Abstract
Undernutrition during pregnancy reduces birth weight and programmes adult phenotype with consequences for life expectancy, but its effects on the phenotype of the placenta, responsible for supplying nutrients for fetal growth, remain largely unknown. Using molecular, morphological and functional analyses, placental phenotype was examined in mice during restriction of dietary intake to 80% of control from day 3 of pregnancy. At day 16, undernutrition reduced placental, but not fetal, weight in association with decreased junctional zone volume and placental expression of glucose transporter Slc2a1. At day 19, both placental and fetal weights were reduced in undernourished mice (91% and 87% of control, respectively, P < 0.01), as were the volume and surface area of the labyrinthine zone responsible for placental nutrient transfer (85% and 86%, respectively, P < 0.03). However, unidirectional materno-fetal clearance of tracer glucose was maintained and methyl-aminoisobutyric acid increased 166% (P < 0.005) per gram of undernourished placenta, relative to controls. This was associated with an 18% and 27% increased placental expression of glucose and system A amino acid transporters Slc2a1 and Slc38a2, respectively, at day 19 (P < 0.04). At both ages, undernutrition decreased expression of the placental specific transcript of the Igf2 gene by 35% (P < 0.01), although methylation of its promoter was unaffected. The placenta, therefore, adapts to help maintain fetal growth when its own growth is compromised by maternal undernutrition. Consequently, placental phenotype is responsive to environmental conditions and may help predict the risk of adult disease programmed in utero.
Introduction
Undernutrition during pregnancy is a major, global cause of infant mortality and morbidity. It is common in many developing countries but can also occur during disease, eating disorders and periods of famine caused by natural disasters and human conflict in any country (Yajnik, 2004; Lartey, 2008; Jobe, 2009). Voluntary restriction of caloric intake during pregnancy is also becoming evident in women from affluent cultures anxious to retain a svelte body image (Davies & Wardle, 1994). In all societies, undernutrition during pregnancy reduces the birth weight of the infant with consequences for its life expectancy (Barker, 1994). Many epidemiological studies in human populations of different age, gender and ethnicity have shown that low birth weight is associated with an increased risk of developing cardiovascular, metabolic and other life-shortening diseases in later life (Barker, 2007). Since size at birth depends primarily on the fetal nutrient supply (Fowden et al. 2008), these epidemiological studies have led to the concept that predisposition to adult disease can arise in utero as a result of tissue programming by poor nutrition during critical periods of prenatal development (Barker, 1994; Fowden et al. 2005; McMillen & Robinson, 2005; Hanson & Gluckman, 2008).
Prenatal nutritional programming has been investigated experimentally in several species using a range of techniques to manipulate fetal nutrient availability. These studies have shown that nutritionally induced intrauterine growth restriction (IUGR) leads to postnatal abnormalities in cardiovascular and metabolic function consistent with the human epidemiological data (McMillen & Robinson, 2005; Fowden et al. 2006b). For instance, in rats, restriction of dietary protein or calorie intake during pregnancy has been shown to alter the structural and functional development of many fetal tissues with physiological consequences for the adult offspring (Fowden et al. 2005, 2006b; McMillen & Robinson, 2005; Hanson & Gluckman, 2008). However, relatively little is known about prenatal nutritional programming of the placenta, the organ primarily responsible for supplying nutrients to the fetus (Fowden et al. 2006b, 2008).
The nutrient supply capacity of the placenta depends upon its size, morphology, blood flow and transporter abundance, all of which can vary naturally (Sibley et al. 1997; Fowden et al. 2008, 2009). In several species, experimental restriction of placental growth has been shown to cause postnatal physiological abnormalities similar to those seen in response to maternal dietary manipulation during pregnancy (McLaren, 1965; Molteni et al. 1978; McMillen & Robinson, 2005; Fowden et al. 2008, 2009). In pregnant rats, feeding an isocaloric low protein diet reduces placental weight and alters placental efficiency, morphology and nutrient transport prior to the onset of IUGR (Lederman & Rosso, 1980; Jansson et al. 2006; Rutland et al. 2007). Changes in placental efficiency and expression of nutrient transporters are also seen when placental growth is restricted naturally and by other experimental treatments (Langdown & Sugden, 2001; Coan et al. 2008a). However, the extent to which maternal caloric restriction influences placental efficiency and nutrient transport capacity remains unknown.
In part, the effects of nutrition on placental development may be mediated through changes in gene expression, particularly of imprinted genes. These genes are expressed monoallelically according to their parental origin and have a disproportionately important influence on placental development (Reik et al. 2003; Coan et al. 2005; Fowden et al. 2006a). Ablation of several imprinted genes, including Igf2, H19 and Grb10, alters the size and efficiency of the mouse placenta in association with changes in placental morphology and expression of nutrient transporter genes in some instances (Constancia et al. 2005; Angiolini et al. 2006; Coan et al. 2008b). In other tissues nutritionally programmed in utero, there are changes in DNA methylation, particularly at promoter regions, which are related to altered gene expression in adulthood (Rees et al. 2000; Hanson & Gluckman, 2008). Yet, to date, few studies have examined the effects of maternal nutrition on expression or methylation of imprinted or other genes regulating growth and nutrient transport in the placenta. Using mice, this study investigated placental adaptations to maternal undernutrition using structural, functional and molecular analyses to quantify the growth and nutrient transfer capacity of the placenta during late gestation.
Methods
Ethical approval
All procedures were carried out in accordance with the UK Home Office regulations under the Animal (Scientific Procedures) Act 1986 and comply with the policies and regulations of The Journal of Physiology (Drummond, 2009).
Animals
One hundred and thirty-six virgin 6- to 8-week-old C57Bl/6J female mice (Harlan, UK) were group-housed and mated overnight with C57Bl/6J males (Harlan, UK). On the morning of the plug, all females were singly housed and fed ad libitum on a 23% casein diet, (RB 23% CASEIN SY P829274, Dietex International). Following adjustments to the single housing required to monitor individual food intake, females were randomly selected on day 3 of pregnancy to either remain ad lib fed (Control, CT, n= 70) or to receive 80% of the control food intake for the same stage of pregnancy (undernourished, UN, n= 66). Ad libitum food intake was measured daily and all mice were housed under dark:light 12 h:12 h conditions with free access to water. The presence of a copulatory plug was designated as day 1.
Experimental procedures
Unidirectional materno-fetal clearance of non-metabolisable radioactive tracers was measured in pregnant mice at D16 (n= 20 CT, n= 31 UN litters) and D19 (n= 39 CT, n= 27 UN litters). Briefly, mice were anaesthetised with an intraperitoneal injection of 10 μl g−1 of fentanyl fluanisone and midazolam solutions in deionised water (1:1:2, respectively, Jansen Animal Health). The maternal jugular vein was exposed and 100 μl of 14C-methyl-d-glucose (NEN NEC-377; specific activity 2.1 GBq mmol−1) or 14C-methyl aminoisobutyric acid (MeAIB) (NEN NEC-671; specific activity 1.86 GBq mmol−1) in physiological saline (0.9% w/v) was injected intravenously, to provide a measure of transplacental transport by facilitated diffusion and active transport, respectively. At specific times < 5 min after tracer injection, a maternal blood sample was taken for the measurement of blood glucose and plasma counts. The mother was then killed by cervical dislocation. Conceptuses were dissected out and the placentas and fetuses weighed after fetal decapitation. At D16 and D19, the mean time from tracer injection to maternal blood sampling was similar in UN and CT mice for both 14C-methyl-d-glucose (D16, CT 1.91 ± 0.32 min, UN 2.13 ± 0.23 min; D19, CT 2.18 ± 0.28 min, UN 1.98 ± 0.30 min, P > 0.05) and MeAIB (D16, CT 2.17 ± 0.24 min, UN 1.87 ± 0.25 min; D19 CT 2.17 ± 0.23 min, UN 2.05 ± 0.26 min, P > 0.05). The remaining 19 litters (n= 11 CT, n= 8 UN) were allowed to deliver spontaneously at term (overnight 20 days). On the morning of birth, litter size and pup weights were recorded. Following decapitation, a small blood sample (≤20 μl) was taken from the severed neck vessels of the pups and used directly for measurement of blood glucose concentrations.
Plasma and tissue counts
Whole fetuses and placentas were minced and then lysed in Biosol (National Diagnostics) at 55°C for either 48 h (D16) or 96 h (D19). Aliquots of maternal plasma and the tissues lysates were counted in a β counter (LKB Wallac 1216 Liquid Scintillation Counter). Fetally accumulated radioactivity and maternal plasma radioactivity were used to calculate clearance in μl min−1 (g placenta)−1 and fetal uptake in fetal d.p.m. (g fetus)−1 (Sibley et al. 2004). Placental uptake was calculated as placental d.p.m. (g placenta)−1.
Biochemical analyses
Blood glucose concentrations were measured immediately using a hand-held glucometer (One Touch, Ortho-Clinical Diagnostics, High Wycombe, UK). At D16 (n= 6 CT, n= 7 UN) and D19 (n= 7 CT, n= 6 UN), the glycogen content of the second heaviest placenta in each litter was measured enzymatically as glucose produced by amyloglucosidase activity in 10 min at 55°C using methodology published in detail previously (Franko et al. 2007).
Stereological analysis of placental structure
The placenta closest to the mean in a litter (D16, 9 litters; D19, 15 litters) was hemisected and one half fixed in 4% paraformaldehyde for paraffin embedding, the other half in 4% glutaraldehyde, for resin embedding. The details of preparation and analysis of tissue have been described in detail previously (Coan et al. 2004). Briefly, paraffin-embedded tissue was completely sectioned at 7 μm, whereas a 1 μm section close to the midline of each resin-embedded placenta was cut. The Computer Assisted Stereological Toolbox (CAST v2.0) was employed to measure the gross and fine structure of the placenta in a systematic random fashion using paraffin sections. Volume densities of the placental compartment were determined by point counting and converted to absolute volumes according to the total number of sections, section thickness and shrinkage, as described previously (Coan et al. 2004).
Detailed structural analysis of the labyrinthine zone was performed using the resin sections. Volume and surface densities of fetal capillaries, maternal blood spaces and trophoblast were measured by counting 200 events for each parameter using point and cycloid arc grids, respectively. The densities were referred back to the absolute volume of the labyrinthine zone to obtain absolute volumes and surface areas. The thickness of the interhaemal membrane was determined by orthogonal intercept lengths, superimposing a line grid to generate random points on the maternal side of the interhaemal membrane, whereby the shortest distance to the nearest fetal capillary boundary was determined. An estimate of the structural capacity of the placenta for diffusion was calculated from the mean of the surface area of the maternal blood spaces and fetal capillaries divided by the harmonic thickness of the interhaemal membrane. This was then multiplied by Krogh's oxygen diffusion constant to obtain a theoretical diffusion capacity for oxygen of UN and CT placentas (Coan et al. 2004).
Quantitative real-time PCR analysis of placental gene expression
Expression levels were analysed by quantitative real-time PCR (7500 Fast Real-Time PCR System, Applied Biosystems) for the growth regulatory genes (Grb10, Igf2), the placenta-specific transcript of Igf2 (Igf2P0), the placental transporter genes of the system A family of amino acid transporters (Slc38a1, Slc38a2, Slc38a4) and for the predominant placental glucose transporters (GLUTs, Slc2a1/GLUT1, Slc2a3/GLUT3) at D16 and D19. Ribonucleic acids (RNAs) were isolated from whole placentas using Tri-reagent and a standard protocol (Sigma). RNAs were reverse transcribed to cDNAs using Multiscribe Reverse Transcriptase with random primers according to the manufacturer's protocol (Applied Biosystems). RNA (100 ng) was converted to cDNA and diluted such that 36 ng cDNA was used per reaction. All cDNA samples were analysed in triplicate with cDNA derived from six placentas per diet group per gestational age and cDNAs derived from a pool of placentas used with 5-fold dilutions to create a standard curve. The following Taqman Gene Expression Assays (Applied Biosystems), and Taqman Universal PCR Master Mix, were used:
Slc38a1, Mm00506391_m1; Slc38a2, Mm00628416_m1; Slc38a4, Mm00459056_m1; Slc2a1, Mm00441473_m1; Slc2a3, Mm00441483_m1; Igf2, Mm00439564_m1; H19, Mm00469706_g1; Grb10, Mm01180444_m1, with optimised, thermocycling PCR conditions of an initial 2 min at 50°C followed by 10 min at 95°C, and then 40 cycles (15 s at 95°C and 1 min at 60°C for extension). Primers for detecting levels of the placenta-specific transcript of Igf2 (P0) are as follows: forward primer CCGAGGCCTGTACCACCTA, reverse primer CCTCGGCTCAGACCTCAGTA, FAM CCGAGGCCTCTGCCACC. The relative standard curve method was used for quantifying levels of gene expression. In order to normalise gene expression levels, transcripts from Sdha, Actb, Tbp and Gapd were compared in order to find the optimal combination. Consequently, genes of interest were normalised to mean expression levels of Gapd, 4352339E, and Tbp, Mm01277045_m1, as described previously (Silver et al. 2006).
Sequenom massARRAY quantitative methylation analysis
Genomic DNA was isolated from D16 and D19 placentas following standard methods. Aliquots of 1.5 μg were converted with sodium bisulphite using the EpiTect kit according to the manufacturer's instructions (Qiagen, UK). Amplification of bisulphite-treated DNA (∼2.5 ng μl−1) was performed using HotStar Taq DNA polymerase (Qiagen, UK) with the following primers and parameters:
Igf2P0 promoter, 5′-AGGAAGAGAGTAGGGTGTTAGGTGATTGTTAGGTG-3′ and 5′-CAGTAATACGACTCACTATAGGGAGAAGGCTTAAATCAATATTAACAACCCCCTCC-3′ (annealing 59°C);
H19-ICR, 5′-AGGAAGAGAGTTTGAGGAGTTTTAAGGTAGAAGGG-3′ and 5′-CAGTAATACGACTCACTATAGGGAGAAGGCTAAACCAAAAAACTTAACTCATTCCC-3′ (annealing 61°C);
Slc38a4 Un1 promoter, 5′-AGGAAGAGAGGGTGGAGTTTAGTTGTTTAGTTGTT-3′ and 5′-CAGTAATACGACTCACTATAGGGAGAAGGCTAATAAACCCCTTCTACCAAAAAAAA-3′ (annealing 52°C)
The PCR programme consisted of 45 cycles of 20 s at 94°C, 30 s at the annealing temperature and 1 min at 72°C. The amplicons of bisulfite PCR were treated with shrimp alkaline phosphatase (SAP) followed by in vitro transcription with T7 RNA polymerase and base-specific cleavage using MassCLEAVE Kit-T7 according to the manufacturer's protocol (Sequenom Inc., CA, USA). The samples were desalted and spotted on a 384-pad SpectroCHIP (Sequenom), followed by spectral acquisition on a MassARRAY Analyzer Compact MALDI-TOF MS (Sequenom). The resultant methylation calls were performed by the EpiTYPER software (Sequenom) to generate quantitative results for each CpG site or an aggregate of two CpG sites. Two independent bisulphite-converted DNAs were analysed per sample. Six placental DNA samples from different litters were used per dietary group at both D16 and D19. The average methylation was calculated as a mean value of the CpG methylation values and expressed as percentage methylation.
Statistical analyses
Values for all data are expressed as mean ±s.e.m. using litter means for analysis of differences with dietary treatment, where appropriate. Significant differences in clearance, structure and gene expression between diets were determined by unpaired t tests (Sigmastat 3.5, Systat Software, Point Richmond, CA, USA). Significant differences between gestational ages within treatment groups were assessed by unpaired t tests.
Results
Biometry
Maternal weights
Mice fed 80% of the ad lib intake were significantly lighter at D16 and D19 of pregnancy than CT mice fed ad lib, although the body weight of the two groups was similar on D1 (Table 1). After removing the gravid uterus, carcases of UN mice were still significantly lighter than those of CT mice (Table 1). Both CT and UN mice weighed more at D19 than D16, yet carcass weight was less at D19 than D16 (Table 1). Litter size did not vary with dietary intake either before (Table 1) or at birth (CT, 6.9 ± 0.5 pups, n= 11 litters; UN, 6.8 ± 0.5 pups, n= 8 litters).
Table 1.
Biometry data and maternal blood glucose concentrations in pregnant mice fed ad libitum (CT), or at 80% of CT food intake (UN) until day 16 or day 19 of pregnancy
| Day 16 |
Day 19 |
|||
|---|---|---|---|---|
| CT | UN | CT | UN | |
| Maternal weight (g) | ||||
| Initial (day 1) | 18.3 ± 0.5 | 18.8 ± 0.3 | 18.2 ± 0.3 | 18.2 ± 0.4 |
| Pregnant, intact | 28.8 ± 0.7 | 27.2 ± 0.4* | 32.4 ± 0.6† | 29.2 ± 0.6*† |
| Pregnant, hysterectomized | 23.1 ± 1.3 | 20.9 ± 0.4* | 21.9 ± 1.2† | 19.1 ± 0.3*† |
| Conceptus weights (mg) | ||||
| Fetus (F) | 403 ± 8 | 409 ± 6 | 1142 ± 14 † | 990 ± 13*† |
| Placenta (P) | 96 ± 2 | 90 ± 1* | 81 ± 1† | 74 ± 1*† |
| F:P weight ratio | 4.2 ± 0.1 | 4.6 ± 0.1* | 14.2 ± 0.2 † | 13.6 ± 0.2*† |
| Litter size | 6.3 ± 0.4 | 6.8 ± 0.3 | 6.6 ± 0.3 | 7.0 ± 0.3 |
| Blood glucose (mmol l−1) | 11.0 ± 0.4 | 8.0 ± 0.7* | 11.4 ± 0.4 | 4.8 ± 0.5*† |
Mean ±s.e.m.
Significant differences (P < 0.05 to P < 0.0001) between mice CT (D16, 20 litters; D19, 37 litters) and UN (D16, 27 litters; D19, 23 litters) for weights and glucose (D16, 20 litters; D19, 37 litters) and UN (D16, 27 litters; D19, 23 litters) assessed by unpaired t test.
Significant differences between D16 and D19 within a dietary group assessed by unpaired t test, P < 0.05 to P < 0.0001.
Offspring and placental weights
Fetuses of UN dams were similar in weight to those of CT mice at D16. However, by D19, UN fetuses were 13% lighter than their CT counterparts (P < 0.01, Table 1). At birth, UN neonates (1080 ± 2 mg, n= 8 litters) weighed 8% less than CT pups (1180 ± 2 mg, n= 11 litters, P < 0.05). Placental weight at D16 was significantly less in UN than CT litters (Table 1). Consequently, placental efficiency measured as grams fetus per gram placenta was significantly higher in UN than CT litters at D16 (Table 1). By D19, UN placentas were significantly lighter and less efficient than CT placentas (Table 1).
Blood glucose concentrations
At both ages, UN dams had a significantly lower blood glucose concentration than CT mice (Table 1). Moreover, as pregnancy advanced, there was a significant reduction in maternal blood glucose levels in UN but not CT animals (Table 1). At birth, UN neonates had significantly lower blood glucose concentrations (1.2 ± 0.1 mmol l−1, n= 8 litters) than CT pups (2.3 ± 0.1 mmol l−1, n= 11 litters, P < 0.01).
Placental transport
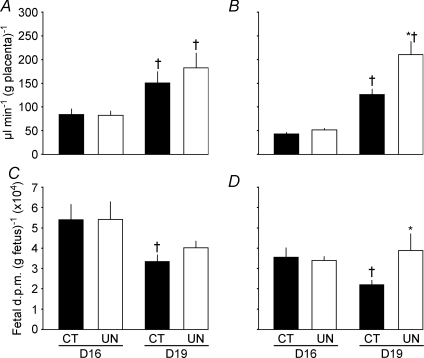
The effects of maternal dietary intake on the unidirectional transplacental clearance of glucose (facilitated diffusion) and amino acid (active transport) depended on the type of transport and gestational age. For 14C-methyl-d-glucose, unidirectional clearance per gram placenta was not significantly different between UN and CT mice at either age and increased to a similar extent between D16 and D19 in both dietary groups (Fig. 1A). At both ages, accumulation of glucose per gram UN fetus matched that of controls (Fig. 1C). In contrast, the effects of undernutrition on transplacental transfer of 14C-methyl-aminoisobutyric acid (MeAIB), an amino acid analogue transferred primarily by the system A family of amino acid transporters, differed with gestational age (Fig. 1B and D). At D16, placental MeAIB clearance was similar in the two dietary groups, whereas at D19 it was 58% greater in UN than CT placentas (P < 0.01, Fig. 1B). Weight-specific fetal accumulation of MeAIB was unaffected by maternal dietary intake at D16 but was significantly greater in UN than CT mice at D19 in association with an ontogenic decrease in MeAIB accumulation per gram CT, but not UN, fetus between D16 and D19 (P < 0.02, Fig. 1D).
Figure 1.
Unidirectional flux (clearance) across the placenta (A and B) and fetal accumulation of tracer per gram of fetus (C and D) of [14C]-methyl-d-glucose (A and C) and [14C]-MeAIB (B and D) in mice fed ad libitum (control, CT, filled columns) and undernourished by feeding 80% of control food intake (UN, open columns) at D16 and D19 of pregnancy. Mean litter clearance ±s.e.m. *Significant differences between CT and UN assessed by unpaired t test, P < 0.05 to P < 0.01. †Significant differences between age within treatment group, unpaired t test, P < 0.05 to P < 0.0001.
Placental morphology
Quantifiable differences in placental morphology existed between UN and CT mice at both ages (Table 2). At D16 and D19, the absolute volume of UN placentas was reduced significantly compared to CT placentas (Table 2), in line with the reduced placental weight (Table 1). At D16, this was associated with a reduced absolute volume and volume fraction of the junctional but not labyrinthine zone of the placenta (Table 2). At D19, growth-restricted UN placentas had a significantly smaller labyrinthine zone volume than their CT counterparts, although junctional zone volumes and volume fractions of the two zones were similar in UN and CT groups (Table 2). Labyrinthine zone volume, therefore, expanded in CT but not UN placentas between D16 and D19 (Table 2).
Table 2.
Stereological analysis of placentas and placental glycogen content from mice fed ad libitum (CT) or at 80% of CT intake (UN) until day 16 or day 19 of pregnancy
| Day 16 |
Day 19 |
|||
|---|---|---|---|---|
| CT | UN | CT | UN | |
| Placental compartment volume (mm3) | ||||
| Placenta | 100 ± 5 | 83 ± 4* | 85 ± 3† | 72 ± 2*† |
| Lz | 41 ± 1 | 41 ± 4 | 52 ± 2† | 44 ± 2* |
| Jz | 43 ± 5 | 27 ± 2* | 20 ± 2† | 17 ± 1† |
| Db | 16 ± 1 | 15 ± 2 | 13 ± 1† | 10 ± 1 |
| Placental compartment volume fraction (%) | ||||
| Lz | 40 ± 1 | 45 ± 2 | 52 ± 2† | 52 ± 2† |
| Jz | 41 ± 1 | 35 ± 1* | 29 ± 2† | 29 ± 2† |
| Db | 24 ± 1 | 25 ± 2 | 23 ± 1 | 22 ± 1 |
| Lz component volume (mm3) | ||||
| FC | 4.4 ± 1 | 5.3 ± 1 | 8.1 ± 1† | 5.9 ± 1* |
| MBS | 7.4 ± 1 | 6.2 ± 1 | 9 ± 1 | 7 ± 0.3* |
| LT | 27 ± 1 | 26 ± 2 | 35 ± 2† | 31 ± 1† |
| Lz component volume fraction (%) | ||||
| FC | 19 ± 2 | 21 ± 2 | 23 ± 1† | 21 ± 1 |
| MBS | 25 ± 1 | 23 ± 2 | 25 ± 1 | 24 ± 1 |
| LT | 54 ± 2 | 54 ± 3 | 55 ± 1 | 57 ± 1 |
| Lz interhaemal membrane surface areas (m2) | ||||
| FC | 11 ± 1 | 11 ± 2 | 17 ± 1† | 14 ± 1 |
| MBS | 17 ± 1 | 15 ± 2 | 24 ± 1† | 22 ± 1† |
| Mean | 14 ± 1 | 13 ± 4 | 21 ± 1† | 18 ± 1* |
| Interhaemal membrane harmonic mean thickness (μm) | ||||
| Th | 4.39 ± 0.26 | 4.71 ± 0.25 | 3.61 ± 0.13† | 3.70 ± 0.20† |
| Theoretical diffusion capacity (mm2 min−1 kPa−1) | 5.7 ± 1.0 | 4.9 ± 1.0 | 9.9 ± 1.0 | 8.6 ± 1.0 |
| Epilabyrinthine component volumes (mm3) | ||||
| GC | 16 ± 2 | 5 ± 1* | 2 ± 0.1† | 2 ± 0.1† |
| Non-GC | 43 ± 3 | 37 ± 3 | 29 ± 4† | 22 ± 1† |
| Epilabyrinthine volume fraction (%) | ||||
| GC | 31 ± 1 | 20 ± 2* | 15 ± 2† | 18 ± 1 |
| Non-GC | 59 ± 1 | 70 ± 2* | 75 ± 2† | 72 ± 1 |
| Glycogen content (mg g−1) | 12.1 ± 0.4 | 10.5 ± 0.4* | 4.4 ± 0.3† | 4.2 ± 0.3† |
Mean ±s.e.m. For stereological measurements: CT (D16, 4 litters; D19, 7 litters), UN (D16, 5 litters; D19, 7 litters). For glycogen content: CT (D16, 6 placentas from 6 litters; D19, 7 placentas from 7 litters), UN (D16, 7 placentas from 7 litters; D19 6 placentas from 6 litters). Abbreviations: Db, decidua basalis; FC, fetal capillaries; GC, glycogen cell; Jz, junctional zone; LT, labyrinthine trophoblast; Lz, labyrinthine zone; MBS, maternal blood spaces; non-GC, non-glycogen cell mass including all other Jz and Db cell types.
Significant differences between CT and UN assessed by unpaired t test, P < 0.005.
Significant differences between D16 and D19 within a dietary group assessed by unpaired t test, P < 0.05 to P < 0.001.
Within the labyrinthine zone, the volumes of the maternal blood spaces, fetal capillaries and labyrinthine trophoblast were similar in UN and CT placentas at D16 (Table 2). However, at D19, both maternal blood space and fetal capillary volume of the UN placentas were significantly less than CT values (Table 2). Fetal capillary volume and the mean surface area of labyrinthine interhaemal membrane increased between D16 and D19 in CT but not UN placentas (Table 2). Consequently, the mean surface area of labyrinthine interhaemal membrane was similar in the two dietary groups at D16 but significantly less in UN than CT placentas at D19 (Table 2). The thickness of the labyrinthine interhaemal membrane did not differ with dietary intake at either age and decreased to the same extent between D16 and D19 in both dietary groups (Table 2). At both ages, the theoretical diffusion capacity of the placenta, a structural measure of interhaemal membrane efficiency for simple diffusion, was similar in UN and CT mice (Table 2).
The glycogen content of the placenta and the total volume and volume fraction of glycogen cells in the junctional zone and decidua were all significantly less in UN than CT placentas at D16 (Table 2). Placental glycogen content and glycogen cell volume decreased between D16 and D19, concurrent with a decrease in glycogen cell mass, resulting in a similar volume for this zone in both dietary groups by D19 (Table 2). Consequently, these values and the volume fraction of glycogen cells in the epilabyrinthine region did not differ significantly with maternal dietary intake at D19 (Table 2).
Placental gene expression
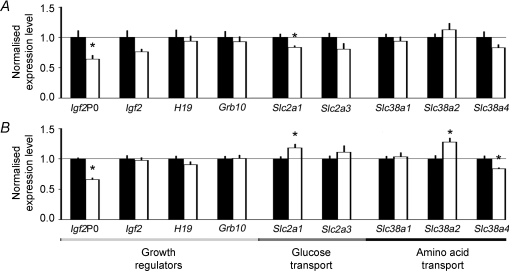
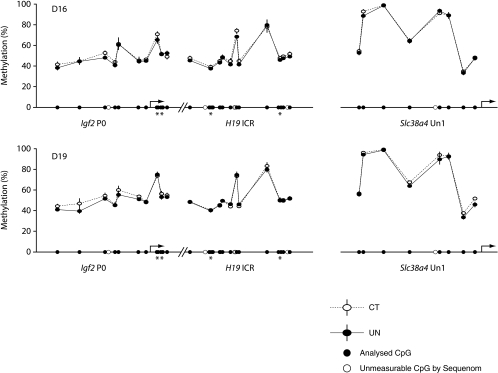
In UN placentas at both ages, expression of the placenta-specific transcript of Igf2, Igf2P0, but not total Igf2 expression, was reduced to ∼65% of controls (Fig. 2). Analysis of the methylation status of the Igf2P0 promoter in UN and CT placentas revealed no significant differences in methylation at either age (Fig. 3). Both H19 gene expression and methylation of the imprinting control region (ICR) of this gene were similar in UN and CT placentas at D16 and D19 (Figs 2 and 3). At both ages, there was no significant difference in Grb10 expression between UN and CT placentas (Fig. 2).
Figure 2.
Normalised real-time PCR transcripts of growth regulatory and nutrient transporter genes in placentas from mice fed ad libitum (control, CT, filled columns) or undernourished (UN, open columns) by feeding 80% of the control food intake at D16 (A) and D19 (B) of pregnancy. Mean ±s.e.m. of the placentas closest the mean for 6 litters for each treatment and gestational age. *Significant differences between treatment groups were assessed by unpaired t test.
Figure 3.
Methylation profile of Igf2P0 promoter, H19-ICR and Slc38a4 Un1 promoter regions in placentas at D16 and D19 from mice fed ad libitum (CT, open symbols, n= 6 at both ages) or undernourished by feeding 80% of the control intake (UN, filled symbols and continuous lines, n= 6 at both ages) measured by Sequenom massARRAY. The results are presented as average of percentage methylation ±s.e.m. for each CpG site or aggregate of two CpG sites (*).
Expression of the facilitated glucose transporter gene Slc2a1/GLUT1 in UN placentas was reduced to 83% of controls at D16 (Fig. 2A). However, by D19, Slc2a1 expression was significantly greater in UN than CT placentas (Fig. 2B). Expression of Slc2a3/GLUT3, the other prominent glucose transporter in the placenta, was similar in the two dietary groups at both ages (Fig. 2A and B). At D16, mRNA transcripts encoding the system A amino acid transporter family, Slc38a1, Slc38a2 and Slc38a4, were expressed at similar levels in UN and CT placentas (Fig. 2A). However, by D19 in UN placentas, expression of Slc38a2 was significantly increased to 127% of controls, whereas Slc38a4 expression was decreased to 83% of controls (Fig. 2B). The methylation status of the Un1 promoter of the Slc38a4 gene, primarily responsible for placental Slc38a4 expression, did not differ between dietary groups at either age (Fig. 3). Expression of Slc38a1 was similar in UN and CT placentas at D19 (Fig. 2B).
Discussion
The results show that the mouse placenta can adapt its phenotype to help maintain fetal growth in late gestation when its own growth is restricted by maternal undernutrition during pregnancy. These adaptations were both morphological and functional, and dependent on gestational age. At D16 before the major fetal growth spurt, UN placentas were more efficient at supporting fetal growth as fetal weight was normal, despite maternal hypoglycaemia and reduced placental mass. In part, this was due to maintained growth of the labyrinthine zone responsible for nutrient transfer relative to other zones in the UN placenta. In contrast, at D19 towards the end of the normal fetal growth spurt, both placental and fetal weights, and the volume and surface area of the labyrinthine zone, were reduced in UN compared to control mice. Despite this, unidirectional materno-fetal clearance of glucose was maintained whilst clearance of MeAIB was higher per gram UN placenta relative to D19 controls, in association with increased placental expression of specific nutrient transporter genes. Placental phenotype is, therefore, responsive to nutritional conditions and adapts to maximise the capacity for materno-fetal nutrient transfer when fetal nutrient availability is compromised by restricting maternal food intake and placental growth.
Since the weight of UN placentas was reduced by 5–10% at both ages and fell to the same extent as in controls between D16 and D19, maternal undernutrition probably limited the main proliferative phase of placental growth that normally occurs before D16 (Georgiades et al. 2002; Coan et al. 2006). In particular, it was growth of the junctional zone that was initially impaired, chiefly from a reduced glycogen cell mass. Preferential development of the labyrinthine zone relative to junctional zone in these conditions may help maintain the fetal nutrient supply and account for the normal fetal weight at D16. However, disproportionate lack of glycogen and other hormone-producing cells in the undernourished junctional zone may have consequences for energy balance and materno-fetal nutrient partitioning nearer term when fetal demands for nutrients are rising most rapidly in absolute terms (Coan et al. 2006). Indeed, this labyrinthine ‘sparing’ effect was not maintained until term as the labyrinthine zone and the surface area for nutrient exchange failed to expand normally in UN placentas between D16 and D19. Since the actual volume of labyrinthine trophoblast in UN placentas increased between D16 and D19 while labyrinthine surface area did not, impaired angiogenesis may be the primary defect in placental development during late gestation in UN dams. The UN placenta is, therefore, not only small but also morphologically abnormal by D19, which, together with the increasing severity of maternal hypoglycaemia, will contribute to the ensuing IUGR.
At both ages, placental growth restriction was accompanied by reduced expression of the labyrinthine specific transcript of the imprinted Igf2 gene Igf2P0 which is known to stimulate placental growth (Constancia et al. 2005; Coan et al. 2008b). In Igf2P0 null mutants, the placenta is growth restricted from D14 and has 50% less labyrinthine surface area by D19 but, like the UN placenta at D16, is more efficient at supporting fetal growth (Constancia et al. 2005). Reduced abundance of Igf2 has also been observed in rat and guinea pig placentas growth restricted through reducing nutrient availability by uterine artery ligation and maternal caloric restriction, respectively (Price et al. 1992; Olausson & Sohlstrom, 2003). However, there was no change in total Igf2 gene expression in UN mouse placentas, which emphasises the importance of the Igf2P0 transcript in regulating placental development in mice. The decrease in placental Igf2P0 expression during undernutrition was not due to changes in methylation of the Igf2P0 promoter or H19-ICR involved in Igf2 imprinting (Reik et al. 2003; Fowden et al. 2006a). In contrast to naturally small placentas (Coan et al. 2008a), the UN placenta maintained normal expression of Grb10, a maternally expressed imprinted gene that inhibits placental growth (Charalambous et al. 2003).
On a weight-specific basis, unidirectional placental clearance and fetal accumulation of tracer glucose were unaffected by restricting dietary intake of pregnant mice, despite the abnormalities in placental morphology. Both the Slc2a1/GLUT1 and Slc2a3/GLUT3 glucose transporters were detected in UN placentas but only Slc2a1 expression differed from control values, consistent with previous findings in other species of greater sensitivity of placental GLUT1 than GLUT3 to nutritional stimuli (Illsley, 2000). Both transporters are required for normal intrauterine development in mice as deletion of either gene results in IUGR (Wang et al. 2006; Ganguly et al. 2007). However, localization of the two transporter proteins in rat placentas suggests that their functions differ with Slc2a1/GLUT1 regulating placental glucose uptake, particularly in the junctional zone, while both transporters are required for transplacental glucose transfer to the fetus (Shin et al. 1997). Thus, at D16, the decreased Slc2a1 expression in UN placentas may limit placental glucose utilization and contribute to the observed growth restriction of the junctional zone. The relatively low glucose requirement of the D16 fetus could then be met by lowering fetal glucose levels to maximise the transplacental glucose concentration gradient regulating net glucose transfer to the fetus. A similar mechanism to aid fetal glucose acquisition is seen in human infants with small placentas (Marconi et al. 1996). In contrast, at D19, Slc2a1 expression was increased in the small UN mouse placenta, as occurs in naturally small placentas and in placentas of mice fed a high fat diet (Coan et al. 2008a; Jones et al. 2009). Although materno-fetal glucose transfer capacity per gram of UN placenta was normal, the net flux of glucose into the UN fetus will be less due to a smaller transplacental glucose concentration gradient caused by the greater degree of maternal hypoglycaemia at D19. Increased Slc2a1 expression in UN placentas at D19 may, therefore, help maintain a glucose supply to feto-placental tissues when more glucose is needed to support the greater fetal mass near term. The switch from decreased Slc2a1 expression at D16 to increased expression at D19 in small UN placentas may, therefore, reflect a mechanism for optimising fetal glucose delivery in relation to fetal glucose demands at different gestational age.
Although UN placentas had a reduced surface area by D19, they transported more MeAIB per gram and had a higher expression of the Slc38a2 amino acid transporter than controls. Similar increases in MeAIB transport and Slc38a2 gene expression have been seen in late gestation in naturally small placentas within litters and in placentas from mice fed a high fat diet (Coan et al. 2008a; Jones et al. 2009). System A amino acid transport is also increased in small placentas of human infants at the lower end of the normal birth weight range (Godfrey et al. 1998). The molecular mechanisms upregulating expression of the system A amino acid transporters in small placentas remain unknown but may involve decreased expression of the Igf2P0 transcript. Certainly, deletion of this transcript increases MeAIB transport per gram mutant placenta and, in preliminary experiments, prevents upregulation of placental MeAIB transport in response to maternal caloric restriction (Constancia et al. 2005; Sferruzzi-Perri et al. 2009).
Paradoxically, Slc38a4/SNAT4 expression was down-regulated in UN placentas at D19. In humans, expression and function of SNAT4 is diminished in term relative to first trimester placentas, which suggests that this transporter may become less important in regulating transplacental amino acid transfer and fetal growth with increasing gestational age (Desforges et al. 2009). Recent preliminary findings in the Slc38a4 knockout mouse suggest that this transporter is directly involved in placental growth and localised predominantly to the junctional zone (G. Kelsey & M. Constancia, unpublished observations). Downregulation of Slc38a4 expression in UN placentas near term may, therefore, contribute to placental growth restriction but, by limiting amino acid uptake into the junctional zone, spare amino acids for transport to the fetus. Overall, the changes in amino acid transporter expression in UN placentas resulted in increased fetal amino acid accumulation at D19. This may help sustain fetal growth in the last days of gestation by providing substrates for tissue accretion and oxidative metabolism. System A-mediated amino acid transport is known to be important for fetal growth in rats near term and amino acid carbon is used oxidatively to meet the energy requirements of hypoglycaemic sheep fetuses during late gestation (Cramer et al. 2002; Regnault et al. 2005). Indeed, UN fetuses gained double the weight of controls between D19 and delivery in the current study, although their body weight was still less than normal at birth.
The nutrient transporter and growth regulatory genes assessed in the current study were selected on the basis of their known role in regulating placental nutrient transfer capacity but are probably only a small subset of the genes altered in expression in the placenta by maternal undernutrition (Constancia et al. 2002, 2005; Coan et al. 2008a; Jones et al. 2009). Recent studies on mice fed an isocaloric low protein diet during the second half of pregnancy have shown the changed expression of over 200 genes in the placenta at day 18 (Gheorghe et al. 2009). However, changes in the expression of imprinted genes are likely to be particularly important in the conflict between maternal and paternal genomes in allocating maternal resources to fetal growth with the mother restraining the paternal drive for fetal nutrient acquisition in an individual pregnancy to distribute resources more equally among all her potential offspring (Constancia & Reik, 2004). Indeed, the current finding that reducing caloric intake during pregnancy decreases the abundance of two paternally expressed, growth-enhancing imprinted genes Igf2P0 and Slc38a4, but has little effect on the abundance of two maternally expressed, growth-restraining imprinted genes Grb10 and H19 in the placenta, is consistent with this concept. At the placental level, the conflict between the parental genomes may be even more pronounced when nutrients are scarce, especially in mice, which re-allocate a significant proportion of maternal body mass to the gravid uterus during late gestation, even in normal nutritional conditions. Since the changes in placental expression of key imprinted genes during undernutrition were not associated with altered methylation of their promoters or known control regions, these nutritional effects must be mediated via other epigenetic mechanisms, such as histone modifications, chromatin remodelling or alterations in the transcriptome.
In summary, small UN placentas adapt their phenotype to better match nutrient supply to fetal demands for growth when nutrient availability is compromised. These adaptations appeared to be primarily morphological at D16 with relative loss of the junctional zone whereas, they were largely functional at D19 with increased expression of two nutrient supply genes. Thus, small mouse placentas appear to respond to fetal demand signals and upregulate expression of nutrient supply genes during late gestation, irrespective of whether placental growth restriction occurs naturally or by manipulation of nutrition and/or gene expression (Constancia et al. 2005; Coan et al. 2008a,b;). These changes will alter the absolute and relative quantities of nutrients supplied to the fetus with implications for growth and functioning of tissues both pre- and postnatally. However, the specific expression pattern of placental nutrient transporter and growth regulatory genes differs with the cause of IUGR. Consequently, placental phenotype, particularly of these genes, may provide a good index of the conditions experienced during intrauterine development and allow the better prediction of the risk of adult disease programmed in utero.
Acknowledgments
The authors would like to thank Nuala Daw, Julie Gautrey, Chris Cardinal and Wendy Cassidy for technical assistance. This study was funded by a grant from the BBSRC. This study was funded by a grant from the BBSRC and MRC CORD.
Glossary
Abbreviations
- CT
control, fed diet ad libitum
- D
days after copulation
- ICR
imprinting control region
- IUGR
intrauterine growth restriction
- MeAIB
methyl-aminoisobutyric acid
- UN
undernourished (fed 80% of CT diet)
Author contributions
P.M.C.: design, analysis and interpretation of data, drafting, revising and final approval. A.L.F.: conception, design, analysis and interpretation of data, drafting, revising and final approval. G.J.B. and M.C.: conception, design, interpretation of data, revising and final approval. O.R.V., S.Y. and S.L.F.: analysis and interpretation of data, revising content and final approval.
References
- Angiolini E, Fowden A, Coan P, Sandovici I, Smith P, Dean W, Burton G, Tycko B, Reik W, Sibley C, Constancia M. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta. 2006;27(Suppl. A):S98–102. doi: 10.1016/j.placenta.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Barker DJP. Mothers, Babies and Disease in Later Life. London: BMJ Publishing Group; 1994. [Google Scholar]
- Barker DJP. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci U S A. 2003;100:8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan PM, Angiolini E, Sandovici I, Burton GJ, Constancia M, Fowden AL. Adaptations in placental nutrient transfer capacity to meet fetal growth demands depend on placental size in mice. J Physiol. 2008a;586:4567–4576. doi: 10.1113/jphysiol.2008.156133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta – a review. Placenta. 2005;26(Suppl. A):S10–20. doi: 10.1016/j.placenta.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Coan PM, Conroy N, Burton GJ, Ferguson-Smith AC. Origin and characteristics of glycogen cells in the developing murine placenta. Dev Dyn. 2006;235:3280–3294. doi: 10.1002/dvdy.20981. [DOI] [PubMed] [Google Scholar]
- Coan PM, Ferguson-Smith AC, Burton GJ. Developmental dynamics of the definitive mouse placenta assessed by stereology. Biol Reprod. 2004;70:1806–1813. doi: 10.1095/biolreprod.103.024166. [DOI] [PubMed] [Google Scholar]
- Coan PM, Fowden AL, Constancia M, Ferguson Smith AC, Burton GJ, Sibley CP. Disproportional effects of Igf2 knockout on placental morphology and diffusional exchange characteristics in the mouse. J Physiol. 2008b;586:5023–5032. doi: 10.1113/jphysiol.2008.157313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, Fowden A. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci U S A. 2005;102:19219–19224. doi: 10.1073/pnas.0504468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, Reik W. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- Constancia M, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- Cramer S, Beveridge M, Kilberg M, Novak D. Physiological importance of system A-mediated amino acid transport to rat fetal development. Am J Physiol Cell Physiol. 2002;282:C153–C160. doi: 10.1152/ajpcell.2002.282.1.C153. [DOI] [PubMed] [Google Scholar]
- Davies K, Wardle J. Body image and dieting in pregnancy. J Psychosom Res. 1994;38:787–799. doi: 10.1016/0022-3999(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Desforges M, Mynett KJ, Jones RL, Greenwood SL, Westwood M, Sibley CP, Glazier JD. The SNAT4 isoform of the system A amino acid transporter is functional in human placental microvillous plasma membrane. J Physiol. 2009;587:61–72. doi: 10.1113/jphysiol.2008.161331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond GB. Reporting ethical matters in The Journal of Physiology: standards and advice. J Physiol. 2009;587:713–719. doi: 10.1113/jphysiol.2008.167387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden A, Sferruzzi-Perri AN, Coan PM, Constancia M, Burton GJ. Placental efficiency and adaptation: endocrine regulation. J Physiol. 2009;587:3459–3472. doi: 10.1113/jphysiol.2009.173013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden AL, Forhead AJ, Coan PM, Burton GJ. The placenta and intrauterine programming. J Neuroendocrinol. 2008;20:439–450. doi: 10.1111/j.1365-2826.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Giussani DA, Forhead AJ. Endocrine and metabolic programming during intrauterine development. Early Hum Dev. 2005;81:723–734. doi: 10.1016/j.earlhumdev.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006a;65(Suppl. 3):50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Ward JW, Wooding FP, Forhead AJ, Constancia M. Programming placental nutrient transport capacity. J Physiol. 2006b;572:5–15. doi: 10.1113/jphysiol.2005.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko KL, Guissani DA, Forhead AJ, Fowden AL. Effects of dexamethasone on the glucogenic capacity of fetal, pregnant and non-pregnant adult sheep. J Endocrinol. 2007;191:67–73. doi: 10.1677/joe.1.07063. [DOI] [PubMed] [Google Scholar]
- Ganguly A, McKnight RA, Raychaudhuri S, Shin BC, Ma Z, Moley K, Devaskar SU. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am J Physiol Endocrinol Metab. 2007;292:E1241–1255. doi: 10.1152/ajpendo.00344.2006. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Ferguson-Smith A, Burton GJ. Comparative developmental anatomy of the murine and human placenta. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- Gheorghe CP, Goyal R, Holweger JD, Longo LD. Placental gene expression responses to maternal protein restriction in the mouse. Placenta. 2009;30:411–417. doi: 10.1016/j.placenta.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey KM, Matthews N, Glazier J, Jackson A, Wilman C, Sibley CP. Neutral amino acid uptake by the microvillous plasma membrane of the human placenta is inversely related to fetal size at birth in normal pregnancy. J Clin Endocrinol Metab. 1998;83:3320–3326. doi: 10.1210/jcem.83.9.5132. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol. 2008;102:90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- Illsley NP. Glucose transporters in the human placenta. Placenta. 2000;21:14–22. doi: 10.1053/plac.1999.0448. [DOI] [PubMed] [Google Scholar]
- Jansson N, Pettersson J, Haafiz A, Ericsson A, Palmberg I, Tranberg M, Ganapathy V, Powell TL, Jansson T. Down-regulation of placental transport of amino acids precedes the development of intrauterine growth restriction in rats fed a low protein diet. J Physiol. 2006;576:935–946. doi: 10.1113/jphysiol.2006.116509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe AH. Maternal eating, infant feeding and growth. J Pediatr. 2009;154:A1. doi: 10.1016/j.jpeds.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Jones HN, Woollett LA, Barbour N, Prasad PD, Powell TL, Jansson T. High-fat diet before and during pregnancy causes marked up-regulation of placental nutrient transport and fetal overgrowth in C57/BL6 mice. FASEB J. 2009;23:271–278. doi: 10.1096/fj.08-116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdown ML, Sugden MC. Enhanced placental GLUT1 and GLUT3 expression in dexamethasone-induced fetal growth retardation. Mol Cell Endocrinol. 2001;185:109–117. doi: 10.1016/s0303-7207(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Lartey A. Maternal and child nutrition in Sub-Saharan Africa; challenges and interventions. Proc Soc Nutr. 2008;67:105–108. doi: 10.1017/S0029665108006083. [DOI] [PubMed] [Google Scholar]
- Lederman SA, Rosso P. Effects of food restriction on fetal and placental growth and maternal body composition. Growth. 1980;44:77–88. [PubMed] [Google Scholar]
- McLaren A. Genetic and environmental effects on foetal and placental growth in mice. J Reprod Fertil. 1965;9:79–98. doi: 10.1530/jrf.0.0090079. [DOI] [PubMed] [Google Scholar]
- McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- Marconi AM, Paolini C, Buscaglia M, Zerbe G, Battaglia FC, Pardi G. The impact of gestational age and fetal growth on the maternal–fetal glucose concentration difference. Obstet Gynecol. 1996;87:937–942. doi: 10.1016/0029-7844(96)00048-8. [DOI] [PubMed] [Google Scholar]
- Molteni RA, Stys SJ, Battaglia FC. Relationship of fetal and placental weight in human beings: fetal/placental weight ratios at various gestational ages and birth weight distributions. J Reprod Med. 1978;21:327–334. [PubMed] [Google Scholar]
- Olausson H, Sohlstrom A. Effects of food restriction and pregnancy on the expression of insulin-like growth factors-I and -II in tissues from guinea pigs. J Endocrinol. 2003;179:437–445. doi: 10.1677/joe.0.1790437. [DOI] [PubMed] [Google Scholar]
- Price WA, Rong L, Stiles AD, D’Ercole AJ. Changes in IGF-I and -II, IGF binding protein and IGF receptor transcript abundance after uterine artery ligation. Pediatr Res. 1992;32:291–295. doi: 10.1203/00006450-199209000-00009. [DOI] [PubMed] [Google Scholar]
- Rees WD, Hay SM, Brown DS, Antipatis C, Palmer RM. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J Nutr. 2000;130:1821–1826. doi: 10.1093/jn/130.7.1821. [DOI] [PubMed] [Google Scholar]
- Regnault TR, Friedman JE, Wilkening RB, Anthony RV, Hay WW., Jr Fetoplacental transport and utilization of amino acids in IUGR – a review. Placenta. 2005;26(Suppl. A):S52–62. doi: 10.1016/j.placenta.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Reik W, Constancia M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, Tycko B, Sibley C. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol. 2003;547:35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutland CS, Latunde-Dada AO, Thorpe A, Plant R, Langley-Evans S, Leach L. Effect of gestational nutrition on vascular integrity in the murine placenta. Placenta. 2007;28:734–742. doi: 10.1016/j.placenta.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Sferruzzi-Perri AN, Coan PM, Vaughan OR, Constancia M, Burton GJ, Fowden AL. Igf2 deficiency modifies placental adaptation to maternal undernutrition. Reprod Sci. 2009;16:165A. [Google Scholar]
- Shin BC, Fujikura K, Suzuki T, Tanaka S, Takata K. Glucose transporter GLUT3 in the rat placental barrier: a possible machinery for the transplacental transfer of glucose. Endocrinology. 1997;138:3997–4004. doi: 10.1210/endo.138.9.5369. [DOI] [PubMed] [Google Scholar]
- Sibley C, Glazier J, D'Souza S. Placental transporter activity and expression in relation to fetal growth. Exp Physiol. 1997;82:389–402. doi: 10.1113/expphysiol.1997.sp004034. [DOI] [PubMed] [Google Scholar]
- Sibley CP, Coan PM, Ferguson-Smith AC, Dean W, Hughes J, Smith P, Reik W, Burton GJ, Fowden AL, Constancia M. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc Natl Acad Sci U S A. 2004;101:8204–8208. doi: 10.1073/pnas.0402508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pascual JM, Yang H, Engelstad K, Mao X, Cheng J, Yoo J, Noebels JL, De Vivo DC. A mouse model for Glut-1 haploinsufficiency. Hum Mol Genet. 2006;15:1169–1179. doi: 10.1093/hmg/ddl032. [DOI] [PubMed] [Google Scholar]
- Yajnik CS. Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr. 2004;134:205–210. doi: 10.1093/jn/134.1.205. [DOI] [PubMed] [Google Scholar]