Abstract
Rapid, sensitive and quantitative assays for proteases are important for drug development and in the diagnosis of disease. Here, an assay for protease activity which uses inductively coupled plasma-mass spectrometry (ICP-MS) detection is described. Peptidic α-chymotrypsin substrates were synthesized containing a lanthanide ion chelate at the N-terminus to provide a distinct elemental tag. A biotin label was appended to the C-terminus of the peptide allowing separation of uncleaved peptide from the enzymatic digestion. The enzyme activity was determined by quantifying the lanthanide ion signal of the peptide cleavage products by ICP-MS. Biotinylated substrates synthesized include Lu-DTPA-Asp-Leu-Leu-Val-Tyr∼Asp-Lys(Biotin) and Lu-DTPA-βAla-βAla-βAla-βAla-Gly-Ser-Ala-Tyr∼Gly-Lys-Arg-Lys(biotin)-amide. Parallel assays with a commercially available fluorogenic substrate (Suc-AAPF-AMC) for α-chymotrypsin were performed for comparison. Using the ICP-MS assay enzyme concentrations as low as 2 pM could be readily detected which was superior to the detection limit of an assay using the α-chymotrypsin fluorogenic substrate (Suc-AAPF-AMC). Furthermore, we demonstrated the use of this approach to detect chymotrypsin activity in HeLa cell lysates.
Keywords: ICP-MS, multiplex, α-chymotrypsin, proteases, lanthanides
Introduction
Proteases are known to be involved in a number of pathological conditions including cancer [1], diseases of the central nervous system [2], cardiac diseases [3] and numerous viral and infectious diseases [4]. The ability of proteases to catalytically and specifically cleave peptide substrates makes them ideal biomarkers for the diagnosis of disease [5]. Investigation of the pathological and physiological functions of proteases requires sensitive assays to detect their activity in complex tissue samples and other biological matrixes. Commonly protease assays use fluorogenic or chromogenic peptide substrates [6-9]. Fluorescence resonance energy transfer (FRET) substrates containing a fluorophore and a quencher attached to opposite ends of the peptide substrate are also a common choice [10, 11]. Other methods for detecting protease activity include assays based on calorimetric [12], amperometric [13], radioactive [14, 15], and chemiluminescent [16] detection. In addition assays based on analytical instruments such as HPLC, GC, MS, NMR or IR spectrometers have also been developed [17]. These methods usually involve the detection of the activity of a single protease or use a single protease substrate in each assay.
The development of multiplexed protease assays which are sensitive, reproducible and have a large dynamic range would be advantageous for applications in medicine and biology [18- 21]. The benefit of being able to quantify the activity of numerous proteases in a given sample is that it allows a multiparametric analysis to identify characteristic patterns of protease activity for diagnostic purposes. Multiplexed protease assays would be especially beneficial when only small quantities of an analyte solution are available. The optical assays commonly applied to detect protease activity are limited in their ability to be used for a multiplex detection. The shortfall of chromogenic and fluorescent methods lie in the broad overlapping absorption and emission wavelengths of the dyes used in these assays which complicate the use of multiple substrates in a single sample. A recent example of a duplex assay employing luminescence energy transfer to quantum dots is a good example of the challenges in multiplexing protease assays based on chromophores [22].
Using ICP-MS and elemental tagging overcomes the shortcomings of optical detection in multiplexed biological assays because signals from elemental tags are well resolved and essentially non-overlapping [23]. ICP-MS is a sensitive analytical tool (sub-part per trillion, or attomole/microlitre detection) which offers analyte quantification with high precision, low detection limits and a large dynamic range (9 orders of magnitude) [24]. The use of antibodies labeled with lanthanide metal tags has been demonstrated to be highly effective for the multiplexed detection of cell surface antigens [23] and in standard immunoaffinity assays [25].
Lanthanide metals have been used as tags in proteases activity assays but these methods exploit the long-lived luminescence of lanthanide ions such as Eu3+ and Tb3+ [26]. There have also been previous reports where elemental tagging has been used to detect proteins [27-30], but to the best of our knowledge this is the first example of using lanthanide ions in conjugation with ICP-MS to develop multiplexed protease assays. α-Chymotrypsin was used as a model enzyme and its activity was measured using a Lu-tagged peptide substrate to demonstrate the potential of elemental assays to detect protease activity.
Experimental Section
Materials
The protected amino acids, HOBt [N-hydroxybenzotriazole] and HBTU [2-(1H-benzotriazole-1-yl)-1,1,3,3- tetramethyl uranium hexafluorophosphate] were purchased from GL Biochem (Shanghai). The biotin labeled lysine [Fmoc-Lys (biotin)-OH] and Fmoc β Ala-OH were purchased from EMD Biosciences. Streptavidin Agarose bead suspension (binding capacity: >85 nmol free biotin/mL) was purchased from EMD Biosciences. α-Chymotrypsin from bovine pancreas Type II was purchased from Sigma and was used without any further purification. α-Chymotrypsin fluorogenic substrate [Suc-AAPF-AMC] was purchased from EMD Biosciences. NovaPEG Rink amide resin (capacity 0.67 mmole/g) was purchased from Nova Biochem. ICP-MS grade Baseline Hydrochloric acid (35%) was purchased from Seastar Chemicals. Iridium standard 1000 mg/L (1000 ppm) was purchased from SpexCertiprep and the lutetium plasma standard was purchased from SpecPure. All other reagents, including the lanthanide salts, buffer salts and solvents were bought from Sigma. All buffers were made using Milli-Q water (Millipore).
Peptide synthesis
Enzyme substrates were synthesized using standard Fmoc SPPS (solid phase peptide synthesis). Peptides were manually synthesized on NovaPEG Rink amide resin (0.67 mmole/g loading, 150 mg). Each amino acid was coupled using HOBt and HBTU to generate the active ester in the presence of DIEA (diisopropylethylamine) using DMF (dimethylformamide) as the solvent (resin: amino acid: HOBt: HBTU: DIEA- 1:4:4:4:8). Removal of the Fmoc group was done using 20% piperidine in DMF (20 minutes). For coupling Fmoc-Lys(biotin) OH, NMP (N- methyl pyrolidone) was used as the solvent. Each residue was coupled for 40 minutes. Deprotection and coupling was confirmed using the TNBS (2,4,6-trinitrobenzene sulfonic acid) test. NHS (N-Hydroxysuccinimide) and HBTU were used to attach DTPA (Diehtylenetriaminepentaacetic acid) to the N-terminus of the peptide. Molar ratios used were, resin: DTPA: DIEA: HBTU: NHS (1: 14: 140:28:28). The DTPA (512 mg) and DIEA (2.15 mL) were added to DMF (4 mL) and the suspension was heated to dissolve the DTPA. HBTU and NHS were dissolved in DMF and added to the DTPA solution after it had cooled to room temperature. The resulting solution was then added to the resin and allowed to couple overnight.
The protected peptide substrates were globally deprotected using TFA (trifluoroacetic acid)/water/TIS (triisopropyl silane) (95: 2.5: 2.5). Following evaporation of 90% of the solvent the peptide was precipitated from the TFA solution in cold diethyl ether.
Purification was carried out using reverse phase HPLC (high-performance liquid chromatography). Solvents used were ACN (acetonitrile) + 0.1% TFA and water + 0.1% TFA. The gradient used to elute substrate 1 was 2% ACN for 5 min, 45 min to 50% ACN, and 60 min to 100% ACN with a flow rate of 10 mL/min. The gradient used to elute substrate 2 was 2% ACN for 2 min, 15 min to 15% ACN, 40 min to 30% ACN, 45 min to 50% ACN, and 50 min to 100% ACN with a flow rate of 10 mL/min.
Loading the substrate with a lanthanide metal
The purified peptides were incubated with excess lanthanide salt (approximately 3 mM) in water at room temperature (10 min). Desalting was done using a C18 column (Bondapak® C18 125 Å, 37-55 μm, 5cc) that was pre-washed with 10 mM ammonium citrate buffer pH 6 (100 mL) and water (100 mL). The peptide was loaded on the pre-washed column. It was washed with 10 mM ammonium citrate buffer pH 6 (100 mL) and water (100 mL), the peptide was eluted in 50% ACN. The solution was frozen and lyophilized.
The lutetium loading of the peptide was confirmed using MALDI-MS (Matrix-assisted laser desorption/ionization-mass spectrometry) and the peptide was quantified by ICP-MS using a lutetium plasma standard (1 ppb)
α-Chymotrypsin ICP-MS assays
α-Chymotrypsin was reconstituted in 1 mM HCl containing 2 mM CaCl2. Aliquots were kept at -20 °C and were used within a week of reconstitution, according to the manufacturer's instructions. The assays were carried out in reaction buffer (100 mM tris pH 8, 10 mM CaCl2, 50 mM NaCl) at 25 °C. The reaction was stopped by diluting the aliquots into 8 M urea (5 μL in 100 μL). Streptavidin agarose bead suspension (70 μL) was added to each sample and was shaken for 30 min at 25 °C. Samples were centrifuged (13,000 rpm, 25 min) and the supernatant was diluted according to substrate and enzyme concentrations used. Blanks were treated identically but did not contain any enzyme. A sample containing the substrate and no enzyme, which was not treated with the streptavidin agarose bead suspension served as a positive control. Triplicate dilutions of the same sample were used to estimate instrumental error which was usually less than 3%. Error bars for graphs plotted represent standard deviation for 3 independent experiments each measured in triplicate.
α-Chymotrypsin fluorescence assay
α-Chymotrypsin fluorogenic substrate [Suc-AAPF-AMC] was used in control fluorescence assays. The assay was run using the same protocol as the ICP-MS assay with the elimination of the streptavidin agarose bead suspension pull down step. The reactions were stopped by diluting the aliquots in 8 M urea (10 μL in 100 μL). Blank samples were run without the enzyme. The fluorescence was measured using a Tecan safire 2 fluorescence plate reader with an excitation wavelength of 380 nm, emission wavelength of 460 nm, integration time was 40 μs, lag time was 0 μs, and bandwidth was 20 nm.
ICP-MS sample preparation
50 μL of diluted assay sample was added to 50 μL Ir standard (1 ppb in 10% HCl). Each sample was run on the ELAN DRCPlusTM (PerkinElmer SCIEX) in triplicate. The lutetium plasma standard, diluted to 1 ppb was used to quantify the samples. Full parameters for the ICP-MS experiments can be found in the supplementary material.
Cell Lysate assays
The cells (HeLa cells) were plated at 50% confluence in 100 mm tissue culture dishes in regular growth media (DMEM, 10% FBS, 2 mM L-glutamine, antibiotics). For Doxorubicin treated cells, Doxorubicin (1 μg/mL) was added in fresh media after 24 hours. A control set of cells was also prepared that were not treated with Doxorubicin. The cells were cultured for 48 hours. Cells were then collected with a scraper and pelleted (500 × g, 10 min). Cells were washed with cold PBS and lysed with cold lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate) on ice with occasional trituration. The cell debris was pelleted by centrifuging (10,000 × g, 10 min). The total protein concentration of the lysates was measured (A280, Nanodrop). Lysates were aliquoted into tubes and stored at -80 °C. Lysates were diluted in reaction buffer (100 mM tris pH 8, 10 mM CaCl2, 50 mM NaCl) to working protein concentrations. Substrate 2 (100 μM) was incubated (37 °C) with lysates (100 μg/mL), the reaction was stopped after 60 min by diluting in 8 M urea (5 μL in 100 μL). Streptavidin agarose bead suspension (70 μL) was added to each sample and was shaken for 30 min at 25 °C. Samples were centrifuged (13,000 rpm, 25 min) and the supernatant was diluted (final dilution was 200 times). A blank control was run that was treated the identically but did not contain lysate. A sample containing the substrate and no lysate, which was not treated with streptavidin agarose bead suspension served as a positive control.
Results and Discussions
Design of the assay
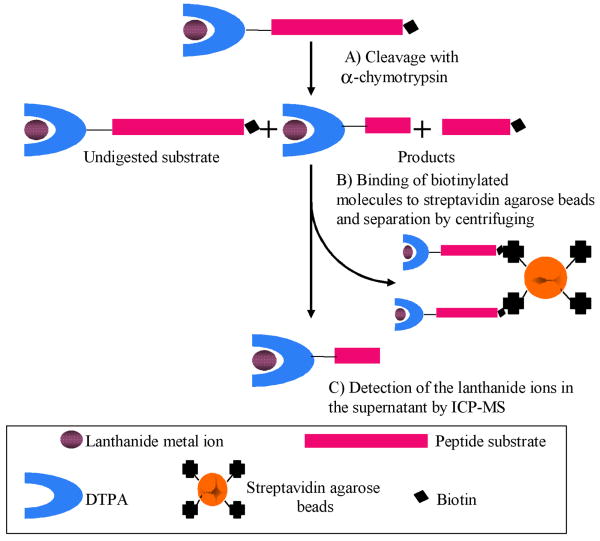
The design of the novel ICP-MS assay for α-chymotrypsin is shown in Figure 1. The assay is based upon a dual labeled peptide substrate that contains an N-terminal lanthanide chelate and a C-terminal biotin tag. The biotin tag was incorporated into the peptide sequence during the solid phase synthesis using the commercially available Fmoc-lys(biotin)-OH building block [31, 32]. Lanthanides are known to form thermodynamically and kinetically stable complexes with diethylenetriaminepentaacetic acid (DTPA)-based ligands and this chelate was coupled to the N-terminus of the peptide substrate [33, 34]. The sequences of the synthesized α-chymotrypsin substrates are shown in Table 1. Substrate 1 was designed based on a commercially available α-chymotrypsin substrate [35]. The lanthanide metal ions were loaded into the DTPA labeled substrates after peptide purification. In the study reported here lutetium ions were chosen as the metal tags but DTPA is a general chelator and would allow for any of the lanthanide ions to be used as the tagging element.
Fig. 1.
Outline of the ICP-MS based protease assay. The protease substrate was digested with α-chymotrypsin. The undigested substrate was separated from the digestion product by adding streptavidin agarose bead suspension followed by centrifuging. The supernatant was analyzed for lutetium signal by ICP-MS after dilution.
Table 1.
Sequences of the α-chymotrypsin substrates used
| Substrate 1 | DTPA-Asp-Leu-Leu-Val-Tyr∼Asp-Lys(biotin)-amide |
| Substrate 2 | DTPA-βAla- βAla-βAla-βAla-Gly-Ser-Ala-Tyr∼Gly-Lys-Arg-Lys(biotin)-amide |
| Fluorogenic substrate | Suc-Ala-Ala-Pro-Phe-(7-amino-4-methylcoumarin) |
Substrate 2 has a spacer of four β-alanine residues [36] between the DTPA-Lu complex and the site of enzymatic cleavage. Since β-alanine does not occur in nature, it is unlikely to be recognized by proteases and serves as a spacer without interfering with the substrate specificity of the enzyme.
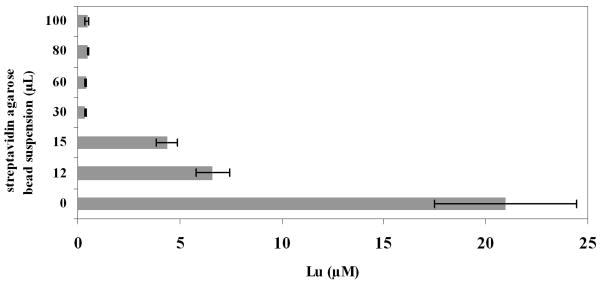
The enzyme assays were run under the optimized conditions for α-chymotrypsin (100 mM tris pH 8, 10 mM CaCl2, 50 mM NaCl at 25 °C). The reactions were quenched with 8 M urea. This quench is expected to denature all proteases while not affecting the coordination chemistry of the DTPA ligand. 8 M urea does not denature streptavidin, thus biotinylated species can be removed from the digestion by a streptavidin agarose bead suspension [37, 38] The amount of streptavidin suspension to effectively remove all of the biotinylated species in the reaction was optimized (Figure 2). It was found that a 50-fold excess of streptavidin agarose, based on commercial loading and total biotin-labeled peptides in the assay, reproducibly removed all the biotin-labeled peptide. After removing any undigested substrate the supernatant was diluted and the metal content was analyzed by ICP-MS. The lanthanide ion concentration of the supernatant is a direct measure of the amount of cleavage product formed by the protease. For blank controls, samples with no enzyme were treated with streptavidin agarose bead suspension to pull down all the substrate. For positive controls, samples not treated with streptavidin agarose bead suspension were used, as this would correspond to the maximum signal that could be obtained upon complete cleavage of substrate.
Fig. 2.
Optimization of the streptavidin agarose bead suspension required in the assay. A 20 μM solution of substrate 2 was diluted 20 times (5 μL into 100 μl 8 M Urea) and different amounts of streptavidin agarose bead suspension (binding capacity: >85 nmol free biotin/mL) was added and incubated for 30 minutes at 25° C with shaking. The supernatant was further diluted (20 times) and analyzed by ICP-MS. The data points represent an average ± 1 standard deviation for three independent experiments measured in triplicate.
Enzymatic studies
The ICP-MS assay protocol was used to digest substrate 1 with α-chymotrypsin. Unexpectedly under a wide range of experimental conditions investigated, less than 10% of substrate 1 was cleaved by the protease. Parallel fluorescence assays with a commonly used commercially available fluorogenic α-chymotrypsin substrate [Suc-AAPF-AMC] were performed as control experiments [39]. A competition assay between substrate 1 and the fluorescent substrate did not show any inhibition of the cleavage of the fluorescent substrate suggesting α-chymotrypsin had little affinity for substrate 1 (Supplementary Figure S1). It is likely that the DTPA-Lu complex was preventing substrate 1 from being cleaved by the enzyme. This could be due to reduced binding of substrate 1 to the enzyme active site or the inhibition of the catalytic step after the Michaelis complex has been formed.
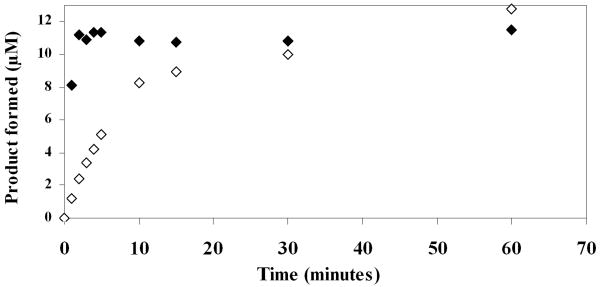
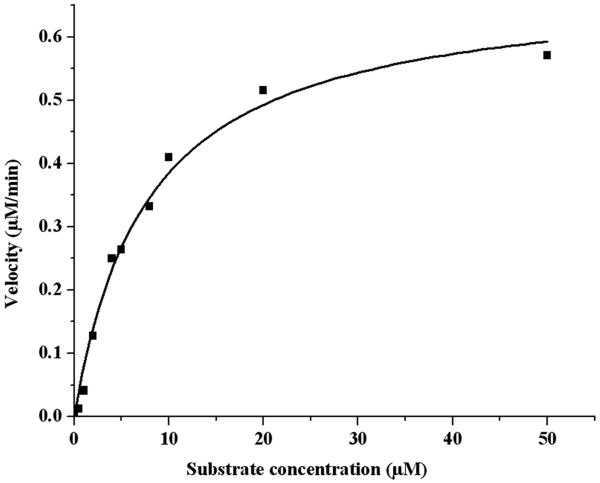
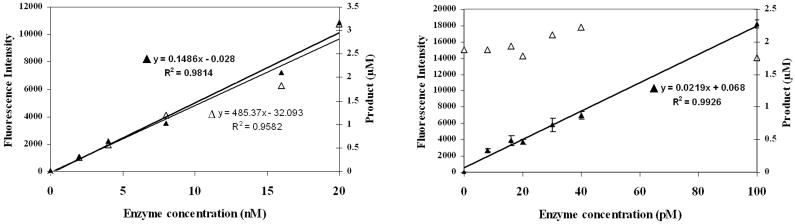
Substrate 2 was designed to have a spacer between the lanthanide tag and the site of enzymatic cleavage, and to further simplify the synthesis and handling of the substrate, a water soluble α-chymotrypsin substrate was chosen [40]. When substrate 2 (10 μM) was incubated with 20 nM and 200 nM enzyme, complete cleavage of substrate was seen in 30 minutes and 3 minutes, respectively (Figure 3). The Km for substrate 2, calculated using the ICP-MS assay, was found to be 7.8 μM and kcat was 0.57 s-1 (Figure 4). These values are comparable to those determined for the control fluorogenic substrate (Km 15 μM, kcat 1.5 s-1) [6] and to those of other chymotrypsin substrates [41- 43]. A linear relationship was observed between the enzyme concentration and product formed in the assay showing that the assay can be used quantitatively (Figure 5). At high enzyme concentrations, from 2 nM to 20 nM, both the fluorescence and the ICP-MS assays showed linear relationships between the product formed and the enzyme concentration in the assay. However, low enzyme concentrations of 8 pM to 100 pM were below the limits of detection for the fluorescence assay and no increase in signal over background (reading obtained at 0 pM enzyme concentration) was observed. In this range, however the ICP-MS assay showed a linear relationship between product formation and enzyme concentration. The error bars in Figure 5 represent a standard deviation of three independent experiments, each measured in triplicate, showing reproducibility of the method. Thus, the ICP-MS based assay protocol is sensitive and can be used to quantify enzyme concentrations in the low picomolar range, where the standard fluorescent substrate did not produce a robust signal. With the ICP-MS assay there was a 7 fold increase in signal above the background at enzyme concentrations as low as 2 pM, whereas under the same conditions, no α-chymotrypsin activity was detected using the fluorescence assay (Supplementary Figure S2). These results validate the use of ICP-MS as a simple, sensitive and reproducible method to detect enzyme activity.
Fig. 3.
α-Chymotrypsin cleavage of substrate 2. Substrate 2 (10 μM) was incubated with α-chymotrypsin 200 nM (◆) or 20 nM (◊) in reaction buffer (100 mM Tris pH 8, 10 mM CaCl2, 50 mM NaCl) at 25 °C. The reaction was stopped by diluting the aliquots in a 20 fold excess of 8 M urea. Samples were processed as described in the experimental methods section.
Fig. 4.
Determination of the Michaelis constants for substrate 2 and α-chymotrypsin. Substrate 2 was incubated in reaction buffer (100 mM Tris pH 8, 10 mM CaCl2, 50 mM NaCl) with α-chymotrypsin (20 nM) for 5 minutes. The reaction was stopped by diluting the aliquots in a 20 fold excess of 8 M urea. Samples were processed as described in the experimental methods section.
Fig. 5.
Comparison of the detection of α-chymotrypsin activity using ICP-MS and fluorescence based assays. A) Substrate 2 (20 μM, ▲) or Suc-AAPF-AMC (20 μM, Δ) was incubated in reaction buffer (100 mM Tris pH 8, 10 mM CaCl2, 50 mM NaCl) with α-chymotrypsin at 25 °C for 5 minutes. B) Substrate 2 (20 μM, ▲) or Suc-AAPF-AMC (20 μM, Δ) was incubated in reaction buffer (100 mM Tris pH 8, 10 mM CaCl2, 50 mM NaCl, 1 mg/ml BSA) with α-chymotrypsin at 25 °C for 60 minutes. The reaction was stopped by diluting the aliquots in a 20 fold excess of 8 M urea. Samples were processed as described in the experimental methods section. The data points represent an average ± 1 standard deviation for three independent experiments measured in triplicate.
Cell Lysate experiments
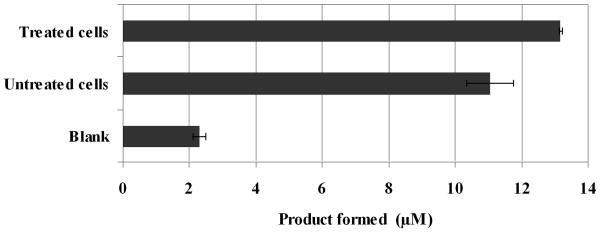
Doxorubicin, which is well known to induce apoptosis, was used to treat Hela cells in culture [18]. Substrate 2 was used to test the response between the doxorubicin-treated and untreated cells for the expression of α-chymotrypsin activity. Lysates from doxorubicin-treated cells show increased α-chymotrypsin activity (Figure 6). This is in agreement with observations from prior studies which suggest that, along with cysteine protease activity such as the caspases, serine protease activity is also increased when apoptosis is induced [18]. The fact that substrate 2 was used in cell lysates, suggests that such metal-tagged protease substrates will be effective for measuring protease activity in complex biological matrixes.
Fig. 6.
Quantitation of α-chymotrypsin activity in HeLa cell lysates. Cell lysates from cells treated or untreated with doxorubicin (1 μg/mL) were incubated with substrate 2 (100 μM) in reaction buffer (100 mM Tris pH 8, 10 mM CaCl2, 50 mM NaCl) for 1 hour. The reaction was stopped by diluting the aliquots in a 20 fold excess of 8 M urea. Samples were processed as described in the experimental methods section. The data points represent an average ± 1 standard deviation for three independent experiments measured in triplicate.
Conclusion
A novel protease assay using ICP-MS as a detection method is presented. The peptide substrates synthesized for α-chymotrypsin showed that the presence of a spacer between the lanthanide ion complex and the site of cleavage played a critical role in the affinity of the substrate for the enzyme. Lu-DTPA-βAla-βAla-βAla-βAla-Gly-Ser-Ala-Tyr∼Gly-Lys-Arg-Lys(biotin)-amide, designed as a α-chymotrypsin substrate, showed high affinity for the enzyme having a Km in the low micromolar range. Futhermore, α-chymotrypsin activity was detected in lysates of cells treated with doxorubicin showing an increase of serine proteolytic activity during the apoptotic cascade. The results show the ICP-MS assay offers a superior sensitivity to the fluorescence assay which is the most common method used to measure protease activity.
The potential for this assay lies in the ability to multiplex the experiment by adding additional peptide substrates synthesized in an analogous manner but tagged with different lanthanide metals. We have used α-chymotrypsin to develop and illustrate this analytical technique, but many other orthogonal protease substrates for enzymes such as the caspases, calpains, MMPs and ADAMs are available for multiplexed assays and our lab is continuing research in this direction.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from NIH grant #GM076127-01A1 for the support of this work. We also thank Professor M. Winnik for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stetler-Stevenson WG, Yu AE. Proteases in invasion: matrix metalloproteinases. Seminars in Cancer biology. 2001;11:143–152. doi: 10.1006/scbi.2000.0365. [DOI] [PubMed] [Google Scholar]
- 2.Molinari F. Extracellular proteases and their inhibitors in genetic diseases of the central nervous system. Hum Mol Genet. 2003;12:195–200. doi: 10.1093/hmg/ddg276. [DOI] [PubMed] [Google Scholar]
- 3.Singh RB. Role of proteases in the pathophysiology of cardiac disease. Mol Cell Biochem. 2004;263:241–256. doi: 10.1023/B:MCBI.0000041865.63445.40. [DOI] [PubMed] [Google Scholar]
- 4.Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends in Microbiology. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 5.Richard I. The genetic and molecular bases of monogenic disorders affecting proteolytic systems. J Med Genet. 2005;42:529–539. doi: 10.1136/jmg.2004.028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman M, Yurewicz E, Patel G. A new fluorogenic substrate for chymotrypsin. Anal Biochem. 1976;70:258–262. doi: 10.1016/s0003-2697(76)80066-8. [DOI] [PubMed] [Google Scholar]
- 7.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. PNAS. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosalia DN. Functional Phenotyping of human plasma using a 361-fluorogenic substrate biosensing microarray. Biotechnol Bioeng. 2006;94:1099–1110. doi: 10.1002/bit.20927. [DOI] [PubMed] [Google Scholar]
- 9.Rlangera BE, Cooper AG, Bendich AJ. On the Heterogeneity of Three-Times-crystallized α-Chymotrypsin. Biochemistry. 1964;3:1880–1883. doi: 10.1021/bi00900a015. [DOI] [PubMed] [Google Scholar]
- 10.Matayoshi ED, Wang GT, Krafft GA, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:754–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 11.Korkmaz B, Attucci S, Juliano MA, Kalupov T, Jourdan ML, Juliano L, Gauthier F. Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nature Protocols. 2008;3:991–1000. doi: 10.1038/nprot.2008.63. [DOI] [PubMed] [Google Scholar]
- 12.Williams BA, Toone EJ. Calorimetric evaluation of enzyme kinetic parameters. J Org Chem. 1993;58:3507–3510. [Google Scholar]
- 13.Ionescu RE. Protease amperometric sensor. Anal Chem. 2006;78:6327–6331. doi: 10.1021/ac060253w. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig R, Lucius R, Mentlein R. A radioactive assay for the degradation of neuropeptide. Biochimie. 1995;77:739–743. doi: 10.1016/0300-9084(96)88191-0. [DOI] [PubMed] [Google Scholar]
- 15.Wormser U, Zbinden G. Characterization of proteolytic systems in human and rat urine. Biochem Biophys Res Commun. 1985;127:191–197. doi: 10.1016/s0006-291x(85)80143-1. [DOI] [PubMed] [Google Scholar]
- 16.Richard JA, Jean Ludovic, Romieu A, Massonneau M, Fraissignes PN, Renard PY. Chemiluminescent probe for the in vitro detection of protease activity. Org Lett. 2007;9:4853–4855. doi: 10.1021/ol702190y. [DOI] [PubMed] [Google Scholar]
- 17.Wahler D, Reymond JL. Novel Methods for Biocatalyst screening. Current Opinion in Chemical Biology. 2001;5:152–158. doi: 10.1016/s1367-5931(00)00184-8. [DOI] [PubMed] [Google Scholar]
- 18.Henares TG, Mizutani F, Sekizawa R, Hisamoto H. Single-drop analysis of various proteases in a cancer cell lysate using a capillary-assembled microchip. Anal Bioanal Chem. 2008;391:2507–2512. doi: 10.1007/s00216-008-2105-x. [DOI] [PubMed] [Google Scholar]
- 19.Kim YP, Oh YH, Oh E, Ko S, Han MK, Kim HS. Energy Transfer-Based Multiplexed Assay of Proteases by Using Gold Nanoparticle and Quantum Dot Conjugates on a Surface. Anal Chem. 2008;80:4634–4641. doi: 10.1021/ac702416e. [DOI] [PubMed] [Google Scholar]
- 20.Medintzi IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot–peptide conjugates. Nature Materials. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 21.Clapp AR, Medintz IL, Uyeda HT, Fisher BR, Goldman ER, Bawendi MG, Mattoussi H. Quantum Dot-Based Multiplexed Fluorescence Resonance Energy Transfer. J Am Chem Soc. 2005;127:18212–18221. doi: 10.1021/ja054630i. [DOI] [PubMed] [Google Scholar]
- 22.Xia Z, Xing Y, So MK, Koh AL, Sinclair R, Rao J. Multiplex Detection of Protease Activity with Quantum Dot Nanosensors Prepared by Intein-Mediated Specific Bioconjugation. Anal Chem. 2008;80:8649–8655. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou XD, Zhang GH, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Polymer-based elemental tags for sensitive Bioassays. Angew Chem Int Ed. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ornatsky OI, Kinach R, Bandura DR, Lou X, Tanner SD, Baranov VI, Nitz M, Winnik MA. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2008;23:463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Careri M, Elviri L, Mangia A. Element-tagged immunoassay with inductively coupled plasma mass spectrometry for multianalyte detection. Anal Bioanal Chem. 2009;393:57–61. doi: 10.1007/s00216-008-2419-8. [DOI] [PubMed] [Google Scholar]
- 26.Karvinen J, Laitala V, Makinen ML, Mulari O, Tamminen J, Hermonen J, Hurskainen P, Haemmila I. Fluorescence quenching- Based assays for hydrolyzing enzymes. Application of time-resolved fluorometry in assays for caspase, helicase, and phosphatase. Anal Chem. 2004;76:1429–1436. doi: 10.1021/ac030234b. [DOI] [PubMed] [Google Scholar]
- 27.Hu S, Zhang S, Hu Z, Xing Z, Zhang X. Detection of multiple proteins on one spot by laser ablation inductively coupled plasma mass spectrometry and application to immunomicroarray with element-tagged antibodies. Anal Chem. 2007;79:923–929. doi: 10.1021/ac061269p. [DOI] [PubMed] [Google Scholar]
- 28.Careri M, Elviri L, Mangia A, Mucchino C. ICP-MS as a novel detection system for quantitative element-tagged immunoassay of hidden peanut allergens in foods. Anal Bioanal Chem. 2007;387:1851–1854. doi: 10.1007/s00216-006-1091-0. [DOI] [PubMed] [Google Scholar]
- 29.Razumienko E, Ornatsky O. Element-tagged immunoassay with ICP-MS detection: Evaluation and comparison to conventional immunoassays. J Immunol Methods. 2008;336:56–63. doi: 10.1016/j.jim.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettmer J. Elemental tagging in inorganic mass spectrometric bioanalysis. Anal Bioanal Chem. 2006;386:7–11. doi: 10.1007/s00216-006-0557-4. [DOI] [PubMed] [Google Scholar]
- 31.Andresen H, Grötzinger C, Zarse K, Birringer M, Hessenius C, Kreuzer OJ, Ehrentreich-Förster E, Bier FF. Peptide microarrays with site-specifically immobilized synthetic peptides for antibody diagnostics. Sens Actuators, B. 2006;113(2):655–663. doi: 10.1016/j.snb.2005.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown AM, George SM, Blume AJ, Dushin RG, Jacobsen JS, Sonnenberg-Reines J. Biotinylated and Cysteine-Modified Peptides as useful reagents for studing the inhibition of Cathepsin G. Anal Chem. 1994;217:139–147. doi: 10.1006/abio.1994.1094. [DOI] [PubMed] [Google Scholar]
- 33.Edwards WB, Fields CG, Anderson CJ, Pajeau TS, Welch MJ, Fields GB. Generally Applicable, Convenient Solid-Phase Synthesis and Receptor Affinities of Octreotide Analogs. J Med Chem. 1994;37:3749–3757. doi: 10.1021/jm00048a011. [DOI] [PubMed] [Google Scholar]
- 34.Moulin C, Amekraz B, Steiner V, Plancque G, Ansoborlo E. Speciation studies on DTPA using the complementary nature of electrospray ionization mass spectrometry and time-resolved laser-induced fluorescence. Appl Spectrosc. 2003;57:1151–1161. doi: 10.1366/00037020360696026. [DOI] [PubMed] [Google Scholar]
- 35.Tsubuki S, Kawasaki H, Saito Y, Miyashita N, Inomata M, Kawashima S. Purification and Characterization of a Z-Leu-Leu-Leu-MCA Degrading Protease Expected to Regulate Neurite Formation: A Novel Catalytic Activity in Proteasome. Biochem Biophys Res Commun. 1993;196:1195–1201. doi: 10.1006/bbrc.1993.2378. [DOI] [PubMed] [Google Scholar]
- 36.Kofoed J, Reymond JL. Identification of protease substrates by combinatorial profiling on TentaGel beads. Chem Commun. 2007:4453–4455. doi: 10.1039/b713595e. [DOI] [PubMed] [Google Scholar]
- 37.Martin CJ, Frazier AR. The urea denaturation of α-chymotyrpsin. J Biol Chem. 1963;238:3869–3875. [PubMed] [Google Scholar]
- 38.Kurzban GP, Bayer EA, Wilchek M, Horowitz PM. The quaternary structure of streptavidin in urea. J Biol Chem. 1991;266:14470–14477. [PubMed] [Google Scholar]
- 39.Oshima G. Interaction of a-Chymotrypsin with Dimyristoyl Phosphatidylcholine Vesicles. J Biochem. 1984;95:1131–1136. doi: 10.1093/oxfordjournals.jbchem.a134701. [DOI] [PubMed] [Google Scholar]
- 40.Nishikata M, Yoshimura Y, Deyama Y, Suzuki K. Continuous assay of protein tyrosine phosphatases based on fluorescence resonance energy transfer. Biochimie. 2006;88:879–886. doi: 10.1016/j.biochi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Bausert JH, Wolfbeis OS, Moser R, Koller E. Fluorometric continous kinetic assay of α-chymotrypsin using new protease substrates possessing long-wave excitation and emission maxima. Anal Biochem. 1988;171:393–397. doi: 10.1016/0003-2697(88)90503-9. [DOI] [PubMed] [Google Scholar]
- 42.Bauer CA, Thompson RC, Blout ER. The activity centers of Streptomyces griseus Protease 3. α-chymotrypsin, and elastase: Enzyme-Substrate interactions close to the scissile bond. Biochemistry. 1976;15:1296–1299. doi: 10.1021/bi00651a020. [DOI] [PubMed] [Google Scholar]
- 43.Case A, Stein RL. Mechanistic origins of the substrate selectivity of serine proteases. Biochemistry. 2003;42:3335–3348. doi: 10.1021/bi020668l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.