Iron is a nutrient that is indispensable for growth of almost all living organisms. The virulence of pathogenic organisms in the mammalian host is related to the availability of iron, therefore, microbial iron acquisition mechanisms are an important determinant of infection potential. Indeed, iron levels regulate 10–20% of all genes present on microbial genomes, including iron acquisition mechanism and virulence factor encoding genes. The association between iron and microbial virulence is well established (121,158). Many infectious diseases, such as gonorrhea, malaria, tuberculosis, and diarrheal infections, depend on the expression of microbial acquisition mechanisms capable of competing with the host’s iron scavenging mechanisms. The availability of iron in the mammalian host is extremely low, therefore, successful pathogens evolved strategies to overcome this limitation. Some pathogens produce high affinity metal chelators capable of competing with the host’s scavenging mechanisms, while others degrade and release mammalian proteins such as transferrin or hemoglobin that bind iron or hemin. Iron overload in the host from genetic predispositions, therapeutic intervention, or nutritional status can also increase the risk of infection by many pathogens, such as Plasmodium falciparum and Mycobacterium tuberculosis (121). Treating the host by administering iron chelators that capture free iron and compete with the acquisition system can limit microbial iron acquisition. However, this treatment also limits the availability of iron for the host, which can also have a damaging effect. Another way to alter iron availability is to interfere with the microbial iron acquisition mechanisms required by the pathogen to survive at specific stages of infection. Therefore, understanding the role of the iron acquisition mechanisms at various stages of bacterial infection may lead to the development of additional interventional strategies.
The oral cavity harbors at least 700 different species of bacteria (1,19,104,105,139). The contribution of many of those species to onset and progression of oral diseases is still to be established. However, several bacterial species are strongly implicated as aetiological agents of periodontal diseases. Among them are members of the Bacteroidetes phylum: Porphyromonas gingivalis, Prevotella intermedia, and Tannerella forsythia ((18,40,105). Of these, P. gingivalis is the most intensively investigated oral bacterium, and significant progress has been made in recent years regarding its iron uptake mechanisms. Because of this, we chose P. gingivalis as a model to describe the role of iron in host-pathogen interactions in the oral cavity.
P. gingivalis is a black pigmented, anaerobic, gram-negative bacterium that is implicated as a prime aetiological agent of initiation and progression of periodontal disease (40,46). As periodontal disease is also associated with increased risk of systemic conditions, such as coronary heart disease and preterm delivery of low birthweight infants (43,50,93,107,119,120,148), the burden of infection caused by P. gingivalis may be higher than previously estimated. The periodontal status of patients was significantly improved following elimination of the bacterium (reviewed in (40,113), however, current therapies still allow for recurrent infections (3,17,125). Currently, the standard therapeutic approach includes mechanical removal of the organism followed by antibiotic treatment. But rapidly increasing resistance to antibiotics and the potential for recurrent infection suggests more targeted and longer-lasting approaches are needed to reduce the levels of P. gingivalis in the oral cavity. To achieve this goal, a thorough knowledge of the biology of P. gingivalis is needed.
Periodontal diseases are triggered by microorganisms and manifest themselves in inflammation, bleeding, and destruction of soft and tooth tissues. P. gingivalis is found in healthy sites such as the buccal mucosa, tongue, tonsils, palate and supragingival dental plaque on the tooth enamel surface (86), but its numbers are significantly elevated in the subgingival environment and especially in diseased areas (92), suggesting the bacterium is well adapted and benefits from the hostile inflammatory state of the host. To survive in the diseased periodontal pocket the pathogen must be able to escape host defenses and acquire sufficient nutrients that will sustain its growth. One such nutrient required for growth of P. gingivalis is iron, and its presence has a profound effect on the bacterium’s virulence. Using a mouse abscess model, the Ebersole group demonstrated that bacteria grown under hemin-limitation were less virulent than their counterparts grown in hemin-excess (64,83,87). In this review, we discuss the mechanisms of iron acquisition by P. gingivalis. We also describe regulatory mechanisms, because excess iron reacts with peroxide to generate hydroxyl radicals that are detrimental to the bacterial cell. P. gingivalis is distantly related to bacteria in which iron acquisition and homeostasis mechanisms are well known, so this review provides novel supportive information with regard to iron acquisition mechanisms in bacteria.
Characteristics of iron
Iron is a transitional metal belonging to group 8 and period 4 of the periodic table. It can exist in reduced form, as ferrous iron (Fe2+) or oxidized form, as ferric iron (Fe3+). Ferrous iron is more soluble in water and is highly reactive, while ferric iron is less soluble (approximately 10−9 M at neutral pH) (9). Pure iron is rarely found in nature, as it readily oxidizes in the presence of oxygen or moisture.
Ferrous iron is an indispensable cofactor for most living organisms. The few exceptions include organisms in iron-depleted environments that evolved the ability to use similar metals such as manganese for catalytic purposes. Iron often exists as heme (composed of iron and protoporphyrin IX) in enzymes that carry out cytochrome catalyzed oxidation reactions, or in proteins transporting oxygen such as hemoglobin, myoglobin, and leghemoglobin. Redox enzymes also contain inorganic iron in the form of iron-sulfur clusters (nitrogenase, hydrogenase, oxidative phosphorylation enzymes). Other non-heme iron proteins include ribonucleotide reductase enzyme that reduces ribose to deoxyribose and thus is required for DNA synthesis (6).
Iron has the ability to be readily oxidized and reduced, which makes it essential to life, but it is also biochemically dangerous at the same time, because free iron is capable of catalyzing the conversion of hydrogen peroxides to free radicals. This reaction is known as the Fenton reaction:
The radicals can react with a variety of cellular components, such as nucleic acids (DNA, RNA), proteins, and lipids (and thus membranes), and thereby cause substantial damage that ultimately may kill the cell. To defend against toxic effects, living organisms use and store iron in the protein-bound forms. Therefore, iron levels must be sufficient for essential functions and low enough to prevent accumulation of free iron, which can damage cells via the highly reactive hydroxyl radicals.
In bacteria, iron is also required for processes such as respiration (complexed with redox enzyme systems), DNA synthesis, and free radical-scavenging mechanisms. Ferritin-like proteins are major intracellular iron storage proteins that protect against DNA damage by sequestering free reactive iron into an unreactive protein-bound form.
Availability of iron in mammalian host
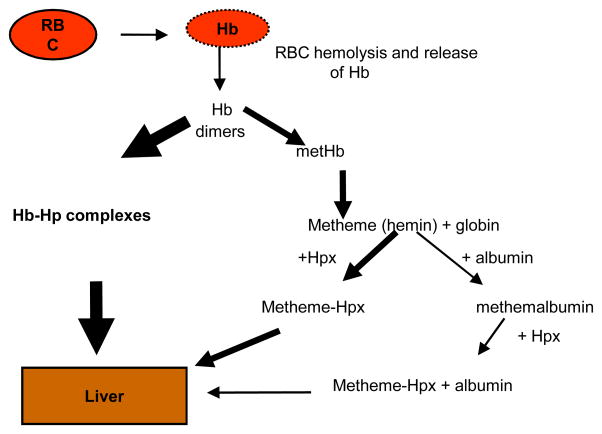
In mammals, approximately 76% of body iron is contained in red blood cells as hemoglobin. The erythrocyte hemoprotein contains iron bound to protoporphyrin IX to form hemin. The availability of hemin in physiological conditions is depicted in Fig. 1. Lysis of erythrocytes releases hemoglobin, which is then rapidly and strongly bound by haptoglobin (affinity less than 10−15 M for the hemoglobin-haptoglobin complexes) (55). In addition, any hemin released from the hemoglobin is rapidly bound by two other host hemin-binding proteins, hemopexin and albumin. These host proteins bind hemin with high affinity (10−12 M for hemin-hemopexin (54) and l0−8 M for hemin-albumin (8). While albumin is a highly abundant protein in serum (35–50 mg/ml), the concentrations of haptoglobin and hemopexin are in the range of 0.8–2.3 mg/ml and 0.4–1.5 mg/ml, respectively. The formation of hemo-protein complexes, however, is dependent on the affinity of the proteins to hemin or hemoproteins. Because haptoglobin has the highest affinity and scavenges hemoglobin very rapidly and tightly, the hemoglobin-haptoglobin complexes are the major hemin source in human hosts. Extracellular free iron is also rapidly bound by transferrin in serum and lactoferrin at mucosal surfaces.
Fig. 1. Availability of hemin in physiological conditions.
Upon vascular trauma, red blood cells (RBC) are released. Following RBC lysis, hemoglobin (Hb) is released. The released hemoglobin is rapidly bound by haptoglobin (Hp) and the complex is transported to the liver. This is the major hemin pathway. Upon depletion of Hp-binding capacity, hemoglobin degrades and the released hemin is bound by a high affinity serum protein, hemopexin (Hpx), and transported to the liver. Any remaining free hemin is bound by a lower affinity protein, albumin. The hemin-albumin complexes can also be bound by hemopexin and the heme-Hpx complexes can be transported to the liver.
Senescent red blood cells are processed by reticulo-endothelial macrophages that in turn provide for 90% of the iron required for erythropoiesis, which is the largest physiological process involving iron. Only approximately 10% of body iron comes from the diet (35). Dietary iron is absorbed by the duodenum and is bound by transferrin, which then transports the metal to various cells. When the iron-binding transferrin capacity is exceeded, iron toxicity may occur. For protection, mammalian hosts had to evolve complex control systems for transport and storage of the nutrient (52). The regulatory mechanisms needed to balance the release of iron from macrophages as well as uptake by enterocytes to fulfill the erythroblastic demand.
As mentioned, the ability of the ferrous form of iron to generate reactive oxygen species and hydroxyl radicals necessitates that any free iron be sequestered to prevent DNA and lipid damage. High affinity binding proteins such as transferrin (found in serum), lactoferrin (found in extracellular secretions), and ferritin (intracellular iron storage protein) (101) bind free iron. However, the majority of iron is sequestered as heme, mainly as hemoglobin. Heme also is found in myoglobin and serves as a prosthetic group for a variety of enzymes, including cytochromes, catalyses, and peroxidases.
The amount of free iron available for bacterial acquisition is extremely low (10−24 M), due to the presence of iron-binding proteins in mammalian blood. This amount may even be lower during infection, as the host downregulates iron availability with disease. Interferon-gamma plays a role in promoting the maturation of macrophages into phagosomes and ultimately in modulation of transferrin synthesis and expression of the transferrin receptor (117). However, iron has an inhibitory effect on interferon-gamma expression and thus reduces the expression of the transferrin receptor and ultimately limits the amount of iron available in the form of iron-loaded transferrin. Additional iron regulation occurs during the course of infectious diseases, for example, the recently-reported siperophore-chelator, lipocalin, complexes with siderophores and interferes directly with iron acquisition by microbial pathogens. Therefore, because chronic inflammation and infection affect the host’s iron metabolism and ultimately the availability of iron, determining the causes or effects of infectious diseases can be difficult. The current treatment for iron deficiency is blood transfusion of the host. However, too much free iron released from erythrocytes during transfusion may promote the growth of bacteria, and, as a result, the patient could die from an infectious disease instead of anemia. Therefore, giving blood transfusions for treatment purposes should be done judiciously.
Bacterial iron transport mechanisms
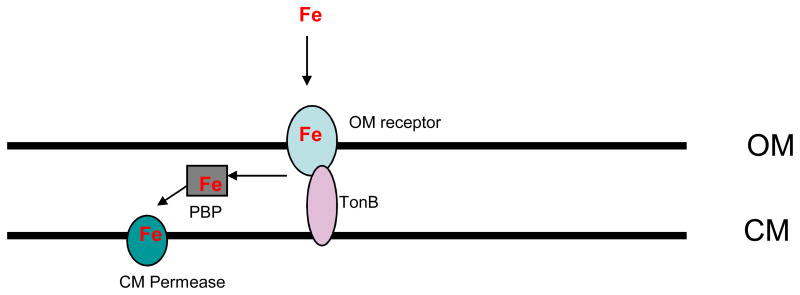
Because in vivo concentrations of free iron are too low (10−24 M) to support the growth of microorganisms, specialized iron acquisition mechanisms capable of extracting the metal from host proteins are needed for bacterial colonization of the human host. Bacteria must be able to acquire the iron from the host scavenging proteins either by proteolytic degradation and/or by expression of high affinity binding receptors capable of wresting the iron from the host hemin-binding proteins (71,98). Bacteria acquire both iron and hemin. Following removal of the metal by bacterial heme oxygenases, the internalized hemin serves as a source of iron. However, hemin is too large to freely diffuse through the bacterial membranes, so bacteria have also developed mechanisms to transport the hemin across the membrane(s). Both, gram-negative and gram-positive bacteria employ similar mechanisms that utilize cell surface receptors and membrane transport systems. In gram-positive bacteria the iron must transverse only one membrane, while in gram-negatives the metal must be transported across two membranes. The receptor-mediated transport across the outer membrane is energy-dependent and is powered by the energy transducing complex composed of three proteins: TonB, ExbB and ExbD (TonB/ExbB/ExbD) complex (13,74,157) (Fig. 2). The periplasmic heme is then bound by a periplasmic heme-binding protein that transports heme to cytoplasmic ATP-dependent permease for subsequent internalization and ultimate degradation into biliverdin, Fe, and carbon monoxide (CO) by heme oxygenase. Similarly, iron is scavenged by surface receptors and transported through the cytoplasmic membrane via ATP-dependent transporters (Fig. 2). It appears the organization of genes encoding iron-uptake systems in operons is common to all bacteria (141,147,161).
Fig. 2. Schematic representation of iron uptake in gram-negative bacteria.
Outer Membrane (OM), cytoplasmic membrane (CM). Iron (Fe) is captured by an outer membrane transporter (OM receptor) and transported through the outer membrane (OM) into periplasm where it is bound by a periplasmic iron binding protein (PBP). PBP then delivers Fe to cytoplasmic permease (CM permease) which transports the metal across the cytoplasmic membrane (CM) into the cytoplasm. The metal transport activity of the OM receptor is energy dependent which is delivered by a TonB protein (energy transducer).
Intracellular pathogens and iron
Some pathogens have evolved to evade the mammalian immune system (e.g., Mycobacterium tuberculosis, Salmonella, Shigella, Listeria monocytogenes and multiple oral bacteria including P. gingivalis, P. intermedia, T. forsythia). These pathogens are intracellular and reside within macrophages, phagosomes, or epithelial cells (36,37,68,160,165). The intracellular and extracellular environments differ in terms of nutrient availability, oxygen tension, temperature and the amount as well as form of available iron. Ferritin is an intracellular iron source and accounts for 23% of intracellular iron (while hemoglobin present in erythrocytes contains approximately 76% of the total body iron content). Intracellular pathogens have to compete for iron with the host’s pH-dependent metal transporter, natural resistance-associated macrophage protein (NRAMP)(15,45). A NRAMP-like mycobacterial metal transporter has been implicated in iron acquisition by M. tuberculosis (2). The differential expression of Shigella’s iron coding genes illustrates the differences between the intra- and extracellular environments; while the siderophore encoding genes are expressed extracellularly, the expression of aerobactin synthesis and ferrichrome receptor genes is elevated in the intracellular environment (160). This differential gene expression provides support that siderophores scavenge iron from extracellular sources.
P. gingivalis and hemin/iron
P. gingivalis requires hemin for growth (122). Analogous to other anaerobes, such as Prevotella ruminicola and Bacteroides fragilis, cell yields of the bacterium in the absence of hemin are extremely low (82). Hemin is required for synthesis of the cytochrome b subunit of P. gingivalis fumarate reductase. Since reduction of fumarate to succinate is the central pathway for generation of metabolic energy in P. gingivalis, it is not surprising that hemin is a critical growth factor for this bacterium (116). P. gingivalis is incapable of synthesizing protoporphyrin IX, the precursor of hemin, due to the lack of the enzymes 5-aminolevulinic acid synthase and porphobilinogen deaminase (116). Therefore, the pathogen requires exogenously supplied hemin to grow. Finally, hemin stored on the surface of P. gingivalis cells in the form of a black pigmentation composed of μ-oxo-heme may provide a protective role against peroxide stress (137).
P. gingivalis’ primary niche is the periodontal pocket bathed in gingival crevicular fluid. The composition of gingival crevicular fluid is similar to that of serum and contains an array of hemoglobin and hemin binding proteins, including haptoglobin, hemopexin, and albumin. Thus, the main source of hemin in gingival crevicular fluid is hemoglobin derived from lysed erythrocytes. Of note, Shizukuishi et al. (126) reported that P. gingivalis uses hemoglobin as a source of iron more effectively than it uses other iron sources. The concentrations of hemoglobin in crevicular fluid may vary depending on the degree of bleeding associated with periodontal tissue destruction, but this has not been precisely defined in the literature. Hemoglobin, as noted previously, is not available to P. gingivalis, because it is rapidly and irreversibly bound by haptoglobin in crevicular fluid. Thus, to overcome the scavenging effect of haptoglobin, P. gingivalis must have effective mechanisms for removal of hemin from the hemoglobin-haptoglobin complex. Other, less abundant hemin sources include hemin-hemopexin complexes and hemin-albumin complexes. However, these are hemoproteins, and thus successful bacterial pathogens must be able to acquire the nutrient from the host scavenging proteins either by proteolytic degradation and/or by expression of high affinity binding receptors capable of sequestering the hemin out of the host hemin-binding proteins. Although P. gingivalis appears to preferentially acquire hemin, inorganic iron, transferrin, and lactoferrin have also been shown to support growth of this organism (69), suggesting different mechanisms of iron acquisition exist in P. gingivalis. Many pathogenic bacteria produce siderophores that are capable of chelating iron from transferrin and lactoferrin (22). However, P. gingivalis lacks a siderophore system, so siderophore-independent mechanisms of iron acquisition must be present in this organism. P. gingivalis has been demonstrated to invade and survive in a variety of mammalian cells (32,36). The iron acquisition mechanisms and sources of iron available in these various environments may differ from the ones playing a role in the extracellular environment.
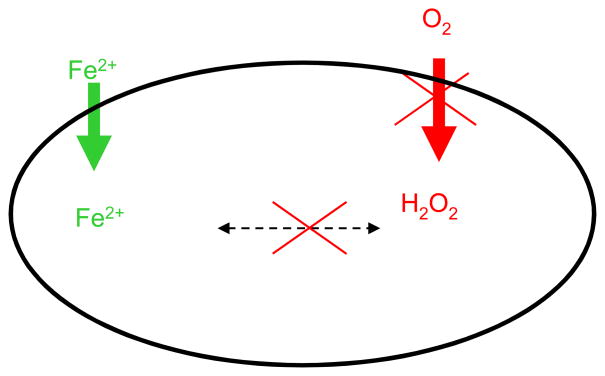
P. gingivalis and the entire Bacteroidetes phylum are quite distant from γ-proteobacteria in which the iron acquisition mechanisms have been intensely investigated. Therefore, the mechanisms of metal acquisition in the Bacteroidetes group may also be different, so further investigation may provide novel insight into iron acquisition in anaerobic bacteria. Iron is more soluble in anaerobic conditions and thus more available for uptake by bacterial cells. Oxidative stress generated in the presence of ferrous iron, on the other hand, may not be as problematic in the absence of oxygen (Fig. 3).
Fig. 3. Iron and oxygen in anaerobic conditions.
Ferrous iron (Fe2+) is more soluble in anaerobic conditions and thus more available for bacterial uptake (the increased iron transport in anaerobic conditions when compared to aerobic one is designated by green arrow). At the same time lack of oxygen in anaerobic conditions leads to reduced oxygen metabolism and thus reduced generation of hydrogen peroxide (H2O2). The reduction in generation of is designated by red arrow. Ultimately, generation of hydroxyl radicals resulting from interaction of iron and peroxide is reduced/or eliminated in the absence of oxygen.
P. gingivalis and hemin/iron acquisition mechanisms
(i) Hemagglutinin/protease family
In conditions where heme is not readily available, the secretion of proteins that bind red blood cells, lyse them, and release hemoglobin is highly advantageous, as it significantly increases the levels of hemin available to pathogens. Therefore, the ability of a microorganism to bind erythrocytes would be expected to increase the iron concentration in the vicinity of the bacterial cells.
P. gingivalis encodes a family of protease/hemagglutinins termed gingipains. Two genes, rgpA (encoding arg-gingipain, RgpA) and kgp (encoding lys-gingipain, Kgp), code for multidomain high molecular weight complexes composed of N-terminal proteolytic domains and C-terminal adhesin domains (24). While the RgpA has specificity for the arginyl peptide bonds (Arg-X specificity) the Kgp is specific for the lysyl bonds (Lys-X specificity). The C-terminally encoded hemagglutinin/adhesin domains were demonstrated to bind a variety of substrates including fibrinogen and fibronectin, as well as, red blood cells (14,23,57,106,124). Another member of the protease/hemagglutinin gene family, hagA (coding for hemagglutinin A), contains four direct repeats that are each 1350 bp in length and code for hemagglutinin/adhesin activities (47). The repeats are highly similar to the hemagglutinin-encoding regions of both gingipain genes: rgpA and kgp. Also, another member of the family, rgpB, codes for a second arg-gingipain (Arg-X specific protease) without the C-terminal hemagglutinin/adhesin domain. The sequence of the rgpB is similar to the protease-encoding portion of the rgpA gene.
Besides the protease associated hemagglutinins, P. gingivalis encodes several other proteins capable of binding red blood cells (95). Hemagglutinins such as hemagglutinin B (HagB) and hemagglutinin C (HagC) were demonstrated to bind erythrocytes (73,110). Also, HagA-like hemagglutinins either encoded by hagA or associated with the cysteine-specific proteases (gingipains) were shown to bind red blood cells (109). Once bound, the erythrocytes are disrupted by hemolysins and hemoglobin is released. Then hemoglobin is bound by hemoglobin-binding proteins such as the hemoglobin receptor (HbR) encoded by the gingipain genes (94). The hemoglobin binding to this receptor was shown to be mediated by the porphyrin IX component of hemoglobin, further demonstrating that HbR is also a hemin receptor (31). This receptor is proposed to function as a high affinity hemophore at the bacterial cell surface that captures hemin from hemoglobin, presumably after hemin is released from the globin, a process mediated by the P. gingivalis proteases (102).
The P. gingivalis gingipains play an indispensable role in the release of hemin from hemoglobin. The first indication of the involvement of the proteases was observation that mutation of the lys-gingipain protease, Kgp, renders the bacterial colonies white when grown on blood agar plates, indicating the protease plays a major role in acquisition of hemin from hemoglobin (75,96). Subsequently, Kgp was demonstrated to play a role in degradation of hemoglobin (75,140). Fujimura et al. (41) showed that the arg-gingipain proteases, Rgps, also play a role in hemoglobin degradation. More recent work by the Smalley et al. group demonstrated the involvement of both proteases in pigment formation in P. gingivalis (135,136,138). The Rgp activity was essential for conversion of oxyhemoglobin into methemoglobin, which then was more susceptible for degradation by the Kgp protease. The human alpha globin chain contains 11 Lys residues and the beta chain has 10 Lys residues, so the globin molecule can be degraded into small peptides suitable for bacterial nutrition. The role of Kgp in the acquisition of amino acids (aa) from hemoglobin is still to be demonstrated. The hemin released from hemoglobin is then bound by the hemin/hemoglobin receptor, HbR, which also plays a role in formation of the μ-oxo bishaem on the surface of P. gingivalis (134).
It is noteworthy that the Curtis et al. laboratory has reported inhibitory activity of hemin on the Arg-X protease (153). Similar results were also obtained in the Lewis laboratory (Lewis et al., unpublished). The Arg-X protease is crucial in the release of hemin from hemoglobin, so the concentrations of hemin could serve as one mechanism to keep the protease activity in check. Ultimately, regulation of hemin availability in this manner could prevent excessive accumulation of the nutrient and avoid possible oxidative stress resulting from the presence of free hemin in the vicinity of the bacterial cell. On the other hand, hemin is required for bacterial growth and increased hemin concentrations correlated with faster growth rates as well as higher proteolytic activities of P. gingivalis cells (83,132). Thus, higher hemin availability may also serve as a signal for increased proteolytic activity of the bacteria ultimately enabling growth.
In addition to a role in the release of hemin from hemoglobin, P. gingivalis proteases may play a role in the release of hemin from hemalbumin (131). Further, gingipains were demonstrated to degrade other host iron proteins such as haptoglobin, hemopexin, and transferrin (140). Thus, protease action appears to significantly contribute to the availability of iron for bacterial uptake.
Hemin uptake mechanisms
Once released from host proteins, the hemin/iron must be transported into a bacterial cell. P. gingivalis is a gram-negative bacterium requiring transport of hemin across two membranes. As mentioned above, genes coding for iron transport functions across the membranes are usually clustered together on the genome. Three such multigenic clusters encoding proteins thought to be involved in the hemin acquisition pathways have been detected in the genome of P. gingivalis W83 (Fig. 3)(95). The first locus, designated ihtABCDE (iron-heme transport), is composed of five open reading frames (ORFs) coding for a TonB-dependent outer membrane receptor (IhtA), lipoprotein (IhtB), periplasmic binding protein (IhtC), permease(IhtD), and cytoplasmic ATP binding protein (IhtE) (30). The IhtAB components resemble a two-component receptor analogous to the transferrin receptor, TbpBA (20,72). The surface exposure and hemin-binding ability of the IhtB protein has been demonstrated, however, the exact contribution of the locus to hemin uptake in P. gingivalis, as well as the specificity of the receptor for hemin sources remains unknown. The second locus has a similar composition in that a TonB-dependent receptor Tla (also known as Tlr for TonB-linked receptor) is followed by a putative ATP-binding cassette hemin transport system, htrABCD (38,130). Direct evidence for the role of the locus in hemin uptake was obtained using a Tla deficient mutant. Growth of this mutant was significantly impaired on low levels of hemin (38). As in the case of the iht locus, the specificity for various hemoproteins or hemin alone is still to be determined.
Our laboratory identified a third hemin uptake locus, designated hmu (77). A portion of the locus was first reported in P. gingivalis 53977 by Karunakaran et al. (61), who demonstrated the presence of a hemin-repressed gene encoding a TonB-dependent outer membrane protein, designated HemR (hemin-regulated receptor). Upstream of hemR, a 429-bp open reading frame (orfl) was located. The two genes were independently transcribed and while hemR was repressed by hemin, orfl was upregulated in the presence of hemin. Using a different P. gingivalis strain, A7436, Simpson et al. (127) demonstrated the presence of a homolog of this locus consisting of hmuY (gene 90% identical to orfl) and hmuR (gene identical at the 5′ end to hemR but differing significantly at the 3′ end). hmuY and hmuR were demonstrated to be co-transcribed and repressed by iron. In addition, Liu et al. (81) has demonstrated that hmuR is also negatively regulated by hemin. Further analysis of the sequences surrounding hmuYR in the genome of P. gingivalis W83 revealed another portion of the locus, designated hmuSTUV, which is also a part of the same operon (77). The hmuSTUV portion of the locus encodes a fusion of a putative cobalamin biosynthesis [CobN/magnesium chelatase (HmuS)], two putative proteins containing multiple predicted transmembrane regions (typical for permeases) (HmuT and HmuU), and putative protein containing an N-terminal transmembrane region (suggesting it is membrane-associated). The structural composition of proteins encoded by the hmu locus is consistent with that of other gram-negative bacterial iron transport systems that are typically composed of an outer membrane receptor, a periplasmic binding protein, inner membrane transporter (permease), and inner membrane-associated protein that can bind and hydrolyze ATP (Fig. 2). A novel feature, when compared to other hemin uptake loci, is the presence of hmuS gene encoding putative chelatase. This finding suggests that the hemin uptake mechanisms in P. gingivalis may also be novel compared to other bacterial iron uptake systems. The hmu locus has been found in most P. gingivalis strains and similar loci are present in other anaerobic bacteria, including B. fragilis, B. thetaiotamicron, and P. intermedia. Growth of Bacteroidetes is dependent on hemin, so it is probable that the P. gingivalis hemin uptake locus, hmu, encodes a universal hemin uptake system in these organisms. As such, the locus may serve as a model system for examination of hemin uptake in the Bacteroidetes phylum of microorganisms.
Further analysis of the locus revealed that the genes within the locus are co-transcribed (77,99). Although expression of all genes is iron repressible, there is differential regulation within the locus resulting in overexpression of the promoter-proximal genes when compared to promoter distal genes. Such differential regulation may account for overexpression of the outer membrane receptor components and thus increase the chances of hemin capture by P. gingivalis. Once captured, the hemin may then be transported through the inner membrane component of the system.
Several studies have demonstrated that the hmu locus plays a role in hemin uptake in P. gingivalis (80,97,100,128)(Lewis et al., unpublished). A mutant deficient in the locus did not grow well in low concentrations of hemin, hemoglobin, and human serum supplemented with hemin as the hemin source. Of note, the mutant failed to grow with hemoglobin-haptoglobin complexes as hemin sources (Lewis et al., unpublished). These data indicate the hmu operon is required for growth of P. gingivalis with hemoglobin-hapoglobin complexes as the hemin source. This is physiologically relevant because released hemoglobin is rapidly bound by a haptoglobin, and thus hemoglobin-haptoglobin complexes rather than free hemoglobin would be expected to be encountered in the mammalian body.
The contribution of the hmu locus to hemin acquisition was also verified by examining the ability of the mutant devoid of the locus to transport radiolabeled hemin. Bacterial cells derived from both iron replete- and iron deplete-conditions were examined. Hemin uptake was elevated by iron depletion, and while in iron depleted conditions the mutant strain showed a four-fold reduced ability to take up hemin. No hemin uptake was observed in the mutant strain in hemin replete conditions (77). On the other hand, no significant differences in hemin binding were observed between the mutant and parental cells derived from hemin-replete or hemin-deplete cells, suggesting there are other components accounting for hemin binding in P. gingivalis (77). It is likely these are non-specific hemin-binding structures.
The contribution of the hmu locus to the binding of hemin and hemoproteins was also examined using spectrophotometric assay or ELISA (80). Reduced binding of hemin, hemoglobin and human albumin-serum complexes was observed in HmuR-deficient mutants. Using Escherichia coli strains overexpressing various forms of the HmuR protein, the contribution of the four amino acid asparagine-proline-aspartic acid-leucine (NPDL) motif to hemin binding was demonstrated (80).
The HmuY protein
Analysis of outer membrane protein profiles demonstrated that 23 kDa protein is the major protein upregulated in iron-deplete conditions (77). N-terminal sequence analysis of a 21 kDa fragment of the protein resulting from processing by the P. gingivalis lys-gingipain protease, Kgp, revealed it was the hemin uptake protein, HmuY. Further, iron-dependent regulation as well as the outer membrane location was confirmed using Western blot analysis with antibody specific to the HmuY protein (77).
The HmuY protein is likely the same as the 26 kDa hemin – repressible protein characterized by Bramanti et al. (11). This protein played a role in hemin acquisition, was surface exposed in hemin limited conditions, and bound hemin (12). The sequence of the protein matched that of the HmuY sequence (66). Further characterization of the HmuY protein demonstrated that it may exist in multimeric form ranging from monomers to tetramers (77). The tetrameric form was shown to be involved in hemin binding (100). Recently, the crystal structure of the hemin-bound HmuY protein was determined (159). The protein was shown to exist in tetrameric form with hemin, and analysis revealed an all-beta fold that mimicked a right hand. Finally, HmuY was shown to bind ATP (99), which supports the slight phosphatase activity exhibited by the protein (89).
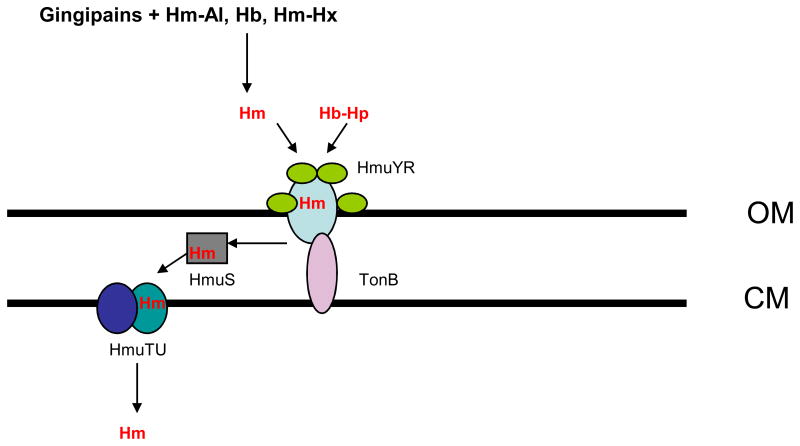
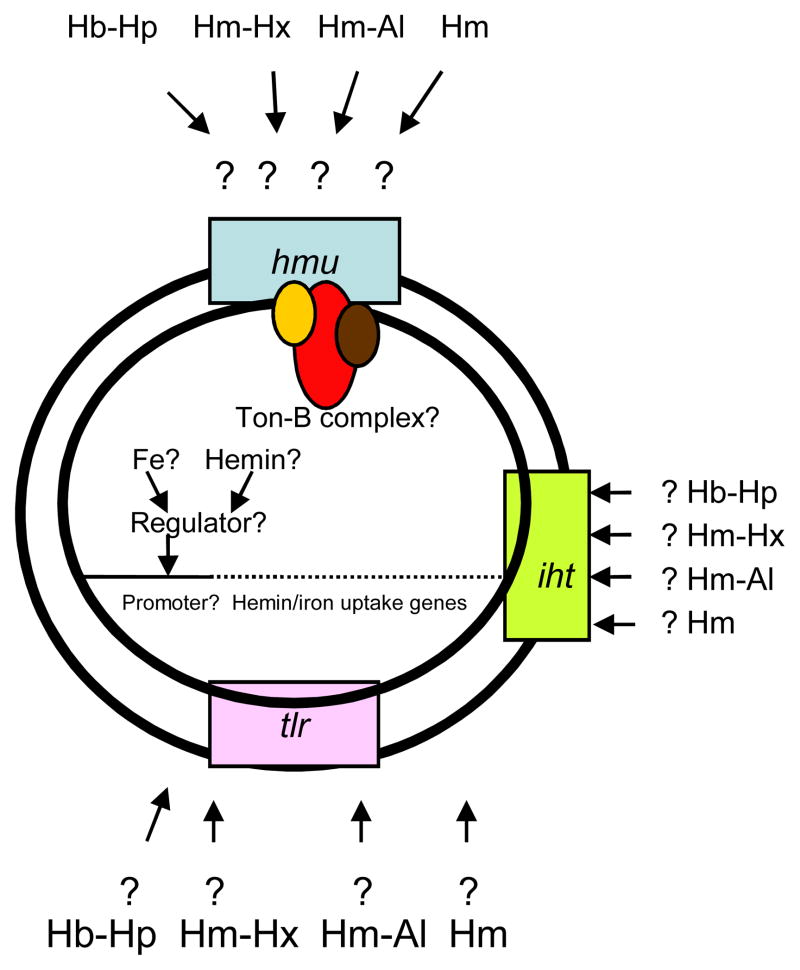
Based on the available data regarding the role of the hmu operon in hemin uptake, the following model can be proposed. The hmu locus appears to play a role in hemin acquisition from hemoglobin-haptoglobin complexes. Such complexes are formed following the release of hemoglobin from lysed erythrocytes, which are then rapidly bound by haptoglobin. Thus, the hemoglobin-haptoglobin complexes are present in the extracellular environment and the hmu-encoded proteins might play a role in iron acquisition by extracellular bacteria. While the outer membrane proteins, HmuY and HmuR, could function as an outer membrane hemin receptor (analogously to the transferrin receptor, TbpBA) (20), we propose the HmuSTUV proteins are involved in processing and transport of the hemin molecule across the inner membrane. The genomic location and structural features of the HmuY and HmuR proteins suggest that they form a two-component HmuYR outer membrane receptor (similar to that found in Neisseria species)(20). The differential expression within the hmu locus results in overexpression of the hmuY-specific transcript when compared with the hmuR transcript. The significantly larger levels of the hmuY transcript may also explain the observation that the HmuY lipoprotein is preferentially observed as the major outer membrane protein in iron deplete conditions (77). Again, this observation mirrors a similar one for the TbpBA receptor, in which the lipoprotein is more abundant than the TonB-dependent receptor (115). In Neisseria, the TbpB protein is also a lipoprotein playing an accessory role in the acquisition of iron from transferrin, while the TbpA protein is a TonB-dependent outer membrane receptor transporting the iron across the outer membrane. Although only the TbpA component is indispensable for iron acquisition, the TbpB lipoprotein increases the efficiency of the iron removal from transferrin (5,21). The exact mechanism of iron removal from transferrin by the TbpBA receptor is still to be determined. As for the P. gingivalis HmuYR receptor, the HmuY protein was demonstrated to bind hemin, while HmuR binds both hemin and hemoglobin (80,99). The affinity of the proteins for hemin, when examined separately, is too low to compete for hemin with the hemoglobin-haptoglobin complexes, however, it is probable that the two proteins work cooperatively when extracting hemin from the hemoglobin-haptoglobin complexes. Establishing this potential will require determination of the HmuY-HmuR interaction using molecular, biochemical, and structural approaches. As the two-component receptors are implicated in acquisition of iron from iron-protein complexes, and the single component receptors play a role in transport of uncomplexed iron, the presence of the two components in the P. gingivalis hemoglobin-haptoglobin receptor would be expected. The hmu locus also contributes to transport of uncomplexed hemin (77,80), Lewis et al., unpublished). The provision of uncomplexed hemin may be aided by the action of the P. gingivalis gingipain proteases, which have been demonstrated to extract hemin from hemoglobin (as described above)(75,135,140). The proteases also were shown to degrade hemalbumin, hemopexin, and haptoglobin (140), which would also be expected to contribute to the release of hemin for uptake by the hmu -encoded hemin transport mechanism. The structural assignments of proteins encoded by the hmuSTUV portion of the locus suggest that this section plays a role in transport of hemin from the periplasmic space and through the inner membrane. The two permeases, HmuS and HmuT, would thus transport hemin across the inner membrane. The role of the putative chelatase, HmuS, is still to be determined, as it appears to be a novel component when compared with hemin uptake loci in other bacteria. The proposed functional assignment of the Hmu proteins is depicted in Fig. 5. Of note, the iron-hemin transport locus, iht, also codes for outer membrane proteins capable of forming two-component receptors and thus may play a significant role in acquisition of heme complexed with host proteins.
Fig. 5. Schematic representation of the predicted role of proteins encoded by the hemin uptake, hmu locus.
Hemoglobin-haptoglobin complexes (Hb-Hp), hemin-hemopexin complexes (Hm-Hx), hemin-albumin complexes (Hm-Al) are degraded by P. gingivalis cysteine proteases, gingipains. The released hemin (Hm) is then captured by the cell surface lipoprotein, HmuY, which delivers it to the outer membrane transporter, HmuR. Alternatively, the HmuY interacts with HmuR and extracts hemin from the Hb-Hp complexes. Hm is transported through the outer membrane (OM) into periplasm by HmuR where it interacts with a novel chelatase-like protein, HmuS delivering it to the cytoplasmic memmebrane (CM). Ultimately, two permease-like proteins, HmuT and HmuU, transport Hm into cytoplasm.
Other TonB proteins, as predicted using the genomic sequence of P. gingivalis W83, include PG0637, PG0899, PG1242, PG1655, and PG1752 (www.oralgen.lanl.gov). Finally, the rag locus coding for TonB-dependent outer receptor (RagA) and outer membrane lipoprotein (RagB) also may play a role in metal acquisition (25). The roles of the various two-component hemin receptors, as well as the hemin uptake loci, are still to be determined.
Energy sources for iron/hemin transport in P. gingivalis
The TonB protein was demonstrated to physically interact with the outer membrane TonB-dependent receptors (129). Several of the P. gingivalis outer membrane hemin receptors, such as the HmuR protein, contain a TonB binding motif (127) and therefore we expect the P. gingivalis receptors also associate with TonB-like proteins. Two putative TonB proteins, PG0707 and PG1912, are encoded on the genome of P. gingivalis W83 (www.oralgen.lanl.gov) (95). PG0707 expression was induced in iron-deplete conditions (Lewis et al., unpublished), indicating that the protein plays a role in iron acquisition. It is noteworthy that the PG0707-encoding locus contains only one novel gene while similar loci found in Eubacteria also contain genes coding for accessory proteins, ExbB and ExbD (70,74). PG1912, on the other hand, is preceded by an ORF (PG1913) coding for a putative DNA-binding regulator with repressor activity (www.oralgen.lanl.gov). Again, no TonB-accessory proteins are encoded by the locus. These observations suggest the energy transduction mechanism in P. gingivalis is different from that observed in Eubacteria.
Non-heme iron acquisition mechanisms
P. gingivalis can also use transferrin as an iron source (126). Transferrin-binding ability that is rapid, reversible, and specific has been demonstrated in this bacterium (146). Transferrin binding is elevated in iron-depleted conditions indicating that expression of the transferrin-binding component is elevated in iron limited conditions. However, the specific transferrin receptor has yet to be identified.
P. gingivalis grows only in conditions where the oxygen concentrations are reduced (34)(Lewis et al., unpublished) and the pathogen prefers very low oxygen concentrations. In the deoxygenated environment, the reduced form of iron (ferrous iron) would be expected. As noted earlier, ferrous iron is more soluble and thus more available for bacterial uptake. Ferrous iron may be transported by the ferrous iron transporter, FeoB. FeoB protein was initially identified in E. coli K12 and characterized as a cytoplasmic membrane ferrous iron transporter (60). Currently, genes encoding homologs of the FeoB protein are shown to be present in a number of bacteria. Also, FeoBs of several bacteria were demonstrated to be required for high affinity ferrous iron uptake (114,152,155). The P. gingivalis W83 genome encodes two proteins whose products exhibited sequence similarity to the FeoB of E. coli K12. FeoB1 encoded by PG1138 is 844 aa in length and shows 52.6% similarity and 39.2% identity to the E. coli FeoB. FeoB2, encoded by PG0930, is 725 aa in length and exhibits 40% similarity and 31% identity with the E. coli FeoB. Interestingly, the specificity of the transporters differs, and while the P. gingivalis FeoB1 transports iron the FeoB2 has been implicated in manganese accumulation (28). To further investigate the role of the P. gingivalis FeoB1 and FeoB2 proteins in divalent metal cation transport, the feoB1 and feoB2 mutants, as well as the wild-type W83 were examined for their ability to acquire 55Fe2+ and 54Mn2+. Iron uptake in P. gingivalis occurred very rapidly and, whereas iron uptake was elevated throughout the experiment in the feoB2 mutant strain (1.5- to 2-fold higher than the parental W83 strain, suggesting FeoB2 is not a major iron transporter in P. gingivalis), the feoB1 mutant lost its ability to transport iron, indicating FeoB1 is a major iron transporter in P. gingivalis (51). Uptake of manganese in P. gingivalis W83, accomplished using 50 nM of extracellular 54Mn2+, suggested the presence of a high-affinity manganese transporter. The feoB2 mutant exhibited a drastic decrease in 54Mn2+ uptake compared to the wild-type strain (approximately 12-fold reduction observed after 20 min of incubation) (51), demonstrating the FeoB2 manganese-transport ability. No reduction in manganese uptake was observed in the FeoB1-deficient mutant, suggesting this protein is not a major transporter of manganese in P. gingivalis (Anaya-Bergman, et al., unpublished).
Interplay of iron and manganese
Metals play a major role in oxidative stress protection. The roles of two metals, iron and manganese, are particularly well known. Manganese has similar characteristics to iron. However, an excess of iron has a detrimental effect on host cells, while the presence of manganese has a protective role. Iron, especially in its reduced form, potentiates oxygen toxicity via the Fenton reaction by converting the less reactive hydrogen peroxide to the more reactive oxygen species, such as hydroxyl radical and ferryl iron. The antioxidant superoxide, on the other hand, by releasing iron from iron-containing molecules, favors the Fenton reaction. Thus, we assume that strict regulation of iron assimilation is required to prevent an excess of free intracellular iron that in turn could lead to oxidative stress in microorganisms. This is especially important for bacteria inhabiting anaerobic environments, because under these conditions iron is in its more reactive reduced form and is more soluble and therefore freely available to microorganisms. Manganese, like iron, is a transitional metal; however, manganese plays a very different role in organisms. Manganese is an essential trace element for most microorganisms and recent research indicates previously unappreciated roles for manganese during infection (166). Manganese has a significant role in oxidative stress protection in bacterial pathogens and manganese homeostasis was shown to play a large role in oxidative stress protection of a variety of bacteria (58,63,143,149,166). It can function as a cofactor of oxidative stress protection enzymes such as superoxide dismutase (SOD)(26,164) as well as non-enzymatic antioxidant (151). Significantly, it has been shown that Mn2+ acquisition by bacteria is linked to virulence in the host. Salmonella enterica encodes two systems, mntH and sitABC, that have been identified as selective Mn2+ transporters. Mutation of both transporters of S. enterica markedly attenuated virulence of the bacteria, and the attenuation was in part dependent on the presence of a functional natural resistance-associated macrophage protein1 (NRAMP1) in the host (10,166). These results suggest that the mechanisms of Mn2+ acquisition may provide new targets for development of strategies to control bacterial infection.
The effects of both manganese and iron on protection or promotion of oxidative stress in P. gingivalis have been investigated by examination of the contribution of FeoBs to peroxide resistance. While the ferrous iron transporter, FeoB1 mutant showed reduced growth inhibition in the presence of peroxide when compared to the parental strain, the manganese transporter, FeoB2 mutant was more sensitive to peroxide stress (51). These results demonstrated that the mutation of manganese transporter imparts increased sensitivity while mutation of the ferrous iron transpoprter imparts increased resistance to oxidative stress. The effects of manganese and the contribution of manganese transporter, FeoB2, to anaerobic and aerobic growth were also examined. While manganese had no effect on growth of P. gingivalis W83 in anaerobic conditions, manganese-dependent differences in P. gingivalis W83 growth were noted when the strain was incubated in the presence of 6% O2. Growth did not occur when manganese was absent. However, supplementing the growth media with 200 nM Mn2+ restored the ability of the bacteria to grow, supporting the requirement for the metal for P. gingivalis growth in the presence of oxygen (51). Analysis of the FeoB2 mutant revealed no manganese-dependent differences in growth in anaerobic conditions; however, addition of 200 nM Mn2+ was not sufficient to restore growth in anaerobic conditions, indicating that the FeoB2 transporter is required for bacterial growth in the presence of oxygen when low manganese concentrations are present. Only high concentrations of manganese (25 μM) were able to partially restore the growth of aerobically-grown P. gingivalis. Similar results were obtained when 25 μM of Mn2+ was added to the culture media (growth of both parental and mutant strains was restored), thus further demonstrating the role of manganese in protection of P. gingivalis from oxidative stress.
Iron storage
As with other living organisms, both eukaryotic and prokaryotic, P. gingivalis needs to regulate intracellular unbound iron pools to avoid metal-mediated oxidative stress damage. The pathogen does this by regulating iron uptake mechanisms, as well as by the action of intracellular storage proteins. Ferritin and ferritin-like Dps (DNA protection during starvation) protein identified in P. gingivalis scavenge free iron and as such may also serve as iron sources in times of iron deficiency (111,154). Expression of ferritin and ferritin-like Dps protein was shown to be oxygen and iron induced (76)(Lewis et al., unpublished), providing support that to the proteins function in protection against iron-mediated oxidative stress.
Role of metals in P. gingivalis – host interaction
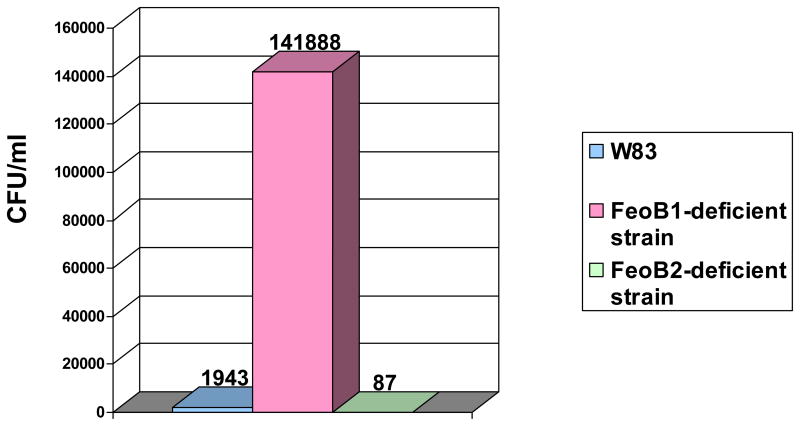
Since all eukaryotic cell types (including endothelial cells, vascular smooth muscle cells, and adventitial fibroblasts) (150) can generate both extracellular and intracellular reactive oxygen species, effective oxidative stress defense mechanisms must be present in P. gingivalis to enable it to survive within the host cells. As described above, iron plays a crucial role in mediating oxidative stress damage due to its reactivity with hydrogen peroxide. Hydrogen peroxide is one of the main weapons used by host cells to combat invading microbes, therefore, the presence of high intracellular iron content would predispose the bacteria to die by host defense mechanisms. Indeed, a ferrous iron transporter (FeoB1)-deficient mutant was demonstrated to have a reduced ability to survive with host cells (Anaya-Bergman et al., unpublished) (Fig. 6). However, in a mouse abscess model the ferrous iron transporter FeoB1 mutant exhibited reduced survival abilities, so the protein seems to be indispensable for iron acquisition in P. gingivalis (28). Together, these data, while contradictory, highlight the importance of examining the role of iron uptake mechanisms at various stages of infection. It is probable that iron deficiencies promote intracellular survival of P. gingivalis, however, iron is needed for bacterial growth.
Fig. 6. Survival of P. gingivalis with host cells.
P. gingivalis strains: parental (W83), ferrous iron transporter FeoB1 mutant, and manganese transporter FeoB2 mutant were examined for their ability to survive within host cells. Bacteria recovered from host cells following infection with parental and FeoB mutant strains were determined by counting colony forming units (CFU). The numbers above the bars represent the mean value derived from an experiment performed in triplicate.
On the other hand, the manganese transporter FeoB2 mutant, which was deficient in the manganese transporter, showed a reduced ability to survive within host cells compared to the parental strain (Fig. 6), supporting the crucial role of manganese in oxidative stress protection (51). Although both strains were able to invade human umbilical vein endothelial cells (HUVECs) to the same extent, an approximately 20-fold reduction in survival rates of the mutant was observed when compared to the parental strain.
Thus, in conclusion, the biological roles of ferrous iron transporter, FeoB1, and manganese transporter, FeoB2, are contrary, which agrees with the roles of iron and manganese in oxidative stress protection. While FeoB2 is required for survival of P. gingivalis in host cells, the presence of FeoB1 has the opposite effect. The properties of FeoB1 and FeoB2 are compared in Table 1.
Table 1.
Comparison of properties of FeoB1 and FeoB2.
| Biological process | FeoB1 | FeoB2 |
|---|---|---|
| Metal Uptake | Fe2+ transporter | Mn2+ transporter |
| Anaerobic Growth | Required | Not required |
| Oxidative Stress Protection | Not required | Required |
| Intracellular Survival in Host Cells | Not required | Required |
| Virulence in Mouse Model | Required | Not required |
In the P. gingivalis–host interaction, the iron uptake components may also play a role in modulating host response in addition to their metal transport capabilities. In support of this suggestion, the hemagglutinins, in addition to binding red blood cells and other host proteins, were shown to bind defensins, a host antimicrobial peptides, and thus interfere with the host’s defense mechanisms. Another hemagglutinin, HagB, was shown to modulate the host’s inflammatory response (167). Furthermore, the hemin-hemoglobin receptor, HbR, binds bone marrow macrophages and induces phosphorylation of ERK (extracellular signal-regulated kinase), p38 [mitogen-activated protein (MAP) kinase], NF-kappaB (nuclear factor kappa-light-chain-enhancer of activated B cells), and Akt (an enzyme that is a member of the serine/threonine-specific protein kinase family) (42). These signaling events lead to inhibition of osteoclastogenesis from bone marrow macrophages. Interestingly, the identity of the N-terminus of the hemin uptake protein, HmuY, to a protein exhibiting fibroblast-activating property indicates that iron acquisition mechanisms may have broader functions than so far identified (89–91). As reported by Mihara et al., HmuY (designated as fibroblast-activating factor, FAF) had significant proliferation-stimulating activity on normal gingival fibroblasts as well as displayed functional similarity to several human-derived growth factors. Furthermore, it had weak phosphatase activity, a property implicated in bone resorption (89). Finally, the HmuR receptor was upregulated in P. gingivalis grown as a three-species biofilm, and a mutant deficient in the protein failed to form three species biofilms indicating the HmuR protein may have adhesive properties in addition to its hemin acquisition functions (59).
The examples described above show the metal transporters have a role in oxidative stress protection of P. gingivalis during host-pathogen interactions. Further, the transporters mediate several pathogenic properties, including inflammation, the host’s innate response, cell proliferation, bone resorption and even biofilm formation. By virtue of being surface exposed, the iron acquisition mechanisms come in direct contact with host cells. It is probable that when induced in iron limited conditions, these components are able to trigger a host response, ultimately leading to inflammation, which is a provision associated with iron. On the other hand, as discussed below, internalization and life in a persistent-type stage could be another alternative.
Iron-dependent transcriptome of P. gingivalis
Iron may affect the transcription of as many as 10–20% of genes encoded on microbial genomes. Work in our laboratory has shown that up to 71 genes had altered expression at >1.5 fold level (Lewis et al., unpublished). The hemin uptake encoding locus, hmu, was significantly upregulated. These data are in agreement with the previously demonstrated iron-dependent regulation of the locus (77). Also, a locus coding for an outer membrane putative lipoprotein and a putative outer membrane TonB-dependent receptor was upregulated. The significant upregulation of the locus suggests that it encodes an iron uptake locus that has yet to be characterized. The predicted functional assignments of the role of the proteins would be in agreement with the structures of two-component receptors (surface lipoprotein and outer membrane TonB-dependent receptor), such as the P. gingivalis HmuYR and IhtBA (29,77) as well as the transferrin and lactoferrin receptors (20,78). Expression of other hemin uptake loci (e.g., tla and iht) was not affected by different iron concentrations. Interestingly, iron levels had no effect on expression of the major ferrous iron transporter, FeoB1 (28), suggesting that regulation of transcription of this gene is iron-independent and therefore novel compared to homologs of the protein found in other organisms.
In agreement with their function as free iron scavengers, genes encoding the iron storage proteins were downregulated. These included genes encoding the ferritin-like protein (Dps) and ferritin (Ftn). While Dps was shown to be required for oxidative stress protection (154), ferritin is indispensable for growth of P. gingivalis in iron-limited conditions (111). Another upregulated gene encoding iron-protein was rbr, coding for rubrerythrin, which is indispensable for protection of P. gingivalis from peroxide stress (144).
Thus, the major theme observed among genes in iron limited conditions was the upregulation of iron uptake mechanisms and downregulation of proteins implicated in iron storage or oxidative stress defense. These trends are not surprising considering the role iron plays in generation of oxidative stress. In the absence of iron, the generation of oxidative stress mediators such as the hydroxyl radicals from peroxide are diminished, thus in agreement with the downregulation of the oxidative stress defenses (Fig. 7). It is interesting to note that the downregulated genes were induced by oxidative stress (76)and are iron-regulated, thus further demonstrating the connection between iron and oxidative stress.
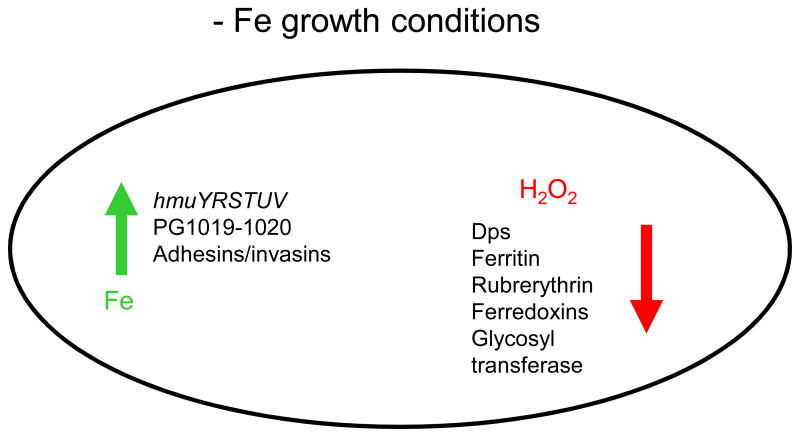
Fig. 7. Opposing gene regulatory functions of iron.
In iron limited (−Fe) conditions, transcription of genes coding for proteins involved in iron uptake mechanisms was increased while expression of oxidative stress response-encoding genes was reduced when compared to iron replete conditions. Green arrow indicates increase in transcription of genes encoding iron transport mechanisms such as the hemin uptake locus (hmu) and putative hemin uptake locus encoded by PG1019–1020, as well as genes coding for adhesins and/or invasins. At the same time reduced transcription of genes coding for mechanisms protecting the cells from peroxide (H2O2) and other sources of oxidative stress was reduced (designated by red arrow). These genes code for: Dps (DNA protection during starvation protein), rubrerythrin, ferritin, and ferredoxin. Also, glycosyl transferase-encoding genes were downregulated.
Iron or heme are required for function of several metabolic enzymes such as the fumarate-reductase, FRD (PG1614-PG1617), ferredoxin-oxidoreductase system (PG1809-PG1813), and proteins of the electron transport system (PG0302-PG0307). Genes coding for those proteins were also observed to be downregulated (Lewis et al., unpublished) which could be perceived as an energy-conservation strategy, because without iron such proteins would be non-functional. The conserved energy would then be used for production of the iron uptake systems.
Iron limitation resulted in upregulation of several genes coding for adhesins/invasions (Lewis et al., unpublished). Two genes coding for leucine rich repeat proteins, PG0350 and PG1374, had elevated expression in iron limited conditions. While the protein encoded by PG0350 was shown to be required for biofilm formation (16,167), the protein encoded by PG1473 was demonstrated to play a role in invasion of KB cells by P. gingivalis W50 (27). Thus, combined with the reduced oxidative stress threat suggested by the reduced expression of the oxidative stress defense mechanisms, the iron-stressed bacteria have a predilection for invasion of host cells or formation of biofilms. This strategy is intuitive because iron-stressed bacteria have lower metabolic capacity and reduced growth, and thus require an environment that would allow them to persist while being protected from host defense mechanisms. In addition, iron stressed bacteria have an increased capacity to survive in host cells when compared with iron-replete cells (Lewis et al., unpublished, Anaya-Bergman et al., unpublished). Indeed, proteomic analysis of intracellular bacteria demonstrated reduced expression of iron uptake mechanisms as well as adhesive proteins thus further supporting the model based on the iron concentrations (162). Finally, nearly half of the regulated genes coded for proteins with unknown function, suggesting novel aspects of the adaptation of P. gingivalis to iron-limitation when compared with other bacteria such as the well known E. coli, may yet be found.
The comparison of iron-dependent regulation to that of the hemin-mediated regulation as reported by Dashper et al. (27) revealed significant overlap among the regulated genes, indicating gene regulation at various hemin levels is iron- and not protoporphyrin IX-mediated. Similar results were obtained when examining the expression of the hmu operon (77).
Mechanisms of iron homeostasis
Generally, in bacteria, the ferric uptake regulator (Fur) is the major iron-dependent regulator that represses transcription initiation of iron uptake-encoding genes in response to an increase in intracellular iron levels (48). In addition to iron uptake genes, Fur also controls expression of numerous iron-containing proteins (39,49). Fur in its apo form is inactive, however, in the presence of iron it forms an iron-bound holo-enzyme. In addition, the presence of iron induces formation of dimers, which then bind to signature sequences present in the vicinity of promoter regions of iron-regulated genes. Interestingly, recent work demonstrated that Fur also downregulates expression of a small RNA (sRNA), the 90 nt sRNA RyhB of E. coli, which plays a role in degradation of mRNAs derived from genes coding for iron-using proteins (85). RyhB was shown to prevent translation of iron-using proteins by pairing with an antisense mRNAs in response to iron depletion (84). A similar mechanism, operating in other organisms including Pseudomonas aeruginosa and Saccharomyces cerevisiae has also been found (88). Finally, extracytoplasmic function (ECF) sigma factors were demonstrated to play a role in bacterial iron homeostasis (108,156).
(i) protein regulators
From the Oralgen database (http://www.oralgen.lanl.gov/), we identified two homologues of iron-dependent regulators in the genome of P. gingivalis W83; a putative ferric uptake regulator; Fur (annotated as PG0425, ferric uptake regulation protein) and Reg (annotated as PG0931, a probable iron-dependent transcriptional repressor). We believe these homologues are candidates to play a role in iron-dependent regulation in this bacterium. Since Fur has been shown to regulate expression of iron and manganese transporters in E. coli and Salmonella typhimurium, so it is probable Fur plays a role in regulation of metal homeostasis in P. gingivalis as well, and may also contribute to oxidative stress defense homeostasis. The P. gingivalis ferric uptake regulator (Fur) has limited similarity to that of the E. coli Fur and thus future structural characterization of the regulator offers the prospect of adding significant new knowledge to our understanding of the mechanism of action of this regulator.
Analysis of the sequence of P. gingivalis W83 revealed the presence of a gene (PG0931) encoding putative DtxR- related metalloregulatory protein, which is located 35 bp upstream of feoB2. We designated the putative regulatory protein Reg. It has been shown that expression of a major manganese transporter, MntH, in Bacillus subtilis is subject to control of a DtxR-related (diphtheria toxin repressor) manganese dependent regulator (MntR) responding to manganese concentration (79). The DtxR-like family of proteins includes metalloregulatory proteins that are widely distributed among many microorganisms and can be divided into three general groups based on the type of metal they respond to. The first group are regulators of iron uptake mechanisms, represented by the Corynebacterium diphtheriae DtxR (123,145). The second group are regulators of Mn2+ transport and include S. enterica MntR (56,62). The third group can respond to both Fe2+ and Mn2+, and include the Staphylococcus epidermidis SirR (53). Comparison of the protein sequence of the P. gingivalis DtxR-like regulator with other DtxR-like regulators reveals that it contains a metal-binding domain that can bind either Mn or Fe, as well as a dimerisation domain. This suggests it has a role in regulation of metal homeostasis. Interestingly, the C-terminal region of Reg is similar to FeoA, which is a small protein, and is probably involved in Fe2+ transport (60). Although this protein has not been found to be fused to previously characterized DtxR-like regulators, it is found at the C-terminus of a variety of putative metal dependent transcriptional regulators of various microorganisms (e.g., Mycobacterium bovis, Deinococcus radiodurans). It is possible the chimeric protein has novel properties when compared with other characterized members of the DtxR family. Furthermore, our preliminary results using microarray analysis revealed that approximately 100 genes were differentially regulated in a mutant devoid of the Reg protein when compared to parental strain (Lewis et al., unpublished). However, whether or not this is an iron or manganese regulator remains to be determined. Currently, confirmation of the direct involvement of those genes in regulation, as well as examination of the metal specificity of the regulator is underway in our laboratory.
Our data suggested that iron-dependent regulation may also be mediated through the oxidative stress regulator (OxyR) (Lewis et al., unpublished). There is limited sequence homology between the P. gingivalis OxyR and the E. coli OxyR, suggesting there may be significant differences in the mechanisms of action of these regulators among bacteria and/or between aerobes and anaerobes. E. coli OxyR was demonstrated to regulate gene expression in response to oxidative stress, with peroxide being the key activator of the regulator (142). The chemical and structural basis of the mechanism is hypothesized to involve the oxidation of cysteine to a cysteine disulfide bridge, which results in conformational changes that induce DNA binding. The P. gingivalis OxyR was shown to respond not only to peroxide, but also to superoxide (33). In E. coli the response to peroxide is mediated by the SoxR protein, but a homolog of this protein is absent in P. gingivalis, so OxyR takes the role of both regulators. Taken together, the differences between E. coli and P. gingivalis at the biological level lead us to infer their mechanisms of response to oxidative stress are distinct and warrant further detailed investigation.
(ii) sRNA-dependent regulation
Even though it has become apparent in E. coli that numerous small RNAs (sRNAs) are made and are important in gene expression regulation, none have been studied in P. gingivalis. To provide high quality annotation for the oral pathogen research community, a computer pipeline was designed to locate potential sRNAs in bacteria by searching for highly conserved regions in the intergenic sequence (IGS) among and within genomes (www.oralgen.lanl.gov). Combining this approach with same-orientation, terminator-pair prediction, a genome-wide survey for putative sRNA-encoding genes in nine oral pathogens was conducted. Using this approach, 650 candidate sRNAs, 94 repeat regions, and 10 riboswitches (including the cobalamin riboswitch) were identified in P. gingivalis. These data are publically available on the Oralgen website (www.oralgen.lanl.gov). The expression and role in regulation of some of the transcripts present in the intergenic regions has been verified (Kim et al., unpublished). Analysis of 28 IGSs verified the presence of transcripts for most of the predicted sRNAs examined in the study and expression of 30% of the sRNAs was oxygen-dependent (the degree of sRNA expression varied with those encoded by IGS 687 and IGS 48 being the most oxygen-dependent). It is noteworthy that expression of many sRNAs was drastically regulated by iron, indicating possible roles of these sRNAs in iron homeostasis. A mutant deficient in one such iron-regulated intergenic transcript, IGS653, showed drastically altered gene expression when compared to the parental strain, suggesting these sequences play a significant role in gene regulation in P. gingivalis. Interestingly, hmuY was among the regulated genes (Lewis et al., unpublished), indicating involvement of the putative sRNA in iron homeostasis in P. gingivalis. Further work is required to characterize the mechanism of action of the sequences encoded by the IGS653. It has been shown, in E. coli at least, that sRNA species act in concert with the host protein designated Hfq (44). It remains to be determined if this also occurs in other bacteria, but it should be pointed out that P. gingivalis does not appear to encode an Hfq orthologue (95). Therefore, it will be important to discover what other peptides, if any, are required for efficient sRNA-mRNA interactions in this bacterium. Thus, novel findings are expected to be derived from examination of the sRNA mechanisms in the Bacteroidetes group.
As already pointed out in the section on the iron-dependent transcriptome, it is noteworthy that although the expression of the major hemin uptake locus, hmu, is iron dependent, the expression of many other iron uptake loci is not iron dependent. Indeed, the regulation of both metal transporters, feoBs (feoB1 and feoB2) expression is unusual and novel when compared to regulation of the ferrous uptake proteins in other bacteria. The feoBs are not regulated by metal as they are in E. coli, but instead are regulated by oxygen and exposure to host cells, respectively (76)(51)(Anaya-Bergman et al., unpublished). Interestingly, sequences capable of forming extensive secondary structures are present within the 5′ untranslated regions of both of the genes (www.oralgen.lanl.gov). Such sequences may play a role in regulation of expression of feoBs, a hypothesis worthy of further investigation, as it suggests novel mechanisms of regulation of metal transporters may be discovered in P. gingivalis.
Secondary structures may also play a regulatory role within multigenic transcriptional units. Differential expression within the hmu operon has been demonstrated (77) that results in higher levels of the promoter-proximal messages encoding for the outer membrane receptor when compared to the promoter-distal messages coding for periplasmic and cytoplasmic components of the transport system. Such differential gene expression may result either from differential transcript expression or differential transcript stability. Several bacterial operons exhibit differential gene expression (e.g., the periplasmic iron transport-encoding fbpABC operon of Neisseria meningitidis (65) or the transferrin receptor-encoding tbpBA operon of Neisseria gonorrhea), and the differences in transcript levels within these operons are due to the presence of sequences capable of forming secondary structures that either interfere with transcription of the sequences or promote stability of the message (115). Analysis of the hmu locus revealed the presence of sequences capable of forming secondary structures following the hmuY and hmuS genes, with calculated energies of dissociation of −16.9 kcalM−1 and −20.8 kcalM−1, respectively (77). Higher temperatures were needed to generate cDNA from transcripts containing these sequences, suggesting they form and possibly play a role in the differential expression observed within the hmu operon.
(iii) sigma factors as iron homeostasis regulators
Finally, sigma factors are components mediating genome-wide regulatory processes. The components bind reversibly to DNA-dependent RNA polymerase (RNAP) to form holoenzyme. This in turn enables binding of the holoenzyme to specific promoters and leads to mRNA synthesis from the promoters. Although the holoenzyme can drive the synthesis of mRNA, this process can also be regulated by several transcriptional regulators; either activators or repressors. The P. gingivalis DNA-dependent RNA polymerase has been reported to differ from that in E. coli (67). The differences were at the protein level, as well as in the ability of the polymerase to recognize E. coli promoters (purified P. gingivalis polymerase bound to P. gingivalis promoters but not to E. coli promoters). Thus we hypothesize that the sigma factors in P. gingivalis will also differ from the well-characterized factors in E. coli, both at the protein level as well as in regards to their target recognition.
To date, the promoters of the Bacteroidetes group have not been well defined and our study indicates there are significant differences in both the consensus sequences as well as in the position of the promoters (promoters of several P. gingivalis genes are located far upstream of the translational start site, e.g., promoters of the hmu, fimA, and feoB’s loci) (77,103,163) (Lewis et al., unpublished). Therefore, further investigation of the promoter regions as well as the transcription process in Bacteroides is warranted. So far, sigma factors have not been characterized in Bacteroidetes and thus our studies of these factors in P. gingivalis could shed light on the transcription process in this group of bacteria. This is supported by our observation that the sigma factors examined in this study share significant similarity with putative sigma factors in other bacteria belonging to the Bacteroidetes phylum.
Analysis of the P. gingivalis W83 genome revealed that as many as eight putative sigma factors may be present in this bacterium. These included: PG0162 (SigG, ECF subfamily), PG0194, PG0536 (RpoD), PG0879.1 (sigma – 24 factor), PG0984 (RpoN, sigma – 54 factor), PG1158, PG1449 (SigG, ECF subfamily), and PG1595 (SigH, ECF subfamily) (results from the Oralgen website, www.oralgen.lanl.gov). While RpoD (sigma – 70), known as the primary sigma factor, and RpoN (sigma – 54) are required for transcription of a wide variety of genes, the remaining sigma factors, also known as alternative sigma factors, direct the transcription of a specific subsets of genes. The presence of multiple alternative sigma factors implies they also may mediate regulation of iron-uptake mechanisms in P. gingivalis. Elevated expression of the PG1158 in iron-limited conditions suggests that the encoded protein may play a role in iron homeostasis in P. gingivalis (Lewis et al., unpublished).
Based on the evidence, we conclude all the mechanisms found in other bacteria also appear to be present in P. gingivalis. However, the roles of the regulatory mechanisms as well as the players involved in regulation still need to be determined.
Pigmentation of P. gingivalis colonies
P. gingivalis forms black-pigmented colonies when grown on blood agar plates (75). The black pigment consists of μ-oxo heme dimers that are generated when oxygen is complexed between two hemin molecules (137). This mechanism may protect P. gingivalis from oxidative stress generated by host cells (133). So far, gingipains were demonstrated to be required for generation of the heme dimer (75,135). However, the release of hemin does not explain the deposition of large amounts of hemin on the cell surface. Genes encoding lipopolysaccharide biosynthesis were recently reported to be required for the pigment formation (118). It is probable that the pigment hemin is non-specifically associated with the bacterial cell surfaces due to charge and hydrophobic interactions.
Hemin and lipopolysaccharide
Lipopolysaccharide is the major bacterial component eliciting response from host cells. The P. gingivalis lipopolysaccharide differs significantly from that of E. coli lipopolysaccharide in the number of phosphate groups as well as lipid A fatty acids positions (112). Furthermore, lipid A heterogeneity between various P. gingivalis strains has been observed and is attributed to the action of acyltransferases, LpxA and LpxD (two proteins catalyzing early stages of lipid A biosynthesis) (7). As it is the lipid A component of lipopolysaccharide that interacts with host cell receptors, differences in the fatty acid composition of lipid A also may lead to differences in virulence of P. gingivalis. It is notable that the lipid A composition was dependent on the hemin concentrations present in culture media (4). Hemin has been demonstrated to have a profound effect on the virulence of P. gingivalis (64,83), and one possible mechanism of action may be through modulation of the lipopolysaccharide structure.
Concluding remarks and future directions
The outcome of the battle for iron between pathogenic bacteria and the host determines the occurrence of infection. In this review, we use P. gingivalis as a model oral organism and describe recent findings regarding iron acquisition mechanisms, with particular focus on novel findings when compared with aerobic counterparts such as the well-studied bacterium Escherichia coli. Several members of the Bacteroidetes phylum are implicated in development and progression of periodontal disease. Considering the phylogenetic relatedness, also underscored by sequence similarity between the various members of the family, knowledge gained using one genus of bacteria can also be applicable for other bacteria belonging to this family. Significant progress has been made in recent years regarding iron acquisition and regulation in P. gingivalis, therefore, this organism was selected as a model system to describe new findings regarding the role of iron in host-pathogen interactions in the oral cavity. P. gingivalis inhabits various niches (supragingival, subgingival, systemic location) and exists both intracellularly and extracellularly. This environmental variation necessitates a variety of mechanisms to allow the organism to adapt to the range of niches. Oxygen concentrations also vary among the niches, thus influencing the availability of iron. Therefore, multiple iron acquisition systems, along with many levels of regulation must be present in P. gingivalis.
P. gingivalis is very distantly related to Eubacterium and iron transport has been thoroughly investigated in this anaerobically growing pathogen. P. gingivalis grows anaerobically or in the presence of low concentrations of oxygen. In anaerobic conditions, iron is more soluble and thus more abundant for bacterial acquisition. Also, in the absence of oxygen, high iron concentrations pose no threat of oxidative stress to the bacterium. Considering this, we expected to uncover novel results in P. gingivalis. Indeed, although some of the mechanisms of iron acquisition and regulation resembled that of other bacteria, many novel findings were also observed. These included the structure of the major hemin uptake locus, hmu, the presence of secondary structures influencing the differential transcription within the locus, the structure of the major hemin binding protein, HmuY, and the mechanisms of iron-dependent regulation, which negatively influenced expression of iron uptake mechanisms and increased the transcription of genes coding for oxidative stress protection mechanisms. It should be noted, that the molecular basis of the regulation appears to involve the oxidative stress response regulator, OxyR, which previously was demonstrated to control expression of oxidative stress mechanisms, but was shown to also mediate iron-dependent regulation in P. gingivalis. The novel findings were made possible by the availability of the genomic sequence as well as other resources for high-throughput gene and protein expression. The nucleotide sequence of the genome of P. gingivalis W83 has already been completed collaboratively at the Forsyth Dental Center and The Institute for Genomic Research (TIGR) and is available to investigators at http://www.pgingivalis.org/. Also, annotated databases are available from the Oral Pathogen Sequence Database at Los Alamos National Laboratory (http://www.stdgen.lanl.gov/oragen/) and the J. Craig Venter Institute’s Comprehensive Microbial Resource (www.jcvi.org). Lastly, P. gingivalis W83 genomic microarrays are available to all interested investigators. Despite the progress made regarding iron acquisition in P. gingivalis, many gaps still exist in our knowledge and understanding of the mechanisms of action. Genomic tools combined with structural analysis of the mechanisms are expected to continue providing further insights into the roles of the various mechanisms of iron acquisition and regulation in P. gingivalis. Finally, the mechanisms of action of the various iron metabolism mechanisms in vivo will need to be illuminated and are expected to provide insights into the requirement of the large number of putative iron uptake mechanisms encoded on the genome of P. gingivalis.
My group and others have established that iron/hemin acquisition is an important feature in P. gingivalis pathogenesis, so it follows that anything that would disrupt iron/hemin acquisition or use represents a novel anti-infective interventional strategy. To my knowledge, there are no antibiotics or vaccines that target iron/hemin homeostasis in any pathogen. Thus, work leading to a cellular and molecular understanding of iron/hemin metabolism in P. gingivalis could produce novel translational approaches that will tip the outcome of the host-bacteria battle for iron in favor of the host, and thus allow both treatment and prevention of adult periodontal disease. In light of increasing antibiotic resistance such novel approaches targeting pathogen-specific essential functions are needed and the knowledge gained from investigating the iron acquisition systems of P. gingivalis could shed light on similar targets in organisms related to P. gingivalis, such as P. intermedia, T. forsythia, B. fragilis, Bacteroides thetaiotaomicron and Capnocytophaga hutchinsonii. Despite the growing concern with infections caused by anaerobic bacteria belonging to the family of Bacteroidetes phylum, little is known about the fundamental biology of these organisms. Understanding the molecular basis of iron homeostasis in Bacteroidetes will be a step forward in understanding the capability of the bacteria to colonize in the host and cause disease.
Fig. 4. Schematic representation of the hemin uptake mechanisms present in P. gingivalis W83.
Three hemin uptake loci designated as hmu, iht, and tlr are present on the genome of P. gingivalis W83. These loci may encode mechanisms that transport may hemin derived from: Hb-Hp (hemoglobin-haptoglobin complexes), Hm-Hx (hemin-hemopexin complexes), Hm-Al (hemin-albumin complexes), or uncomplexed Hm (hemin). The question marks denote areas yet to be investigated. Loci encoding the energy transducer proteins, TonB complex, are also present in P. gingivalis W83. Finally, the regulator of the various loci and the role of iron (Fe) and hemin in this process still remain to be defined.
Acknowledgments
Work on hemin and iron uptake mechanisms in P. gingivalis was supported by NIH-NIDCR grants K22DE014180 to J.P. Lewis, R01DE018039 to J. P. Lewis and R01DE016124 to J. P. Lewis.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agranoff D, I, Monahan M, Mangan JA, Butcher PD, Krishna S. Mycobacterium tuberculosis expresses a novel pH-dependent divalent cation transporter belonging to the Nramp family. J Exp Med. 1999;190:717–724. doi: 10.1084/jem.190.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ainamo J, Ainamo A. Risk assessment of recurrence of disease during supportive periodontal care. Epidemiological considerations. J Clin Periodontol. 1996;23:232–239. doi: 10.1111/j.1600-051x.1996.tb02082.x. [DOI] [PubMed] [Google Scholar]
- 4.Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect Immun. 2006;74:4474–4485. doi: 10.1128/IAI.01924-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JE, Sparling PF, Cornelissen CN. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J Bacteriol. 1994;176:3162–3170. doi: 10.1128/jb.176.11.3162-3170.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 7.Bainbridge BW, Karimi-Naser L, Reife R, Blethen F, Ernst RK, Darveau RP. Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis. J Bacteriol. 2008;190:4549–4558. doi: 10.1128/JB.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaven GH, Chen SH, d’ AA, Gratzer WB. A spectroscopic study of the haemin--human-serum-albumin system. Eur J Biochem. 1974;41:539–546. doi: 10.1111/j.1432-1033.1974.tb03295.x. [DOI] [PubMed] [Google Scholar]
- 9.Boukhalfa H, Crumbliss AL. Chemical aspects of siderophore mediated iron transport. Biometals. 2002;15:325–339. doi: 10.1023/a:1020218608266. [DOI] [PubMed] [Google Scholar]
- 10.Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar typhimurium. Infect Immun. 2002;70:6032–6042. doi: 10.1128/IAI.70.11.6032-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bramanti TE, Holt SC. Effect of porphyrins and host iron transport proteins on outer membrane protein expression in Porphyromonas (Bacteroides) gingivalis: identification of a novel 26 kDa hemin-repressible surface protein. Microb Pathog. 1992;13:61–73. doi: 10.1016/0882-4010(92)90032-j. [DOI] [PubMed] [Google Scholar]
- 12.Bramanti TE, Holt SC. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 14.Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003;4:409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 15.Canonne-Hergaux F, Gruenheid S, Govoni G, Gros P. The Nramp1 protein and its role in resistance to infection and macrophage function. Proc Assoc Am Physicians. 1999;111:283–289. doi: 10.1046/j.1525-1381.1999.99236.x. [DOI] [PubMed] [Google Scholar]
- 16.Capestany CA, Kuboniwa M, Jung IY, Park Y, Tribble GD, Lamont RJ. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun. 2006;74:3002–3005. doi: 10.1128/IAI.74.5.3002-3005.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi JI, Nakagawa T, Yamada S, Takazoe I, Okuda K. Clinical, microbiological and immunological studies on recurrent periodontal disease. J Clin Periodontol. 1990;17:426–434. [PubMed] [Google Scholar]
- 18.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombo AP, Haffajee AD, Dewhirst FE, Paster BJ, Smith CM, Cugini MA, Socransky SS. Clinical and microbiological features of refractory periodontitis subjects. J Clin Periodontol. 1998;25:169–180. doi: 10.1111/j.1600-051x.1998.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 20.Cornelissen CN. Transferrin-iron uptake by Gram-negative bacteria. Front Biosci. 2003;8:d836–d847. doi: 10.2741/1076. [DOI] [PubMed] [Google Scholar]
- 21.Cornelissen CN, Biswas GD, Tsai J, Paruchuri DK, Thompson SA, Sparling PF. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crosa JH. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis MA. Analysis of the protease and adhesin domains of the PrpRI of Porphyromonas gingivalis. J Periodontal Res. 1997;32:133–139. doi: 10.1111/j.1600-0765.1997.tb01394.x. [DOI] [PubMed] [Google Scholar]
- 24.Curtis MA, duse-Opoku J, Rangarajan M. Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med. 2001;12:192–216. doi: 10.1177/10454411010120030101. [DOI] [PubMed] [Google Scholar]
- 25.Curtis MA, Hanley SA, duse-Opoku J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J Periodontal Res. 1999;34:400–405. doi: 10.1111/j.1600-0765.1999.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 26.D’Mello RA, Langford PR, Kroll JS. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect Immun. 1997;65:2700–2706. doi: 10.1128/iai.65.7.2700-2706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, Walsh KA, Slakeski N, Chen D, Lissel JP, Butler CA, O’Brien-Simpson NM, Barr IG, Reynolds EC. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J Bacteriol. 2009;191:1044–1055. doi: 10.1128/JB.01270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dashper SG, Butler CA, Lissel JP, Paolini RA, Hoffmann B, Veith PD, O’Brien-Simpson NM, Snelgrove SL, Tsiros JT, Reynolds EC. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J Biol Chem. 2005;280:28095–28102. doi: 10.1074/jbc.M503896200. [DOI] [PubMed] [Google Scholar]
- 29.Dashper SG, Hendtlass A, Slakeski N, Jackson C, Cross KJ, Brownfield L, Hamilton R, Barr I, Reynolds EC. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dashper SG, Hendtlass A, Slakeski N, Jackson C, Cross KJ, Brownfield L, Hamilton R, Barr I, Reynolds EC. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeCarlo AA, Paramaesvaran M, Yun PL, Collyer C, Hunter N. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J Bacteriol. 1999;181:3784–3791. doi: 10.1128/jb.181.12.3784-3791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66:5337–5343. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol. 2006;188:2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz PI, Zilm PS, Wasinger V, Corthals GL, Rogers AH. Studies on NADH oxidase and alkyl hydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19:137–143. doi: 10.1111/j.0902-0055.2004.00120.x. [DOI] [PubMed] [Google Scholar]
- 35.Doherty CP. Host-pathogen interactions: the role of iron. J Nutr. 2007;137:1341–1344. doi: 10.1093/jn/137.5.1341. [DOI] [PubMed] [Google Scholar]
- 36.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Invasion of human coronary artery cells by periodontal pathogens. Infect Immun. 1999;67:5792–5798. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorn BR, Leung KL, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun. 1998;66:6054–6057. doi: 10.1128/iai.66.12.6054-6057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.duse-Opoku J, Slaney JM, Rangarajan M, Muir J, Young KA, Curtis MA. The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escolar L, Perez-Martin J, de V L. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ezzo PJ, Cutler CW. Microorganisms as risk indicators for periodontal disease. Periodontol 2000. 2003;32:24–35. doi: 10.1046/j.0906-6713.2003.03203.x. [DOI] [PubMed] [Google Scholar]
- 41.Fujimura S, Hirai K, Shibata Y, Nakayama K, Nakamura T. Comparative properties of envelope-associated arginine-gingipains and lysine-gingipain of Porphyromonas gingivalis. FEMS Microbiol Lett. 1998;163:173–179. doi: 10.1111/j.1574-6968.1998.tb13042.x. [DOI] [PubMed] [Google Scholar]
- 42.Fujimura Y, Hotokezaka H, Ohara N, Naito M, Sakai E, Yoshimura M, Narita Y, Kitaura H, Yoshida N, Nakayama K. The hemoglobin receptor protein of Porphyromonas gingivalis inhibits receptor activator NF-kappaB ligand-induced osteoclastogenesis from bone marrow macrophages. Infect Immun. 2006;74:2544–2551. doi: 10.1128/IAI.74.5.2544-2551.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia RI, Henshaw MM, Krall EA. Relationship between periodontal disease and systemic health. Periodontol 2000. 2001;25:21–36. doi: 10.1034/j.1600-0757.2001.22250103.x. [DOI] [PubMed] [Google Scholar]
- 44.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 45.Gruenheid S, Canonne-Hergaux F, Gauthier S, Hackam DJ, Grinstein S, Gros P. The iron transport protein NRAMP2 is an integral membrane glycoprotein that colocalizes with transferrin in recycling endosomes. J Exp Med. 1999;189:831–841. doi: 10.1084/jem.189.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 47.Han N, Whitlock J, Progulske-Fox A. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect Immun. 1996;64:4000–4007. doi: 10.1128/iai.64.10.4000-4007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 49.Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 50.Haraszthy VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. Identification of periodontal pathogens in atheromatous plaques. J Periodontol. 2000;71:1554–1560. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- 51.He J, Miyazaki H, Anaya C, Yu F, Yeudall WA, Lewis JP. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect Immun. 2006;74:4214–4223. doi: 10.1128/IAI.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 53.Hill PJ, Cockayne A, Landers P, Morrissey JA, Sims CM, Williams P. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect Immun. 1998;66:4123–4129. doi: 10.1128/iai.66.9.4123-4129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrkal Z, Vodrazka Z, Kalousek I. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur J Biochem. 1974;43:73–78. doi: 10.1111/j.1432-1033.1974.tb03386.x. [DOI] [PubMed] [Google Scholar]
- 55.Hwang PK, Greer J. Interaction between hemoglobin subunits in the hemoglobin. haptoglobin complex. J Biol Chem. 1980;255:3038–3041. [PubMed] [Google Scholar]
- 56.Ikeda JS, Janakiraman A, Kehres DG, Maguire ME, Slauch JM. Transcriptional regulation of sitABCD of Salmonella enterica serovar typhimurium by MntR and Fur. J Bacteriol. 2005;187:912–922. doi: 10.1128/JB.187.3.912-922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 58.Jakubovics NS, Jenkinson HF. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology. 2001;147:1709–1718. doi: 10.1099/00221287-147-7-1709. [DOI] [PubMed] [Google Scholar]
- 59.James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, Demuth DR, Lamont RJ. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. Regulation of Salmonella enterica serovar typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+) J Bacteriol. 2002;184:3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 64.Kesavalu L, Holt SC, Ebersole JL. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol Immunol. 2003;18:226–233. doi: 10.1034/j.1399-302x.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 65.Khun HH, Deved V, Wong H, Lee BC. fbpABC gene cluster in Neisseria meningitidis is transcribed as an operon. Infect Immun. 2000;68:7166–7171. doi: 10.1128/iai.68.12.7166-7171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim SJ, Chu L, Holt SC. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 67.Klimpel KW, V, Clark L. The RNA polymerases of Porphyromonas gingivalis and Fusobacterium nucleatum are unrelated to the RNA polymerase of Escherichia coli. J Dent Res. 1990;69:1567–1572. doi: 10.1177/00220345900690090601. [DOI] [PubMed] [Google Scholar]
- 68.Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larsen RA, Myers PS, Skare JT, Seachord CL, Darveau RP, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee BC. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 72.Legrain M, Mazarin V, Irwin SW, Bouchon B, Quentin-Millet MJ, Jacobs E, Schryvers AB. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene. 1993;130:73–80. doi: 10.1016/0378-1119(93)90348-7. [DOI] [PubMed] [Google Scholar]
- 73.Lepine G, Progulske-Fox A. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:65–78. doi: 10.1111/j.1399-302x.1996.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 74.Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 75.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis JP, Iyer D, naya-Bergman C. Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced accumulation of lactate. Microbiology. 2009 doi: 10.1099/mic.0.027953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewis JP, Plata K, Yu F, Rosato A, Anaya C. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152:3367–3382. doi: 10.1099/mic.0.29011-0. [DOI] [PubMed] [Google Scholar]
- 78.Lewis LA, Rohde K, Gipson M, Behrens B, Gray E, Toth SI, Roe BA, Dyer DW. Identification and molecular analysis of lbpBA, which encodes the two-component meningococcal lactoferrin receptor. Infect Immun. 1998;66:3017–3023. doi: 10.1128/iai.66.6.3017-3023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lieser SA, Davis TC, Helmann JD, Cohen SM. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry. 2003;42:12634–12642. doi: 10.1021/bi0350248. [DOI] [PubMed] [Google Scholar]
- 80.Liu X, Olczak T, Guo HC, Dixon DW, Genco CA. Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect Immun. 2006;74:1222–1232. doi: 10.1128/IAI.74.2.1222-1232.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X, Sroka A, Potempa J, Genco CA. Coordinate expression of the Porphyromonas gingivalis lysine-specific gingipain proteinase, Kgp, arginine-specific gingipain proteinase, RgpA, and the heme/hemoglobin receptor, HmuR. Biol Chem. 2004;385:1049–1057. doi: 10.1515/BC.2004.136. [DOI] [PubMed] [Google Scholar]
- 82.Macy JM, Probst I. The biology of gastrointestinal Bacteroides. Annu Rev Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- 83.Marsh PD, McDermid AS, McKee AS, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology. 1994;140(Pt 4):861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 84.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 86.Matto J, Saarela M, Alaluusua S, Oja V, Jousimies-Somer H, Asikainen S. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J Clin Microbiol. 1998;36:157–160. doi: 10.1128/jcm.36.1.157-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKee AS, McDermid AS, Baskerville A, Dowsett AB, Ellwood DC, Marsh PD. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect Immun. 1986;52:349–355. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mercier A, Pelletier B, Labbe S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mihara J, Holt SC. Purification and characterization of fibroblast-activating factor isolated from Porphyromonas gingivalis W50. Infect Immun. 1993;61:588–595. doi: 10.1128/iai.61.2.588-595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mihara J, Miyazawa Y, Holt SC. Modulation of growth and function of human gingival fibroblasts by fibroblast-activating factor derived from Porphyromonas gingivalis W50. Infect Immun. 1993;61:596–601. doi: 10.1128/iai.61.2.596-601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mihara J, Yoneda T, Holt SC. Role of Porphyromonas gingivalis-derived fibroblast-activating factor in bone resorption. Infect Immun. 1993;61:3562–3564. doi: 10.1128/iai.61.8.3562-3564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–739. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 93.Mustapha IZ, Debrey S, Oladubu M, Ugarte R. Markers of systemic bacterial exposure in periodontal disease and cardiovascular disease risk: a systematic review and meta-analysis. J Periodontol. 2007;78:2289–2302. doi: 10.1902/jop.2007.070140. [DOI] [PubMed] [Google Scholar]
- 94.Nakayama K, Ratnayake DB, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 95.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 97.Olczak T, Dixon DW, Genco CA. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olczak T, Simpson W, Liu X, Genco CA. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 99.Olczak T, Siudeja K, Olczak M. Purification and initial characterization of a novel Porphyromonas gingivalis HmuY protein expressed in Escherichia coli and insect cells. Protein Expr Purif. 2006;49:299–306. doi: 10.1016/j.pep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 100.Olczak T, Sroka A, Potempa J, Olczak M. Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch Microbiol. 2008;189:197–210. doi: 10.1007/s00203-007-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Otto BR, Verweij-van Vught AM, MacLaren DM. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 102.Paramaesvaran M, Nguyen KA, Caldon E, McDonald JA, Najdi S, Gonzaga G, Langley DB, DeCarlo A, Crossley MJ, Hunter N, Collyer CA. Porphyrin-mediated cell surface heme capture from hemoglobin by Porphyromonas gingivalis. J Bacteriol. 2003;185:2528–2537. doi: 10.1128/JB.185.8.2528-2537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park Y, Xie H, Lamont RJ. Transcriptional organization of the Porphyromonas gingivalis fimA locus. FEMS Microbiol Lett. 2007;273:103–108. doi: 10.1111/j.1574-6968.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- 104.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 106.Pike RN, Potempa J, McGraw W, Coetzer TH, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Polyzos NP, I, Polyzos P, Mauri D, Tzioras S, Tsappi M, Cortinovis I, Casazza G. Effect of periodontal disease treatment during pregnancy on preterm birth incidence: a metaanalysis of randomized trials. Am J Obstet Gynecol. 2009;200:225–232. doi: 10.1016/j.ajog.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 108.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front Biosci. 2003;8:d661–d686. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 109.Progulske-Fox A, Tumwasorn S, Holt SC. The expression and function of a Bacteroides gingivalis hemagglutinin gene in Escherichia coli. Oral Microbiol Immunol. 1989;4:121–131. doi: 10.1111/j.1399-302x.1989.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 110.Progulske-Fox A, Tumwasorn S, Lepine G, Whitlock J, Savett D, Ferretti JJ, Banas JA. The cloning, expression and sequence analysis of a second Porphyromonas gingivalis gene that codes for a protein involved in hemagglutination. Oral Microbiol Immunol. 1995;10:311–318. doi: 10.1111/j.1399-302x.1995.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 111.Ratnayake DB, Wai SN, Shi Y, Amako K, Nakayama H, Nakayama K. Ferritin from the obligate anaerobe Porphyromonas gingivalis: purification, gene cloning and mutant studies. Microbiology. 2000;146(Pt 5):1119–1127. doi: 10.1099/00221287-146-5-1119. [DOI] [PubMed] [Google Scholar]
- 112.Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, Howald WN, Darveau RP. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006;8:857–868. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 113.Renvert S, Dahlen G, Wikstrom M. Treatment of periodontal disease based on microbiological diagnosis. Relation between microbiological and clinical parameters during 5 years. J Periodontol. 1996;67:562–571. doi: 10.1902/jop.1996.67.6.562. [DOI] [PubMed] [Google Scholar]
- 114.Robey M, Cianciotto NP. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun. 2002;70:5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ronpirin C, Jerse AE, Cornelissen CN. Gonococcal genes encoding transferrin-binding proteins A and B are arranged in a bicistronic operon but are subject to differential expression. Infect Immun. 2001;69:6336–6347. doi: 10.1128/IAI.69.10.6336-6347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roper JM, Raux E, Brindley AA, Schubert HL, Gharbia SE, Shah HN, Warren MJ. The enigma of cobalamin (Vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J Biol Chem. 2000;275:40316–40323. doi: 10.1074/jbc.M007146200. [DOI] [PubMed] [Google Scholar]
- 117.Ryu SY, Jeong KS, Kang BN, Park SJ, Yoon WK, Kim SH, Kim TH. Modulation of transferrin synthesis, transferrin receptor expression, iNOS expression and NO production in mouse macrophages by cytokines, either alone or in combination. Anticancer Res. 2000;20:3331–3338. [PubMed] [Google Scholar]
- 118.Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology. 2009;155:1282–1293. doi: 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- 119.Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann Periodontol. 2003;8:38–53. doi: 10.1902/annals.2003.8.1.38. [DOI] [PubMed] [Google Scholar]
- 120.Scannapieco FA, Bush RB, Paju S. Periodontal disease as a risk factor for adverse pregnancy outcomes. A systematic review. Ann Periodontol. 2003;8:70–78. doi: 10.1902/annals.2003.8.1.70. [DOI] [PubMed] [Google Scholar]
- 121.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 122.Schifferle RE, Shostad SA, Bayers-Thering MT, Dyer DW, Neiders ME. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J Endod. 1996;22:352–355. doi: 10.1016/S0099-2399(96)80216-0. [DOI] [PubMed] [Google Scholar]
- 123.Schmitt MP, Twiddy EM, Holmes RK. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci U S A. 1992;89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 125.Shiloah J, Patters MR. Repopulation of periodontal pockets by microbial pathogens in the absence of supportive therapy. J Periodontol. 1996;67:130–139. doi: 10.1902/jop.1996.67.2.130. [DOI] [PubMed] [Google Scholar]
- 126.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;131:313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 127.Simpson W, Olczak T, Genco CA. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Simpsonv W, Olczak T, Genco CA. Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim Pol. 2004;51:253–262. [PubMed] [Google Scholar]
- 129.Skare JT, Ahmer BM, Seachord CL, Darveau RP, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 130.Slakeski N, Dashper SG, Cook P, Poon C, Moore C, Reynolds EC. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol. 2000;15:388–392. doi: 10.1034/j.1399-302x.2000.150609.x. [DOI] [PubMed] [Google Scholar]
- 131.Smalley JW, Birss AJ. Albumin and hemalbumin degradation by Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:254–258. doi: 10.1111/j.1399-302x.1997.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 132.Smalley JW, Birss AJ, McKee AS, Marsh PD. Haemin-restriction influences haemin-binding, haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalis W50. FEMS Microbiol Lett. 1991;69:63–67. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 133.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 134.Smalley JW, Birss AJ, Szmigielski B, Potempa J. The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo bishaem formation from monomeric iron(III) protoporphyrin IX. Microbiology. 2006;152:1839–1845. doi: 10.1099/mic.0.28835-0. [DOI] [PubMed] [Google Scholar]
- 135.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Sequential action of R- and K-specific gingipains of Porphyromonas gingivalis in the generation of the haem-containing pigment from oxyhaemoglobin. Arch Biochem Biophys. 2007;465:44–49. doi: 10.1016/j.abb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 136.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Mechanism of methaemoglobin breakdown by the lysine-specific gingipain of the periodontal pathogen Porphyromonas gingivalis. Biol Chem. 2008;389:1235–1238. doi: 10.1515/BC.2008.XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smalley JW, Silver J, Marsh PJ, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331(Pt 3):681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smalley JW, Thomas MF, Birss AJ, Withnall R, Silver J. A combination of both arginine- and lysine-specific gingipain activity of Porphyromonas gingivalis is necessary for the generation of the micro-oxo bishaem-containing pigment from haemoglobin. Biochem J. 2004;379:833–840. doi: 10.1042/BJ20031221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 140.Sroka A, Sztukowska M, Potempa J, Travis J, Genco CA. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J Bacteriol. 2001;183:5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 142.Storz G, Tartaglia LA, Ames BN. The OxyR regulon. Antonie Van Leeuwenhoek. 1990;58:157–161. doi: 10.1007/BF00548927. [DOI] [PubMed] [Google Scholar]
- 143.Supek F, Supekova L, Nelson H, Nelson N. Function of metal-ion homeostasis in the cell division cycle, mitochondrial protein processing, sensitivity to mycobacterial infection and brain function. J Exp Biol. 1997;200:321–330. doi: 10.1242/jeb.200.2.321. [DOI] [PubMed] [Google Scholar]
- 144.Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM., Jr Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol. 2002;44:479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- 145.Tao X, Schiering N, Zeng HY, Ringe D, Murphy JR. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 146.Tazaki K, Inoshita E, Amano A, Hanioka T, Tamagawa H, Shizukuishi S. Interaction of Porphyromonas gingivalis with transferrin. FEMS Microbiol Lett. 1995;131:161–166. doi: 10.1016/0378-1097(95)00253-2. [DOI] [PubMed] [Google Scholar]
- 147.Thompson JM, Jones HA, Perry RD. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tonetti MS. Periodontitis and risk for atherosclerosis: an update on intervention trials. J Clin Periodontol. 2009;36(Suppl 10):15–19. doi: 10.1111/j.1600-051X.2009.01417.x. [DOI] [PubMed] [Google Scholar]
- 149.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 150.Touyz RM, Schiffrin EL. Reactive oxygen species in vascular biology: implications in hypertension. Histochem Cell Biol. 2004;122:339–352. doi: 10.1007/s00418-004-0696-7. [DOI] [PubMed] [Google Scholar]
- 151.Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol Microbiol. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 152.Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.US, Harper F, Curtis MA. Haemin inhibits the trypsin-like enzyme activity of Porphyromonas gingivalis W83. FEMS Microbiol Lett. 1990;60:169–172. doi: 10.1111/j.1574-6968.1990.tb03883.x. [DOI] [PubMed] [Google Scholar]
- 154.Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun. 2003;71:1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Velayudhan J, Hughes NJ, McColm AA, Bagshaw J, Clayton CL, Andrews SC, Kelly DJ. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol Microbiol. 2000;37:274–286. doi: 10.1046/j.1365-2958.2000.01987.x. [DOI] [PubMed] [Google Scholar]
- 156.Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol. 2002;45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 157.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 158.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 159.Wojtowicz H, Guevara T, Tallant C, Olczak M, Sroka A, Potempa J, Sola M, Olczak T, Gomis-Ruth FX. Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathog. 2009;5:e1000419. doi: 10.1371/journal.ppat.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wyckoff EE, Boulette ML, Payne SM. Genetics and environmental regulation of Shigella iron transport systems. Biometals. 2009;22:43–51. doi: 10.1007/s10534-008-9188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wyckoff EE, Duncan D, Torres AG, Mills M, Maase K, Payne SM. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 162.Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, Hackett M. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie H, Lamont RJ. Promoter architecture of the Porphyromonas gingivalis fimbrillin gene. Infect Immun. 1999;67:3227–3235. doi: 10.1128/iai.67.7.3227-3235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:2819–2826. doi: 10.1128/iai.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zaharik ML, Finlay BB. Mn2+ and bacterial pathogenesis. Front Biosci. 2004;9:1035–1042. doi: 10.2741/1317. [DOI] [PubMed] [Google Scholar]
- 167.Zhang P, Martin M, Michalek SM, Katz J. Role of mitogen-activated protein kinases and NF-kappaB in the regulation of proinflammatory and anti-inflammatory cytokines by Porphyromonas gingivalis hemagglutinin B. Infect Immun. 2005;73:3990–3998. doi: 10.1128/IAI.73.7.3990-3998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]