Abstract
The 7-methylguanosine cap added to the 5′ end of mRNA is essential for efficient gene expression and cell viability. Methylation of the guanosine cap is necessary for the translation of most cellular mRNAs in all eukaryotic organisms in which it has been investigated. In some experimental systems, cap methylation has also been demonstrated to promote transcription, splicing, polyadenylation and nuclear export of mRNA. The present review discusses how the 7-methylguanosine cap is synthesized by cellular enzymes, the impact that the 7-methylguanosine cap has on biological processes, and how the mRNA cap methylation reaction is regulated.
Keywords: gene regulation, mRNA cap methylation, translation, transcription
Abbreviations: CBC, cap-binding complex; CBP, cap-binding protein; CDK, cyclin-dependent kinase; CE, capping enzyme; CTD, C-terminal domain; DSIF, DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity inducing factor; eIF, eukaryotic initiation factor; NELF, negative elongation factor; P-TEFb, positive transcription elongation factor b; RNGTT, RNA guanylyltransferase and 5′ triphosphatase; RNMT, RNA (guanine-7-) methyltransferase; SAH, S-adenosylhomocysteine; SAHH, SAH hydrolase; SAM, S-adenosylmethionine; TFIIH, transcription factor IIH; UTR, untranslated region; xSAHH, Xenopus laevis SAHH
INTRODUCTION
Eukaryotic mRNA is modified by the addition of the 7-methylguanosine ‘cap’ to the first transcribed nucleotide. From yeast to humans, this modification is necessary for efficient gene expression and cell viability. The 7-methylguanosine cap is required for the translation of the majority of mRNAs, and it has also been reported to stabilize mRNA against attack by exonucleases and to promote transcription, splicing, polyadenylation and nuclear export of mRNA. Formation of the 7-methylguanosine cap occurs as the substrate mRNA is being transcribed and is catalysed by enzymes which are recruited to RNA polymerase II. Cellular proteins which regulate mRNA cap methylation are likely to function either by regulating the recruitment of the cap methyltransferase to RNA polymerase II, or by regulating the cap methyltransferase activity. Several cellular factors have now been found which regulate mRNA cap methylation and these are discussed in the present review. Viruses also have mechanisms to ensure efficient mRNA cap methylation. Since this is out of the scope of the present review, readers are directed two other excellent reviews which discuss regulation of cap methylation by viral proteins [1,2]. Furichi and Shatkin [2] also provide a comprehensive history of the discovery of the mRNA 7-methylguanosine cap.
CREATION OF THE 7-METHYLGUANOSINE CAP
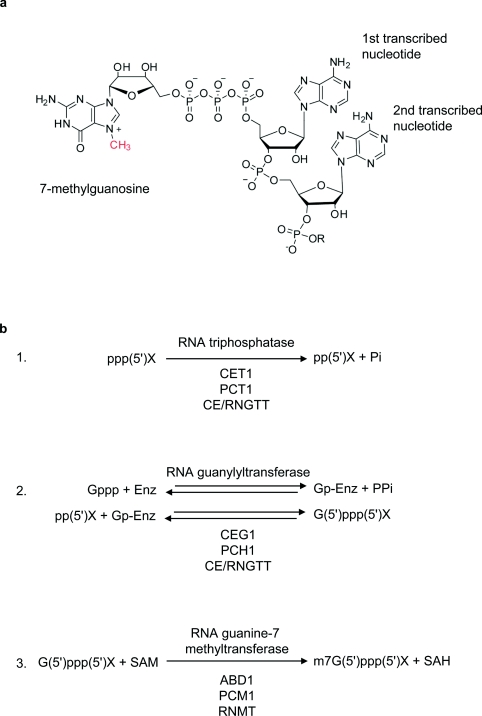
The 7-methylguanosine cap is joined to the first transcribed nucleotide via the 5′ hydroxyl group, through a triphosphate linkage, to produce m7G(5′)ppp(5′)X, where m7G is 7-methylguanosine, p is a phosphate group and X is the first transcribed nucleotide (Figure 1a) [2,3]. This 5′–5′ linkage is in contrast with the 3′–5′ phosphodiester bond, which links nucleotides in transcribed RNA. The 7-methylguanosine cap is formed by the action of three enzymes (Figure 1b). (i) The 5′ triphosphate group on nascent RNA, ppp(5′)X, is hydrolysed by an RNA 5′ triphosphatase to produce diphosphate–RNA, pp(5′)X. (ii) A guanylyltransferase catalyses the addition of GMP to the diphosphate–RNA to produce the guanosine cap, G(5′)ppp(5′)X via a two-step reversible reaction. Initially, the guanylyltransferase reacts with the α-phosphate of GTP, forming an enzyme–GMP intermediate, Gp-Enz, and releasing pyrophosphate, PPi. The GMP is transferred from Gp-Enz to pp(5′)X to create G(5′)ppp(5′)X, and the enzyme is regenerated. Finally, (iii) an RNMT [RNA (guanine-7-) methyltransferase] catalyses the methylation of the guanosine cap at the N-7 position to produce the 7-methylguanosine cap, m7G(5′)ppp(5′)X. SAM (S-adenosylmethionine) is used as the methyl donor.
Figure 1. Synthesis of the 7-methylguanosine cap.
(a) The 7-methylguanosine cap. The methyl group of 7-methylguanosine is indicated in red. (b) Reactions that synthesize the 7-methylguanosine cap. The synthesis is described in detail in the text. Enzymes which catalyse reactions are indicated above the reactions, and the names of the enzymes from S. cerevisiae, S. pombe and Homo sapiens are indicated below the reactions.
Addition of the 7-methylguanosine cap is often referred to as ‘capping’, and the enzymes that catalyse the reactions are referred to as the ‘capping enzymes’. In the present review, the processes of guanosine cap addition and guanosine cap methylation are discussed distinctly and referred to as ‘capping’ and ‘cap methylation’ respectively.
Much of the characterization of the biochemical pathway which produces the 7-methylguanosine cap has been carried out in the yeast species Saccharomyces cerevisiae and Schizosaccharomyces pombe, or in vitro using the enzymes encoded by these organisms. The three enzymic activities required to synthesize the 7-methylguanosine cap are present as three separate proteins in S. cerevisiae and S. pombe. Cet1p and Pct1 are the triphosphatases, Ceg1p and Pch1 are the guanylyltransferases, and Abd1 and Pcm1 are the RNMTs in S. cerevisiae and S. pombe respectively [4–6]. In mammals and other metazoa, the triphosphatase and guanylytransferase activities are found in the same polypeptide, called CE (capping enzyme) or RNGTT (RNA guanylyltransferase and 5′ triphosphatase) [7–10]. The mammalian RNMT is a distinct protein [8,10,11]. The guanylyltransferases and cap methyltransferases are conserved in structure and mode of action from yeast to metazoa, whereas the RNA triphosphatases are quite different in structure and mode of action in yeast and metazoa [1].
Higher organisms may have evolved to have the triphosphatase and guanylyltransferase on one polypeptide to permit efficient co-ordination of the reactions required for guanosine cap addition. Some viruses, including vaccinia virus, encode a single polypeptide containing the triphosphatase, guanylyltransferase and methyltransferase, thus permitting efficient capping and cap methylation [1]. The fact that higher organisms have retained a distinct guanosine capping enzyme (CE/RNGTT) and RNMT permits differential regulation of guanosine capping and cap methylation [12,13].
PROCESSES DEPENDENT ON CAP METHYLATION
The 7-methylguanosine cap has been demonstrated to influence several steps of gene expression, including transcription, splicing, polyadenylation, nuclear export of mRNA, translation and mRNA stability, and these are discussed individually below. For some of these processes, the role of the 7-methylguanosine cap and the unmethylated guanosine cap have been compared, whereas for other processes, only the role of the 7-methylguanosine cap has been investigated.
Transcription elongation
mRNA capping and cap methylation occur ‘co-transcriptionally’, that is, as the RNA is being transcribed. Furthermore, generation of the 7-methylguanosine cap and the enzymes which catalyse this process promote transcription elongation. This is discussed in more detail below in the discussion of the mechanism of mRNA cap methylation.
Splicing
RNA splicing is dependent on the 7-methylguanosine cap. In vitro, the splicing reaction has been demonstrated to be dependent on the substrate RNA having a 7-methylguanosine cap and could be inhibited by free 7-methylguanosine [14,15]. Splicing of the first intron was found to be particularly dependent on the presence of a 7-methylguanosine cap [16]. These findings were consolidated in vivo by work carried out in Xenopus laevis oocytes, where microinjected non-guanosine-capped mRNA, A(5′)ppp(5′)G, was spliced less efficiently than 7-methylguanosine-capped mRNA, m7G(5′)ppp(5′)X. The 7-methylguanosine cap promoted splicing predominantly on the 5′ proximal intron [17]. Use of temperature-sensitive mutants of the guanylyltransferase Ceg1 have allowed rapid inhibition of capping in S. cerevisiae [18–20]. Inhibition of capping led to a defect in splicing, but the severity was dependent on the Ceg1 mutation and was gene-specific. The proportion of genes that are dependent on the 7-methylguanosine cap for splicing is not known for either yeast or mammals.
The dependency of splicing on the 7-methylguanosine cap is mediated by the heterodimeric CBC (cap-binding complex), which consists of CBP80 (cap-binding protein 80) and CBP20. Immunodepletion of this complex from HeLa cell extracts inhibited in vitro splicing reactions [21]. Microinjection of X. laevis oocytes with antibodies raised against CBP20 inhibited CBP20 interaction with the mRNA cap and inhibited splicing [22]. CBC interacts with splicing complex components and is required for efficient recognition of the 5′-splice site on the 5′-proximal intron [23,24]. These findings support the model that splice sites are defined by interactions which span exons [25].
mRNA translation
From yeast to humans, the 7-methylguanosine cap is necessary for the translation of most mRNAs, with the exception of a subset of mRNAs translated by internal ribosome entry [26]. Initially, methylation of the guanosine cap was demonstrated to be necessary for efficient translation in cell-free systems [27,28]. In vivo, mRNA injected into X. laevis oocytes was translated more efficiently if it contained a 7-methylguanosine cap [29,30]. In S. cerevisiae, temperature-sensitive mutants of Abd1 were synthesized, which facilitated rapid inhibition of Abd1 in vivo. This resulted in a rapid and almost complete loss of protein synthesis, consistent with mRNA cap methylation being essential for the efficient translation of most mRNAs [31].
Cap-dependent translation initiates with eIF4F (eukaryotic initiation factor 4F) complex formation on the 7-methylguanosine cap. This is required for the 5′ end of mRNA to be recruited to the 43S initiation complex containing the 40S ribosomal subunit, associated initiation factors and the initiator tRNA tRNAiMet [32]. The first mRNA-dependent step of translation is the binding of the eIF4F complex to the 7-methylguanosine cap, which is mediated by the eIF4E subunit. eIF4E has a significantly higher affinity for the 7-methylguanosine cap than the unmethylated guanosine cap, and this has been hypothesized to prevent free cellular GTP from interfering with translation initiation [33].
mRNA polyadenylation
mRNA is modified at the 3′ end by endonucleolytic cleavage and addition of the poly(A) tail [34]. In vitro, the endonucleolytic cleavage step is dependent on the CBC [35]. A recent study used chromatin immunoprecipitations to demonstrate that the mammalian capping enzyme RNGTT/CE and the mammalian cap methyltransferase RNMT are localized not only at the 5′ end of the gene but are also found throughout the gene, including a significant fraction at the 3′ end of the gene [36]. Potentially, the capping enzyme and cap methyltransferase could influence polyadenylation at this stage.
In S. cerevisiae, when capping was inhibited rapidly using temperature-sensitive mutants of the capping enzyme Ceg1, polyadenylation of CHY2 mRNA was unaffected [18]. The length of poly(A) tails on total mRNA was investigated and was also not significantly altered following inhibition of capping [18]. Therefore, in S. cerevisiae, polyadenylation of mRNA appears to be largely independent of the 7-methylguanosine cap.
Nuclear export of mRNA
Nuclear export of a subset of mRNAs is dependent on the 7-methylguanosine cap. Nuclear export of microinjected mRNA in X. laevis oocytes could be inhibited by 7-methylguanosinecapped RNA, but not uncapped mRNA [37]. Export of snRNAs (small nuclear RNAs) was also shown to be dependent on the CBC in X. laevis oocytes [22]. However, inactivation of temperature-sensitive mutants of the guanylyltransferase Ceg1 in S. cerevisiae did not alter the distribution of mRNA between the nucleus and cytoplasm, and therefore the 7-methylguanosine cap is not required for the export of the majority of mRNAs in this organism [18].
mRNA stability
The presence of a 7-methylguanosine cap can protect mRNA from degradation. When RNA was microinjected into X. laevis oocytes or incubated in mammalian cell extracts, 7-methylguanosine and unmethylated guanosine-capped mRNA was significantly more stable than uncapped mRNA [38,39]. Subsequently, the 7-methylguanosine cap was found to protect mRNA from degradation by exonuclease activity [40]. Use of temperature-sensitive mutants of the S. cerevisiae guanylyltransferase Ceg1 facilitated rapid inhibition of guanosine capping in vivo [18–20]. Although some mRNAs were rapidly degraded following loss of Ceg1 activity, others were relatively stable and therefore the necessity for a guanosine cap to stabilize mRNA may be dependent on the RNA sequence, secondary structure, or some other gene-specific factor. It is also a possibility that mRNA stability is more dependent on the guanosine cap in higher organsims.
Methylation of the mRNA guanosine cap also has the potential to indirectly regulate mRNA stability. The capping reaction catalysed by the guanylyltransferase is reversible, and the product of the back reaction, uncapped mRNA, is degraded rapidly by exonucleases (Figure 1b) [38,40]. Once the guanosine cap is methylated, it is no longer a substrate for the guanylyltransferase, and, since cap methylation appears to be irreversible, it indirectly stabilizes the RNA. Current data, however, suggest that, in vivo, the 5′ end of mRNA only encounters a significant concentration of guanylyltransferase during transcription (see below), and therefore mRNA cap methylation may not be required to inhibit the guanosine cap removal catalysed by the guanylyltransferase [41]. Experiments carried out in S. cerevisiae and X. laevis oocytes also indicate that cap methylation plays a relatively minor role in mRNA stability. Unmethylated-capped mRNA and 7-methylguanosine-capped mRNA were equivalently stable when injected into X. laevis oocytes [38]. However, these cells are unusual since they appear to lack decapping activity, and therefore these results may not be able to be extrapolated to all vertebrate cells. Inhibition of the S. cerevisiae cap methyltransferase Abd1 in vivo also resulted in a relatively small fall in the mRNA levels of the genes investigated [20,31]. In contrast, as described previously, inhibition of the S. cerevisiae guanylyltransferase Ceg1 resulted in almost complete loss of the same mRNAs over the same time course [20,31].
RNMT CATALYSES THE CAP METHYLATION REACTION
Methylation of the guanosine cap on the N-7 position is catalysed by the enzyme RNMT. Mammalian RNA guanine-7 cap methyltransferase activities were first isolated from rat liver and HeLa cells based on in vitro cap methylation assays [42,43]. The first cellular cap methyltransferase to be identified was the RNMT from S. cerevisiae, ABD1, on the basis of sequence homology to the cap methyltransferase domain found in the vaccinia virus D1 subunit [6]. The cap methyltransferases from S. cerevisiae and mammals appear to be isolated as monomers and are able to function effectively as such [6,42,44]. The S. pombe cap methyltransferase Pcm1 is isolated in a complex with the guanylyltransferase and P-TEFb (positive transcription elongation factor b), but these factors have not been reported to alter the activity of Pcm1 [45].
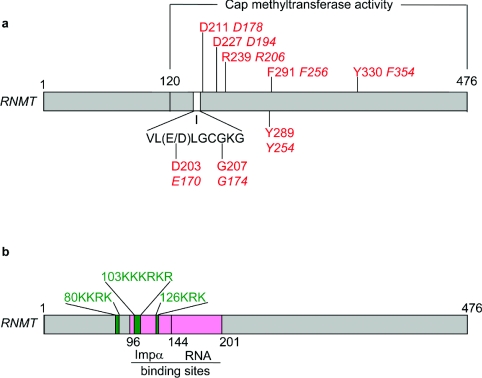
Following the identification of ABD1 as a cap methyltransferase, it was demonstrated to be required for cell growth, providing the first genetic evidence that mRNA cap methylation has an essential function in cells [6]. Subsequently the human cap methyltransferase RNMT was also found to be required for mammalian cell survival [46,47]. Since both RNMT and ABD1 are essential genes, there is not another pleiotropic cap methyltransferase that can compensate for loss of RNMT or ABD1. The cellular cap methyltransferases are highly conserved, including the SAM-binding site/motif I, VLD/ELGCGKG, which is essential for catalysis (Figure 2a) [8,10,11,44,48]. In particular, the C-terminus of RNMT and Abd1 are highly homologous, and the C-terminus of RNMT can substitute for ABD1 in S. cerevisiae [44]. Furthermore, the entire S. cerevisiae capping apparatus, that is, the RNA triphosphatase CET1, the guanylyltransferase CEG1, and the cap methyltransferase ABD1, can be replaced by the mammalian bifunctional capping enzyme CE/RNGTT and the cap methyltransferase RNMT [44]. The N-terminal 110 amino acids of Abd1 and the N-terminal 120 amino acids of the human cap methyltransferase RNMT are not essential for in vitro cap methyltransferase activity or for cell growth and survival (Figure 2a) [44,46,49]. At present, the function of the N-terminus of the cellular cap methyltransferases is not known.
Figure 2. RNMT functional domains.
(a) The minimal domain required for cap methyltransferase activity is indicated (residues 120–476). The SAM-binding domain/motif I, VL(E/D)LGCGKG, is shown. Residues essential for cap methyltransferase activity and cell viability are indicated in red: residues found in the human mRNA cap methytransferase RNMT are indicated in normal type; and those found in the S. cerevisiae cap methyltransferase ABD1 are indicated in italics. (b) Nuclear localization signals are indicated in green. Importin α (Impα)- and RNA-binding sites are indicated in pink.
When ABD1 was first demonstrated to be an essential gene, it was not clear whether this was due to the cap methyltransferase activity of the enzyme or some other hypothetical function of the protein. Subsequently, a series of point mutations were made in ABD1 (and in the analogous residues in RNMT) (Figure 2a). All point mutants that inhibited the enzymic activity were essential for cell growth and survival, strengthening the hypothesis that RNMT activity is essential for cell survival (Figure 2a) [31,44,48,49]. Interestingly, some point mutations in regions that are identical from yeast to humans were not found to be essential for cap methylation or yeast viability [31]. It remains to be seen whether such regions are required under specific growth conditions.
Other residues which are essential for RNMT function in vivo are those which ensure correct localization [46] (Figure 2b). RNMT is thought to function predominantly, if not entirely, in the nucleus at the initiation of transcription, and was localized to the nucleus by use of an RNMT–GFP (where GFP is green fluorescent protein) fusion protein [50]. The nuclear localization of RNMT is critical for its function. A truncation mutant (human RNMT residues 144–476), which localizes to the cytosol, was enzymically active but unable to rescue cells deficient in RNMT [46]. Three alternate nuclear localization signals were identified in human RNMT, K80KRK, K103KRKR and K126RK.
All discussion of the human cap methyltransferase RNMT refers to the dominant isoform in the present review. However, three splice variant cDNAs transcribed from the RNMT gene, designated hCMT1a, b and c, were cloned from human colon mucosa and transcripts for all three variants were expressed in a variety of tissues [8]. hCMT1c is the predominant isoform of RNMT discussed in the present review and is 476 amino acids long. hCMT1a encodes the same protein as hCMT1c, but has a different 5′- and 3′-UTR (untranslated region). hCMT1b encodes a distinct protein which is 504 amino acids long; it has the same N-terminal 464 amino acids as hCMT1c and differs thereafter. Recombinant hCMT1a/c was demonstrated to be active in an in vitro RNA methyltransferase assay, whereas hCM1b was not. Since the different RNMT isoforms have different UTRs, the potential exists for them to be differentially regulated, and changing the ratio of active hCMT1a/c isoforms to inactive hCMT1b isoform could potentially regulate cellular cap methyltransferase activity. However, this remains speculation at this stage, since hCMT1b has not yet been demonstrated to be expressed at the protein level.
mRNA CAP METHYLATION OCCURS CO-TRANSCRIPTIONALLY
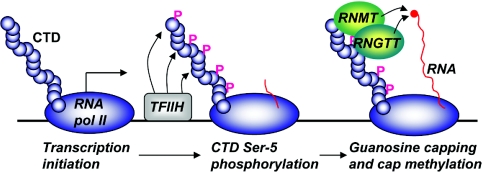
As discussed above, mRNA cap methylation occurs ‘co-transcriptionally’, i.e. as the RNA to which it is added is being transcribed (Figure 3). The 5′-end of nascent RNA is guanosine capped, and the guanosine cap is methylated as the RNA emerges from RNA polymerase II, in the vicinity of the RNA polymerase II large subunit CTD (C-terminal domain) [41,51,52]. The CTD contains approx. 50 repeats (depending on the species) of the consensus sequence YSTPSPS and can be phosphorylated on multiple residues, with Ser2 and Ser5 phosphorylation being the most comprehensively characterized [53,54]. The CTD forms a docking site for the enzymes and factors which mediate RNA modifications, including mRNA cap methylation, splicing and polyadenylation. The CTD residues are phosphorylated and dephosphorylated during distinct stages of gene transcription, and the different patterns of phosphorylation are thought to form a ‘CTD code’ that dictates which enzymes and factors are recruited, retained or removed from the transcribing polymerase [41,51,52]. Thus RNA modifications are correctly co-ordinated in space and time.
Figure 3. mRNA cap methylation occurs co-transcriptionally.
RNA polymerase II is recruited to chromatin with the CTD hypophosphorylated. At the initiation of transcription, TFIIH phosphorylates CTD on Ser5. Enzymes which catalyse the formation of the 7-methylguanosine cap, RNGTT and RNMT, are recruited to the phosphorylated CTD, proximal to their substrate, the 5′ end of nascent RNA. An animation of this Figure is available at http://www.BiochemJ.org/bj/425/0295/bj4250295add.htm.
RNA polymerase II is recruited to chromatin with the CTD unphosphorylated [53,54]. During transcription initiation, the CTD is phosphorylated on Ser5 by CDK7 (cyclin-dependent kinase 7), which is a subunit of the basic transcription factor TFIIH (transcription factor IIH). At this stage, the polymerase pauses under the influence of DSIF [DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity inducing factor] and NELF (negative elongation factor). The enzymes which create the 7-methylguanosine cap in yeast and mammals were found to be recruited preferentially to the RNA polymerase II CTD when it is phosphorylated on Ser5 [7,55–57]. The guanylyltransferases also bind to Spt5, a component of DSIF, or the analogous complex in yeast, Spt4-Spt5 [58–60]. Thus the guanosine-capping enzymes and the cap methyltransferase are recruited to their substrate, the 5′ end of nascent RNA, as it emerges from the RNA polymerase II complex. Furthermore, the guanylyltransferase is also stimulated by interactions with Ser5-phosphorylated CTD and Spt5 [59,61]. Guanosine capping was demonstrated to be enhanced on RNA that is being actively transcribed compared with free RNA [12]. However, stimulation of capping on actively transcribed RNA was only partially dependent on the CTD [12]. This suggests that the capping enzyme may also be recruited by another domain of RNA polymerase II or another component of the transcription apparatus.
mRNA CAP METHYLATION-DEPENDENT TRANSCRIPTION
Shortly after transcription initiates, RNA polymerase II ‘pauses’ and transcription stops. Release from this arrest into productive transcription elongation is now recognized to be a major point in the control of transcription [62]. The guanylyltransferases and methyltransferases are recruited to RNA polymerase II during pausing, and release from pausing into transcription elongation is partially dependent on these enzymes.
One of the initial findings that suggested a relationship between cap methylation and RNA polymerase II-dependent transcription was that, in vitro, SAH (S-adenosylhomocysteine), an inhibitor of methylation reactions, was found to inhibit the initiation of transcription by RNA polymerase II [63]. SAH did not inhibit transcription elongation, or the activity, of purified RNA polymerase II, and therefore this implied that a methylation reaction which occurred early on during the transcription of a gene was coupled to transcription initiation. Subsequently, the mammalian capping enzyme CE/RNGTT was found to relieve transcriptional repression mediated by NELF [64]. At least in vitro, catalytically inactive CE/RNGTT mutants could also relieve repression mediated by NELF, equivalently to the wild-type enzyme [64]. This implies that it is the CE/RNGTT protein itself, rather than the process of guanosine capping, or the guanosine cap, that relieves repression mediated by NELF. The relationship between guanosine capping and transcription elongation was also observed in vivo when mutations in the yeast capping enzyme Ceg1 caused defects in transcription elongation [65].
Inactivation and mutation of the S. cerevisiae cap methyltransferase Abd1 also inhibited RNA polymerase II recruitment to promoters and movement through the gene in vivo [66]. Similar to what was observed for capping enzyme-stimulated transcription, an Abd1 mutant defective for cap methyltransferase activity could promote RNA polymerase II recruitment to most of the genes investigated, suggesting that it is the cap methyltransferase protein itself, rather than the process of cap methylation or the 7-methylguanosine cap, which stimulates transcription [66].
Transcription elongation is dependent on the P-TEFb [67]. P-TEFb, which contains the kinase CDK9, phosphorylates the RNA polymerase II CTD on Ser2 and relieves repression mediated by DSIF/NELF, thus promoting transcription elongation. In S. pombe, CDK9 was isolated in a complex with all three enzymes required for cap methylation, i.e. the triphosphatase Pct1, the guanosine-capping enzyme Pch1 and the cap methyltransferase Pcm1 [45,68]. The interaction between the S. pombe cap methyltransferase Pcm1 and CDK9 is such that all detectable Pcm1 can be cleared from cell extracts by an immunoprecipitation with CDK9 [69]. Pcm1 was found to mediate recruitment of P-TEFb to chromatin and CTD Ser2 phosphorylation [69,70]. In Caenorhabditis elegans, a similiar relationship between the capping enzyme and P-TEFb is implied by the finding that inhibition of expression of the capping enzyme Cel-1 resulted in loss of CTD Ser2 phosphorylation, without loss of CDK9 expression [71]. It remains to be seen in S. cerevisiae and mammalian systems, where such an avid interaction between CDK9 and the cap methyltransferase has not been documented, whether P-TEFb recruitment is also dependent on the cap methyltransferase.
Although the experiments described above demonstrate that the guanosine-capping enzymes and cap methyltransferases promote transcription elongation, they do not appear to be required, at least for transcription of a subset of genes in S. cerevisiae. Following inhibition of the capping enzyme Ceg1 in S. cerevisiae, transcription of at least some genes occurred independently of capping [20]. Furthermore, following inhibition of the S. cerevisiae methyltransferase Abd1, a subset of genes did not exhibit loss of recruitment of RNA polymerase II [66]. In another study, inhibition of Abd1 using temperature-sensitive mutants lead to a minor loss in transcript level, which paralleled a loss in rRNA expression, which is not predicted to be directly dependent on cap methylation [31]. These studies suggest that the role of the capping enzymes and cap methyltransferases in transcription elongation is gene-specific in S. cerevisiae, and this may be the case in other species too.
REGULATION OF mRNA CAP METHYLATION
The mRNA guanosine cap is methylated predominantly, if not entirely, as RNA polymerase II is paused, shortly after the initiation of transcription. As described above, at this stage the CTD of RNA polymerase II is phosphorylated on Ser5, and this forms a docking site for the mRNA cap methyltransferase (Figure 3). Known regulators of mRNA cap methylation either increase the activity of the cap methyltransferase or regulate recruitment of the cap methyltransferase to RNA polymerase II. The regulators of cap methylation described below have typically only been studied in one species or a few closely related ones. However, due to the high degree of homology between all the cap methyltransferases, and the fact that the mammalian cap methyltransferase can functionally substitute for the S. cerevisiae cap methyltransferase in vivo, it is likely that the regulators function with some equivalence in all species in which they are expressed [44].
SAM
Certain Abd1 temperature-sensitive mutants cause a severe growth defect at the restrictive temperature, and a screen for suppressors of this phenotype identified SAM synthase, the enzyme which synthesizes the methyl donor SAM as a critical activator of cap methyltransferase activity [31]. Furthermore, the growth defect was also suppressed in a dose-dependent manner by incubation in SAM, which indicates that it is levels of SAM rather than the SAM synthase protein itself that directly stimulates Abd1 activity.
cdc34
The same screen for suppressors of temperature-sensitive mutants of Abd1 also identified a ubiquitin-conjugating enzyme, cdc34 [31]. Mutations that affect the ubiquitin-conjugating activity of cdc34 or its interaction with the E3–SCF (where SCF is Skp1/cullin/F-box) complex inhibited rescue of the growth defect, correlating the functional interaction of cdc34 with Abd1 with its ubiquitin-conjugating activity. cdc34 did not affect ABD1 transcript levels (or SAM synthase transcript levels, as discussed above). Many possible scenarios exist for how cdc34 may regulate Abd1. cdc34 may positively regulate Abd1 either by directly ubiquitating Abd1 or a co-activator, thus increasing their activity. Alternatively, cdc34 may ubiquitinate a repressor of Adb1 and target it for degradation.
Importin α
The nuclear transporter importin α was found to interact directly with RNMT in a yeast two-hybrid interaction screen [50]. Importin α enhanced RNMT binding to capped mRNA and enhanced RNMT activity in vitro. Importin α forms a complex with importin β and transports nuclear localization signal-containing proteins to the nucleus [72]. On encountering a high concentration of RanGTP in the nucleus, this complex dissociates, releasing the transported proteins. Importin β alone had no effect on in vitro RNMT activity, but inhibited importin α binding to RNMT and enhancement of RNMT activity. RanGTP inhibited this action of importin β. Therefore importin α stimulation of RNMT probably predominates in the nucleus where RanGTP levels are high and importin α and β are dissociated.
SIGNALLING PATHWAYS THAT REGULATE mRNA CAP METHYLATION
The potential also exists for mRNA cap methylation to be regulated in response to cellular signalling pathways. Currently, few examples exist and the details are limited.
Myc
The proto-oncogenes c-Myc and N-Myc (neuroblastoma-derived Myc) are up-regulated in response to growth factors and are essential for the proliferative response. The Myc proteins are weak transcription factors which typically up-regulate mRNA levels by 2-fold. Expression of the Myc proteins was also demonstrated to up-regulate mRNA cap methylation on specific target mRNAs [73,74]. Using antibodies raised against 7-methylguanosine to purify 7-methylguanosine-capped mRNA, c-Myc was found to up-regulate mRNA cap methylation on its transcriptional targets by up to 8-fold. Significantly, c-Myc was found to up-regulate cap methylation more than transcription for most transcriptional target genes investigated, resulting in greater c-Myc-induced gene expression than had been previously recognized. Myc proteins could also induce mRNA cap methylation on mRNAs that were not transcriptional targets of Myc.
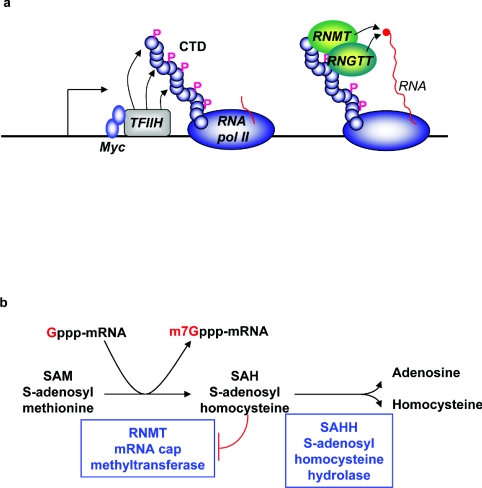
Although Myc is a transcription factor, Myc-induced up-regulation of mRNA cap methylation is not simply a result of increased transcription of components of the cap methylation apparatus. Myc mutants lacking transcriptional activity could up-regulate mRNA cap methylation of certain transcripts, implying that Myc proteins have a relatively direct effect on mRNA cap methylation. Myc binds close to transcription initiation sites [75,76], and was found to bind directly to components of TFIIH, to enhance recruitment of TFIIH to initiation sites, and to increase RNA polymerase II phosphorylation on Ser5 of the CTD (Figure 4a) [73]. Since RNMT binds to Ser5-phosphorylated CTD [57], Myc-induced RNA polymerase II phosphorylation is likely to increase the length of time that RNMT is recruited to genes, thus increasing the probability of an mRNA guanosine cap being methylated. It has yet to be proven, however, that Myc promotes increased RNMT recruitment, and the exact mechanism of Myc-induced cap methylation is unresolved.
Figure 4. Up-regulation of mRNA cap methylation.
(a) Myc binds to DNA proximal to transcription initation sites. Myc binds to TFIIH subunits, promotes TFIIH recruitment to chromatin and enhances CTD phosphorylation. Then RNGTT and RNMT bind to phosphorylated CTD and catalyse formation of the 7-methylguanosine cap. (b) SAM is the methyl donor for the cap methylation reaction catalysed by the mRNA cap methyltransferase RNMT. SAH is the byproduct of the reaction, which inhibits RNMT. SAHH hydrolyses SAH to adenosine and homocysteine, thus relieving repression of the RNMT.
Although immunoprecipitations carried out with anti7-methylguanosine antibodies can indicate the relative level of cap methylation of a gene product, they cannot determine the absolute level of cap methylation. However, the fact that cap methylation can be up-regulated more than transcription, or independently of transcription, means that pools of mRNAs are present in the cell which are unmethylated on the guanosine cap. If cap methylation can only occur at the initiation of transcription, then such unmethylated mRNAs will be weakly translated until degraded. If, however, mRNA cap methylation could be regulated post-transcriptionally, then unmethylated mRNAs will represent a dormant pool that could potentially be activated by cap methylation. Although mRNA can be cap methylated in cell extracts in vitro [42], physiological examples of post-transcriptional cap methylation have not been reported.
E2F1
E2F1 was also demonstrated to up-regulate mRNA cap methylation for a subset of its transcriptional targets [74]. E2F1 is a transcription factor which promotes cell proliferation and has a number of functional similarities to the Myc proteins, including binding proximal to transcription initiation sites [77]. Presumably E2F1 regulates cap methylation using a similar mechanism to c-Myc.
SAHH (SAH hydrolase)
The enzyme SAHH (also known as AHCY) has the potential to regulate the majority of methylation reactions, including mRNA cap methylation. SAH is a by-product of cellular methylation reactions that inhibits methyltransferases (Figure 4b) [78]. SAHH hydrolyses SAH, thus relieving repression of methyltransferases (Figure 4b). mRNA cap methylation has been proposed to be up-regulated by the X. laevis SAHH xSAHH during X. laevis oocyte gastrulation [79,80]. During gastrulation, xSAHH is gradually translocated from the cytoplasm to the nucleus, coinciding with the increase in zygotic mRNA synthesis [79]. In support of a role for SAHH in co-transcriptional mRNA cap methylation, a fraction of xSAHH was found to co-localize with RNA polymerase II on nascent transcripts [79], and xSAHH could be immunoprecipitated with the X. laevis cap methyltransferase [80]. It is worth noting that an increase in cap methyltransferase activity was observed during X. laevis maturation, although the mechanism was not explored, and it is not clear whether it was dependent on a change in xSAHH activity [30].
In mammalian cell systems, SAHH was identified as a Myc target gene that was up-regulated by increased transcription and cap methylation [81]. The up-regulation of SAHH was necessary to permit Myc-induced cap methylation but not transcription. Myc-induced SAHH up-regulation was also necessary for Myc-induced protein synthesis and cell proliferation, thus correlating cap methylation with the core Myc functions. Although SAHH has the potential to regulate all methylation reactions, only Mycinduced cap methyation was SAHH-dependent, whereas Myc-induced total RNA, DNA and protein methylation were not. This suggests that either the RNMT is accutely sensitive to SAH levels, or that the local concentration of SAH rises to significant levels around sites of mRNA cap methylation.
DEMETHYLASES AND DECAPPING ENZYMES?
The major mechanism by which the mRNA cap methylation is reversed is ‘decapping’, which is the removal of the entire 7-methylguanosine cap [82–84]. There are two major decapping pathways that initiate following repression of translation. Dcp2 and its associated proteins hydrolyse 7-methylguanosine-capped mRNA, producing m7GDP and monophosphate-RNA, which is degraded rapidly in a 5′ to 3′ direction by exonucleases. DcpS hydrolyses the 7-methylguanosine structure following degradation of the mRNA from the 3′ end, releasing m7GMP, and therefore is described as a scavenger enzyme. Decapping can be regulated at several stages, which can affect all, or a specific subset of, mRNAs [82–84].
Decapping removes the entire 7-methylguanosine cap. Currently, demethylation of 7-methylguanosine has not been described and, to our knowledge, there is no experimental evidence to suggest that such a demethylase will be identified.
CONCLUSION
The 7-methylguanosine cap is essential for mRNA translation and cell viability from yeast to mammals. It also has a role in transcription elongation, mRNA stability and degradation, and mediates other RNA processing events, including splicing, poly(A) tail addition and nuclear export. The effect of the 7-methylguanosine cap on these processes varies in different experimental systems, which raises the possibility that the 7-methylguanosine cap has gene-specific, cell-lineage-specific and species-specific roles. Given the recent observations that mRNA cap methylation can be regulated by cellular transcription factors, the precise role of the 7-methylguanosine cap in has become even more intriguing.
Online data
ACKNOWLEDGEMENTS
I thank Martin Bushell, Sander Van Kasteren, Tristan Henderson, Rebecca Bounds, Gavin Preston and Evelyn Henderson for their contributions to this review.
References
- 1.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell. Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y., Shatkin A. J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 4.Shibagaki Y., Itoh N., Yamada H., Nagata S., Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 5.Tsukamoto T., Shibagaki Y., Imajoh-Ohmi S., Murakoshi T., Suzuki M., Nakamura A., Gotoh H., Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 6.Mao X., Schwer B., Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue Z., Maldonado E., Pillutla R., Cho H., Reinberg D., Shatkin A. J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukamoto T., Shibagaki Y., Niikura Y., Mizumoto K. Cloning and characterization of three human cDNAs encoding mRNA (guanine-7-)-methyltransferase, an mRNA cap methylase. Biochem. Biophys. Res. Commun. 1998;251:27–34. doi: 10.1006/bbrc.1998.9402. [DOI] [PubMed] [Google Scholar]
- 9.Yamada-Okabe T., Doi R., Shimmi O., Arisawa M., Yamada-Okabe H. Isolation and characterization of a human cDNA for mRNA 5′-capping enzyme. Nucleic Acids Res. 1998;26:1700–1706. doi: 10.1093/nar/26.7.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillutla R. C., Shimamoto A., Furuichi Y., Shatkin A. J. Human mRNA capping enzyme (RNGTT) and cap methyltransferase (RNMT) map to 6q16 and 18p11.22-p11.23, respectively. Genomics. 1998;54:351–353. doi: 10.1006/geno.1998.5604. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa K., Nagase T., Nakajima D., Seki N., Ohira M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. VIII. 78 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1997;4:307–313. doi: 10.1093/dnares/4.5.307. [DOI] [PubMed] [Google Scholar]
- 12.Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 13.Cole M. D., Cowling V. H. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat. Rev. Mol. Cell. Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 15.Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno M., Sakamoto H., Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue K., Ohno M., Sakamoto H., Shimura Y. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev. 1989;3:1472–1479. doi: 10.1101/gad.3.9.1472. [DOI] [PubMed] [Google Scholar]
- 18.Fresco L. D., Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B., Shuman S. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA. 1996;2:574–583. [PMC free article] [PubMed] [Google Scholar]
- 20.Schwer B., Mao X., Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I. W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 22.Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I. W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 23.Lewis J. D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I. W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 24.Colot H. V., Stutz F., Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 26.Spriggs K. A., Stoneley M., Bushell M., Willis A. E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 27.Muthukrishnan S., Both G. W., Furuichi Y., Shatkin A. J. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 28.Both G. W., Banerjee A. K., Shatkin A. J. Methylation-dependent translation of viral messenger RNAs in vitro. Proc. Natl. Acad. Sci. U.S.A. 1975;72:1189–1193. doi: 10.1073/pnas.72.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond D. R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillian-Daniel D. L., Gray N. K., Astrom J., Barkoff A., Wickens M. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol. Cell. Biol. 1998;18:6152–6163. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer B., Saha N., Mao X., Chen H. W., Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gingras A. C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 33.von der Haar T., Gross J. D., Wagner G., McCarthy J. E. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 34.Shatkin A. J., Manley J. L. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 35.Flaherty S. M., Fortes P., Izaurralde E., Mattaj I. W., Gilmartin G. M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover-Cutter K., Kim S., Espinosa J., Bentley D. L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarmolowski A., Boelens W. C., Izaurralde E., Mattaj I. W. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuichi Y., LaFiandra A., Shatkin A. J. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 39.Shimotohno K., Kodama Y., Hashimoto J., Miura K. I. Importance of 5′-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1977;74:2734–2738. doi: 10.1073/pnas.74.7.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murthy K. G., Park P., Manley J. L. A nuclear micrococcal-sensitive, ATP-dependent exoribonuclease degrades uncapped but not capped RNA substrates. Nucleic Acids Res. 1991;19:2685–2692. doi: 10.1093/nar/19.10.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bentley D. L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Ensinger M. J., Moss B. Modification of the 5′ terminus of mRNA by an RNA (guanine-7-)-methyltransferase from HeLa cells. J. Biol. Chem. 1976;251:5283–5291. [PubMed] [Google Scholar]
- 43.Mizumoto K., Lipmann F. Transmethylation and transguanylylation in 5′-RNA capping system isolated from rat liver nuclei. Proc. Natl. Acad. Sci. U.S.A. 1979;76:4961–4965. doi: 10.1073/pnas.76.10.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha N., Schwer B., Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- 45.Pei Y., Du H., Singer J., Stamour C., Granitto S., Shuman S., Fisher R. P. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol. Cell. Biol. 2006;26:777–788. doi: 10.1128/MCB.26.3.777-788.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shafer B., Chu C., Shatkin A. J. Human mRNA cap methyltransferase: alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol. Cell. Biol. 2005;25:2644–2649. doi: 10.1128/MCB.25.7.2644-2649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu C., Shatkin A. J. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol. Cell. Biol. 2008;28:5829–5836. doi: 10.1128/MCB.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S. P., Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 49.Mao X., Schwer B., Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol. Cell. Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen Y., Shatkin A. J. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-α. Genes Dev. 2000;14:2944–2949. doi: 10.1101/gad.848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phatnani H. P., Greenleaf A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 52.Moore M. J., Proudfoot N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Chapman R. D., Heidemann M., Hintermair C., Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Egloff S., Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 55.McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D. L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho C. K., Sriskanda V., McCracken S., Bentley D., Schwer B., Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 57.Pillutla R. C., Yue Z., Maldonado E., Shatkin A. J. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 1998;273:21443–21446. doi: 10.1074/jbc.273.34.21443. [DOI] [PubMed] [Google Scholar]
- 58.Pei Y., Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 59.Wen Y., Shatkin A. J. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindstrom D. L., Squazzo S. L., Muster N., Burckin T. A., Wachter K. C., Emigh C. A., McCleery J. A., Yates J. R., 3rd, Hartzog G. A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho C. K., Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 62.Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jove R., Manley J. L. Transcription initiation by RNA polymerase II is inhibited by S-adenosylhomocysteine. Proc. Natl. Acad. Sci. U.S.A. 1982;79:5842–5846. doi: 10.1073/pnas.79.19.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandal S. S., Chu C., Wada T., Handa H., Shatkin A. J., Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H. J., Jeong S. H., Heo J. H., Jeong S. J., Kim S. T., Youn H. D., Han J. W., Lee H. W., Cho E. J. mRNA capping enzyme activity is coupled to an early transcription elongation. Mol. Cell. Biol. 2004;24:6184–6193. doi: 10.1128/MCB.24.14.6184-6193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroeder S. C., Zorio D. A., Schwer B., Shuman S., Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 67.Peterlin B. M., Price D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 68.Pei Y., Schwer B., Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- 69.Viladevall L., St Amour C. V., Rosebrock A., Schneider S., Zhang C., Allen J. J., Shokat K. M., Schwer B., Leatherwood J. K., Fisher R. P. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guiguen A., Soutourina J., Dewez M., Tafforeau L., Dieu M., Raes M., Vandenhaute J., Werner M., Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;26:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takagi T., Walker A. K., Sawa C., Diehn F., Takase Y., Blackwell T. K., Buratowski S. The Caenorhabditis elegans mRNA 5′-capping enzyme. In vitro and in vivo characterization. J. Biol. Chem. 2003;278:14174–14184. doi: 10.1074/jbc.M212101200. [DOI] [PubMed] [Google Scholar]
- 72.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell. Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 73.Cowling V. H., Cole M. D. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole M. D., Cowling V. H. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28:1169–1175. doi: 10.1038/onc.2008.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell. Biol. 2005;25:3401–3410. doi: 10.1128/MCB.25.9.3401-3410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown S. J., Cole M. D., Erives A. J. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics. 2008;9:442. doi: 10.1186/1471-2164-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang P. K., Gordon R. K., Tal J., Zeng G. C., Doctor B. P., Pardhasaradhi K., McCann P. P. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 79.Radomski N., Kaufmann C., Dreyer C. Nuclear accumulation of S-adenosylhomocysteine hydrolase in transcriptionally active cells during development of Xenopus laevis. Mol. Biol. Cell. 1999;10:4283–4298. doi: 10.1091/mbc.10.12.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Radomski N., Barreto G., Kaufmann C., Yokoska J., Mizumoto K., Dreyer C. Interaction of S-adenosylhomocysteine hydrolase of Xenopus laevis with mRNA(guanine-7-)methyltransferase: implication on its nuclear compartmentalisation and on cap methylation of hnRNA. Biochim. Biophys. Acta. 2002;1590:93–102. doi: 10.1016/s0167-4889(02)00205-7. [DOI] [PubMed] [Google Scholar]
- 81.Fernandez-Sanchez M. E., Gonatopoulos-Pournatzis T., Preston G., Lawlor M. A., Cowling V. H. S-Adenosyl homocysteine hydrolase (SAHH) is required for Myc-induced mRNA cap methylation, protein synthesis and cell proliferation. Mol. Cell. Biol. 2009;29:6182–6191. doi: 10.1128/MCB.00973-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franks T. M., Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simon E., Camier S., Seraphin B. New insights into the control of mRNA decapping. Trends Biochem. Sci. 2006;31:241–243. doi: 10.1016/j.tibs.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Liu H., Kiledjian M. Decapping the message: a beginning or an end. Biochem. Soc. Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.