Abstract
Background information. The BOR (branchio-oto-renal) syndrome is a dominant disorder most commonly caused by mutations in the EYA1 (Eyes Absent 1) gene. Symptoms commonly include deafness and renal anomalies.
Results. We have used the embryos of the frog Xenopus laevis as an animal model for early ear development to examine the effects of different EYA1 mutations. Four eya1 mRNAs encoding proteins correlated with congenital anomalies in human were injected into early stage embryos. We show that the expression of mutations associated with BOR, even in the presence of normal levels of endogenous eya1 mRNA, leads to morphologically abnormal ear development as measured by overall otic vesicle size, establishment of sensory tissue and otic innervation. The molecular consequences of mutant eya1 expression were assessed by QPCR (quantitative PCR) analysis and in situ hybridization. Embryos expressing mutant eya1 showed altered levels of multiple genes (six1, dach, neuroD, ngnr-1 and nt3) important for normal ear development.
Conclusions. These studies lend support to the hypothesis that dominant-negative effects of EYA1 mutations may have a role in the pathogenesis of BOR.
Keywords: branchio-oto-renal (BOR), disease model, Eyes Absent 1 (EYA1) gene, otic development, Xenopus
Abbreviations: BOR, branchio-oto-renal; DACH, dachshund; DIG, digoxigenin; DMED, dimethylethylenediamine; EYA1, Eyes Absent 1; GFP, green fluorescent protein; GST, glutathione transferase; HAD, haloacid dehalogenase; HRP, horseradish peroxidase; QPCR, quantitative PCR; RT–PCR, reverse transcription–PCR; six, sine oculis; spcd, spinal cord; THF, tetrahydrofuran
Introduction
The BOR (branchio-oto-renal) syndrome in humans is an autosomal dominant disorder characterized by branchial cleft abnormalities, otic developmental defects and renal malformations. All of the causes of BOR remain to be determined but mutations in several different genes have been implicated. To date, autosomal dominant mutations in the EYA1 (Eyes Absent 1) gene (Abdelhak et al., 1997b) are the most common genetic cause of BOR. EYA1 is the human homologue of the Drosophila gene eya (eyes absent), in which null mutations result in eyeless fly embryos due to apoptotic loss of eye disc cells (Bonini et al., 1993). Subsequent studies reported homologues of the eya gene in vertebrates (Duncan et al., 1997) and the rescue of the Drosophila phenotype with copies of human and murine EYA1 confirmed functional similarity (Bui et al., 2000).
The multiple members of the EYA gene family have a relatively divergent N-terminal activation domain coupled with a conserved C-terminal EYA domain. Although EYA1 protein does not bind directly to DNA, the EYA domain interacts with other DNA-binding proteins. Transcription factors SIX (sine oculis) and DACH (dachshund) are among the proteins that can form a complex with EYA1, and translocation of EYA1 from the cytoplasm into the nucleus seems to be SIX dependent (Ohto et al., 1999). EYA1 is a member of the phosphatase subfamily of the HAD (haloacid dehalogenase) superfamily and can act as a protein tyrosine phosphatase (Rayapureddi et al., 2003; Tootle et al., 2003); mutations that compromise this enzymatic activity alter its ability to serve as a transcription cofactor (Tootle et al., 2003; Mutsuddi et al., 2005).
Tracking the developmental consequences of EYA1 mutations has taken advantage of previous studies in a number of model organisms including studies in Xenopus laevis. Initial characterization of the Xenopus homologue of EYA1 revealed 85% identity at the amino acid level with the human form of the protein (David et al., 2001). Xenopus eya1 expression begins during early gastrulation and continues throughout development. Early embryonic expression is similar to that seen during mammalian development, including expression in all neurogenic placodes and placodally derived structures with the exception of the lens (David et al., 2001). Of particular relevance to the work reported here is the expression of eya1 in the otic placode and the developing ear (Bane et al., 2005). Recently, Schlosser et al. (2008) probed early developmental defects that arose after morpholino-targeted reduction of eya1 protein. They came to the conclusion that eya1 was required for normal pre-placodal ectoderm and placodal neuron formation. Their studies were consistent with conclusions drawn from studies on Eya1 in mutant mice (Zou et al., 2004, 2008).
The human phenotype associated with different EYA1 mutations can vary, and over eighty mutations have been reported (Orten et al., 2008). We note that as more detailed information about the EYA1 gene became available, current numerical designations found in the Human Gene Mutation Database no longer match historical designations. In this paper, we chose to study mutations representative of a range of phenotypes. For example, E363K was originally designated E330K (Azuma et al., 2000) and is associated with ocular defects without BOR; G426S [originally G393S (Azuma et al., 2000)] is associated with BOR and ocular defects; R440Q [originally R407Q (Kumar et al., 1997) and one of the most common BOR mutations (Orten et al., 2008)] and L505R [originally L472R (Abdelhak et al., 1997a)] are associated with BOR without ocular defects. Others have examined these mutations in an attempt to understand the molecular mechanisms that lead to syndromic consequences. Although the results are not uniformly consistent they do provide some interesting observations. For example, in vitro and tissue culture experiments with E363K, G426S and L505R showed that these mutant proteins preserve their ability to translocate to the nucleus (Buller et al., 2001) in a Six protein-dependent manner although E358K and L505R fail to bind directly to Six1 (Buller et al., 2001; Rayapureddi and Hegde, 2006). This suggests that an additional intermediate may be required for Six1-dependent transport of Eya1 to the nucleus. When assayed as GST (glutathione transferase) fusions E363K, G426S and L505R mutants have reduced phosphatase activity; however, when G426S was analysed in its non-fusion form it had wild-type phosphatase activity (Mutsuddi et al., 2005; Rayapureddi and Hegde, 2006). These findings indicate that phosphatase activity alone may not guarantee normal function or, alternatively, that factors that affect folding or association with other proteins (as may occur with the GST fusions) may mask phosphatase activity. Studies in Drosophila (Mutsuddi et al., 2005) concluded that E363K and G426S could induce ectopic eye structures but that E363K, G426S and L505R mutations were incapable of fully rescuing an eya-null mutation. However, all of these mutations compromised the transcriptional activation of a tested target gene.
We are particularly interested in how EYA1 mutations alter early ear development and use Xenopus embryos as a model. These embryos offer several advantages. The first is that mRNA encoding mutant forms of eya1 can be directly injected into embryos. mRNA injection endows embryos with the capacity to produce the mutant protein without having to create and maintain stocks of animals with the mutations that are being studied. Secondly, for bilateral structures like ears, one-sided injections are easily performed by injecting only one of the two blastomeres at the two-cell stage. This allows one side of the embryo to serve as a control, while the other side is used to examine changes in development. Finally, unlike mammals, fertilization and development are external in Xenopus, simplifying observation of the embryos.
As noted above, BOR is an autosomal dominant disorder. Haploinsufficiency is one of the mechanisms by which genetic mutations result in autosomal dominant disorders. The BOR/EYA1 pathway has been proposed to fall into this class (Zhang et al., 2004; Orten et al., 2008). Although it is clear that reducing the amount of EYA1 product leads to developmental defects (Schlosser et al., 2008; Zou et al., 2008), the possibility that these mutations also act in a dominant-negative fashion remains an open question. The experiments described here explore whether the BOR-causing mutations mediate their effect solely via haploinsufficiency or whether a dominant-negative effect is also possible. We also examine whether early, but developmentally transient, expression of aberrant eya1 protein irrevocably alters otic development. We report here that the presence of mRNA encoding mutant eya1 compromises the normal formation of the otic system by affecting the structure, innervation and gene expression.
Results
Expression of eya1 mutants after injection of mRNA into Xenopus embryos
Three of the four eya1 mutations used in the present study are associated with the BOR syndrome in humans. Figure 1 shows the alignment of human and the slightly smaller Xenopus protein in the region where the mutations occur. As described in the Materials and methods section, an 11-amino-acid T7 epitope was incorporated into the N-terminal region of eya1 proteins encoded by mRNAs used in the present study. We chose this epitope to distinguish the protein made by injected mRNA from that made by endogenous eya1 mRNA, and because the antibodies recognizing the T7 epitope have low background reactivity to proteins made during Xenopus embryogenesis. In Figure 2, we provide evidence that mRNA for all of the proteins being used in the present study were translated after injection into Xenopus embryos. Developmental staging (Nieuwkoop and Faber, 1967) and landmarks of otic development (Schlosser and Northcutt, 2000; Bever et al., 2003; Bane et al., 2005; Quick and Serrano, 2005) noted in this manuscript include stage 12 (mid-gastrula), stage 16 (mid-neural fold), stage 23 (the first slight depression of the otic placode) and stage 27 (otic vesicle closed). From stage 32 to stage 40, the ear grows, hair cells begin to develop and the medial and lateral aspects of the sensory regions of the ear become distinct. By stage 47 the membranous labyrinth of the ear is almost fully formed although the development of auditory components continues through metamorphosis (Fritzsch, 1988).
Figure 1. Comparison of the partial amino acid sequence of human and Xenopus eya1.
Amino acid substitutions associated with BOR and ocular defects are in larger boldface font. Italic letters indicate conserved motifs characteristic of HAD family proteins. (This Figure is modified from Rayapureddi and Hegde, 2006.) Mutation designations for human (hEYA1) as cited in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) are above the aligned sequences, and the corresponding numbering for Xenopus (Xeya1) is below them. OD, ocular defects.
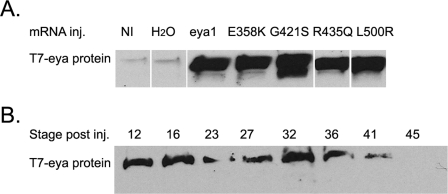
Figure 2. Expression of eya1 after injection of mRNA encoding eya1 proteins into Xenopus embryos.
Embryos were injected (inj.) with 250 ng of mRNA, and protein isolated from two embryos was loaded on to each lane. (A) A Western blot that shows that mRNAs encoding eya1 and four mutant variants produce detectable protein in stage 12 embryos. Anti-T7 antibody was used to detect proteins. (B) A Western blot that shows that eya1 protein made from injected mRNA persists through stage 41. Anti-T7 antibody was used to detect proteins. NI, non-injected control.
All of the proteins could be detected using an anti-eya1 antibody provided by Dr Gerhard Schlosser (Martin Ryan Institute, National University of Ireland, Galway, Ireland; results not shown); however, the production of protein from injected mRNA was more easily detected by reactivity to the T7 epitope. Proteins were readily detected by stage 12 (Figure 2A), approximately the time the endogenous eya1 gene is expressed (David et al., 2001). When we compared the levels of protein expressed in Figure 2(A), we detected a less than 2-fold variation in expression. We detected the expression of protein from injected mRNA for many hours. For example in Figure 2(B), eya1 protein in embryos injected with mRNA was detected through to stage 41. We do note that the mRNA injected in these assays is distributed throughout the embryo, whereas endogenous eya1 mRNA is spatially restricted (David et al., 2001).
mRNA encoding epitope-tagged wild-type eya1 produces functional protein
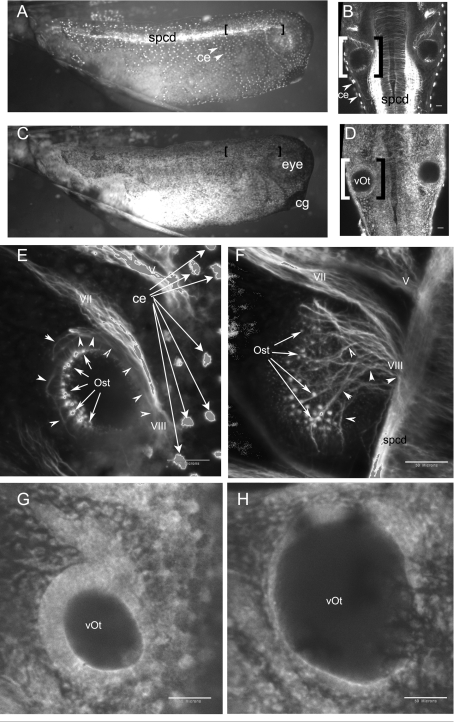
We next established that the epitope-tagged protein made by injected mRNA was capable of directing eya1-controlled processes. In the studies presented here, we routinely assayed otic development using embryos fixed for whole mount analysis at stages 32 and 40. For morphological assessment each embryo was exposed to antibodies to detect acetylated α-tubulin to mark sensory and neural tissue. All embryos were also treated with phalloidin to detect F-actin to provide a more global view of surrounding tissues. An example of the approach is seen in Figures 3(A)–3(E) and 3(G) (images from stage 32) and Figures 3(F) and 3(H) (the otic section of stage 40). Tubulin is readily seen (Figures 3A, 3B, 3E and 3F) in the spcd (spinal cord), developing nerves (Figures 3E and 3F, cranial nerves V, VII and VIII), sensory patches (Figures 3E and 3F, labelled Ost) and the neurites matching the sensory patches to the VIII cranial nerve (arrowheads in Figures 3E and 3F). Acetylated tubulin is also present in ciliated epidermal cells (ce) that are not relevant for our study on otic development. Bracketed areas in Figures 3(A) and 3(C) indicate the regions that were imaged for Figures 3(B) and 3(D). Bracketed regions in Figures 3(B) and 3(D) indicate the region enlarged in Figures 3(E) and 3(G). F-actin detection with phalloidin allows a strong contrast between the cells that form the otic vesicle and the hollow chamber within (marked vOt) in Figures 3(D), 3(G) and 3(H). Because otic sensory tissue is derived from otic epithelium any changes in development of the otic vesicle might compromise sensory tissue development. When examining treated embryos for developmental defects, we use images like those in Figure 3 to assess the size of the otic vesicle, the approximate number and distribution of sensory patches and the connections of the sensory patches to the VIII cranial nerve.
Figure 3. Detection of otic development in Xenopus embryos.
Otic development was monitored by fluorescence using whole mount immunohistochemistry to detect acetylated tubulin (A, B, E, F) and F-actin using phalloidin (C, D, G, H). (A–E, G) These Figures show progressively increased magnification of a stage 32 embryo, with bracketed regions indicating the region shown in the next magnification. (A, C) These Figures were visualized using a fluorescent microscope and are shown in lateral view with heads on the right. (B, D) These Figures were visualized through the dorsal–ventral plane using a confocal microscope with the anterior at the top of the images. (E–H) These Figures are confocal sections through the dorsal–lateral prospect. The anterior of the embryo is towards the top of each image. (F, H) These Figures are the otic region from a stage 40 embryo. The spcd, ciliated epithelial cells (ce), otic sensory tissue (Ost), neurites associated with otic sensory tissue (arrowheads) and cranial nerves (V, VII and VIII) are noted on panels detecting tubulin; the otic vesicle (vOt), eye and cement gland (cg) are noted on panels detecting F-actin. Scale bars in (B, D, E, F, G, H), 50 μm.
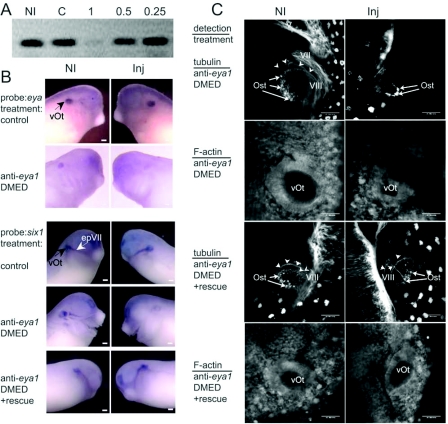
Functionality of T7-epitope-tagged eya1 proteins derived from injected mRNA was tested in trials that attempted to rescue embryos depleted of endogenous eya1 via antisense oligonucleotides. There are several possible approaches that use oligonucleotides to alter protein production of a specific transcript (Dagle and Weeks, 2001). Schlosser et al. (2008) used morpholino antisense oligonucleotides to investigate eya1 loss. Most commonly, morpholinos work by blocking translation, so evidence of their action must be determined by analysis of protein loss. In our alternative approach, we used oligonucleotides that have blocks of DMED (dimethylethylenediamine)-modified internucleoside linkages near their 3′- and 5′-ends that stabilize the oligonucleotide and also have a series of at least five phosphodiester linkages to direct targeted RNaseH-mediated degradation of endogenous mRNA (Dagle et al., 2000). For the eya1 mRNA we found that 1 ng of a DMED anti-eya1 oligonucleotide reduced the endogenous level of mRNA by over 90% (Figure 4A), while a control oligonucleotide leaves the mRNA levels unaltered. We next injected 1 ng of oligonucleotide into one blastomere of a two-cell embryo. In Figure 4(B), loss of eya1 mRNA was shown by in situ hybridization where the antisense oligonucleotide injected side of the embryo has dramatically less signal. Consistent with the results of Schlosser et al. (2008) in Xenopus and Zou et al. (2004, 2008) in mouse we noted that loss of eya1 mRNA alters the expression of other otic genes. For example, in Figure 3(B), the side of the embryo lacking eya1 mRNA also has reduced six1 mRNA levels. Significant for the present study, the expression of six1 is restored by the injection of oligonucleotide-resistant T7-tagged eya1 mRNA. Rescue mRNA resistance results from replacement of several of the nucleotides that make the endogenous mRNA complementary to the oligonucleotide with sequence encoding the epitope tag. The reduction of eya1 mRNA compromises the formation of the otocyst as shown in the confocal sections of a stage 32 embryo (Figure 3C). These sections show the anterior portion of the otocyst near the top of the image and cut through the embryo in the dorsal–lateral plane. The side of the embryo that has reduced levels of eya1 mRNA had a much smaller region of otic sensory tissue, no visible neurite outgrowth (arrowheads in Figure 4) from the sensory tissue and no visible establishment of the VIII cranial nerve. Phalloidin staining allows the identification of a very small otocyst (vOt) compared with the control non-injected side of the embryo. As in our examination of six1 expression, normal otocyst development was rescued using wild-type T7-tagged eya1 mRNA. Thus the epitope-tagged proteins used in the present study are expected to accurately reflect eya1 protein activity.
Figure 4. Oligonucleotide-mediated loss of eya1 and rescue by injection of eya1 mRNA.
(A) DMED-modified oligonucleotides complementary to Xenopus eya1 reduce the levels of eya1 mRNA. Eya1 mRNA was assayed from non-injected embryos (NI), embryos injected with 1 ng of a control oligonucleotide (C) or 1, 0.5 or 0.25 ng of anti-eya1 oligonucleotide. (B) Injection of 1 ng of anti-eya1 oligonucleotide into one side of the embryo reduces the level of endogenous eya1 and six1 mRNAs shown by in situ hybridization. (C) Reduction of eya1 correlates with inability to form a proper otocyst, as assayed by whole mount immunohistochemical detection of tubulin and F-actin by confocal microscopy along the dorsal–lateral plane. Both six1 expression and normal otocyst formation are rescued by co-injection of oligonucleotide with 250 pg of rescue mRNA encoding T7–eya1 protein. The otic vesicle (vOt) and facial epibranchial placode (epVII) are identified in (B). In (C), otic sensory tissue (Ost), cranial nerves (V, VII and VIII) and neurites leading to the VIII nerve (arrowheads) are labelled. Scale bars on in situ hybridization images, 200 μm; scale bars on confocal images, 50 μm. Inj. injected.
Developmental consequences of injection of mRNA encoding eya1 proteins
Our analysis of the consequences of injecting mRNA encoding different eya1 proteins assessed toxicity and general effects on normal development. Injection of mRNA into one or two cell embryos can cause unintended consequences due to ectopic or overexpression of protein. In Table 1 the survival of non-injected, GFP (green fluorescent protein) mRNA (used as a reporter of the distribution on injected material) or embryos injected with eya1/GFP mix was tallied. From fertilization through to stage 32, embryos injected with GFP or eya1/GFP mRNA had similar survival rates. After stage 32, embryos injected with mutant forms of eya1 exhibited lower survival rates and had a number of morphological defects. Commonly seen defects included minor delays in reaching stage appropriate landmarks compared with controls (delayed development), shortened body axis and abnormal dorsal bending. Using stage 32 embryos to assess treatment groups, we made several comparative measurements to give a quantitative summary of the defects. Table 2 shows the results of two separate trials where ten embryos in each treatment group were measured for overall length, and for three measurements that focused on the embryonic head, looking at the distance between the cement gland and the middle of the eye, the cement gland and the ear and the diameter of the eye.
Table 1. Survival of Xenopus embryos after injection treatment.
Embryos [except non-injected controls (NI)] were injected with a mix of indicated eya1 mRNA and GFP mRNAs. Four different trials were included in the analysis. All survival analyses started with the indicated number of embryos at stage 10 (early gastrula stage).
| Survival (%) | |||||||
|---|---|---|---|---|---|---|---|
| Stage | NI | eya1 | E358K | G421S | L500R | R435Q | GFP |
| 32 | 97 | 77 | 84 | 82 | 79 | 74 | 73 |
| 40 | 96 | 65 | 60 | 50 | 54 | 49 | 65 |
| 47 | 96 | 62 | 53 | 38 | 40 | 35 | 62 |
| Embryos analysed (n) | 210 | 276 | 250 | 251 | 287 | 281 | 273 |
Table 2. Changes in body dimensions correlated with injection of eya1 mRNAs.
Embryos from two different matings were assayed at approximately stage 32 of development. Ten embryos in each treatment group were measured. *Average measurements with P<0.05. CG, cement gland.
| Treatment | Body length (mm) (S.D.) | CG to eye (mm) (S.D.) | CG to ear (mm) (S.D.) | Eye diameter (mm) (S.D.) |
|---|---|---|---|---|
| Non-injected no. 1 | 5.50 (0.22) | 0.76 (0.06) | 1.27 (0.08) | 0.45 (0.06) |
| eya1 no. 1 | 5.59 (0.30) | 0.86 (0.08) | 1.25 (0.09) | 0.43 (0.04) |
| E358K no. 1 | 5.39 (0.32) | 0.78 (0.10) | 1.26 (0.11) | 0.41 (0.05) |
| G421S no. 1 | 4.58* (0.93) | 0.73 (0.11) | 1.15* (0.10) | 0.36* (0.03) |
| R435Q no. 1 | 4.68* (0.58) | 0.76 (0.07) | 1.18* (0.09) | 0.41 (0.05) |
| L500R no. 1 | 4.33* (1.10) | 0.77 (0.09) | 1.17* (0.05) | 0.36* (0.05) |
| Non-injected no. 2 | 5.43 (0.34) | 0.72 (0.09) | 1.17 (0.11) | 0.42 (0.03) |
| eya1 no. 2 | 5.63 (0.29) | 0.83* (0.05) | 1.29* (0.12) | 0.42 (0.04) |
| E358K no. 2 | 5.28 (0.49) | 0.79 (0.07) | 1.21 (0.08) | 0.39* (0.05) |
| G421S no. 2 | 4.51* (0.83) | 0.78 (0.09) | 1.22 (0.13) | 0.38* (0.04) |
| R435Q no. 2 | 4.49* (1.01) | 0.78 (0.07) | 1.22 (0.13) | 0.40 (0.06) |
| L500R no. 2 | 4.36* (1.08) | 0.71 (0.08) | 1.20 (0.10) | 0.38 (0.06) |
These values were analysed using the Student's t test comparing non-injected embryos within each treatment group. The analysis supported the visual conclusion that three of the mutants, G421S, R435Q and L500R, stunted overall growth, while wild-type eya1 and E358K did not. Not obvious on visual inspection, but statistically significant in both trials, reduced eye diameter was seen with the G421S mutation. In one of the two trials, we also noted reduced eye diameter in groups injected with the E358K and L500R mutants. Although E358K and G421S are associated with optical defects in humans, L500R is not, so the relevance of this minor alteration in eye size is unclear. In one trial there was a slight but statistically significant increase in the size of embryonic heads (as judged from the distance from the cement gland to the eye or ear) in embryos injected with wild-type eya1 mRNA.
Embryos expressing eya1 mutations have abnormal otic development
To analyse the influence of mutant forms of eya1 on ear development, we injected one blastomere of two-cell embryos with mRNA encoding eya1.
Ear development was monitored by whole mount immunohistochemistry of stage 32 (Figure 5) and 40 embryos (Figure 6) followed by confocal microscopy. By stage 32 it is relatively easy to identify the otocyst as well as the early establishment of sensory hair cells and innervation (Figure 5; see Supplementary Figures S1 and S2 at http://www.biolcell.org/boc/102/boc1020277add.htm).
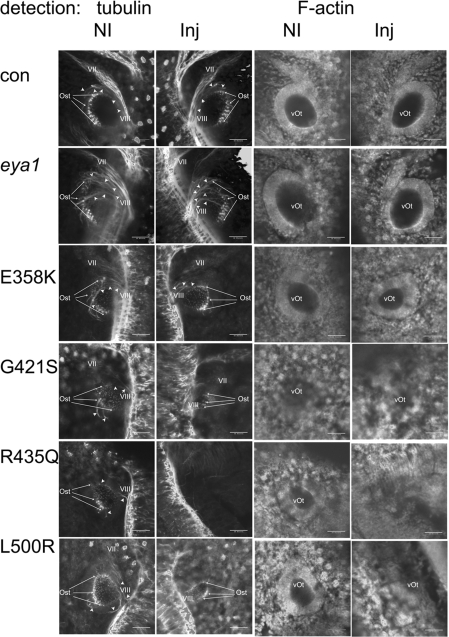
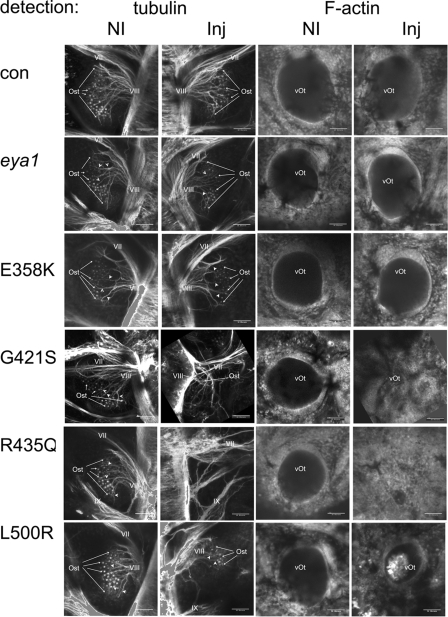
Figure 5. Stage 32 otic development is disrupted by eya1 mutants linked to BOR.
Stage 32 embryos were imaged using a confocal microscope from the dorsal–lateral prospect. The anterior of the embryo is towards the top of each image. Dual panels on the left show tubulin distribution; dual panels on the right detect F-actin. In each dual set, the non-injected side (NI) of the embryo is on the left and the injected side (Inj) is on the right. Material injected into the right side of the embryo is indicated for each row of panels. In panels detecting tubulin, the cranial nerves (VII and VIII) are labelled when visible. The label for VIII is placed as close to the attachment to the spcd as possible. Otic sensory tissue (Ost) is pointed out using arrows that help indicate the most anterior and posterior extents of this tissue type. Arrowheads indicate neurites that are associated with sensory tissue and VIII. The otic vesicle (vOt) is labelled where visible on panels showing F-actin. Scale bars, 50 μm. (Note: the right panel for non-injected controls are also shown enlarged in Figures 3E and 3G.) con, control.
Figure 6. Stage 40 otic development is disrupted by eya1 mutants linked to BOR.
Stage 40 embryos were imaged using a confocal microscope from the dorsal–lateral prospect. The anterior of the embryo is towards the top of each image. Dual panels on the left show tubulin distribution; dual panels on the right show F-actin. In each dual set, the non-injected side (NI) of the embryo is on the left and the injected side (Inj) is on the right. Material injected into the right side of the embryo is indicated for each row of panels. In panels detecting tubulin the cranial nerves (VII, VIII and IX) are labelled when visible. The label for VIII is placed as close to the attachment to the spcd as possible. Otic sensory tissue (Ost) is pointed out using arrows that help indicate the most anterior and posterior extent of this tissue type and arrowheads show examples of neurites connecting to the VIII nerve. The otic vesicle (vOt) is labelled where visible on panels showing F-actin. Scale bars, 50 μm. (Note: the right panel for non-injected controls are also shown enlarged in Figures 3F and 3H). con, control.
In Figures 5 and 6, paired images of treated embryos show the non-injected side on the left and the injected side on the right viewed along the dorsal–lateral plane of the embryo. When embryos were analysed using phalloidin to detect F-actin, the size and the integrity of the otic vesicle could be assessed. Expression of eya1 or the E358K mutant did not alter the size of the otic vesicle. However, overexpression of the three mutations linked to BOR resulted in reduced otic vesicle size and disordered tissue surrounding the vesicle (Figure 5, G421S and L500R; and Figure 6, L500R). In some cases (Figure 5, R435Q, and Figure 6, G421S and R435Q), no otic vesicle could be detected. Direct measurements were made on 20 embryos from each treatment group. As shown in Table 3, the defects were not completely penetrant; approx. 50% of embryos injected with BOR-associated mutations had otocysts at least 10% smaller than controls. In Table 3, changes in otocyst size were scored compared with controls as normal (less than 10% different), mild (10–30% smaller), moderate (30–50% smaller) and severe (>50% smaller). Notably, the largest group of the affected embryos had otocysts less than half the size of controls.
Table 3. Changes in otocyst dimensions correlated with injection of eya1 mRNAs.
n=20. *P<0.05.
| Left or non-injected | Right or injected | Change (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Treatment | Average otocyst size (μm2) | S.D. | Average otocyst size (μm2) | S.D. | Normal | Mild | Medium | Severe |
| Non-injected | 6836 | 992 | 6880 | 1247 | 100 | 0 | 0 | 0 |
| eya1 | 6510 | 2714 | 6576 | 2713 | 95 | 5 | 0 | 0 |
| E358K | 6020 | 1051 | 5961 | 1016 | 95 | 5 | 0 | 0 |
| G421S | 5353 | 844 | 4037* | 2189 | 55 | 10 | 10 | 15 |
| R435Q | 6411 | 1922 | 4038* | 2820 | 55 | 5 | 5 | 35 |
| L500R | 6053 | 2193 | 4145* | 2493 | 50 | 10 | 10 | 30 |
The effect of the eya1 mutations on the development of the sensory system was examined by detection of acetylated-α-tubulin staining. The vestibulocochlear nerve (VIII cranial nerve) could be identified in most cases, although some examples of embryos treated with R435Q (Figures 5 and 6) and G421S and L500R (results not shown) failed to form a detectable VIII cranial nerve. The developing sensorial regions of the otocyst appear as punctate staining within the otocyst and neurite projections from sensory regions were detected (arrowheads in Figures 5 and 6). For the three BOR mutations, there was less otic sensorial tissue formation (Figures 5 and 6 and Supplementary Figure S2) and some embryos (R345Q, Figures 5 and 6) had no discernible otic sensory tissue. The reduction of sensorial tissue always correlated with a reduction in the size of the otic vesicle. Developmental defects in embryos expressing BOR mutant protein persist, as can be seen in Figure 6 where stage 40 embryos were assayed. Thus the defects observed at stage 32 were not simply due to delayed development since they were not corrected when the nervous system was assayed at stage 40.
We noted a tendency for injection of any of the eya1 mutants to slow otic development when treated embryos were compared with non-treated siblings at stage 32 (Figure 5). For example, comparing the non-injected sides of G421S, R435Q or L500R with control embryos, the sensory patches are smaller and neurite outgrowth is less obvious. However, for each of these mutants, the injected side of the embryo was significantly more altered than the non-injected side. The observed bilateral effect may be due to diffusion of some of the injected mRNA before full membrane closure at the two-cell stage to the non-injected side. Alternatively, a few cells may have migrated across axial boundaries during development (Wetts and Fraser, 1989).
Expression of eya1 mutant protein reduces the normal expression of developmentally regulated genes
As presented above, the expression of BOR-related mutations of eya1 leads to developmental defects. We next examined whether the expression of mutant proteins was reflected by changes in gene expression. Based on morphological observations we concentrated on the three mutations linked to BOR, G421S, R435Q and L500R. First, mRNAs expressing the eya1 proteins were injected into one-cell embryos. In these assays embryos were harvested at stage 25, a time in development after the otic placode has formed but before the vesicle completely closes at stage 27. By stage 25, under normal conditions, the trigeminal ganglia have already formed and the neurotrophic genes nt3 and bdnf and the neuronal determination and differentiation genes ngnr-1 and neuroD (Schlosser and Northcutt, 2000) are all being expressed in the developing ear. Tbx1 expression is already active in the embryo; however, expression in the ear does not occur until stage 28 (Ataliotis et al., 2005). Although these genes are not limited to the ear, their regulated expression is critical for normal otic development. We isolated RNA from the heads of stage 25 embryos and assayed the levels of endogenous eya1 (endoeya1 in Table 4) expression and levels of the other mRNAs (Table 4). None of the mRNAs injected significantly altered the expression of endogenous eya1 consistent with the conclusion that eya1 is not autoregulated. Injection of mRNA encoding wild-type eya1 leads to increased levels of all of the genes tested except tbx1. Conversely, expression of any of the mutants leads to a decrease in levels of the downstream genes tested except tbx1.
Table 4. QPCR analysis of selected genes in embryos injected with mRNA expressing eya1 mutant protein.
Data shown were collected from three different trials, each performed in triplicate. *P<0.01.
| mRNA level in treated embryos/mRNA level in control embryos | ||||||
|---|---|---|---|---|---|---|
| Treatment | Endo eya1 | NT3 | NeuroD | Ngn1 | Tbx1 | BDNF |
| eya1 | 1±0.3 | 2.8±0.6* | 3.7±0.9* | 3.2±0.5* | 1±0.2 | 2.2±0.2* |
| G421S | 1.2±0.4 | 0.4±0.1* | 0.3±0.05* | 0.3±0.08* | 0.9±0.2 | 0.2±0.07* |
| R435Q | 1.3±0.4 | 0.3±0.07* | 0.3±0.1* | 0.4±0.02* | 0.9±0.1 | 0.3±0.2* |
| L500R | 0.9±0.3 | 0.3±0.1* | 0.4±0.02* | 0.4±0.03* | 0.8±0.2 | 0.4±0.09* |
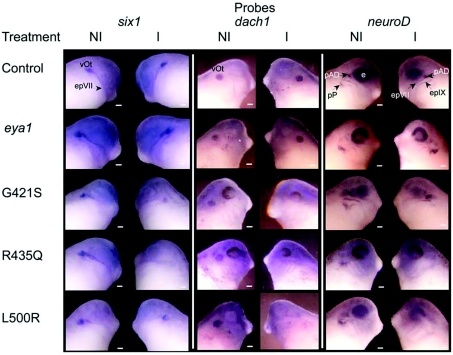
We confirmed the spatial effect on the expression of key otic genes by in situ hybridization. Eya1 protein works in complex with six1 and dach. For in situ analysis we returned to a protocol where we injected only one cell of two cell embryos, allowing the uninjected side to serve as the control. At least 30 embryos in each treatment group were assayed. Reduced expression like that seen in Figure 7 was noted in 14/30 G421S-, 15/40 R435Q- and 15/36 L500R-treated embryos. A similar analysis was performed on transcripts shown to have reduced expression by QPCR (quantitative PCR). NeuroD expression is shown in Figure 7 as representative of those trials. In situ analysis shows that levels of neuroD transcripts were reduced in the region corresponding to the developing ear. These results are consistent with the failure to properly regulate a normal programme for otic development when mutant eya1 protein is present.
Figure 7. Expression of eya1 mutations that cause BOR in humans reduces otic expression of six1, dach and neuroD.
One cell of a two-cell stage embryo was injected with mRNA encoding Eya1 protein. Stage 28 embryos were assayed for otic gene expression by in situ hybridization. In each pair of panels, the non-injected side (NI) of the embryo is shown on the left and the side injected with mRNA (I) is on the right. The otic vesicle (vOt), facial epibranchial placode (epVII and epIX), glossopharyngeal epibranchial placode, anterior lateral line placode (pAD), posterior lateral line placode (pP) and the eye (e) are noted. Scale bars, 200 μm.
Discussion
Examining the early developmental consequences of genes that lead to congenital defects can be very difficult in mammalian systems. For processes where similar developmental patterning occurs in other organisms, a great deal can be learned by taking advantage of the experimental accessibility of those organisms. In these studies we have used the developing embryos of the frog X. laevis to examine the effects of mutations in the EYA1 gene on early development of the ear.
The work presented here took advantage of the ease with which oligonucleotides and mRNA encoding eya1 or its mutant forms could be introduced into early embryos. Others (Zhang et al., 2004; Schlosser et al., 2008; Zou et al., 2004, 2008) have reported previously that loss or reduction of Eya1 protein alters ear development. Consistent with their studies, we found that oligonucleotide-directed reduction of eya1 protein expression inhibits the formation of the otic placode. We further showed that defects resulting from oligonucleotide-mediated reduction of eya1 mRNA were rescued by injection of exogenous eya1 encoding mRNA. Previous studies and ours support the conclusion that reduced level of EYA1 protein (i.e. haploinsufficiency) results in developmental abnormalities consistent with BOR.
The reduction trials suggest that normal levels of eya1 protein are necessary, but are they sufficient to ensure normal otic development when mutant eya1 protein is added to the mix? We supplemented the normal pool of eya1 mRNA with either wild-type or mutant encoding mRNA to look for dominant-negative effects. Expression of wild-type eya1 protein and its expression outside its usual expression domains had few overt consequences. These results are consistent with the requirement of additional tissue-specific proteins that work with eya1 to execute its function. Schlosser et al. (2008) reported that overexpression of eya1 protein in early Xenopus embryos can block the expression of differentiation genes. Close examination of the embryos we injected with mRNA encoding wild-type eya1 protein detected only minor morphological differences between these embryos and non-injected controls. Although it is clear from our analysis that more eya1 protein was produced from injected mRNA than was endogenously generated, the eya1 protein from the injected mRNA was spread across many tissues. In our studies we injected 250 pg of mRNA into either whole embryos or into one cell of a two-cell embryo. If we estimate the volume of a one-cell embryo to be 1 μl, the highest concentration of injected mRNA in our studies would be 500 pg/μl. Schlosser et al. (2008) reported injecting 500–1000 pg into single blastomeres at the 2–16-cell stage. Their approach would yield blastomeres with between 2 and 64 times more eya1 mRNA than in our studies. Thus it may not be surprising that the expression we generated may have been below the threshold needed for the overexpression phenotypes noted by Schlosser et al. (2008).
How did the expression of different mutant forms of eya1 affect development? The mildest morphological effects were noted with the E358K mutation where only subtle, sporadic changes in eye size and slight developmental delays were observed. The lack of a clear phenotype from the E358K mutation is interesting, as in vitro studies have shown that the E358K mutation compromised phosphatase activity (Mutsuddi et al., 2005), failed to interact with Six1 in co-immunoprecipitation assays (Buller et al., 2001) but was capable of a partial, although incomplete, rescue of an eya-null fly (Mutsuddi et al., 2005). As in humans, our studies did not detect otic defects with E358K. The other three mutations that were tested are all associated with deafness and lead to more recognizable defects. Otic development and overall survival was compromised. We noted some general defects including stunted growth and slowed development that have been seen by others who have injected a variety of mRNAs into Xenopus embryos. These defects may be due to the toxicity related to the amount of mRNA injected or ectopic expression of protein. However, we observed fewer defects when injecting mRNA for GFP, wild-type eya1 or the E358K variant than when injecting mRNA for the BOR-associated mutations. The general developmental disruptions may also have been caused by alteration of eya1 regulation of muscle development along the dorsal axis consistent with the roles seen by others for Eya1 and its transcriptional partner Six (Fougerousse et al., 2002; Grifone et al., 2004, 2007).
The consequences of the BOR-associated mutations were readily apparent. In the most obvious cases the embryos fail to form an otocyst when mRNA-producing mutant protein was injected. As has been shown in other studies on Eya1 (Furuya et al., 2005; Schlosser et al., 2008), the effects were not completely penetrant; however, the expression of these mutations can have dramatic consequences. Notably, these effects occur even when embryos have normal levels of endogenous eya1 mRNA that can be translated into protein. These findings indicate that normal levels of wild-type eya1 are not sufficient to ensure normal otic development when these BOR-associated proteins are also present and that the BOR mutations act in a dominant-negative fashion.
Because eya1 acts with six1 and dach to regulate transcription, we also examined the effect of expression of each of the mutations on a panel of genes that have been identified as being dependent on or downstream targets of Eya1 (Zou et al., 2004; Friedman et al., 2005), including nt3, neuroD, ngnr-1, bdnf and tbx1. Using QPCR, we analysed the levels of mRNA that corresponds to these factors. We showed that overexpression of wild-type eya1 in embryos leads to up-regulation of nt3, neuroD, ngnr-1 and bdnf although morphological changes were not evident in the assays we used. It is possible that a more exhaustive analysis of hair cell sensory neurons in later stages would reveal such changes. As indicated earlier, our results are not identical with the overexpression results reported by Schlosser et al. (2008) who found that overexpression of eya1 repressed the expression of genes required for neurogenesis and neural differentiation when activated slightly later in development. With regard to their studies and to studies on dosage effects of Eya1 during mouse auditory development (Zou et al., 2008), perhaps the absolute amount, the transient nature of expression of injected mRNAs or the different stages of development examined explain some of the differences. Interestingly, the endogenous expression of eya1 was not changed and tbx1 expression was also unaffected. Tbx1 expression in the ear normally begins later than the stage we tested. Tbx1 has been reported to be activated in response to Eya1 (Friedman et al., 2005); thus the supplemented level of eya1 that increased the expression of some genes (nt3, neuroD, ngnr-1 and bdnf) was not sufficient to precociously activate tbx1.
All of the BOR mutant proteins resulted in lower expression of the neurotrophins and neural differentiation genes tested. However, the BOR mutant proteins also lowered levels six1 and dach mRNA in the otic region, indicating that the expression of other early regulators of the developmental program of the ear were also altered. The reduced expression of six1 protein may make a significant contribution to the disruption of downstream gene activation. In addition, although direct binding of eya1 and six1 may not be needed to translocate eya1 into the nucleus (Buller et al., 2001), in the absence of six protein, eya1 tends to remain in the cytoplasm (Ohto et al., 1999). Therefore one consequence of lower levels of six1 protein may be to reduce the total amount of eya1 protein that translocates into the nucleus.
The morphological consequences seen using Xenopus provide the impetus to refine these studies using some of the efficient methods recently developed to generate transgenic embryos to more closely mimic the normal levels and distribution of eya1 gene expression (Allen and Weeks, 2005; Ogino et al., 2006; Pan et al., 2006) as a rapid screen for the early developmental consequences of EYA1 mutations found in humans.
Finally, we conclude that therapies to correct EYA1 mutations are unlikely to be effective by simply restoring levels of functional EYA1 protein to normal levels. It will be critical to also eliminate the expression of the mutant form of the protein.
Materials and methods
Animals
X. laevis were purchased from Xenopus I (Ann Arbor, MI, U.S.A.). Embryos were obtained after hCG (human chorionic gonadotropin)-induced egg laying and in vitro fertilization using standard techniques and staged by the method of Nieuwkoop and Faber (1967). All animal protocols were reviewed and approved by the University of Iowa animal care and use committee.
Oligonucleotide-mediated degradation of Eya1
Modified oligonucleotides were synthesized using H-phosphonate chemistry on an Expedite™ synthesizer (Applied Biosystems, Foster City, CA, U.S.A.) (Dagle and Weeks, 2000; Dagle et al., 2000). All reagents used for automated DNA synthesis were obtained from Glen Research (Sterling, VA, U.S.A.). To generate unmodified phosphodiester bonds, hydrogen phosphonate diesters were oxidized for 4 min with freshly prepared 5% iodine in THF (tetrahydrofuran)/pyridine/water (15:2:2) and then for 3 min with the same solution diluted 1:1 with 8% triethylamine in THF/water (43:3). Oxidative amidation of hydrogen phosphonate diesters was performed manually by using a 10% solution of DMED (Aldrich, Milwaukee, WI, U.S.A.) in anhydrous CCl4. Further processing and purification of oligonucleotides using reverse-phase HPLC and gradient elution with acetylnitrile as the mobile phase were performed as previously described (Dagle and Weeks, 2000; Dagle et al., 2000). After Sephadex G-25 column chromatography, oligonucleotides were dissolved in sterile water and quantified by UV spectroscopy. The sequence of the oligonucleotide used to degrade endogenous eya1 mRNA was A+T+T+T+C C A T A G A C C+T+A+G+A with the sequence complementary to the 17 nucleotides surrounding the AUG (the complement of AUG is the boldface CAT of the oligonucleotide) of the eya1 mRNA of the GenBank® Nucleotide Sequence Database depositions (AF352029.1 and AF352028.1 for Xenopus eya1). Frogs used for antisense oligonucleotide trials were screened to ensure that there were no polymorphisms in the targeted area by isolating genomic DNA by buccal swab followed by PCR amplification and sequencing of a genomic fragment starting 109 nt 5′ and ending 230 nt 3′ of the target sequence. Oligonucleotide effectiveness and dose response was tested using RNA isolated from injected embryos as a substrate for RT–PCR (reverse transcription–PCR). Rescue of oligonucleotide inhibition was accomplished using the T7-epitope-tagged eya1 mRNA described below. The T7 tag interrupts the homology sufficiently to render the rescue mRNA refractory to oligonucleotide-mediated degradation.
Plasmid construction
Full-length cDNAs corresponding to Xenopus eya1 were generated using stage 47 embryonic RNA. Eya1 cDNA was prepared by PCR amplification and subcloned into the plasmid pBGFP/RN3P (Zernicka-Goetz et al., 1996), replacing the GFP gene in the plasmid with eya1 at the EcoRI site. The resulting plasmid was named pBEya1FLexp. Four eya1 mutations, E358K, G421S, L500R and R435Q, were generated by PCR-based site-directed mutagenesis. Amplified fragments were subcloned into pBEya1FLexp by replacement of the following restriction fragments. E358K fragment was subcloned between the PpuMI and NotI sites of pBEya1FLexp, G421S and R435Q were subcloned between two BglII sites, and R435Q was subcloned between HpaI and NotI sites. All eya1 encoding constructs used in the present study also contain an in frame T7 epitope at the N-terminus of the protein. pBEya1FLexp and its derivative constructs allow in vitro expression of eya1 mRNA using T3 polymerase. All constructs were confirmed by sequencing and by in vitro transcription/translation using a coupled transcription translation system (the TNT system and restriction enzymes were purchased from Promega, Madison, WI, U.S.A.) followed by Western-blot analysis.
Embryo culture and microinjection
Capped mRNA of full-length eya1 or eya1 mutations were generated in vitro using the mMessage Machine T3 transcription kit (Ambion). All eya1 encoding plasmids were linearized with SfiI for in vitro transcription. mRNA was injected into a one-cell embryo or one blastomere of two-cell embryos. A 10 nl portion of the solution containing 25 pg/nl of mRNA was injected in each embryo. GFP mRNA (100 pg/nl) was co-injected as a tracer in experiments where one blastomere of a two-cell embryo was injected. Embryos were grown to the desired stage at 18°C. Embryos co-injected with GFP mRNA were pre-sorted according to whether GFP expression was restricted to the left or right side.
Whole mount immunohistochemistry
Detailed methods can be found in Kolker et al. (2000) and Bane et al. (2005) but, briefly, Xenopus embryos were fixed in Dent's fixative [80% (v/v) methanol and (v/v) 20% DMSO] and stored at –20°C until processed. Rehydrated specimens were rinsed in PBS-DT (PBS, 1% Tween 20, 1% DMSO and 0.02% NaN3), and blocked with PBS-DT with 0.1 M glycine, 2% (w/v) non-fat dried skimmed milk powder and 1% BSA for 4 h at room temperature (22°C). The primary antibody, anti-acetylated α-tubulin (6-11B-1; Sigma, St. Louis, MO, U.S.A.), was diluted 1:50 with the block solution. Incubation with the primary antibody was carried out overnight at 4°C. Samples were rinsed ten times over 8 h with PBS-DT. The secondary antibody, anti-mouse IgG2b (Sigma), was diluted 1:200 in block solution and allowed to incubate overnight at 4°C. Samples were rinsed as described for primary antibody treatment and subsequently dehydrated in an ethanol series. For double labelling with phalloidin, embryos were rinsed in methanol, and incubated for 5 h in 3.3 μM phalloidin–FITC (Sigma) diluted in methanol. Embryos were then rinsed twice in methanol and stored overnight at –20°C. Whole mount embryos were viewed on a Bio-Rad MRC-1024 confocal microscope (Bio-Rad, Hercules, CA, U.S.A.) equipped with a krypton/argon laser. Confocal Assistant 4.02, ImageJ and Adobe Photoshop 7.0 were used for image processing.
In situ hybridization
In situ hybridization was carried out essentially as described by Harland (1991). Embryos were fixed at the appropriate stage of development using MEMFA (0.1 M Mops, 2 mM EGTA, 1 mM MgSO4 and 0.4% paraformaldehyde) and rehydrated in TTW (200 mM NaCl, 50 mM Tris, pH 7.4, and 0.1% Tween 20). DIG (digoxigenin)-labelled probes for eya1, six1, dach and neuroD were generated from plasmids containing cDNAs of these transcripts using an Ambion Megascript kit with DIG RNA labelling mix. Probes were hybridized with fixed embryos using hybridization buffer [50% formamide, 5 SSC (1× SSC is 0.15 M NaCl/0.015 M sodium citrate), 1 mg/ml total yeast RNA, 1× Denhardt's, 0.1% Tween 20 and 5 mM EDTA] and, after extensive washing in 0.2 × SSC, were incubated with alkaline phosphatase-conjugated anti-DIG antibody (1:5000; Roche) in MAB [100 mM maleic acid, 150 mM NaCl, pH 7.5, and 0.02 g/ml BMBR (Boehringer Mannheim) blocking reagent (Roche, Mannheim, Germany)] overnight at 4°C. After extensive washing at room temperature with 100 mM Tris (pH 9.5), 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20 and 1 mM levamisol to remove unbound antibody, colorimetric detection of hybrids was accomplished using 0.3 mg/ml of Nitro Blue Tetrazolium and 0.2 mg/ml of BCIP (5-bromo-4-chloroindol-3-yl phosphate) in the washing solution. Colour reactions were stopped using 100% methanol.
Western blotting
Protein extracts were made by homogenization of fresh Xenopus embryos using 20 μl of homogenization buffer (0.1 M NaCl, 1% Triton X-100, 1 mM PMSF and 20 mM Tris/HCl, pH 7.6) per embryo. Protein extracts were mixed with bromoacetate (final concentration 0.067 M) prior to separation on SDS gels. Protein extracted from two embryos was loaded on to each well of a 4–20% precast gel (Bio-Rad). Protein was transferred from gels to nitrocellulose membranes using semidry transfer (Owl, Cambridge, MA, U.S.A.). Membranes were blocked using 10% non-fat dried skimmed milk powder in TBS (20 mM Tris, pH 7.6, and 0.137 M NaCl) for at least 4 h at room temperature. T7. Tag antibody (1:5000; Novagen, EMD Chemicals, Gibbstown, NJ, U.S.A.) or eya1 antibody (1:2000, guinea-pig anti-Xenopus eya1, a gift from Dr Gerhard Schlosser) were used as primary antibodies. Anti-mouse IgG1 HRP (horseradish peroxidase; 1:2000, Sigma) was used as a secondary antibody for T7.Tag antibody and anti-guinea-pig IgG HRP (1:2000; Sigma) for eya1 antibody. The presence of eya1 protein was detected using an ECL (enhanced chemiluminescence; SuperSignal West Pico Chemiluminescent Substrate; Pierce, Rockford, IL, U.S.A.) reaction. When (in Figure 2) Adobe Photoshop was used to reorder lanes from a single gel after digital capture, the reordered lanes were separated by a clear break.
Statistical analysis
Treated embryos were measured for body length, distance between the cement gland and the eye, the cement gland and the ear and the diameter of the eye. Values for each treatment group were compared with the non-injected controls using the Student's t test (two-tailed, assuming two-sample equal variance). Confocal analysis through the dorsal–ventral axis identified and measured the widest diameter of the otocyst. Average otocyst size for each treatment group was used to calculate significant variation using a paired, two-tailed Student's t test. Single embryo analysis separated BOR-causing mutations into four groups: non-affected (less than 10% difference), mild (10–30% difference in otocyst size), moderate (30–50% difference in otocyst size) and severe (>50% difference in otocyst size).
Quantitative RT–PCR
RNA isolated from the heads of stage 25 embryos was used for quantitative RT–PCR analysis. Embryonic heads were separated from the rest of the embryo by cutting just posterior from the otic vesicle and ventrally in front of the developing gut. Fresh tissue was placed in RNAlater (Ambion, Austin, TX, U.S.A.) and stored overnight at 4°C. Total RNA was isolated by using an RNAqueous isolation kit (Ambion). The kit incorporates a DNase step to ensure that isolated RNA is DNA-free. Approx. 300 ng of total RNA was isolated per head. We used 1 μg of total RNA to make cDNA in a total reaction volume of 20 μl. The cDNA reaction was diluted 5-fold to 100 μl. A 5 μl portion of diluted cDNA was assayed in triplicate for each QPCR. Sequences selected for analysis by QPCR were: tbx1, bdnf, nt3, neuroD and ngnr-1. Primers for real-time PCR were designed using Primer 3 software. Amplified cDNA fragment sizes ranged between 50 and 250 bp and were tested prior to QPCR analysis to ensure that they generated a single amplification product. Sequences for primers were identified from GenBank® at NCBI (National Center for Biotechnology Information). Primers used were: endogenous eya1: forward: cgatggctggagggttat, reverse: gtctactatgtgggctggcta; total eya1: forward: cctaccccttcttcccaaac, reverse: atccaccttccgtcttgatg; nt3: forward: aagcaacggcgatacaaatc, reverse: tacaagggaggaggttccaa; tbx1: forward: gctaaaggggctcagtggat, reverse: tggagttcaggatgatgtgg; ngnr-1: forward: gctgctgaagtgcgaatacc, reverse: gtgaagaggaggaggacacg; neuroD: forward: atgaaggggatgaggaggag, reverse: cttttgggtttctggtcgtc; bdnf: forward: gcataggtggtctcggtgtc, reverse: cttgctttcctcgctctgtc; 28S rRNA: forward: ggtgttgacgcgatgtgatttctg, reverse: tagatgacgaggcatttggcatcc.
To determine relative gene expression, the Ct (threshold cycle value) of each candidate was normalized to the Ct of the PCR product for the housekeeping gene 28S rRNA. QPCR was performed in a 96-well format using Power SYBR Green Master Mix (Applied Biosystems) on a 7500 Real Time PCR system (Applied Biosystems). All the reactions were done in triplicate. Reaction conditions used were as follows: stage 1: 50°C for 2 min; stage 2: 95°C for 10 min; stage 3; 95°C for 15 s, 60°C for 1 min. Stage 3 was repeated for 40 cycles. Fold changes between treatment groups and non-injected controls were analysed using a two-tailed Student's t test.
Online data
Acknowledgements
We thank Dr Gerhard Schlosser for providing anti-eya1 antibody. We acknowledge technical contributions from Ben Sutti, Jenna Van Rybroek and Craig Fett. We also acknowledge helpful discussions with Dr Richard Smith (University of Iowa) and the members of his laboratory and we thank Dr Bernd Fritzsch (University of Iowa) for his insightful comments on neuroanatomy and on the manuscript. Confocal microscopy was performed at the University of Iowa Central Microscopy Research.
Funding
This work was supported by the National Institutes of Health [grant number NIH ROI-DC007481 (to J.M. and D.L.W.)].
References
- Abdelhak S., Kalatzis V., Heilig R., Compain S., Samson D., Vincent C., Levi-Acobas F., Cruaud C., Le Merrer M., Mathieu M., et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum. Mol. Genet. 1997a;6:2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- Abdelhak S., Kalatzis V., Heilig R., Compain S., Samson D., Vincent C., Weil D., Cruaud C., Sahly I., Leibovici M., et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 1997b;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Allen B. G., Weeks D. L. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataliotis P., Ivins S., Mohun T. J., Scambler P. J. XTbx1 is a transcriptional activator involved in head and pharyngeal arch development in Xenopus laevis. Dev. Dyn. 2005;232:979–991. doi: 10.1002/dvdy.20276. [DOI] [PubMed] [Google Scholar]
- Azuma N., Hirakiyama A., Inoue T., Asaka A., Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum. Mol. Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- Bane B. C., Van Rybroek J. M., Kolker S. J., Weeks D. L., Manaligod J. M. EYA1 expression in the developing inner ear. Ann. Otol. Rhinol. Laryngol. 2005;114:853–858. doi: 10.1177/000348940511401108. [DOI] [PubMed] [Google Scholar]
- Bever M. M., Jean Y. Y., Fekete D. M. Three-dimensional morphology of inner ear development in Xenopus laevis. Dev. Dyn. 2003;227:422–430. doi: 10.1002/dvdy.10316. [DOI] [PubMed] [Google Scholar]
- Bonini N. M., Leiserson W. M., Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bui Q. T., Zimmerman J. E., Liu H., Bonini N. M. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics. 2000;155:709–720. doi: 10.1093/genetics/155.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller C., Xu X., Marquis V., Schwanke R., Xu P. X. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum. Mol. Genet. 2001;10:2775–2781. doi: 10.1093/hmg/10.24.2775. [DOI] [PubMed] [Google Scholar]
- Dagle J. M., Littig J. L., Sutherland L. B., Weeks D. L. Targeted elimination of zygotic messages in Xenopus laevis embryos by modified oligonucleotides possessing terminal cationic linkages. Nucleic Acids Res. 2000;28:2153–2157. doi: 10.1093/nar/28.10.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagle J. M., Weeks D. L. Selective degradation of targeted mRNAs using partially modified oligonucleotides. Methods Enzymol. 2000;313:420–436. doi: 10.1016/s0076-6879(00)13026-5. [DOI] [PubMed] [Google Scholar]
- Dagle J. M., Weeks D. L. Oligonucleotide-based strategies to reduce gene expression. Differentiation. 2001;69:75–82. doi: 10.1046/j.1432-0436.2001.690201.x. [DOI] [PubMed] [Google Scholar]
- David R., Ahrens K., Wedlich D., Schlosser G. Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech. Dev. 2001;103:189–192. doi: 10.1016/s0925-4773(01)00355-0. [DOI] [PubMed] [Google Scholar]
- Duncan M. K., Kos L., Jenkins N. A., Gilbert D. J., Copeland N. G., Tomarev S. I. Eyes absent: a gene family found in several metazoan phyla. Mamm. Genome. 1997;8:479–485. doi: 10.1007/s003359900480. [DOI] [PubMed] [Google Scholar]
- Fougerousse F., Durand M., Lopez S., Suel L., Demignon J., Thornton C., Ozaki H., Kawakami K., Barbet P., Beckmann J. S., Maire P. Six and Eya expression during human somitogenesis and MyoD gene family activation. J. Muscle Res. Cell Motil. 2002;23:255–264. doi: 10.1023/a:1020990825644. [DOI] [PubMed] [Google Scholar]
- Friedman R. A., Makmura L., Biesiada E., Wang X., Keithley E. M. Eya1 acts upstream of Tbx1, Neurogenin 1, NeuroD and the neurotrophins BDNF and NT-3 during inner ear development. Mech. Dev. 2005;122:625–634. doi: 10.1016/j.mod.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. The amphibian octavo-lateralis system and its regressive and progressive evolution. Acta Biol. Hung. 1988;39:305–322. [PubMed] [Google Scholar]
- Furuya M., Qadota H., Chisholm A. D., Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev. Biol. 2005;286:452–463. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Grifone R., Laclef C., Spitz F., Lopez S., Demignon J., Guidotti J. E., Kawakami K., Xu P. X., Kelly R., Petrof B. J., et al. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P. X., Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Harland R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Kolker S. J., Tajchman U., Weeks D. L. Confocal imaging of early heart development in Xenopus laevis. Dev. Biol. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Deffenbacher K., Cremers C. W., Van Camp G., Kimberling W. J. Branchio-oto-renal syndrome: identification of novel mutations, molecular characterization, mutation distribution, and prospects for genetic testing. Genet. Test. 1997;1:243–251. doi: 10.1089/gte.1997.1.243. [DOI] [PubMed] [Google Scholar]
- Mutsuddi M., Chaffee B., Cassidy J., Silver S. J., Tootle T. L., Rebay I. Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics. 2005;170:687–695. doi: 10.1534/genetics.104.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. 2nd edn. Amsterdam: North Holland Publishing Company; 1967. Normal Table of Xenopus laevis (Daudin) [Google Scholar]
- Ogino H., McConnell W. B., Grainger R. M. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech. Dev. 2006;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ohto H., Kamada S., Tago K., Tominaga S. I., Ozaki H., Sato S., Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orten D. J., Fischer S. M., Sorensen J. L., Radhakrishna U., Cremers C. W., Marres H. A., Van Camp G., Welch K. O., Smith R. J., Kimberling W. J. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum. Mutat. 2008;29:537–544. doi: 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- Pan F. C., Chen Y., Loeber J., Henningfeld K., Pieler T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev. Dyn. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- Quick Q. A., Serrano E. E. Inner ear formation during the early larval development of Xenopus laevis. Dev. Dyn. 2005;234:791–801. doi: 10.1002/dvdy.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapureddi J. P., Hegde R. S. Branchio-oto-renal syndrome associated mutations in Eyes Absent 1 result in loss of phosphatase activity. FEBS Lett. 2006;580:3853–3859. doi: 10.1016/j.febslet.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Rayapureddi J. P., Kattamuri C., Steinmetz B. D., Frankfort B. J., Ostrin E. J., Mardon G., Hegde R. S. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Schlosser G., Awtry T., Brugmann S. A., Jensen E. D., Neilson K., Ruan G., Stammler A., Voelker D., Yan B., Zhang C., et al. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G., Northcutt R. G. Development of neurogenic placodes in Xenopus laevis. J. Comp. Neurol. 2000;418:121–146. [PubMed] [Google Scholar]
- Tootle T. L., Silver S. J., Davies E. L., Newman V., Latek R. R., Mills I. A., Selengut J. D., Parlikar B. E., Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Wetts R., Fraser S. E. Slow intermixing of cells during Xenopus embryogenesis contributes to the consistency of the blastomere fate map. Development. 1989;105:9–15. doi: 10.1242/dev.105.1.9. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Pines J., Ryan K., Siemering K. R., Haseloff J., Evans M. J., Gurdon J. B. An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development. 1996;122:3719–3724. doi: 10.1242/dev.122.12.3719. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Knosp B. M., Maconochie M., Friedman R. A., Smith R. J. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J. Assoc. Res. Otolaryngol. 2004;5:295–304. doi: 10.1007/s10162-004-4044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Silvius D., Fritzsch B., Xu P. X. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Erickson C., Kim E. H., Jin D., Fritzsch B., Xu P. X. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum. Mol. Genet. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.