Abstract
Mouse and human ES (embryonic stem) cells display unusual proliferative properties and can produce pluripotent stem cells indefinitely. Both processes might be important for maintaining the ‘stemness’ of ES cells; however, little is known about how the cell-cycle fate is regulated in ES cells. Oct-4, a master switch of pluripotency, plays an important role in maintaining the pluripotent state of ES cells and may prevent the expression of genes activated during differentiation. Using ZHBTc4 ES cells, we have investigated the effect of Oct-4 on ES cell-cycle control, and we found that Oct-4 down-regulation in ES cells inhibits proliferation by blocking cell-cycle progression in G0/G1. Deletion analysis of the functional domains of Oct-4 indicates that the overall integrity of the Oct-4 functional domains is important for the stimulation of S-phase entry. We also show in the present study that the p21 gene is a target for Oct-4 repression. Furthermore, p21 protein levels were repressed by Oct-4 and were induced by the down-regulation of Oct-4 in ZHBTc4 ES cells. Therefore the down-regulation of p21 by Oct-4 may contribute to the maintenance of ES cell proliferation.
Keywords: cell-cycle control, differentiation, enbryonic stem cell, G1 cell-cycle arrest, Oct-4, p21, self-renewal
Abbreviations: AP, alkaline phosphatase; CDK, cyclin-dependent kinase; CDKI, CDK inhibitor; ES, embryonic stem; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; EGFP, enhanced GFP; HEK, human embryonic kidney; ICM, inner cell mass; PI, propidium iodide; PKM2, pyruvate kinase isozyme type M2; RT, reverse transcription
INTRODUCTION
Mouse and human ES (embryonic stem) cells display unusual proliferative properties, which are achieved by symmetric cell division, while maintaining their pluripotency. These properties can be regulated by transcriptional control in the nucleus through extracellular signals [1]. The oct-4 gene, also referred to as oct-3 and oct-3/4, encodes a nuclear protein that belongs to a family of transcription factors containing the POU DNA-binding domain [2–7]. Oct-4 is normally expressed in the pluripotent stem cells of pregastrulation embryos, including oocytes, early-cleavage-stage embryos and the ICM (inner cell mass) of the blastocyst [3,4,8,9]. Its expression is down-regulated during differentiation, and knockout of oct-4 causes early lethality in mice due to the absence of an ICM [10]. These results suggest that Oct-4 plays a pivotal role in mammalian development [11] and in the self-renewal of ES cells [12]. During human development, Oct-4 is expressed at least until the blastocyst stage in which it regulates gene expression [13].
Transcriptional regulation by Oct-4 is complex. In ES cells, the octamer sequence motif (5′-ATGCAAAT-3′) is active irrespective of its distance from the site of transcriptional initiation [2,14]. However, in differentiated cells, Oct-4 can transactivate its targets only when the octamer motif is in a proximal position [3,15,16]. If the octamer motif is at a distal site, the protein requires stem-cell-specific bridging factors that link it to the transcription initiation site [15]. A number of factors such as Sox2, HMG (high-mobility group), E7, E1A and EWS are known to influence the ability of Oct-4 to act as an activator or repressor [15,17–21]. In addition, the physical association of Oct-4 with PKM2 (pyruvate kinase isozyme type M2) was documented recently, suggesting that PKM2 may also play a role in regulating Oct-4 [22].
Proliferation of ES cells is achieved through self-renewal, and should be regulated by controlling the cell cycle. In comparison with self-renewal, little is known about the cell-cycle regulation of ES cells by Oct-4. Although Oct-4 functions as a master switch during differentiation by regulating cells that have pluripotent potential, or can develop such potential, unfortunately no Oct-4 target gene involved in ES cell-cycle regulation has been identified. As a first step towards investigating how the oct-4 gene product contributes to the cell-cycle regulation of ES cells, we analysed its potential to regulate the process of self-renewal and control the cell cycle. We found that down-regulation of oct-4 expression led to the growth arrest of ES cells by blocking cell-cycle progression in G0/G1. Deletion analysis of the functional domains of Oct-4 indicated that the overall integrity of the Oct-4 functional domains is important for the stimulation of S-phase entry. We also show in the present study that a CDKI [CDK (cyclin-dependent kinase) inhibitor] gene, p21, is a target for repression by Oct-4. Furthermore, an increase in p21 protein levels was induced by the down-regulation of oct-4 gene expression in ZHBTc4 ES cells. Therefore the down-regulation of p21 by Oct-4 may contribute to the maintenance of ES cell proliferation.
MATERIALS AND METHODS
Materials and general methods
Restriction endonucleases, calf intestinal AP (alkaline phosphatase), the Klenow fragment of DNA polymerase I and T4 DNA ligase were purchased from New England Biolabs. [α-32P]dCTP 3000 (Ci/mmol) was obtained from PerkinElmer. Preparation of plasmid DNA, restriction enzyme digestion, agarose gel electrophoresis of DNA, DNA ligation, bacterial transformations and SDS/PAGE of proteins were carried out using standard methods [23].
Constructs
The EGFP [enhanced GFP (green fluorescent protein)]-fusion Oct-4 plasmids, pCAG-IP/FLAG-Oct-4–EGFP, pCAG-IP/FLAG-Oct-4(ΔC)–EGFP, pCAG-IP/FLAG-Oct-4(ΔN)–EGFP, pCAG-IP/FLAG-Oct-4(POU)–EGFP and pCAG-IP/EGFP, were generated through the following steps. For pCAG-IP/FLAG-Oct-4–EGFP, the Oct-4 cDNA was amplified from pcDNA3/Oct-4 [17] using PCR with the primers 5′-Oct-4 (2) (5′- GATCGGATCCCGCTGGACACCTGGCTTC-3′, a BamHI site is underlined) and 3′-Oct-4 (352) (5′-GATCGAATTCGGTTTGAATGCATGGGAG-3′, an EcoRI site is underlined), digested with BamHI and EcoRI, and cloned into the same site of pCMV-Tag2A (Stratagene) to generate pCMV-Tag2A-Oct-4. FLAG-tagged Oct-4 was amplified from pCMV-Tag2A-Oct-4 by PCR using the primers 5′-FLAG (5′-GATCCTCGAGATGGATTACAAGGATGAC-3′, a XhoI site is underlined) and 3′-Oct-4 (352), digested with XhoI and EcoRI, and cloned into the same sites of pEGFP-N1 (Takara Bio Company) to generate pEGFP-N1-FLAG-Oct-4. The plasmid pEGFP-N1-FLAG-Oct-4 was digested with XhoI and NotI and cloned into the same sites of the pCAG-IP vector. For pCAG-IP/FLAG-Oct-4(ΔC)–EGFP, the C-terminal truncated Oct-4 fragment was amplified from pcDNA3/Oct-4 using PCR with the primers 5′-Oct-4 (2) and 3′-Oct-4 (282) (5′-GATCGAATTCGACTTGATCTTTTGCCCT-3′, an EcoRI site is underlined), digested with BamHI and EcoRI, and cloned into the same site of pCMV-Tag2A to generate pCMV-Tag2A-Oct-4(ΔC). The FLAG-tagged Oct-4(ΔC) was then amplified from pCMV-Tag2A-Oct-4(ΔC) using PCR with the primers 5′-FLAG and 3′-Oct-4 (282), digested with XhoI and EcoRI, and cloned into the same sites of pEGFP-N1 to generate pEGFP-N1-FLAG-Oct-4(ΔC). The plasmid pEGFP-N1-FLAG-Oct-4(ΔC) was digested with XhoI and NotI, and cloned into the same sites of the pCAG-IP vector. For pCAG-IP/FLAG-Oct-4(ΔN)–EGFP, the N-terminal truncated Oct-4 fragment was amplified from pcDNA3/Oct-4 using the PCR using primers 5′-Oct-4 (127) (5′-GATCGGATCCCGAGGAGTCCCAGGACAT-3′, a BamHI site is underlined) and 3′-Oct-4 (352), digested with BamHI and EcoRI, and cloned into the same site of pCMV-Tag2A to generate pCMV-Tag2A-Oct-4(ΔN). The FLAG-tagged Oct-4(ΔN) was amplified from pCMV-Tag2A-Oct-4(ΔN) using PCR with primers 5′-FLAG and 3′-Oct-4 (352), digested with XhoI and EcoRI, and cloned into the same sites of pEGFP-N1 to generate pEGFP-N1-FLAG-Oct-4(ΔN). The plasmid pEGFP-N1-FLAG-Oct-4(ΔN) was digested with XhoI and NotI, and cloned into the same sites of the pCAG-IP vector. For the plasmid pCAG-IP/FLAG-Oct-4(POU)–EGFP, the Oct-4 POU fragment was amplified from pcDNA3/Oct-4 by PCR using the primers 5′-Oct-4 (127) and 3′-Oct-4 (282), digested with BamHI and EcoRI, and cloned into the same site of pCMV-Tag2A to generate pCMV-Tag2A-Oct-4(POU). FLAG-tagged Oct-4(POU) was amplified from pCMV-Tag2A-Oct-4(POU) by PCR using the primers 5′-FLAG and 3′-Oct-4 (282), digested with XhoI and EcoRI, and cloned into the same sites of pEGFP-N1 to generate pEGFP-N1-FLAG-Oct-4(POU). The plasmid pEGFP-N1-FLAG-Oct-4(POU) was digested with XhoI and NotI, and cloned into the same sites of pCAG-IP vector. The description of plasmid pCAG-IP/EGFP has been described previously [24].
To generate pcDNA3/p21, full-length p21 cDNA was amplified from IRAV-6A9 by PCR using the primer 5′-p21 (1) (5′-GATCCTCGAGATGTCCAATCCTGGTGAT-3′, an XhoI site is underlined) and primer 3′-p21 (159) (5′-GATCGCGGCCGCTCAGGGTTTTCTCTTGCA-3′, a NotI site is underlined), digested with XhoI and NotI, and cloned into the same sites of pCAG-IP vector to generate pCAG-IP/p21. The plasmid pCAG-IP/p21 was digested with XhoI, blunted with Klenow fragment, and redigested with NotI. The excised fragment was ligated into the EcoRV and NotI sites of pcDNA3 (Invitrogen) to generate pcDNA3/p21.
Cell culture, cell growth curve and population-doubling time
ZHBTc4 ES cells were grown as described previously [24,25]. HEK (human embryonic kidney)-293T cells were maintained in DMEM (Dulbecco's modified Eagle's medium) with 10% heat inactivated fetal calf serum (Gibco-BRL), 100 units/ml penicillin and 100 μg/ml streptomycin. An equal number of ZHBTc4 cells (1×105 cells) were plated in duplicate in 0.2% gelatine-coated 60-mm dishes and cultured with or without doxycycline (1 μg/ml). Cells were harvested at days 1, 2 and 3 after plating. The total number of cells was counted in each individual plate with a haemocytometer, and the mean was calculated. The population-doubling time was estimated by the linear regression between log cell number and culture time.
Western blot analysis
Western blot analysis was performed using anti-Oct-4 (C-10; Santa Cruz Biotechnology), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase, V-18; Santa Cruz Biotechnology), anti-p21 (F5; Santa Cruz Biotechnology), anti-p53 (CM5; Novocastra Immunohistochemistry) or anti-actin (I-19; Santa Cruz Biotechnology) antibodies, and reactive bands were detected by chemiluminescence using the Western Lightening system (PerkinElmer Life Sciences).
Cell-cycle analysis
ZHBTc4 ES cells were harvested at 72 h after doxycycline treatment, washed twice in PBS, and were fixed in 70% ethanol at −20 °C for 20 min. After a further two washes in PBS, the fixed cells were resuspended in PBS containing 100 μg/ml RNase A and incubated at 37 °C for 30 min. The cells were stained with PI (propidium iodide) solution (33 μg/ml PI containing 10% Nonidet P40) for 30 min. They were then analysed by flow cytometry (FACSCalibur; BD Biosciences).
Establishment of the ZHBTc4 ES cell line expressing Oct-4 truncation mutants
ZHBTc4 ES cell lines stably expressing Oct-4 truncation mutants were established as described previously [26]. Briefly, to generate stably expressing ES cell lines, pCAG-IP/EGFP, pCAG-IP/FLAG-Oct-4–EGFP, pCAG-IP/FLAG–Oct-4(ΔC)–EGFP, pCAG-IP/FLAG-Oct-4(ΔN)–EGFP and pCAG-IP/FLAG-Oct-4(POU)–EGFP were linearized with PvuI and 6 μg of each was transfected into ZHBTc4 ES cells (1×107) using a MicroPorator (Digital Bio Technology). At 48 h post-electroporation, puromycin (Sigma) was added to a final concentration of 1 μg/ml to select clones carrying stably integrated plasmid DNA. After selection of transfected ZHBTc4 cells, monoclonal cell lines were isolated by picking individual puromycin-resistant colonies.
AP staining, immunocytochemistry and RT (reverse transcription)–PCR
AP was stained with an AP staining kit (Sigma) as described previously [24]. For immunocytochemistry, ZHBTc4 ES cells expressing Oct-4 deletion mutants were washed in PBS, fixed for 30 min at 37 °C with 3.7% paraformaldehyde solution (3.7% formaldehyde and 0.18% Triton X-100), and stained with the following antibodies: anti-SSEA-1 (480; Santa Cruz Biotechnology), anti-Nanog (ab21603; Abcam) or anti-Sox-2 (Y-17; Santa Cruz Biotechnology). After washing with PBS, the plates or slides were incubated with TRITC (tetramethylrhodamine β-isothiocyanate)-conjugated secondary antibody (Sigma). Fluorescence was detected with a fluorescence microscope (IX71; Olympus) equipped with a DP71 digital camera (Olympus).
Total RNA was prepared from ZHBTc4 ES cells expressing Oct-4 deletion mutants using an RNeasy mini kit (Qiagen) with on-column DNase treatment, and mRNA was purified with an Oligodex-dT mRNA mini kit (Qiagen) followed by cDNA synthesis using a Superscript first-strand synthesis system for RT–PCR (Invitrogen), according to the manufacturer's instructions. RT–PCRs for the Oct-4 downstream target genes and differentiation marker genes were performed with gene-specific primer sets as described previously [12,24].
Northern blot analysis
For Northern blotting, total cellular RNAs were prepared using TRIzol® reagent (Invitogen) and aliquots (10 μg/lane) were separated on 1.5% agarose/formaldehyde gels. RNA was transferred on to Hybond nylon membranes (Amersham Biosciences) and cross-linked to the membrane in a GS Gene Linker UV Chamber (Bio-Rad). The EcoRI- and NotI (0.48 kb)-digested DNA fragments from mouse p21 cDNA were gel purified and labelled with 32P using a Prime-It II Random Primer Labeling kit (Stratagene). Hybridizations were carried out overnight using the radiolabelled probe in ExpressHyb Solution (Clontech) at 68 °C. The blots were washed twice at 68 °C with 2×SSC buffer (1×SSC is 0.15 M NaCl/0.0015 M sodium citrate)/0.1% SDS and radiolabelled bands were visualized by autoradiography.
Reporter gene assays
ZHBTc4 or HEK-293T cells were transiently transfected with plasmids by electroporation using a MicroPorator (Digital Bio Technology) or VivaMagic Reagent (Vivagen) according to the manufacturer's instructions. Luciferase assays were performed with the dual-luciferase assay system, in accordance with the manufacturer's protocol (Promega). Renilla luciferase activities were used to normalize the transfection efficiency.
RESULTS
Oct-4 down-regulation inhibits ES cell proliferation
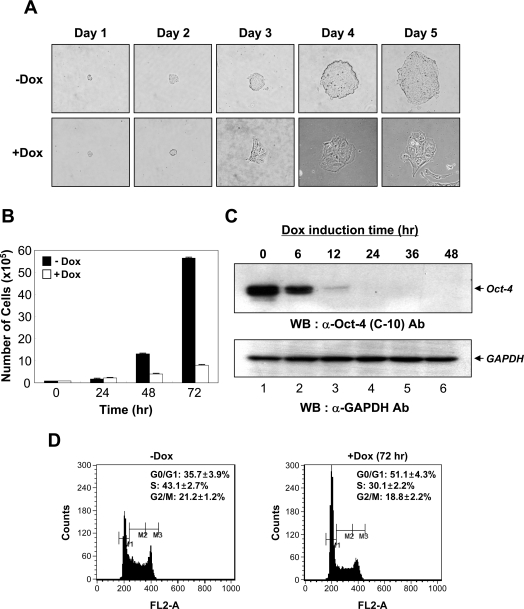
ES cells have remarkable abilities for self-renewal and can produce pluripotent daughter cells indefinitely [27]. Both processes might be important for maintaining the stemness of ES cells; however, little is known about the regulation of the cell-cycle fate in ES cells. To investigate the molecular mechanisms involved in ES cell proliferation, we studied the effect of Oct-4, a key regulator of pluripotency, on ES cell-cycle control using ZHBTc4 ES cells. These ES cells have both endogenous alleles of oct-4 inactivated by gene targeting and contain the Tet (tetracycline)-off oct-4 regulatory system [12]. The cells were colonized and grown like normal undifferentiated ES cells under the maintenance of Oct-4 transgene expression (Figure 1A, upper panels); however, upon treatment with doxycycline, a tetracycline analogue, as previously reported [12,24], definite morphological changes towards differentiation occurred as time elapsed. The cells flattened out and acquired enlarged nuclei and cytoplasm 3 days after treatment with doxycycline (Figure 1A, lower panels).
Figure 1. Growth inhibition of ES cells by the down-regulation of Oct-4.
(A) Time-dependent effects of the down-regulation of Oct-4 on ES cell growth and colony morphology. ZHBTc4 ES cells (3×103) were plated on to 0.2% gelatin-coated 35-mm dishes and treated without (labelled as −Dox) or with 1 μg/ml doxycycline (labelled as +Dox) for 5 days. Cells were monitored daily using an inverted phase-contrast microscope (IX71; Olympus). (B) Effects of Oct-4 down-regulation on the proliferation of ZHBTc4 ES cells. ZHBTc4 ES cells were seeded at 1×105 cells per 0.2% gelatin-coated 60-mm dish and grown in ES cell medium supplemented with (+Dox) or without (−Dox) doxycycline. Cells were counted at 24 h intervals with a haemocytometer for a total of 72 h. Values are means±S.D. Three independent experiments were performed, all of which gave similar results. (C) Western blot analysis of Oct-4 down-regulation in ZHBTc4 ES cells following doxycycline treatment. ZHBTc4 ES cells were treated with 1 μg/ml doxycycline and were extracted 0, 6, 12, 24, 36 and 48 h after doxycycline treatment. The extracted proteins were resolved by SDS/PAGE (12% gels), transferred on to a PVDF membrane, and immunoblotted with anti-Oct-4 antibody (C-10; Santa Cruz Biotechnology) (upper panel). The expression level of GAPDH was used as an internal control (bottom panel). (D) Effects of Oct-4 down-regulation on cell-cycle progression. The DNA contents of ZHBTc4 ES cells cultured in the absence (−Dox) or presence (+Dox) of doxycycline for 72 h were measured by fluorescence with PI staining. M1, M2 and M3 represent gating for G0/G1, S and G2/M populations respectively. Values given are means±S.D. Three independent experiments gave similar results. Ab, antibody; WB, Western blot.
To determine whether the down-regulation of Oct-4 gene expression could alter ES cell growth kinetics, the ZHBTc4 ES cells were cultured in the absence (−Dox) or presence (+Dox) of doxycycline. The growth property of ZHBTc4 ES cells with inhibited Oct-4 expression was markedly different from those expressing Oct-4. The doubling time of ZHBTc4 ES cells, cultured in the presence of doxycycline, was increased from 11.95 h (the doubling time of ZHBTc4 ES cells cultured in the absence of doxycycline) to 24.75 h, which indicated that down-regulation of Oct-4 in ES cells significantly altered ES cell growth (Figure 1B).
The addition of doxycycline to ZHBTc4 ES cells resulted in the rapid repression of Oct-4 transgene expression, as determined by Western blotting of total cell extracts (Figure 1C, upper panel). Expression of the Oct-4 transgene in ZHBTc4 ES cells can be completely turned off within 24 h. Approximately equal amounts of total protein were examined for each of the cell extracts (Figure 1C, lower panel).
Inhibition of proliferation is typically associated with cell-cycle arrest. To investigate this possibility, ZHBTc4 ES cells were cultured in the absence (−Dox) or presence (+Dox) of doxycycline for 72 h. The DNA contents were then analysed by PI staining and flow cytometry. The down-regulation of Oct-4 expression induced a substantial accumulation of ES cells in the G0/G1-phase of the cell cycle (Figure 1D, right-hand panel), when compared with control ES cells (Figure 1D, left-hand panel). Down-regulation of Oct-4 led to a 43% increase (from 35.7% to 51.1%) in the number of ES cells in the G0/G1-phase of the cell cycle. We found that Oct-4 down-regulation reduced the S-phase population by 30% (from 43.1% to 30.1%). Thus Oct-4 down-regulation in ES cells inhibits cellular proliferation by blocking cell-cycle progression in G0/G1.
The overall integrity of the Oct-4 functional domains is necessary to achieve its full potential for self-renewal potential
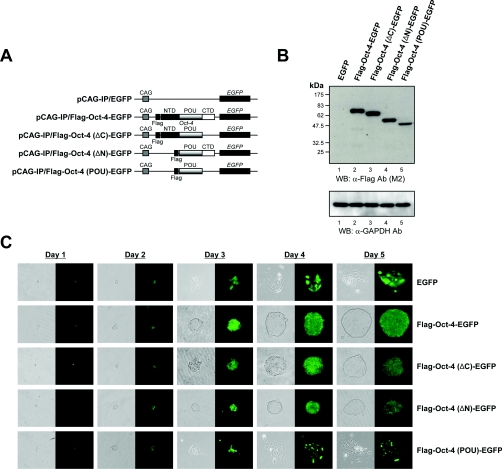
Oct-4 contains N-terminal and C-terminal transactivation domains, as well as a POU DNA-binding domain [28]. To define the critical region(s) within Oct-4 that are required for ES cell self-renewal, the pCAG-IP/EGFP, pCAG-IP/FLAG-Oct-4–EGFP, pCAG-IP/FLAG-Oct-4(ΔC)–EGFP, pCAG-IP/FLAG-Oct-4(ΔN)–EGFP and pCAG-IP/FLAG-Oct-4(POU)–EGFP constructs (Figure 2A) were generated by the PCR method. We constructed plasmids expressing GFP fusions of the Oct-4 truncation mutants in order to monitor the expression of Oct-4 proteins. After selection of pCAG-IP/EGFP, pCAG-IP/FLAG-Oct-4–EGFP, pCAG-IP/Flag-Oct-4(ΔC)–EGFP, pCAG-IP/FLAG-Oct-4(ΔN)–EGFP or pCAG-IP/FLAG-Oct-4(POU)–EGFP-transfected ZHBTc4 cells, 3×103 stable transfectants were seeded on to 35-mm dishes containing 1 μg/ml doxycycline to repress the tetracycline-repressible mouse Oct-4 transgene. The amounts of Oct-4 or Oct-4 deletion mutant proteins in the different clones were determined by Western blot analysis (Figure 2B). Although no Oct-4 protein could be detected in ZHBTc4 ES cells stably transfected with pCAG-IP/EGFP (Figure 2B, lane 1), the Oct-4 or Oct-4 truncation mutant proteins were expressed at similar levels in all clonally derived ES cell lines used in the present study (Figure 2B, lanes 2–5). Consistent with previous reports [12,24,25], the Oct-4-transfected ZHBTc4 ES cells were able to maintain their self-renewal ability and stem-cell phenotype (Figure 2C, second panels). As a control, ZHBTc4 ES cells stably transfected with EGFP and cultured in the presence of doxycycline underwent differentiation (Figure 2C, top panels). The behaviour of the N-terminal domain and/or the C-terminal domain deletion mutants of Oct-4 were also investigated. Interestingly, the ZHBTc4 ES cells expressing FLAG-tagged Oct-4(ΔC)–EGFP or FLAG-tagged Oct-4(ΔN)–EGFP proteins were able to maintain their self-renewal ability and stem-cell phenotype, but the sizes of the colonies were smaller than those of the ZHBTc4 ES cells expressing full-length Oct-4 (Figure 2C, third and fourth panels). When FLAG-tagged Oct-4(POU)–EGFP was stably introduced into ZHBTc4 ES cells, it failed to maintain stem-cell renewal and underwent differentiation (Figure 2C, bottom panels).
Figure 2. Different abilities of the Oct-4 deletion mutants to confer self-renewal.
(A) Schematic representation of the Oct-4–EGFP fusion construct and its derivatives with deleted domains. The functional domains located within Oct-4 are indicated as NTD (N-terminal domain), POU (POU DNA-binding domain) and CTD (C-terminal domain). (B) Immunoblot analysis of the expression of Oct-4 deletion mutants in stably transfected ZHBTc4 cells. ZHBTc4 ES cells were stably transfected with pCAG-IP/EGFP (labelled as EGFP; lane 1), pCAG-IP/FLAG-Oct-4–EGFP (Flag-Oct-4-EGFP; lane 2), pCAG-IP/FLAG-Oct-4(ΔC)–EGFP [Flag-Oct-4(ΔC)-EGFP; lane 3], pCAG-IP/FLAG-Oct-4(ΔN)–EGFP [Flag-Oct-4(ΔN)-EGFP; lane 4] or pCAG-IP/FLAG-Oct-4(POU)–EGFP [Flag-Oct-4(POU)-EGFP; lane 5], and total cell lysates were fractionated by SDS/PAGE (12% gels) and visualized by Western blotting with anti-FLAG (M2; Sigma) or anti-GAPDH (V-18; Santa Cruz Biotechnology) antibodies. The position of the prestained molecular-mass marker (New England Biolabs) is indicated to the left of the gel (molecular mass in kDa). (C) Self-renewal potentials of Oct-4 deletion mutants. ZHBTc4 ES cells stably transfected with pCAG-IP/EGFP (EGFP), pCAG-IP/FLAG-Oct-4–EGFP (Flag-Oct-4-EGFP), pCAG-IP/FLAG-Oct-4(ΔC)–EGFP [Flag-Oct-4(ΔC)-EGFP], pCAG-IP/FLAG-Oct-4(ΔN)–EGFP [Flag-Oct-4(ΔN)EGFP] or pCAG-IP/FLAG-Oct-4(POU)–EGFP [Flag-Oct-4(POU)-EGFP] were cultured in the presence of doxycycline. Phase-contrast (left-hand panels, grey colour) and fluorescence views (right-hand panels, green colour) are shown. Ab, antibody; WB, Western blot.
The N-terminal and C-terminal domains of Oct-4 are important for stimulation of S-phase entry
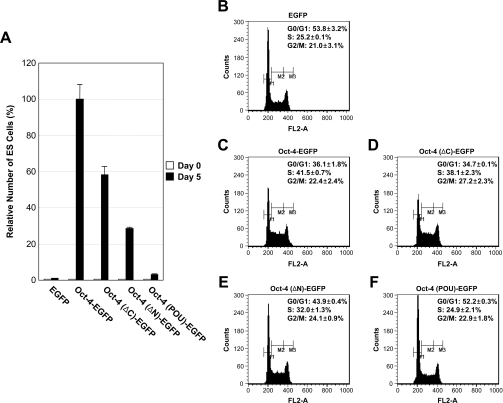
To investigate the functional region(s) of the Oct-4 protein required for the stimulation of ES cell proliferation, the inhibitory potential of ZHBTc4 ES cells expressing Oct-4 deletion mutants was analysed by cell counting. Deletion of the Oct-4 C-terminal domain [Oct-4(ΔC)–EGFP] produced a polypeptide with approx. 40% reduced ES cell proliferation ability relative to intact Oct-4 (Oct-4–EGFP) (Figure 3A). Deletion of amino acids from the N-terminus of Oct-4 [Oct-4(ΔN)–EGFP] further reduced the ES cell proliferation properties (by approx. 70%), whereas ZHBTc4 ES cells expressing Oct-4(POU)–EGFP failed to stimulate ES cell proliferation. Differences in ES cell proliferation potential among the various Oct-4 deletion mutants were not due to differences in protein amounts since Western blot analysis revealed equivalent amounts of protein produced from each construct (Figure 2B).
Figure 3. Functional regions of Oct-4 required for stimulation of S-phase entry of ES cells.
(A) Relative growth rate of ZHBTc4 ES cells expressing Oct-4 deletion mutants. Cells (3×103) were plated on to 0.2% gelatin-coated 35-mm dishes and counted at 5 days with a haemocytometer. Relative self-renewal ability values are shown to the right, with the value obtained by Oct-4 taken as 100%. Each bar represents the average values of two experiments. Three independent experiments gave similar results. (B–F) Effects of Oct-4 deletion mutants on ES cell-cycle progression. DNA contents of ZHBTc4 ES cells expressing EGFP (B), Oct-4–EGFP (C), Oct-4(ΔC)–EGFP (D), Oct-4(ΔN)–EGFP (E) or Oct-4(POU)–EGFP (F) were measured by fluorescence with PI staining. M1, M2 and M3 represent gating for G0/G1, S and G2/M populations respectively. Values shown are means±S.D. Three independent experiments gave similar results.
To investigate the cell-cycle distribution, the DNA content of ZHBTc4 ES cells expressing Oct-4 deletion mutants was analysed by PI staining and flow cytometry (Figures 3B–3F). Compared with ZHBTc4 ES cells expressing intact Oct-4 (Figure 3C), deletion of the N-terminal or C-terminal domain of Oct-4 expression reduced the number of cells in S-phase (Figures 3D and 3E). As can be seen in Figure 3(F), the deletion of both the N-terminal and C-terminal domains of Oct-4 [Oct-4(POU)–EGFP] resulted in a significant increase in the number of cells in the G0/G1 fraction that was accompanied by a decrease in the number of S-phase cells, indicating the failure of ES cell-cycle progression. We also performed the same experiments using different clones and obtained similar results (J. Lee and J. Kim, unpublished work). These results indicated that the overall integrity of Oct-4 is necessary for the full proliferation potential of ES cells.
Different abilities of Oct-4 deletion mutants to maintain the undifferentiated state
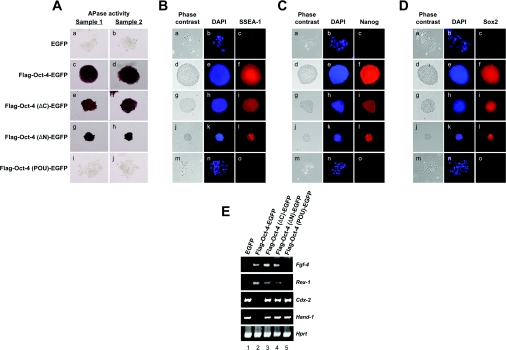
The small colony size and reduced proliferation abilities of doxycycline-treated ZHBTc4 ES cells expressing Oct-4 N- or C-terminal deletion mutants (Figure 2C) suggested change or collapse to maintain the undifferentiated state of the ES cells. To verify this hypothesis, we examined known molecular markers of undifferentiated ES cells. To stain for AP activity (a marker for pluripotent cells of embryonic origin) [29], colonies were tested for staining the cells for AP. ZHBTc4 ES cells expressing FLAG-tagged Oct-4(ΔC)–EGFP or FLAG-tagged Oct-4(ΔN)–EGFP proteins contained AP activity (Figure 4A, panels e–h) comparable with that in ZHBTc4 ES cells expressing full-length Oct-4 (panels c and d). However, the clones expressing FLAG-Oct-4(POU)–EGFP (panels i and j) or EGFP alone (panels a and b) had lost this characteristic, which pointed to a failure to maintain the undifferentiated state.
Figure 4. Characterization of ZHBTc4 ES cells expressing Oct-4 deletion mutants.
(A) Expression of AP. AP activity was assessed in ZHBTc4 ES cells expressing Oct-4 deletion mutants. The expression vectors used in this experiment were the same as those described in Figure 2(A). (B) Expression of SSEA-1. The expression of SSEA-1 in ZHBTc4 ES cells expressing Oct-4 deletion mutants was analysed with an anti-SSEA-1 antibody (480; Santa Cruz Biotechnology). The expression vectors used in this experiment were the same as those described in in Figure 2(A). (C) Expression of Nanog. The expression of Nanog in ZHBTc4 ES cells expressing Oct-4 deletion mutants was analysed with an anti-Nanog antibody (ab21603; Abcam). The expression vectors used in this experiment were the same as those described in Figure 2(A). (D) Expression of Sox2. The expression of Sox2 in ZHBTc4 ES cells expressing Oct-4 deletion mutants was analysed with an anti-Sox2 antibody (Y-17; Santa Cruz Biotechnology). The expression vectors used in this experiment were the same as those described in Figure 2(A). (E) Expression of Oct-4 downstream target genes and lineage-specific markers. RT–PCR analyses of Fgf-4, Rex-1, Cdx-2 and Hand-1 mRNAs were performed in ZHBTc4 ES cells expressing Oct-4 deletion mutants. Hprt was used as a control to qualify the RT–PCR results. Following amplification, an aliquot of each product was analysed by staining the gel with ethidium bromide. The ES cell lines from which the input RNAs used in the RT reactions were derived are shown above the panel. Three independent experiments gave similar results.
We also examined other known molecular markers of undifferentiated ES cells, such as SSEA-1 [30], Nanog [31,32] and Sox2 [33]. Consistent with the results from staining for AP activity, ZHBTc4 ES cells expressing FLAG-tagged Oct-4(ΔC)–EGFP or FLAG-tagged Oct-4(ΔN)–EGFP proteins were positive for immunostaining against SSEA-1 (Figure 4B, panels g–l), Nanog (Figure 4C, panels g–l) or Sox2 (Figure 4D, panels g–l), comparable with that seen in ZHBTc4 ES cells expressing full-length Oct-4 (Figures 4B–4D, panels d–f). However, the clones expressing FLAG-Oct-4(POU)–EGFP (Figures 4B–4D, panels m–o) or EGFP alone (Figures 4B–4D, panels a–c) had lost these characteristics.
To investigate further the features of the ZHBTc4 ES cells expressing these mutants, we investigated the expression level of known Oct-4 downstream target genes such as Fgf-4 [34] and Rex-1 [35] by RT–PCR. As shown in Figure 4(E), the expression of the two Oct-4 downstream target genes was detected in tetracycline-treated ZHBTc4 ES cells stably transfected with FLAG-Oct-4–EGFP (lane 2), FLAG-Oct-4(ΔC)–EGFP (lane 3) or FLAG-Oct-4(ΔN)–EGFP (lane 4) cDNAs.
We also evaluated the expression of differentiated cell markers. As shown in Figure 4(E), the expression of Cdx-2 mRNA, which is implicated in trophoblast differentiation [36], was detected in ZHBTc4 ES cells expressing vector (lane 1) or FLAG-Oct-4(POU)–EGFP (lane 5). Unexpectedly, the expression of Cdx-2 mRNA was also detected in ZHBTc4 ES cells expressing FLAG-Oct-4(ΔC)–EGFP (lane 3) and FLAG-Oct-4(ΔN)–EGFP (lane 4), which were also positive for the undifferentiated markers. However, the Cdx-2 gene was not detectable in ZHBTc4 ES cells expressing FLAG-tagged Oct-4 (lane 2). Hand-1 mRNA, a marker of trophoblast differentiation [37], also appeared in ZHBTc4 ES cells expressing FLAG-Oct-4(ΔC)–EGFP (lane 3) and FLAG-Oct-4(ΔN)–EGFP (lane 4), indicating the induction of differentiation programmes. Taken together, these properties all suggested that ES cells, expressing full-length Oct-4, expanded normally and remained in an undifferentiated state, whereas Oct-4(POU)-expressing ES cells did not. Interestingly, the status of the Oct-4(ΔC)- or Oct-4(ΔN)-expressing ES cells seems to be intermediate between the undifferentiated and differentiated states.
Oct-4 down-regulation induces expression of p21 in ZHBTc4 ES cells
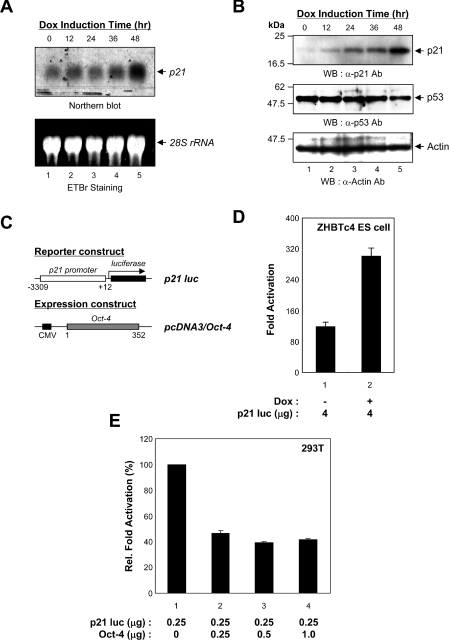
It has been well-characterized that the G1-S transition is mainly regulated by G1 regulatory molecules [38]. Among these regulatory molecules, p21 is a member of the universal CDKIs and plays several critical roles in the regulation of the G1-S transition [39]. To investigate whether Oct-4 down-regulation results in elevated p21 levels in ES cells, we examined p21 levels in ZHBTc4 ES cells. In this system, the addition of doxycycline to the growth medium of ZHBTc4 ES cells resulted in the rapid repression of Oct-4 expression, as determined by Western blot analysis of total cell extracts (Figure 1C). To examine the effects of the down-regulation of Oct-4 expression on endogenous p21 gene expression, Northern blot analysis was performed with total RNA, isolated from these cells in the presence of doxycycline. The results revealed that p21 transcript levels were increased upon down-regulation of oct-4 expression (Figure 5A). We also investigated the induction of p21 protein when Oct-4 expression was down-regulated. Western blot analysis showed almost no signal for p21 protein when Oct-4 is expressed normally (Figure 5B, lane 1). However, down-regulation of Oct-4 induced p21 protein expression, and a strong induction of p21 was observed at 48 h after the addition of the doxycycline (Figure 5B). It has been well demonstrated that p21 expression is regulated at the transcriptional level by both p53-dependent and -independent mechanisms [40]; therefore we decided to investigate the levels of p53 protein in ZHBTc4 ES cells. The analysis of ES cell extracts by immunoblotting, carried out at various time intervals, demonstrated that the level of p53 protein was slightly down-regulated by Oct-4 down-regulation (Figure 5B, middle panel).
Figure 5. Up-regulation of p21 gene expression in ZHBTc4 ES cells down-regulating Oct-4.
(A) Northern blot analysis of p21 mRNA expression. ZHBTc4 ES cells were harvested for the preparation of total RNA at 0, 12, 24, 36 and 48 h following doxycycline treatment. Total RNA was fractionated on a 6% formaldehyde/1.5% agarose gel, transferred on to a nylon membrane, and probed with mouse p21 cDNA as described in the Materials and methods section (upper panel). The ethidium bromide staining of the agarose gel used for the Northern blotting is shown to demonstrate that equal amounts of total RNA were loaded into each lane (lower panel). Arrows indicate the position of migration of the respective RNAs. Three independent experiments gave similar results. (B) Western blot analysis of p21 expression. Total cell extracts were prepared from ZHBTc4 ES cells at 0, 12, 24, 36 and 48 h following doxycycline treatment. The extracts were resolved by SDS/PAGE (15% gel), transferred on to a PVDF membrane, and immunoblotted with anti-p21 (F5; Santa Cruz Biotechnology), anti-p53 (CM5; Novocastra Immunohistochemistry) or anti-actin (I-19; Santa Cruz Biotechnology) antibodies. The positions of the prestained molecular-mass markers (New England Biolabs) are indicated to the left of the gel (molecular mass in kDa). Three independent experiments gave similar results. (C) Schematic representation of the reporter plasmid and Oct-4 expression vector. The p21 luciferase reporter plasmid contains a mouse genomic DNA sequence from nucleotides −3309 to +12. The mouse p21 promoter is indicated by an open box and the luciferase gene is indicated by a solid box. The expression vector driving the production of mouse Oct-4 is also represented. (D) Transcriptional activation of the p21 promoter by down-regulation of Oct-4 in ZHBTc4 ES cells. ZHBTc4 ES cells were transfected with the p21 luciferase reporter plasmid and cultured in the absence (−Dox, lane 1) or presence (+Dox, lane 2) of doxycycline. Firefly luciferase activity was normalized with Renilla luciferase activity to correct for transfection efficiencies. Each transfection was performed at least three times independently, and values are means±S.D. Fold-induction is relative to the empty vector. (E) Transcriptional repression of the p21 promoter by Oct-4. Co-transfections of HEK-293T cells were performed with 0.25 μg of reporter plasmid and 0, 0.25, 0.5 or 1.0 μg of pcDNA3/Oct-4 expression plasmid. Individual transfected DNAs were adjusted to contain equal amounts of total DNA by the addition of the empty expression vector pcDNA3. The average relative transactivation and standard error is presented, with the relative transactivation values obtained with pcDNA3/Oct-4 taken as 100%. The values shown are taken from three independent experiments.
To demonstrate whether the p21 promoter could mediate a transcriptional response to Oct-4, the p21 promoter region was cloned upstream of the firefly luciferase reporter gene (Figure 5C). A CMV (cytomegalovirus)-based expression vector driving the production of Oct-4 was also generated. ZHBTc4 ES cells were transfected with the p21–luciferase reporter plasmid and cultured for 48 h in the absence (Figure 5D, lane 1) or presence (lane 2) of doxycycline. The introduction of p21–luciferase into ZHBTc4 ES cells, cultured in the absence of doxycycline, activated reporter gene transcription 118-fold. However, in the absence of Oct-4 (+Dox), reporter gene activity was increased up to 301-fold, indicating that the down-regulation of Oct-4 in ES cells causes an increases in p21 gene expression. To investigate Oct-4-mediated inhibition of p21 gene expression further, reporter and expression vectors were then introduced into HEK-293T cells and assayed for the induction of luciferase activity. The introduction of increasing amounts of pcDNA3/Oct-4 to a transfection containing fixed amounts of p21–luciferase reporter plasmid led to a reduction in luciferase activity (Figure 5E). These results suggested that Oct-4 might repress p21 promoter activity.
DISCUSSION
The mechanisms by which the ES cell cycle is controlled and sustained remain obscure. It is well known that Oct-4 is an essential transcriptional regulator of genes involved in maintaining the undifferentiated pluripotent state of ES cells, but little has been established as to whether the ES cell-cycle fate is regulated by this protein. In the present study, we have characterized the effect of Oct-4 on ES cell-cycle control using ZHBTc4 ES cells. Previous studies have shown that the length of G1-phase in ES cells is much shorter than in somatic cells [41–43], and in the present study we found that Oct-4 down-regulation in ES cells inhibits cellular proliferation by blocking cell-cycle progression in G0/G1. Deletion analysis of the functional domains of Oct-4 indicated that both the N-terminal and C-terminal transactivation domains of Oct-4 are important for the stimulation of S-phase entry. In addition, we also showed in the present study that the p21 gene is a target for repression by Oct-4 in ZHBTc4 ES cells. Our findings demonstrate that the down-regulation of p21 by Oct-4 may contribute to the maintenance of ES cell proliferation.
Oct-4 is a central mediator of the undifferentiated pluripotent state of ES cells. It was previously reported that maintaining Oct-4 activity within a certain range appears to be critical for stem cell self-renewal, with any increase or decrease triggering differentiation to endoderm/mesoderm or trophectoderm respectively [12]. Although Oct-4 possesses two separate transactivation domains, Oct-4 deletion mutants lacking either the N-terminal domain or the C-terminal domain are able to maintain the stem cell phenotype [25]. Consistent with this result, ZHBTc4 ES cells expressing FLAG-tagged Oct-4(ΔC)–EGFP or FLAG-tagged Oct-4(ΔN)–EGFP proteins were positive for undifferentiated stem cell markers, such as AP, SSEA-1, Nanog, Sox2, Fgf-4 and Rex-1 (Figure 4). However, these ES cells showed smaller colony sizes and were also positive for the expression of markers of differentiated cells, indicating that neither the N-terminal nor the C-terminal domain deletion mutant of the Oct-4 protein is sufficient to maintain the stem cell phenotype. ES cells expressing Oct-4 deletion mutants lacking either the N-terminus or the C-terminus were positive both for the undifferentiated stem cell markers and for markers of differentiated cells (Figure 4). It would be interesting, therefore, to see whether such ES cells are capable of long-term self-renewal and are pluripotent in vitro and in vivo.
Cell-cycle progression is controlled by the CDKs, and p21 is a CDKI that inhibits G1 CDK–cyclin complexes [44–46]. In the present study, the down-regulation of Oct-4 in ES cells induced p21 to a remarkable degree (Figure 5), in a time-dependent manner, both at the mRNA and protein levels. It is well known that p21 expression is regulated largely at the transcriptional level by a p53-dependent mechanism at the transcriptional level [46], which suggests that Oct-4 may regulate p21 expression through the p53 tumour suppressor gene. However, although ZHBTc4 ES cells did express p53, the down-regulation of Oct-4 did not significantly change p53 protein levels (Figure 5B). Having demonstrated that p21 expression is down-regulated by Oct-4 in ES cells, we attempted to determine whether this resulted from a direct effect of Oct-4 with regulatory element(s) in p21. As shown in Figure 5, the down-regulation of Oct-4 in ES cells increased p21 promoter activity (Figure 5D) and the introduction of increasing amounts of Oct-4 led to a reduction in p21 promoter activity (up to 40% of promoter activity) in differentiated cells (HEK-293T cell line; Figure 5E), suggesting that Oct-4 directly represses p21 promoter activity. However, p21 expression can also be regulated post-translationally by both ubiquitin-dependent and -independent proteasome-mediated degradation [47–49]. Thus it would be interesting to determine whether Oct-4 is able to control p21 stability in ES cells and induce pluripotent stem (iPS) cells.
The expression of Oct-4 in doxycyclin-treated ZHBTc4 ES cells led to a consistent increase in the number of cells in S-phase, an effect that was not observed following the expression of EGFP (Figure 3). Consistent with these results, the down-regulation of Oct-4 in ES cells also led to an increase in the number of cells in G0/G1-phase (Figure 1D). To clarify the contribution of the different functional domains of the Oct-4 protein to ES cell-cycle progression, we generated ES cell lines carrying functional domain deletion mutants of Oct-4 (Figure 2A). Deletion of the N-terminal or C-terminal domain of Oct-4 reduced the number of cells in S-phase (Figure 3), indicating that the overall integrity of Oct-4 is required for its ability to stimulate cell-cycle progression in ES cells.
In conclusion, it is likely that Oct-4 contributes towards maintaining cells in an undifferentiated, pluripotent or totipotent state in two ways: (i) by activating certain key genes and (ii) by silencing others. The findings of the present study provide additional evidence that Oct-4 plays a key role in regulating the ES cell cycle positively, as well as maintaining ES cell pluripotency. Therefore Oct-4-mediated p21 gene down-regulation may be a key mechanism for the stimulation of cell-cycle progression of the ICM.
AUTHOR CONTRIBUTIONS
Jungwoon Lee and Jungho Kim designed and performed the research, and wrote and edited the manuscript. Yeorim Go and Inyoung Kang performed some parts of the experiments and Yong-Mahn Han analysed the data. All authors discussed the results and commented on the paper.
ACKNOWLEDGEMENTS
We thank Dr Hitoshi Niwa (RIKEN Center for Developmental Biology, Kobe, Japan) for providing ZHBTc4 ES cells and the pCAG-IP vector.
FUNDING
This work was supported by the Stem Cell Research Center of the 21st Century Frontier Research Program funded by the Ministry of Education, Science, and Technology, Republic of Korea [grant number SC-2211]; the BioGreen21 Program, Rural Development Administration, Republic of Korea [grant number 20070501034009]; the Priority Research Centers Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology [grant number 2009-0093822]. J. L., Y. G. and I. K. were recipients of a research fellowship BK21 from the Ministry of Education and Human Resources Development, and J. K. was a recipient of the SRG programme [grant number 200811026.1] of Sogang University.
References
- 1.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- 3.Scholer H. R., Dressler G. R., Balling R., Rohdewohld H., Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 5.Goto T., Adjaye J., Rodeck C. H., Monk M. Identification of genes expressed in human primordial germ cells at the time of entry of the female germ line into meiosis. Mol. Hum. Reprod. 1999;5:851–860. doi: 10.1093/molehr/5.9.851. [DOI] [PubMed] [Google Scholar]
- 6.Hansis C., Grifo J. A., Krey L. C. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol. Hum. Reprod. 2000;6:999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- 7.Burdon T., Smith A., Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 8.Scholer H. R., Ruppert S., Suzuki N., Chowdhury K., Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 9.Yeom Y. I., Ha H. S., Balling R., Scholer H. R., Artzt K. Structure, expression and chromosomal location of the Oct-4 gene. Mech. Dev. 1991;35:171–179. doi: 10.1016/0925-4773(91)90016-y. [DOI] [PubMed] [Google Scholar]
- 10.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 11.Pesce M., Wang X., Wolgemuth D. J., Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H., Miyazaki J., Smith A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Rahman B., Fiddler M., Rappolee D., Pergament E. Expression of transcription regulating genes in human preimplantation embryos. Hum. Reprod. 1995;10:2787–2792. doi: 10.1093/oxfordjournals.humrep.a135792. [DOI] [PubMed] [Google Scholar]
- 14.Scholer H. R., Balling R., Hatzopoulos A. K., Suzuki N., Gruss P. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 1989;8:2551–2557. doi: 10.1002/j.1460-2075.1989.tb08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholer H. R. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- 16.Brehm A., Ohbo K., Scholer H. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol. Cell. Biol. 1997;17:154–162. doi: 10.1128/mcb.17.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J., Rhee B. K., Bae G. Y., Han Y. M., Kim J. Stimulation of Oct-4 activity by Ewing's sarcoma protein. Stem Cells. 2005;23:738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- 18.Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butteroni C., De Felici M., Scholer H. R., Pesce M. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 2000;304:529–540. doi: 10.1006/jmbi.2000.4238. [DOI] [PubMed] [Google Scholar]
- 20.Brehm A., Ohbo K., Zwerschke W., Botquin V., Jansen-Durr P., Scholer H. R. Synergism with germ line transcription factor Oct-4: viral oncoproteins share the ability to mimic a stem cell-specific activity. Mol. Cell. Biol. 1999;19:2635–2643. doi: 10.1128/mcb.19.4.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholer H. R., Ciesiolka T., Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Kim H. K., Han Y. M., Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Russell D. W. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 24.Lee J., Kim H. K., Rho J. Y., Han Y. M., Kim J. The human OCT-4 isoforms differ in their ability to confer self-renewal. J. Biol. Chem. 2006;281:33554–33565. doi: 10.1074/jbc.M603937200. [DOI] [PubMed] [Google Scholar]
- 25.Niwa H., Masui S., Chambers I., Smith A. G., Miyazaki J. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J., Kim J. Y., Kang I. Y., Kim H. K., Han Y. M., Kim J. The EWS-Oct-4 fusion gene encodes a transforming gene. Biochem. J. 2007;406:519–526. doi: 10.1042/BJ20070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odorico J. S., Kaufman D. S., Thomson J. A. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 28.Pesce M., Scholer H. R. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 29.Evans M. J., Kaufman M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 30.Koestenbauer S., Zech N. H., Juch H., Vanderzwalmen P., Schoonjans L., Dohr G. Embryonic stem cells: similarities and differences between human and murine embryonic stem cells. Am. J. Reprod. Immunol. 2006;55:169–180. doi: 10.1111/j.1600-0897.2005.00354.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambrosetti D. C., Basilico C., Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Shushan E., Thompson J. R., Gudas L. J., Bergman Y. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 1998;18:1866–1878. doi: 10.1128/mcb.18.4.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck F., Erler T., Russell A., James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- 37.Cserjesi P., Brown D., Lyons G. E., Olson E. N. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 38.Sherr C. J. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 39.Sherr C. J., Roberts J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 40.Gartel A. L., Tyner A. L. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 41.Neganova I., Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker K. A., Ghule P. N., Therrien J. A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 43.Fluckiger A. C., Marcy G., Marchand M., Negre D., Cosset F. L., Mitalipov S., Wolf D., Savatier P., Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24:547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 45.Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 46.Gartel A. L., Tyner A. L. Transcriptional regulation of the p21(WAF1/CIP1) gene. Exp. Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 47.Maki C. G., Howley P. M. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 49.Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 α-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]