Abstract
The Comparative Toxicogenomics Database is a public resource that promotes understanding about the effects of environmental chemicals on human health. Currently, CTD describes over 184,000 molecular interactions for more than 5,100 chemicals and 16,300 genes/proteins. We have leveraged this dataset of chemical-gene relationships to compute similarity indices following the statistical method of the Jaccard index. These scores are used to produce lists of comparable genes (“GeneComps”) or chemicals (“ChemComps”) based on shared toxicogenomic profiles. GeneComps and ChemComps are now provided for every curated gene and chemical in CTD. ChemComps are particularly significant because they provide a way to group chemicals based upon their biological effects, instead of their physical or structural properties. These metrics provide a novel way to view and classify genes and chemicals and will help advance testable hypotheses about environmental chemical-genedisease networks.
Availability
CTD is freely available at http://ctd.mdibl.org/
Keywords: gene, chemical, toxicogenomic, database, curation
Background
The Comparative Toxicogenomics Database (CTD) is a public resource that promotes understanding about the effects of environmental chemicals on human health [1]. CTD biocurators manually curate interactions from the scientific literature in a structured format using controlled vocabularies and ontologies for chemicals, genes, diseases, molecular interactions, and organisms [2,3] . These datasets can be used to explore relationships and also to generate novel, testable hypotheses about chemical-gene-disease pathways. Finding chemicals and genes with similar interaction profiles could promote alternative methods for classifying chemicals and help identify additional members of interaction networks. There are many ways to discover and organize related genes and proteins: sequence similarity, co-expression analysis, shared protein-protein interactions, and common biomarkers for a disease. In addition, the Gene Ontology (an annotation vocabulary used to define gene products by molecular function, biological process, and cellular localization) can be data-mined to produce lists of comparable genes [4]. In contrast, criterion for finding similar chemicals has been largely restricted to the physical properties (e.g., molecular weight, atomic elements, boiling point, molar volume, etc.) or atomic structure of compounds [5]. A relatively new approach, however, is to classify chemicals based upon their effect on mRNA expression detected by microarrays [6,7]. While this method and the development ofextensive chemical vocabularies [8] and new ontologies [9] may further help organize compounds, comparative analysis among chemicals with similar gene interaction profiles (beyond mRNA expression) is still lacking.
At CTD we developed a simple approach to discover analogous genes and chemicals based upon shared chemical-gene interaction profiles, which we call GeneComps and ChemComps for comparable genes and comparable chemicals, respectively. CTD biocurators manually curate the literature and annotate over 50 different types of chemical-gene molecular interactions, including, among others, effects on mRNA expression, protein expression, phosphorylation, activity, localization, degradation, metabolic processing, transport, and promoter methylation. Every gene in CTD has a profile of chemicals with which it interacts, and likewise every chemical has a profile of genes with which it interacts. These extended, detailed, and more complete interaction profiles essentially define a “footprint” for a gene or chemical that can be leveraged to discover and cluster genes and chemicals.
Methodology
We used the data available in CTD in September 2009, which included 184,646 chemical-gene interactions for 5,124 chemicals and 16,308 genes. Similarity indices were computed for chemicals (ChemComps) and genes (GeneComps). The degree of similarity was estimated using a modification of the Jaccard index, whose value ranges between 0 and 1 [10]. The index is the ratio of the number of elements in the intersection of two sets (number of shared interactions between two chemicals or genes) divided by the number of elements in the union of two sets (number of combined interactions between two chemicals or genes).
Utility
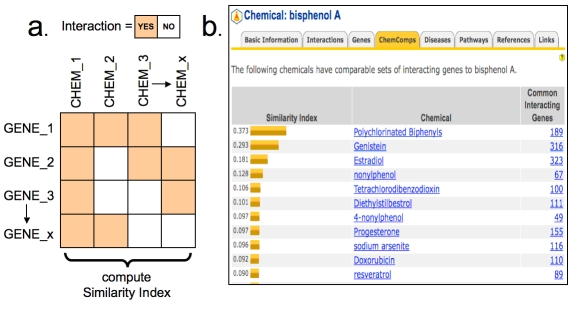
CTD computes statistics that reflect the degree of similarity between the gene interaction profiles of each curated chemical and generates a list of ChemComps (Figure 1a). Likewise, chemical interaction profiles are compared between each curated gene to produce GeneComps. ChemComps and GeneComps provide a simple approach to view chemicals and genes that share interaction profiles. ChemComps especially provide a novel way to classify and organize chemicals based upon biological effects, which can be considered a molecular signature or footprint. Every curated chemical in CTD now includes a ChemComps data tab that lists the top 20 comparable chemicals based upon their ranked similarity index derived from their interaction profile. For example, the chemical bisphenol A (a plastic additive) has curated interactions to 473 genes in CTD. ChemComps lists comparable chemicals that share the most number of interactions with those 473 genes to produce a ranked list that includes polychlorinated biphenyls, genistein, and estradiol (Figure 1b), suggesting that bisphenol A shares many of the networks common to these three chemicals. Similarly, GeneComps are displayed on curated gene pages and lists the genes that share a chemical profile.
Figure 1.
A contingency table of chemical-gene interactions computes the similarity indices (a) to rank comparable chemicals for a user's molecule of interest, such as bisphenol A, which are listed under the ChemComps data tab in CTD (b).
ChemComps and GeneComps are datasets that allow researchers to view and cluster chemicals and genes with similar biological activities. This new metric at CTD provides researchers with additional predictive information that will help construct novel, testable hypotheses about chemical-gene-disease pathways.
Future development
Currently, the similarity indices used to generate GeneComps and ChemComps are derived exclusively by looking for either a “yes” or “no” binary interaction relationship between a gene and chemical (Figure 1a). However, CTD biocurators capture many details about the interactions between these two types of molecules, such as “chemical Y decreases the phosphorylation of protein B” or “protein C results in chemical resistance to chemical Z”, etc. [2,3]. These specific details might be leveraged to derive more qualitative similarity indices, such as finding comparable chemicals that increase vs. decrease the phosphorylation of a protein, or increase vs. decrease the methylation of a gene's promoter.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences [R01ES015065 to APD, CGM, CAS, MCR, TCW and CJM; and P42ES007373 to THH] and the Idea Network of Biomedical Research Excellence program of the National Center for Research Resources [RR-016463 to CJM] of the National Institutes of Health.
Footnotes
Citation:Davis et al, Bioinformation 4(4): 173-174 (2009)
References
- 1.Mattingly CJ, et al. Toxicol Sci. 2006;92:587. doi: 10.1093/toxsci/kfl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis AP, et al. BMC Med Genomics. 2008;1:48. doi: 10.1186/1755-8794-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis AP, et al. Nucleic Acids Res. 2009;37:D786. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M, et al. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheridan RP, Kearsley SK. Drug Discov Today. 2002;7:903. doi: 10.1016/s1359-6446(02)02411-x. [DOI] [PubMed] [Google Scholar]
- 6.Lamb J. Nat Rev Cancer. 2007;7:54. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 7.Williams-Devane CR, et al. Toxicol Sci. 2009;109:358. doi: 10.1093/toxsci/kfp061. [DOI] [PubMed] [Google Scholar]
- 8. http://www.nlm.nih.gov/mesh/
- 9.Degtyarenko K, et al. Nucleic Acids Res. 2008;36:D344. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://en.wikipedia.org/