SYP-1, despite the absence of homolog pairing (not shown). These sun-1(jf18); dlc-1(RNAi) germlines also showed extensive mitotic defects and disorganized nuclei, likely due to a synergistic effect of reduced SUN-1 and dynein function.
Loss of SUN-1 function bypasses the dependence of SC assembly on dynein
As described above, dynein function is normally required for SC assembly, which we attribute to a role in ‘licensing’ SC initiation (see Discussion). Conversely, reduction of SUN-1 function through RNAi or the jf18 mutation results in promiscuous, non-homologous synapsis, suggesting that SUN-1 inhibits premature synapsis initiation. We hypothesized that dynein is required to overcome an inhibitory effect of SUN-1 on synapsis initiation. If so, loss of both SUN-1 and dynein function should lead to promiscuous synapsis. We carried out epistasis analysis by performing dlc-1 RNAi in sun-1(jf18) animals at 20° C (Figure 6B). This resulted in extensive non-homologous synapsis, indistinguishable from the sun-1(jf18) phenotype. This confirms that dynein acts through SUN-1 to promote synapsis, and thereby reveals that dynein is not simply required in a non-specific capacity to promote SC formation; e.g., by transporting SC components to the nucleus.
Previous studies have shown that loss of HTP-1, a component of the axial elements of the SC, also results in a truncated leptotene/zygotene stage and promiscuous, non-homologous synapsis (Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). We tested the interdependence of SUN-1 and HTP-1 localization and function. First, we observed that small SUN-1 patches are clearly observed in htp-1(gk174) mutants, despite the brevity of the transition zone (Figure S14A). Conversely, HTP-1 loaded normally in sun-1(jf18) mutants (data not shown), indicating that neither component is dependent on the other for its normal localization. Second, loss of htp-1 eliminated the dependence of SC polymerization on dynein (Figure S14B), indicating that SUN-1 and HTP-1 are both required to impose a synapsis barrier that is overcome through dynein activity. More details of this analysis are reported in the Supplemental Results.
ZYG-12 promotes homolog pairing and prevents promiscuous synapsis
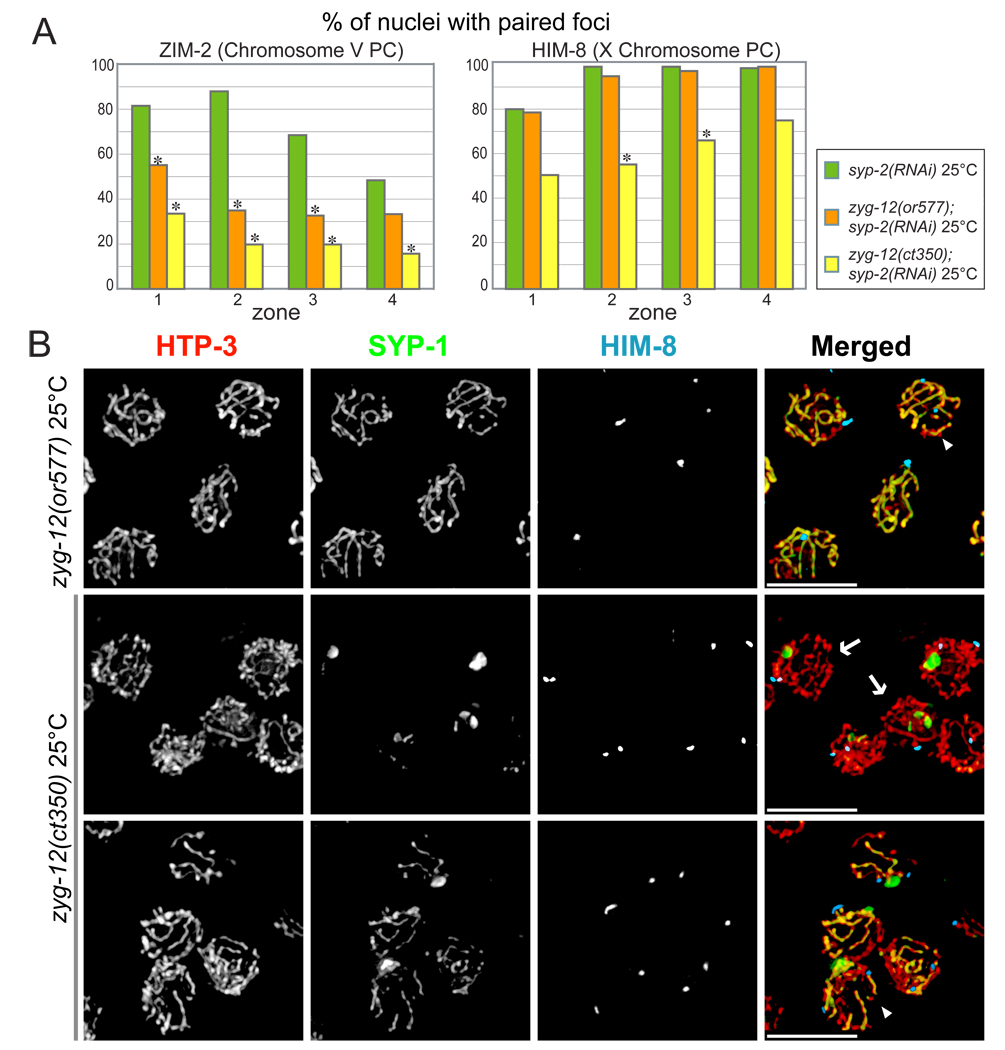
Although two different temperature-sensitive alleles of zyg-12, or577 and ct350, have similar effects in mitotic cells (Malone et al., 2003), their meiotic effects showed interesting differences. ct350 disrupts ZYG-12 self-association as well as its interaction with dynein at elevated temperature in a yeast two-hybrid assay, while or577 disrupts self-association but retains interaction with dynein (Malone et al., 2003). Because both mutant alleles resulted in some non-homologous synapsis (Figure 7B), chromosome pairing at the PCs was quantified under conditions in which synapsis was prevented by RNAi of syp-2. After 6 hours at the restrictive temperature, zyg-12(ct350) severely reduced pairing of the X and V PCs, whereas zyg-12(or577) more subtly reduced V PC pairing and did not perceptibly affect X chromosome PC pairing (Figure 7A).
Figure 7. Mutation of zyg-12 results in reduced homolog pairing and aberrant synapsis.
(A) Pairing of HIM-8 and ZIM-2 foci were scored after shifting to 25° C for 6 hours. ZIM-2 pairing was reduced in both zyg-12(or577) and zyg-12(ct350) hermaphrodites, while HIM-8 pairing was reduced only in zyg-12(ct350). Asterisks indicate a difference from controls at p<0.03. (B) Nuclei from the pachytene region stained for SYP-1(green), HTP-3 (red), and HIM-8 (blue). Animals were fixed after 6 hours at restrictive temperature (25° C). A mixture of unsynapsed, homologously synapsed and non-homologously synapsed chromosomes were observed, with more extensive asynapsis in the zyg-12(ct350) mutant. The arrowhead in the top right panel indicates a nucleus with non-homologous synapsis. The images of zyg-12(ct350) include nuclei with paired HIM-8 foci and SYP-1 polycomplex (arrows, middle row), and non-homologous synapsis (arrowhead, bottom row).
We also examined synapsis in zyg-12ts mutants under the same experimental conditions (25° C for 6 hours, but without syp-2 RNAi). In zyg-12(ct350) hermaphrodites, most early prophase nuclei failed to load SYP-1 onto chromosomes and instead accumulated SYP-1-containing polycomplexes (Figure 7B). A minor fraction of nuclei in zyg-12(ct350) animals loaded stretches of SYP-1, primarily between non-homologous chromosomes. By contrast, in zyg-12(or577) animals, most nuclei loaded stretches of SYP-1 between both homologous and non-homologous chromosomes, with only a small fraction of chromosome lacking SYP-1 staining (Figure 7B). This difference may in part reflect reduced dynein interaction with ZYG-12ct350, based on our results that dynein motor function is required to initiate homologous synapsis.
The observation that both missense alleles of zyg-12 are partially proficient for homolog pairing suggests that they preserve some interaction with microtubules. They also result in smaller, more numerous NE patches (Figure S6), which could reflect reduced interaction with the microtubule cytoskeleton and/or reduced self-association of the mutant proteins.
Discussion
The role of the NE and cytoskeleton in homologous interactions
A fundamental question addressed in this study is how chromosome pairing is coordinated with synapsis so that the SC forms selectively between homologous chromosomes. Previous work from our lab has implicated PCs, special regions on C. elegans chromosomes, in both pairing and synapsis, and suggested that they might participate in coordinating these processes. Here we show that these sites are connected to cytoplasmic dynein and microtubules by a dynamic bridge that includes the SUN/KASH pair, SUN-1 and ZYG-12. Disruption of microtubules inhibited chromosome pairing, and the resulting unpaired chromosomes did not undergo synapsis. By contrast, when dynein was depleted, chromosomes failed to synapse, despite pairing with their homologs at their PCs. When sun-1 or zyg-12 was mutated, homologous chromosomes did not pair properly and synapsed promiscuously. Taken together, these data indicate that the connections between PCs, NE components, and the cytoskeleton actively coordinate pairing and synapsis so that SC assembly is restricted to homologously paired chromosomes.
One interpretation of these results might be that dynein is required to extend pairing from the PCs to the entire chromosome – in other words, to mediate homolog alignment - which is in turn required for SC polymerization. This would be consistent with evidence from S. pombe that dynein-mediated motion facilitates interhomolog interactions along chromosome arms (Ding et al., 2004). While dynein may indeed facilitate alignment in C. elegans, two major lines of evidence argue against this as an explanation of its essential role in SC polymerization. First, extensive chromosome alignment is not required for SC polymerization, as shown by analysis of mutations in sun-1 (Penkner et al. 2007 and this work), zyg-12 (this work), and htp-1 (Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). Second, regions distant from PCs do not achieve stable alignment in the absence of SC polymerization (Colaiacovo et al., 2003; MacQueen et al., 2002). These findings, together with the data presented here, indicate that dynein promotes SC polymerization primarily by overcoming an inhibitory mechanism that acts at the NE, rather than by bringing chromosomes into alignment, and that full alignment is achieved only through the process of synapsis.
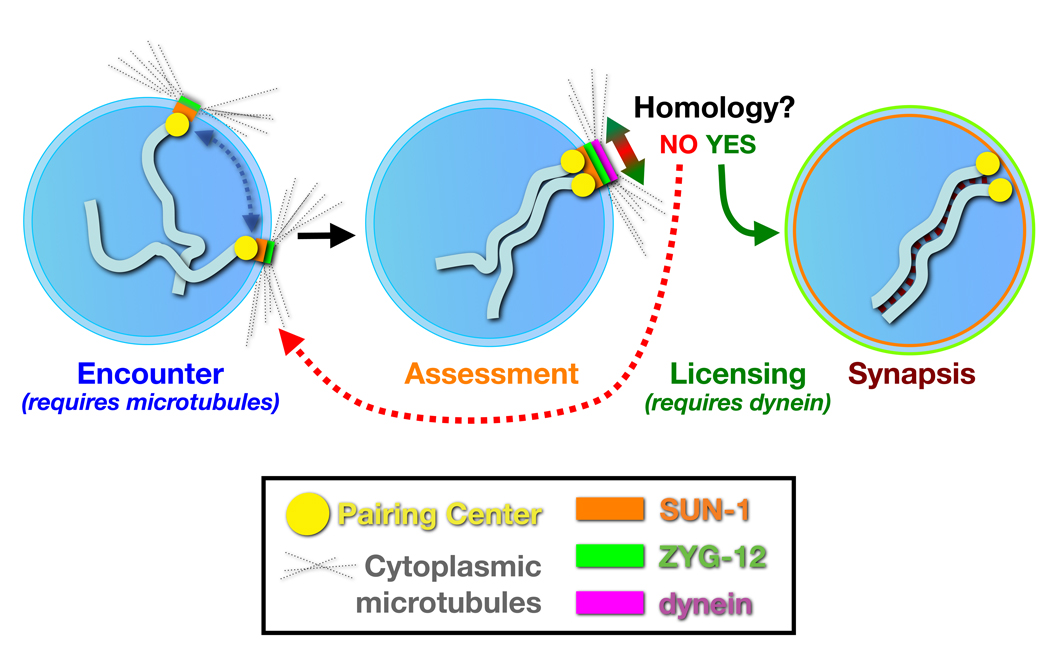
We propose a model in which dynein is required to exert forces that oppose, rather than promote, associations between chromosomes (Figure 8). Non-homologous chromosomes are readily separated by these forces, but homologous chromosomes have sufficient affinity to resist, resulting in mechanical strain that acts through the NE components ZYG-12 and SUN-1. The resulting tension may induce a conformational change or other mechanochemical signal that triggers the initiation of synapsis at the NE attachment sites, i.e., the PCs. This model can explain why dynein is essential for synapsis, and how it might contribute to the efficiency of homolog pairing at PCs without being strictly required for this pairing.
Figure 8. A model for homolog pairing and synapsis.
A nucleus containing a single pair of chromosomes is shown for simplicity. In early meiosis, the NE proteins SUN-1 (orange) and ZYG-12 (green) concentrate by association with individual PCs. Motion of the patches and associated chromosomes is facilitated by microtubules, and promotes encounters between chromosomes. In addition to mediating chromosome encounters, the NE bridge components SUN-1 and ZYG-12 also collaborate to inhibit initiation of synapsis between transiently associated chromosomes. Homology between associated chromosomes is assessed by dynein, acting tangentially at the NE surface. Non-homologous chromosomes normally separate rapidly. However, when dynein exerts force opposing the association between homologous PCs, the resulting tension overcomes a thermodynamic barrier to license synapsis initiation; once initiated, the SC polymerizes processively along the chromosomes, completing their alignment.
Previous studies have indicated that once initiated, SC formation in C. elegans is highly processive and insensitive to homology (MacQueen et al., 2005; Hillers et al., 2003). This implies that synapsis initiation is likely to be a tightly regulated event, with large kinetic and/or thermodynamic barriers that must be overcome. Together with circumstantial evidence that PCs act as major sites of synapsis initiation (MacQueen et al., 2005), it seems likely that an inhibitory mechanism might be specifically imposed at these sites. When SUN-1 function was reduced, the dependence of SC polymerization on dynein was obviated. This implies that a key function of the dynein motor complex is to overcome the synapsis-inhibiting activity of SUN-1 and ZYG-12, which interact with PCs. We note that the persistence of unsynapsed but paired chromosomes in animals with reduced dynein function suggests that synapsis initiation is not solely regulated by a kinetic barrier, as suggested by a model proposed in (MacQueen et al., 2005).
Several important questions remain about the details of the pairing and synapsis initiation mechanisms that transpire at the NE. The nature of the affinity between homologous chromosomes, which must underlie the ability of the cell to differentiate between homologous and non-homologous interactions, remains unknown. In budding and fission yeast, and to a lesser degree in plants and vertebrates, there is good evidence that this recognition is based at least in part upon the recombination machinery, and specifically on strand-invasion intermediates between homologous chromosomes. However, in C. elegans and Drosophila, and likely in diverse other organisms, robust homolog pairing occurs in the absence of double-strand breaks and RAD-51, suggesting that discrimination between like and unlike chromosomes occurs without interchromosomal base-pairing (Reviewed by Bhalla and Dernburg, 2008). The model we have proposed can accommodate diverse mechanisms of homology recognition, as long as cytoskeletal forces during meiosis are tuned to produce forces sufficient to separate non-homologous chromosomes without disrupting appropriate, homologous interactions.
The molecular motors involved in prophase chromosome dynamics also remain to be elucidated. While the dynein motor complex clearly plays a key role, particularly in synapsis initiation, homologs were still able to pair under the most extreme dynein knockdown conditions that we have been able to engineer, suggesting that chromosome pairing may be facilitated by other microtubule motors. Future work will help to fill in many of these crucial gaps in our understanding of meiotic chromosome dynamics.
Conservation and variation in the meiotic roles of cytoskeletal components
The general mechanism of connecting chromosomes to the cytoskeleton via a bridge of NE proteins during meiosis appears to be widespread. While in most organisms this attachment is mediated through telomeres, in C. elegans the PCs appear to have acquired this role. Several lines of evidence indicate that the telomeric bouquet and PC-mediated NE association are evolutionary variants of the same fundamental mechanism. First, SUN-1 homologs have been implicated in telomere attachment in diverse species: in S. pombe, Sad1p/Kms1p play an analogous role to SUN-1/ZYG-12 in C. elegans, by connecting chromosomes to microtubules and dynein. Mps3, the Sad1/SUN-1 homolog in budding yeast, is also required for telomere attachment and clustering during meiosis (Conrad et al., 2007); and Sun1 was recently shown to be required for fertility, telomere attachment to the NE, and efficient synapsis during meiosis in mice (Ding et al., 2007). Second, microtubules appear to play a key role in telomere-mediated meiotic chromosome dynamics in a wide variety of organisms, including S. pombe, mice, rye, and wheat (Corredor and Naranjo, 2007; Cowan and Cande, 2002; Cowan et al., 2002; Ding et al., 1998; Tepperberg et al., 1997; Yamamoto et al., 2001). Here we have demonstrated a role for microtubules in C. elegans acting through the NE and PCs. S. cerevisiae, in which telomere motion is mediated through actin dynamics, presents an interesting, and so far unique, exception.
While key NE components appear to be conserved, the more chromosome-proximal components implicated in NE attachment of meiotic chromosomes are thus far specific to particular evolutionary lineages. The HIM-8/ZIM family in C. elegans, Ndj1 in S. cerevisiae, and Bqt1/Bqt2 in S. pombe all appear to play analogous roles in linking chromosome sites to the NE during meiosis, but are not homologous to each other.
Limited evidence from other species is consistent with a role for chromosome-NE attachment in “ assessing ” local homology to license synapsis – for example, in wheat, homology near telomeres, but not elsewhere on the chromosomes, determines the choice of synaptic partner (Corredor et al., 2007), and synapsis has been reported to initiate near telomeres in a variety of other species. In S. pombe, mutations in bouquet components result in reduced interhomolog recombination and increased recombination between ectopic regions (Davis and Smith, 2006; Niwa et al., 2000), indicating that a primary role of telomere attachment and motion is to enhance the selective association between homologs. In budding yeast, the role of telomere-NE associations and actin-mediated chromosome dynamics is more enigmatic, largely because synapsis and recombination are only subtly affected by mutations that disrupt telomere attachment or movement. Proposed functions have included disrupting ectopic interactions between non-homologous chromosomes, facilitating synapsis by resolving topological entanglements, promoting the completion of recombination, and regulating crossover distribution (Conrad et al., 2008; Kosaka et al., 2008; Koszul et al., 2008; Scherthan et al., 2007; Wanat, 2008). In C. elegans, where homologous pairing and synapsis can be fully uncoupled from recombination (Dernburg et al., 1998), our evidence indicates that the primary roles of the attachment and movement are to promote homolog pairing and to coordinate pairing and synapsis.
Experimental Procedures
Worm strains, genetics and culture conditions
The wild-type C.elegans strain was Bristol N2. All experiments were performed at 20° C under standard laboratory conditions, unless otherwise noted. sun-1(ok1281) was isolated by The C. elegans Knockout Consortium. Sequencing revealed a 581bp-deletion that removes bases 258–838 of the coding sequence and a small intron, resulting in a frame shift right after the breakpoint. This truncated version of SUN-1 retains only the first 85 aa and lacks the predicted transmembrane region (aa109–129). sun-1(gk199) contains a 435bp deletion that spans the promoter, the predicted start codon and part of the predicted transmembrane domain. Immunofluorescence detection of SUN-1 was eliminated by both deletion alleles (see cytological methods, below).
Cytological methods
Sample preparation, immunofluorescence and FISH were performed essentially as in (MacQueen et al., 2005). A polyclonal SUN-1 antibody was raised against a Hisx6-tagged protein fragment corresponding to aa 231–473, purified from E. coli under denaturing conditions. A second polyclonal antibody against SUN-1 aa135–234 was generated using Genomic Antibody Technology™ by Strategic Diagnostics, Inc. Commercial antibodies used in this study included Alexa Fluor 488 rabbit anti-GFP (Invitrogen), monoclonal mouse anti-AFP (MP Biomedicals), and fluorescent secondary antibodies from Invitrogen and Jackson Immunoresearch. Polyclonal antibodies not generated in our lab were graciously provided by Anne Villeneuve (SYP-1), Monique Zetka (HIM-3), Tony Hyman (DLI-1), Chris Malone (ZYG-12), Yossi Gruenbaum (LMN-1), Pierre Gonczy (DHC-1, LIS-1), and Ahna Skop (DNC-1). Except for the 3D-SIM images (Figure 2, Movies S1), all images were acquired with a DeltaVision RT microscope (Applied Precision), using a 100× 1.35 or 1.40 NA objective. Image deconvolution, display, projection, and analysis were performed using the softWoRx package.
Feeding RNAi and drug treatment
For RNAi of all genes other than sun-1, clones from the Ahringer laboratory (Fraser et al., 2000) were used. For sun-1, a PCR product spanning the first 324 bases of coding sequence was cloned into pDONORdT7 and transformed into HT115 (Timmons et al., 2001). Appropriate bacterial strain(s) and empty vector controls were cultured overnight in 20 ml LB + appropriate antibiotics, spun down, and resuspended in 1ml of LB + antibiotics. 70ul of the RNAi bacteria was spread onto 60-mm NGM plates containing 1mM IPTG + antibiotics, and dsRNAs were induced overnight at 37° C. Just before RNAi bacteria were seeded, additional antibiotics (50µl of 50mg/ml stock solution per plate) and IPTG (50 µl of 1M stock solution per plate) were top spread to enhance propagation of bacteria carrying plasmids as well as expression of dsRNAs. For dlc-1 RNAi, either wild type (N2) or dhc-1(or195) L4 animals were placed on freshly prepared RNAi plates, incubated for 24 hours at 15° C, shifted to 25° C for 24 hours, and immediately dissected for cytological analysis. For sun-1 RNAi, wild type (N2) adults were put onto RNAi plates, and their F1 progeny were dissected for cytological analysis. Colchicine treatment was carried out by injecting a solution of 100mM colchicine and 10mM Cy3-dUTP (GE Life Sciences) in injection buffer directly into the distal gonad of young adult worms (16–18 hours post-L4). Injected animals were kept on food at 20° C and dissected 6 hours after injection for cytological analysis. Delivery of the drug(s) into individual gonads was verified by robust Cy3 incorporation and, when colchicine was included, the presence of ≥10 metaphase-arrested nuclei in the premeiotic region. HTI-286 was a generous gift of Tito Fojo and cryptophycin was kindly provided by Eva Nogales.
Quantification of chromosome pairing
Pairing at the PC regions of Chromosomes X and V were measured in samples stained with HIM-8 and ZIM 2 antibodies, essentially as described by Phillips et al. 2005 using a distance threshold of 0.5 µm. To analyze zyg-12ts mutants, wild type (N2), zyg-12(or577) and zyg-12(ct350) adults were transferred to syp-2 RNAi plates, and their F1 progeny were transferred to fresh RNAi plates. F1 progeny at 24 hours post-L4 were shifted to 25° C for 6 hours, dissected, and stained. Loss of SYP-2 function was verified by the absence of SYP-1 immunofluorescence. Worms were kept at the permissive temperature (15° C) unless otherwise mentioned. At least three germlines were scored for each genotype. The total number of nuclei scored for zones 1, 2, 3, and 4, respectively, was as follows: syp-2(RNAi): 157, 191, 187, 153; zyg-12(or577): 214, 209, 205, 156; zyg-12(ct350): 132, 167, 174, 143. For dynein knockdown, wild type (N2) or dhc-1(or195) animals at the L4 stage were picked onto empty vector or dlc-1 RNAi plates, kept at 15° C for 24 hours, shifted to 25° C for 24 hours and stained with HIM-8 and ZIM 2 antibodies. At least three germlines were scored, including 55, 82, 93, and 52 nuclei in wild type (N2) animals, and 73, 96, 115, and 103 nuclei in dhc-1(or195); dlc-1(RNAi) animals. Following colchicine injection, pairing of HIM-8 foci was scored in nuclei with both Cy3 and SYP-1 fluorescence. 5 germlines/378 nuclei from animals injected with Cy3-dUTP only were scored, and 5 germlines/394 nuclei for colchicine-injected gonads. All statistical comparisons were done using Student’s t-test.
Supplementary Material
Acknowledgments
This work was supported by graduate research fellowships from JSPS (A.S.) and the NSF (C.M.P., D.J.W and R.K.), by a research grant from the Keck Laboratory for Advanced Microscopy at UCSF to P.M.C, and by research grants from the American Cancer Society (RSG-07-187-01-GMC) and NIH/NIGMS (R01 GM065591) to A.F.D. We are grateful to John Sedat for the use of the OMX microscope, and to Kent McDonald for expert assistance with EM sample preparation. We thank Chris Malone, Miles Pfaff, Verena Jantsch, Tito Fojo, Eva Nogales, Bruce Bowerman, Yossi Gruenbaum, Pierre Gonczy, Ahna Skop, Anthony Hyman, Anne Villeneuve, and the CGC for providing antibodies, strains, reagents, and technical assistance. We thank Dan Starr, Aaron Severson, Anne Villeneuve, and members of the Dernburg lab for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor E, Lukaszewski AJ, Pachon P, Allen DC, Naranjo T. Terminal regions of wheat chromosomes select their pairing partners in meiosis. Genetics. 2007;177:699–706. doi: 10.1534/genetics.107.078121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor E, Naranjo T. Effect of colchicine and telocentric chromosome conformation on centromere and telomere dynamics at meiotic prophase I in wheat-rye additions. Chromosome Res. 2007;15:231–245. doi: 10.1007/s10577-006-1117-7. [DOI] [PubMed] [Google Scholar]
- Couteau F, Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Cande WZ. Meiotic telomere clustering is inhibited by colchicine but does not require cytoplasmic microtubules. J Cell Sci. 2002;115:3747–3756. doi: 10.1242/jcs.00055. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Carlton PM, Cande WZ. Reorganization and polarization of the meiotic bouquet-stage cell can be uncoupled from telomere clustering. J Cell Sci. 2002;115:3757–3766. doi: 10.1242/jcs.00054. [DOI] [PubMed] [Google Scholar]
- Davis L, Smith GR. The meiotic bouquet promotes homolog interactions and restricts ectopic recombination in Schizosaccharomyces pombe. Genetics. 2006;174:167–177. doi: 10.1534/genetics.106.059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci U S A. 2004;101:6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008;4(9):e1000196. doi: 10.1371/journal.pgen.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Krishnamurthy G, Kaplan J, Zask A, Smith RP, Ellestad G. Probing the interaction of HTI-286 with tubulin using a stilbene analogue. J Am Chem Soc. 2004;126:9898–9899. doi: 10.1021/ja048619e. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11 doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Zhang X. Mechanism of action of cryptophycin. Interaction with the Vinca alkaloid domain of tubulin. J Biol Chem. 1996;271:6192–6198. doi: 10.1074/jbc.271.11.6192. [DOI] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Tepperberg JH, Moses MJ, Nath J. Colchicine effects on meiosis in the male mouse. I. Meiotic prophase: synaptic arrest, univalents, loss of damaged spermatocytes and a possible checkpoint at pachytene. Chromosoma. 1997;106:183–192. doi: 10.1007/s004120050238. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wanat J, Kim K, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4(9):e1000188. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–3946. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186 doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubovych I, Doundoulakis T, Harran PG, Roth MG. A missense mutation in Caenorhabditis elegans prohibitin 2 confers an atypical multidrug resistance. Proc Natl Acad Sci U S A. 2006;103:15523–15528. doi: 10.1073/pnas.0607338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.