Abstract
Hematopoietic stem cell (HSC) differentiation is regulated by cell-intrinsic and extrinsic cues. In addition to transcriptional regulation, post-translational regulation may also control HSC differentiation. To test this hypothesis, we visualized ubiquitin-regulated protein stability of a single transcription factor, c-Myc. The stability of c-Myc protein was instructive of HSC quiescence and c-Myc protein abundance was controlled by the ubiquitin ligase Fbw7. Fine changes in stability of c-Myc protein regulated the HSC “gene expression signature”. Using whole genome genomic approaches, we identified specific regulators of HSC function that are directly controlled by c-Myc binding, however adult HSCs and embryonic stem cells sense and interpret distinctly c-Myc regulated gene expression. These studies show a ubiquitin ligase substrate pair can orchestrate the molecular program of HSC differentiation.
Hematopoietic stem cell (HSC) self-renewal and quiescence are controlled by a highly orchestrated integration of environmental signals, most of them originating from the stem cell niche1, 2. Long term HSCs (LT-HSCs) reside at the top of the developmental pyramid as they possess the ability to self-renew and sustain hematopoiesis3. This fraction of HSCs remains largely quiescent or even dormant throughout the lifetime of an organism. Cells that receive differentiation-promoting niche signals are able to generate multi-potent progenitors (MPPs), cells that have diminished self-renewal and can enter further differentiation routes that will lead them to generate progenitors of the myeloerythroid or lymphoid lineages.
Intense experimentation during the last two decades suggested that tight control of HSC differentiation is controlled by the interplay of a handful genetic and epigenetic regulators of gene transcription4, 5. However, transcriptional control is unlikely to provide the complete answer to the puzzle of stem cell differentiation. It is thus intriguing to suggest that post-transcriptional or even post-translational regulation plays an essential role. The emergence of microRNA function in both embryonic and adult stem cell biology is one example6. Another emerging paradigm of post-translational modification is mono- or polyubiquitination of protein substrates leading to alteration of target half-life or modification of activation status. Ubiquitination is performed by large enzymatic complexes that include ubiquitin activating and conjugating components as well as adapters that dictate substrate specificity7, 8. One of the most important and well-characterized outcomes of protein ubiquitination is targeting to and subsequent degradation by the proteasome. By controlling protein stability and abundance, ubiquitin ligases regulate distinct biological processes, including cell cycle entry and progression9.
Previous studies have suggested that ubiquitination, proteosomal degradation and protein stability could also control stem cell function in different organisms10–13. These studies introduced the intriguing hypothesis that fine-tuning of the half-life, stability and abundance of key regulators by the ubiquitin–proteasome machinery could control HSC function, specifically, self-renewal and differentiation. Testing this hypothesis in vivo is a challenging task as it requires quantitative assessment of substrate abundance in small stem cell and progenitor subsets. To overcome this limitation, we utilized gene-targeted mice in which relative protein abundance can be studied in vivo using flow cytometry and microscopy. As a model ubiquitin-substrate, we selected the transcription factor c-Myc, a well-known oncogene and developmental regulator14, 15. c-Myc expression and function has been suggested to be important for HSC differentiation and more specifically for HSC niche retention and survival16, 17. However, the molecular mechanism by which c-Myc controls HSC function is largely unknown. For example, similar amounts of Myc mRNA are detected in HSC and differentiated progenitors16. This finding introduces the hypothesis that c-Myc functions in stem and progenitor cells are more likely to be controlled post-translationally rather than at the level of transcription. Moreover, several studies have shown that c-Myc protein stability is controlled by the ubiquitin system. At least three distinct E3 ligases (Skp2, Huwe1 and Fbw7)18–20 are involved its regulation.
We demonstrate here that relative abundance of nuclear c-Myc protein was instructive of HSC quiescence and self-renewal status, and c-Myc stability in HSC was controlled by a single E3 ubiquitin ligase, Fbw7. Deletion of Fbw7 led to the overexpression of c-Myc protein in single HSC cells and reduction of c-Myc abundance rescued the Fbw7−/− HSC phenotype. Moreover, we demonstrate that the HSC gene expression signature was controlled by fine changes in relative stability of c-Myc protein and identify specific gene-regulators of HSC quiescence and self-renewal. Whole genome chromatin immunoprecipitation experiments identify genes that are directly regulated by c-Myc promoter-binding and transcriptional activity. Finally, we contrast adult HSC with in vitro self-renewing embryonic stem cells (ESCs). We demonstrate that each stem cell type is able to sense and interpret c-Myc abundance and c-Myc-regulated gene expression in distinct ways. Our studies offer the first example of an ubiquitin ligase–substrate pair able to orchestrate the molecular program of adult mammalian stem cell differentiation.
RESULTS
c-Myc protein abundance in single HSC and progenitors
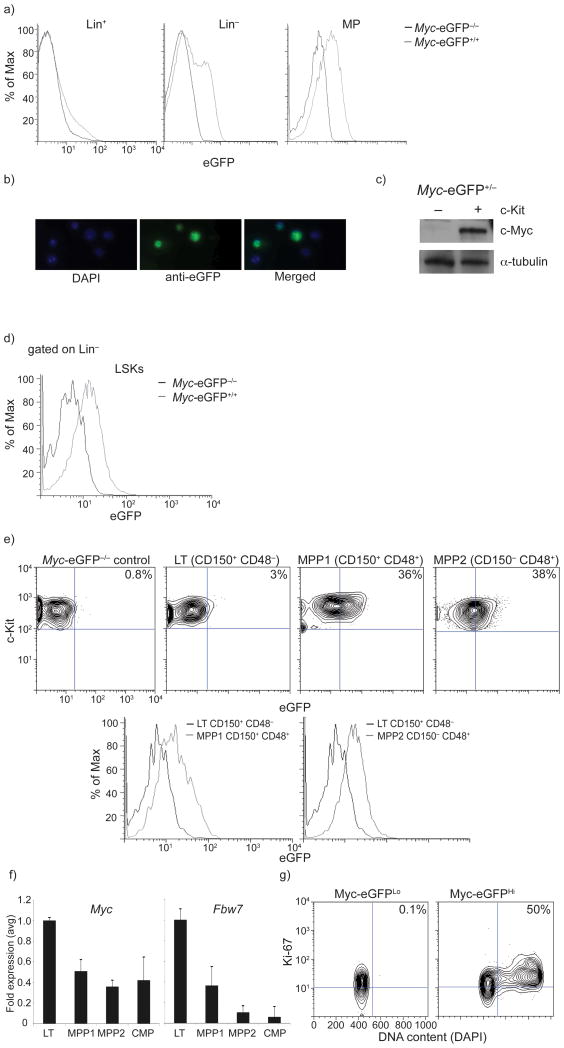
To visualize c-Myc protein abundance in vivo, we took advantage of a recently generated animal model21, in which the eGFP reporter replaced the start codon of Myc within the second exon in the Myc locus, generating a functional N-terminally tagged c-Myc protein that faithfully mirrored the expression of endogenous protein. MyceGFP/eGFP homozygous mice were viable, with no alterations in the kinetics of hematopoietic differentiation or immune function. MyceGFP was not expressed in lineage-positive (Lin+), mature bone marrow (BM) cells, but its expression became evident when we focused on Lin− progenitors (Fig. 1a–c). The highest expression of c-Myc (Myc-eGFP) was detected in the myeloerythroid progenitor fraction (MP), a cell population that originates directly from Lin−, Sca-1+, c-Kit+ (LSK) cells and is actively proliferating. To further validate the faithful expression of the c-Myc-eGFP fusion, we purified wild-type Lin− c-Kit+ progenitors and Lin+ cells from whole bone marrow. Endogenous c-Myc protein was only expressed in the progenitor fraction (Fig. 1d). These experiments showed that c-Myc protein expression was detectable in the LSK fraction and peaked at the MP stage in adult bone marrow.
Figure 1. Visualization of c-Myc protein abundance in early hematopoiesis.
a) Levels of c-Myc-eGFP expression during early hematopoiesis in Lineage+, Lineage−, LSK (Lineage−, c-Kit+, Sca1+) and MP (lineage−, c-Kit+, Sca1) BM subsets. n=6 mice b) Immuno-fluorescence tracing of nuclear c-Myc-eGFP expression in Lineage− bone marrow progenitors. c) Western blot demonstrating presence of c-Myc protein in c-Kit+ and absence in c-Kit− subset of adult bone marrow. d) Overlay histogram depicting c-Myc-eGFP protein expression within LSK cells. e) c-Myc-eGFP expression in eGFP−/−control, LT-HSC and MPP (1, 2) populations. f) qRT-PCR quantification of Myc and Fbw7 mRNA expression during HSC differentiation. g) Levels of c-Myc-eGFP protein correlate with the cell cycle status of LSK cells. Sorted c-Myc-eGFPHi or c-Myc-eGFPLo LSK cells were analyzed using Ki-67 and DAPI staining. Plots are a representation of at least 3 independent experiments.
c-Myc protein correlates with HSC differentiation
We noticed that LSK cells, a population containing HSCs and multipotential progenitors, possessed both a c-Myc-eGFP+ and a c-Myc-eGFP− fraction (Fig. 1e). HSCs differentiate through intermediate stages, generating at least two subsets of multipotential progenitors (MPP1 and MPP2)22. Only long-term (LT)-HSCs have the ability to self-renew and are largely quiescent or even dormant22. Therefore, we subdivided the LSK fraction using both SLAM markers (CD150 and CD48) (Fig. 1f and Supplementary Figure 1) and Flt-3 with CD34 (not shown). A substantial increase in c-Myc protein (Fig. 1f) correlated perfectly with differentiation stage as, LT-HSCs were almost (>95%) exclusively c-Myc-eGFP− whereas the MPP1 fraction was the first subset with detectable c-Myc protein and further increases c-Myc protein abundance occurred in more differentiated CD150−CD48+ MPP2 cells. To examine whether the c-Myc expression changes that we observed were independent of transcriptional control, we compared Myc mRNA expression in the different populations. Myc transcription was not elevated as LT-HSCs differentiated (Fig. 1g), suggesting that the protein changes that we identified were largely due to post-transcriptional mechanisms. These data connect relative c-Myc protein abundance to specific stages of HSC differentiation.
Effects of extrinsic stress signals on c-Myc expression
We then sought to determine whether environmental cues, for example stress or tissue injury, could feedback on LSK proliferation and c-Myc protein abundance. Initially, we discovered that c-Myc protein abundance was not only suggestive of the differentiation stage but also dictated cell cycle status. Indeed, c-Myc-eGFPlo LSK cells were exclusively in G0/G1 phases and c-Myc-eGFPhi cells were actively cycling (Fig. 1h). Moreover, as tissue injury can induce exit of HSCs from quiescence, we used the chemotherapeutic agent fluorouracil (5-FU) to induce myeloablation, a process that normally activates quiescent HSCs23. Consistent with this notion, a 2–3 fold increase of LSK numbers from 5-FU treated MyceGFP/+ mice had entered cell cycle 2 days post treatment when compared to non-treated controls. Most importantly, these cells purified from 5-FU treated animals demonstrated substantially increased amounts of c-Myc protein (Supplementary Fig. 2). These experiments show that extrinsic stress signals can affect c-Myc protein expression in primitive hematopoietic progenitors, illustrating an intriguing feedback regulation.
c-Myc protein expression predicts self-renewal status
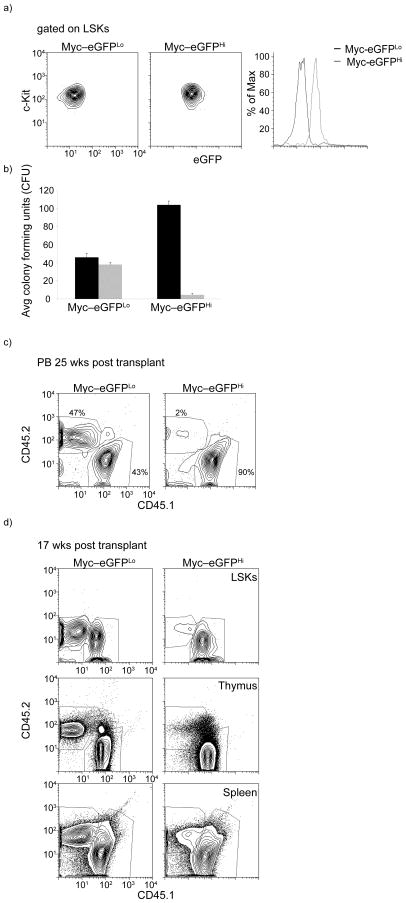
These observations suggested that c-Myc protein abundance (but not mRNA) could dictate the potential of HSCs to self-renew and influence their ability to differentiate. To test this hypothesis we used fluorescence activated cell sorting (FACS) to divide the LSK population into c-Myc-eGFPhi and c-Myc-eGFPlo cells (Fig. 2a). To achieve an estimate of the differentiation capacity of these two populations, we seeded each subset on colony forming unit (CFU) assays using cytokine-supplemented methylcellulose media. The first plating showed that the c-Myc-eGFPhi fraction had an increased (two to three fold) capacity to generate colonies, however, replating of the sorted c-Myc-eGFPhi expressing LSKs yielded a total inability in colony formation, reflecting a putative loss in in vitro self-renewing capacity. In contrast, the c-Myc-eGFPlo population efficiently generated colonies after the first replating, retaining characteristics of bona fide self-renewing HSCs (Fig. 2b). To support these observations with in vivo studies, competitive reconstitutions were performed using 1000 LSK cells purified from both fractions (expressing the marker CD45.2). In agreement with the in vitro studies, c-Myc-eGFPlo cells had the ability to efficiently compete (50:50) and generate all mature lineages as well as stem and progenitor cells 25 weeks after transplantation (Fig. 2c,d). On the other hand, c-Myc-eGFPhi cells were less efficient (6–10% that of the c-Myc-eGFPlo population) in generating long-term chimerism, suggesting that this population is largely devoid of LT-HSC activity (Fig. 2c,d). A previous study suggested that aging could affect differentiation ability of HSCs due to aberrant DNA repair24. As all the experiments described above were performed using 6–10-wk-old animals we asked whether the c-Myc-dependent regulation of HSC self-renewal is a central control mechanism in adult HSCs independently of HSC age. We repeated these experiments using aged (>60-wk-old) HSC. As shown in Supplemental Figure 3a, old and young HSCs demonstrate identical c-Myc-eGFP profiles. Colony assays and bone marrow transplantation assays further proved that relative abundance of c-Myc protein dictate LSK long-term reconstitution ability (Supplementary Figure 3b and data not shown). These findings support the notion that the abundance of c-Myc protein expression can predict the HSC stage and that upon c-Myc protein stabilization HSC cells lose their ability to self-renew.
Figure 2. c-Myc protein levels correlate with loss of HSC self-renewal.
a) FACS-dependent separation of c-Myc-eGFPHi and c-Myc-eGFPLo LSK populations. b) CFU in vitro cultures using sorted c-Myc-eGFPHi and c-Myc-eGFPLo cells. In black: First plating of LSKs, in grey: second plating. Error bars indicate standard deviation (Std) (n=3 mice in 3 independent experiments). c) Peripheral blood chimerism in competitive reconstitution assays at 25 weeks post transplant (n=3 mice). d) Relative chimerism of sorted c-Myc-eGFPHi and c-Myc-eGFPLo LSKs (CD45.2) in the bone marrow (LSK subset), thymus and spleen (d) 17 weeks post transplantation (n=3 mice).
Fbw7 regulates c-Myc protein abundance in adult HSC
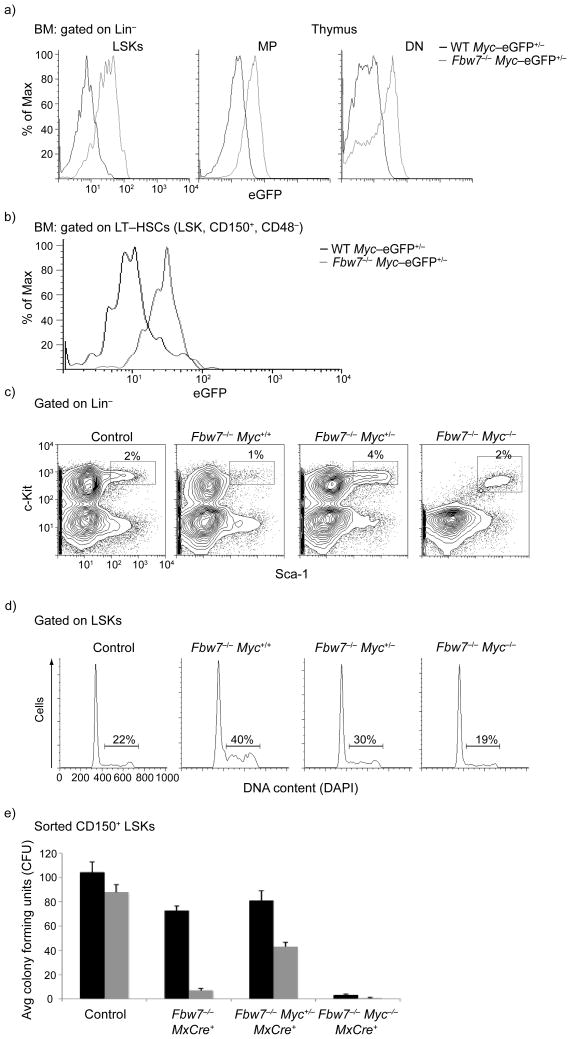
These observations suggested that c-Myc-regulated adult HSC self-renewal could be controlled by a post-transcriptional and likely a post-translational mechanism of regulation. To start probing this hypothesis, we studied the expression pattern of three enzymes, members of the ubiquitin-proteasome system (UPS) that have been previously suggested as putative regulators of c-Myc protein stability (Huwe1, Skp2, Fbw7)25. One of these genes, Fbw7, demonstrated a reciprocal expression pattern when compared to c-Myc-eGFP protein expression (Fig. 1g). To further examine whether Fbw7 controls c-Myc protein stability in HSCs, we generated Mx1-Cre+Fbw7flox/floxMyceGFP/eGFP mice. Mx1 is an type I interferon (IFN)-inducible promoter26 that is activated by intra-peritoneal injection of polyI-polyC resulting in inducible expression of Cre-recombinase and subsequent deletion of the Fbw7 locus. We reasoned that if c-Myc is a bona fide substrate of Fbw7 in HSCs then deletion should significantly stabilize the c-Myc protein, an effect that will correspond to an elevated eGFP mean fluorescence intensity. Consistent with this hypothesis, c-Myc-eGFP abundance in polyI-polyC injected Mx1-Cre+Fbw7flox/floxMyceGFP/eGFP LSK cells was substantially higher than their control, polyI-polyC injected Mx1-Cre+Fbw7+/+MyceGFP/eGFP counterparts (Fig. 3a). More importantly, c-Myc protein was more stable in the LT-HSC compartment as defined by the SLAM markers CD150 and CD48 upon deletion of Fbw7. (Fig. 3b). Such stabilization was also evident in both the MP subset and in thymic CD4−CD8− double negative (DN) T cell progenitors. Moreover, there were no significant differences on the abundance of Myc mRNA expression between the different genotypes (not shown). Thus, these data provide in vivo support of the interaction between c-Myc protein stability and Fbw7 enzymatic function in HSCs.
Figure 3. c-Myc protein stability in HSC is controlled by the ubiquitin ligase Fbw7.
a) FACS analysis showing detection of c-Myc-eGFP in different progenitor populations. LSK, MP and thymic DN (CD4−8−) subsets are shown. b) FACS analysis showing stabilization of c-Myc-eGFP protein in LT-HSC subset (LSK, CD150+, CD48−) upon Fbw7 deletion c-d) Deletion of a single Myc allele significantly rescues the Fbw7−/− HSC phenotype. FACS profiles of bone marrow LSKs (c), cell cycle status of LSKs (d) and methylcellulose assays (CD150+ LSKs) (e) are shown. Black: first plating, Grey: second plating. Error bars indicate standard deviation (Std) (n=5 mice). Plots are a representation of at least 3 independent experiments.
Reduction of c-Myc rescues Fbw7−/− HSC phenotypes
To further prove the relevance of c-Myc stabilization in Fbw7−/− LSK cells, we attempted to rescue the Fbw7 deficiency-induced HSC phenotype by decreasing the c-Myc protein expression14. As total c-Myc deficiency completely abrogates HSC differentiation16, 17 (Fig. 3e), we generated Fbw7−/− LSK cells that lack a single copy of c-Myc (Mx1-Cre+Fbw7flox/floxMycflox/+). Deletion of a single Myc allele rescued LSK numbers, cell cycle status and in vitro self-renewal capacity (Fig. 3c–e). The combined data illustrated in Fig. 2 and Fig. 3 suggest that c-Myc protein stability in HSCs is largely controlled by the activity of the Fbw7 ligase. These results are consistent with our studies that were unable to detect stabilization of additional Fbw7 substrates (Notch1/3, c-jun, cyclin E) in Fbw7−/− hematopoietic progenitors (data not shown and 12).
Fbw7–c-Myc interplay controls LT-HSC differentiation
The previous observations led us to predict that Fbw7 deletion should decrease LT-HSC absolute cell numbers and, conversely, Myc deletion should – at least phenotypically – expand the HSC subset. To test this hypothesis, we initially induced Fbw7 deletion using the Mx1-Cre model (Mx1-Cre+Fbw7flox/flox mice). Deletion of Fbw7 rapidly led to a significant decrease in LSK frequency (Supplemental Fig. 4a). Likewise, within the LSK subset, there was an obvious decline in the frequency and total cell number of CD150+ CD48− LT-HSC subset (Supplemental Fig. 4b,c) suggesting that Fbw7 deletion promotes the LT-HSC to MPP developmental transition and explaining the loss of LT-HSC activity observed in Fbw7−/− animals. On the other hand, inducible deletion of Myc lead to an opposite phenotype characterized by the accumulation of CD150hi, Flt3−, CD34− LT-HSC-like cells (Supplemental Fig. 4d–g). These results further support in vivo the role of the antagonistic interplay between the ubiquitin ligase (Fbw7) and substrate in the differentiation and self-renewal capacity of HSCs.
c-Myc abundance controls the molecular program of HSC
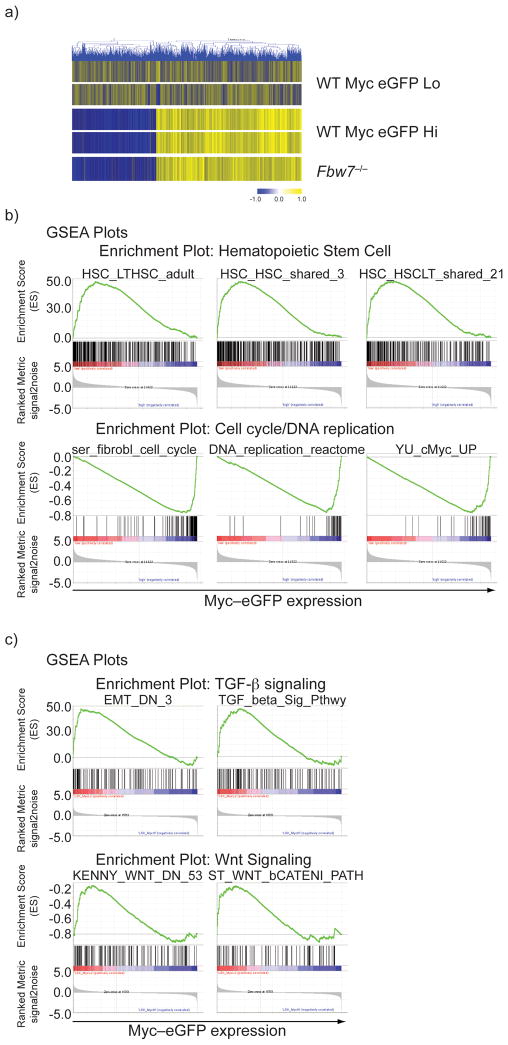
To further understand the mechanistic impact of c-Myc protein stabilization in HSCs, we purified c-Myc-eGFPhi and c-Myc-eGFPlo subsets and used whole-genome transcriptome analysis to define the gene expression signatures of each population. A substantial alteration of the gene expression program accompanies the transition from c-Myc-eGFPlo to c-Myc-eGFPhi stages (Fig. 4a). Indeed, stabilization of c-Myc protein correlated with upregulation of cell cycle (including Ccnd1, CcnA2, Bub1, Aurka, Skp2, Cdk2), transcription (including Narg1, Tcerg1, Ilf3, E2F2, E2F3) and epigenetic (including Ezh1, Suv39h2) regulators. Consequently, a large number of genes known to regulate HSC self-renewal (including Evi1, Eya1, Meis1, Hoxa5/9, Hoxb3, Cdkn1c, Egr1) were significantly down-regulated in the c-Myc–eGFPhi subset. Further analysis has revealed a striking overlap of gene expression signatures between c-Myc–eGFPhi and Fbw7−/− LSKs (Fig. 4a) underlining the close biological connection between the two molecules and further explaining the loss of self-renewal capacity of Fbw7−/− HSCs. Further studies using the Gene Set Enrichment Analysis (GSEA) tool27 provided additional information on shared gene clusters between the c-Myc-eGFPlo versus c-Myc-eGFPhi signature and publicly available datasets. A sizable correlation exists between the c-Myc-eGFPlo gene signature and several published HSC-related datasets (Fig. 4b). Conversely, the c-Myc-eGFPhi signature was significantly enriched in datasets presenting genes that positively control cell cycle progression, DNA replication and c-Myc-induced genes in different cell types28–30. Similarly, KEGG pathway/DAVID database analysis showed significant enrichments associated with cell cycle, DNA replication, and metabolism in the c-Myc-eGFPhi population (not shown). Interestingly, further GSEA analysis revealed significant enrichment of TGF-β and Wnt–β-catenin pathway-regulated genes in the c-Myc-eGFPlo signature (Fig. 4c and Supplementary Fig. 5). Both pathways have been implicated in the regulation of HSC differentiation and self-renewal31, 32. These studies correlate the relative abundance of a single transcription factor to the gene expression signature that defines HSC quiescence and self-renewal.
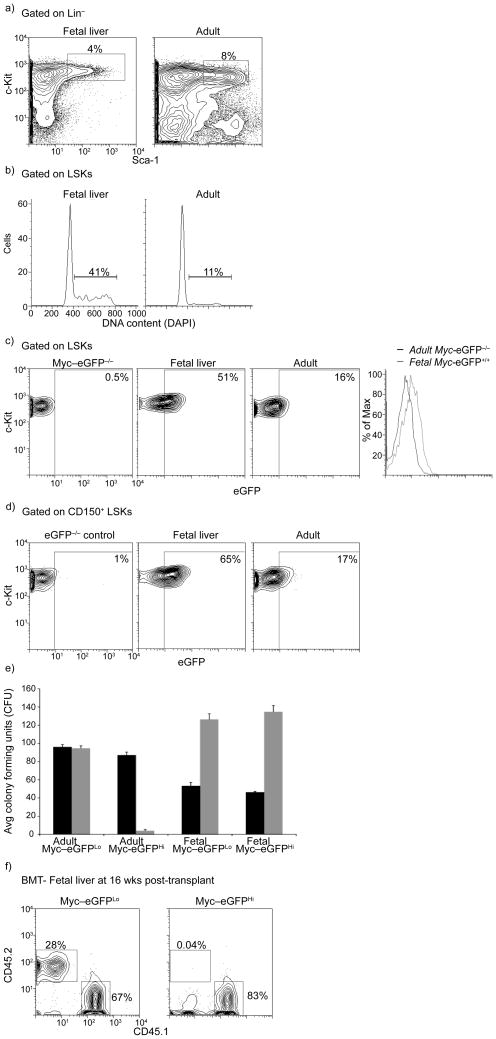
Figure 4. The role of the c-Myc:Fbw7 interaction in fetal liver stem and progenitor cells.
a) FACS staining defining LSK cells in e.d.14.5 fetal liver and adult (6wk old) bone marrow. b) Cell cycle status of fetal and adult LSK cells. c) Levels of c-Myc-eGFP protein expression in fetal and adult LSK cells. The overlay histogram shows induction of c-Myc protein expression in fetal LSKs. d) Levels of c-Myc-eGFP protein expression in CD150+ LSKs. e) Methylcellulose culture using the indicated cell populations purified from the fetal liver (fetal) or the bone marrow (adult). Black: first plating, Grey: second plating. Error bars indicate standard deviation (Std) (n= 6 mice). f) c-Myc-eGFPLo but not c-Myc-eGFPHi expressing fetal liver LSK subsets show chimerism in the peripheral blood 20wks post transplant in competitive reconstitution assays. CD45.2+ cells are donor-derived cells in the peripheral blood (n= 6 mice). Plots are a representation of at least 3 independent experiments.
Identification of genes directly controlled by c-Myc
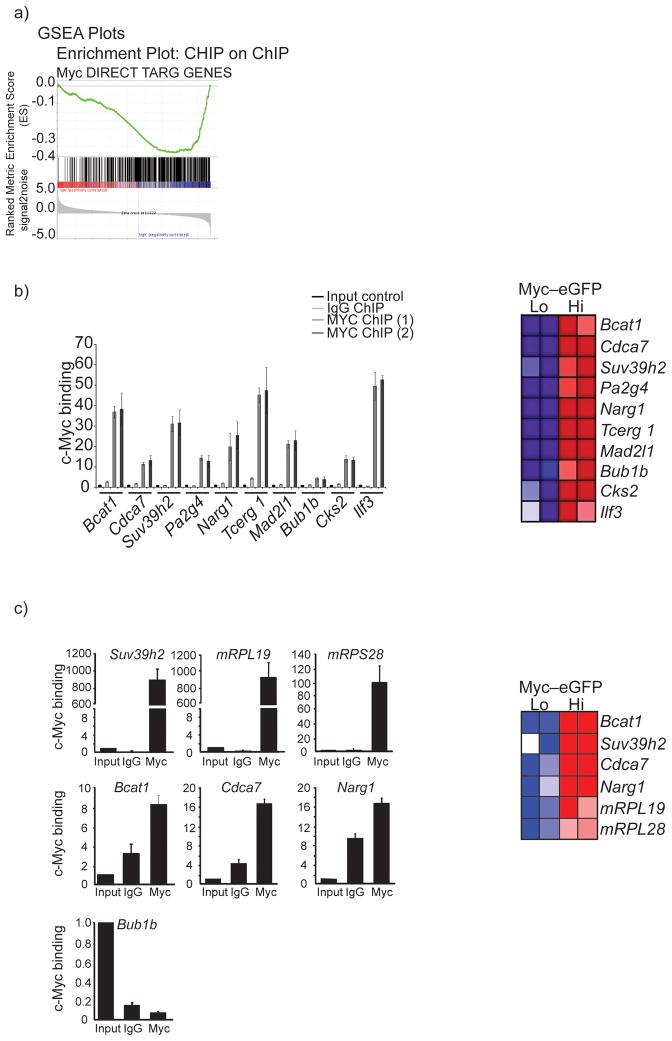
These results are of unique significance as they suggest that c-Myc protein stability is a central regulator of gene expression programs that influence HSC self-renewal, transcription and cell cycle entry. To further explore the mechanism of c-Myc-controlled gene regulation we have compared the identified microarray signatures to our whole genome Chromatin Immunoprecipitation (ChIP2) datasets that contain loci occupied by c-Myc factors in c-Myc-expressing leukemia (T-ALL) cells33. This comparison demonstrated a significant enrichment for genes overexpressed in the c-Myc-eGFPhi subset within the TALL Chip2 dataset (Fig. 5a). The majority of these genes also belonged to functional categories related to cell cycle progression, transcription, translation and epigenetic control (Fig. 5 and Supplemental Figure 6). To demonstrate the validity of these whole-genome wide ChIP2 studies we have selected ten genes previously suggested to play important roles in the regulation of cell cycle progression, gene translation and transcription. Using this model-gene subset we performed conventional ChIP. As predicted, the c-Myc binding sites on the promoters of all these genes were indeed occupied by the c-Myc factor (Fig. 5b).
Figure 5. c-Myc protein abundance directly controls the molecular program of stem cell differentiation, cell cycle entry and self-renewal.
a) Heat map demonstrating relative gene expression signatures of c-Myc-eGFPHi and c-Myc-eGFPLo subsets. A direct comparison to the relative gene expression in Fbw7−/− LSKs is also shown. b) Representative GSEA profiles showing positive correlation of the c-Myc-eGFPLo signature to stem cell gene sets and negative correlation to cell cycle and DNA replication gene sets. c) Representative GSEA profiles showing a positive correlation between both TGF-β and Wnt signaling pathways with c-Myc-eGFPLo gene expression data set.
As the presented database was generated using human c-Myc-expressing hematopoietic tumor (leukemia) cells we sought to further validate these studies using mouse c-Kit-expressing stem and progenitor cells. We were able to demonstrate that the genes bound by c-Myc illustrated in Fig. 5b were also transcriptionally regulated by c-Myc in BM HSCs and progenitor cells (Fig. 5b,c and Supplementary Fig. 7). These genes were expressed in wild-type HSCs, highly expressed in the c-Myc-eGFPhi subset and silenced in HSCs purified from compound Myc−/− mice (Myc and Mycn)16. To further rule out the possibility that present findings were biased due to the ChIP database used, we selected a group of genes that are overexpressed in the c-Myc-eGFPhi LSK subset and also have highly conserved (between mouse and human) putative c-Myc binding sites. We performed conventional ChIP using highly purified bone marrow Lin− c-Myc-eGFPhi progenitors and showed that a significant portion of these genes promoters were indeed bound by c-Myc factors (Fig. 5c). Collectively, these studies demonstrate a mechanistic correlation between c-Myc protein abundance, binding on regulatory DNA sites and transcriptional control of HSC differentiation.
Fbw7:c-Myc axis in fetal liver hematopoietic cells
We next sought to test the importance of the c-Myc stability in a stem cell population closely related to adult HSCs, namely fetal liver HSCs. Although very similar phenotypically and functionally, fetal HSCs actively expand in number in the fetal liver during embryonic days 12.5–14.5 (ref. 34). Thus, we hypothesized that fetal HSCs would show higher amounts of c-Myc protein than their adult counterparts. Our experiments (Fig. 6) have validated this hypothesis. Initially, almost three-fold more fetal LSK cells were in cycle at any given moment, in agreement with previous findings (Fig 6b). Also, abundance of c-Myc protein (in E13.5–14.5 MyceGFP/+ embryos) was substantially higher in fetal total and CD150+ LSKs when compared to adult LSKs (Fig 6c–d).
Figure 6. Genes over-expressed in c-Myc-EGFPHi cells are directly bound by the c-Myc transcription factor.
a) GSEA analysis showing a positive correlation of c-Myc-eGFPHi gene expression profiles to a direct c-Myc target Chip-on-chip dataset. b) ChIP assay and Heat map of selected genes that are over-expressed in the c-Myc-eGFPHi cells. Red color: gene upregulation, Blue color: gene down-regulation. Two individual chromatin IPs using T-ALL cell line genomic DNA are shown. c) ChIP assay of selected genes using genomic DNA from purified Lineage− c-KiteGFP+ bone marrow progenitor cells. Also, Heat Map showing over expression of selected genes in the c-Myc-eGFPHi LSK subset. Error bars indicate standard deviation (std) from 3 independent experiments.
Interestingly, in vitro CFU assays using purified c-Myc-eGFPhi and c-Myc-eGFPlo fetal HSCs revealed no differential ability to generate hematopoietic colonies in primary, secondary or tertiary plating assays (Fig 6e). These findings were consistent with transcriptome studies which showed very similar gene expression signatures between the c-Myc-eGFPhi and c-Myc-eGFPlo fetal LSK subsets, in contrast to the well defined and largely distinct signatures illustrated using their adult counterparts (Fig. 6f and Supplemental Fig. 8). However, long-term competitive reconstitution assays using the fetal liver c-Myc-eGFPhi and c-Myc-eGFPlo LSK subsets demonstrated that when introduced in an adult environment c-Myc-eGFPhi cells were unable to produce long-term chimerism, in agreement with the results obtained using their adult counterparts (Fig. 6e).
Dynamic regulation of Fbw7 and c-Myc in ESCs
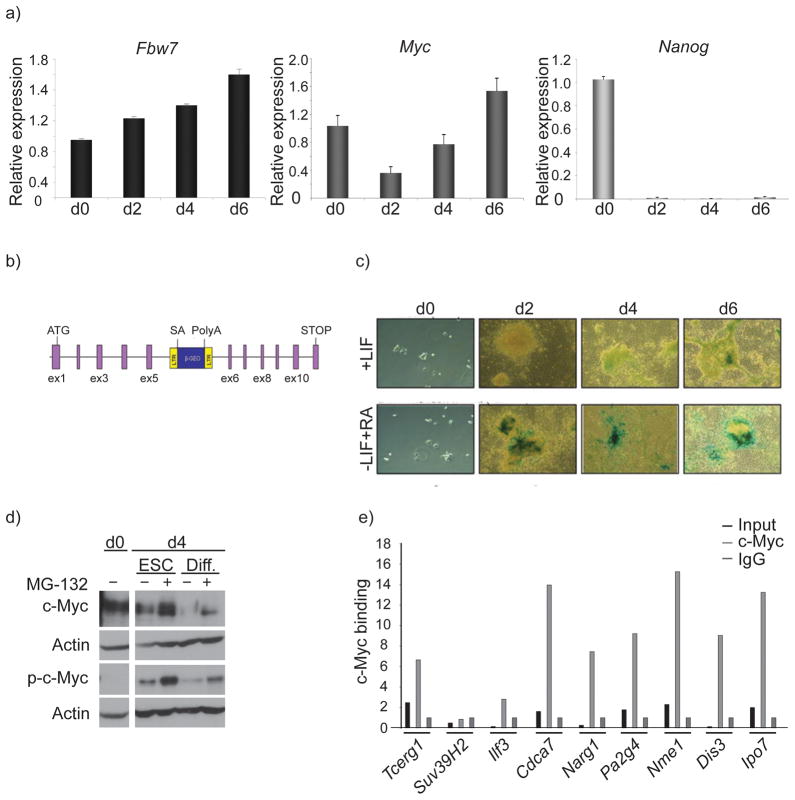
To further probe the universality of the Fbw7–c-Myc interaction in stem cell self-renewal we turned to the study of embryonic stem cells (ESCs), a stem cell type with properties fundamentally distinct from adult and quiescent HSC. ESC are unique among stem cells as they can differentiate into all three germ layers and self-renew extensively in culture35. In terms of cell cycle regulation, ESCs proliferate rapidly, display a shortened G1 phase and possess a constitutively active cyclinE-CDK2. Furthermore, ESCs express low amounts of the D-type cyclins and have almost no CDK4 kinase activity36. It was thus tempting to suggest that Fbw7 could be dispensable for the self-renewal of ESCs. Indeed, our initial expression studies supported this notion as Fbw7 was expressed less in self-renewing ESCs and was upregulated as differentiation was induced (Fig. 7a). To further prove these qPCR studies we used a trapped ESC line, in which a LacZ reporter cassette was introduced in the Fbw7 locus disrupting expression monoallelically (Fig. 7b). Fbw7LacZ expression was almost undetectable in self-renewing ESCs and was subsequently induced upon ESC differentiation (Fig. 7c). Although there were minimal changes in Myc transcript abundance between self-renewing and differentiating ESCs (Fig. 7a), c-Myc protein expression, on the other hand, was readily detectable in self-renewing ESCs and became down-regulated upon induction of differentiation (Fig. 7d), confirming multiple previous studies which connected c-Myc expression to ESC pluripotency37, 38. ChIP studies showed that c-Myc was not only expressed but was also detectable on the promoters of selected predicted gene targets (Fig. 7e).
Figure 7. Patterns of Fbw7 and c-Myc expression in mouse embryonic stem cells.
a) mRNA transcript expression levels of Fbw7 (black), Myc (dark grey) and Nanog (light grey) in self-renewing (+LIF) and differentiating (−LIF, +RA) mouse ESC. Different days of differentiation (d0–6) are shown), b) Schematic diagram of Fbw7 gene-trap cassette. c) LacZ staining in Murine ESCs containing the Fbw7 gene trap cassette depicting upregulation of Fbw7 expression as Murine ESCs differentiate via the removal of LIF and the addition of Retinoic Acid (RA). d) Western blot showing stabilization of both phospho- (T58) and total-c-Myc in self-renewing and differentiating Murine ESCs treated with 20uM of proteosome inhibitor MG-132. e) ChIP assay was carried out using the specific regulators shown in Fig 6 revealing substantial enrichment upon c-Myc immunoprecipitation in Murine ESCs. Error bars indicate standard deviation (std) from 3 independent experiments.
Fbw7 is dispensable for self-renewal of ESCs
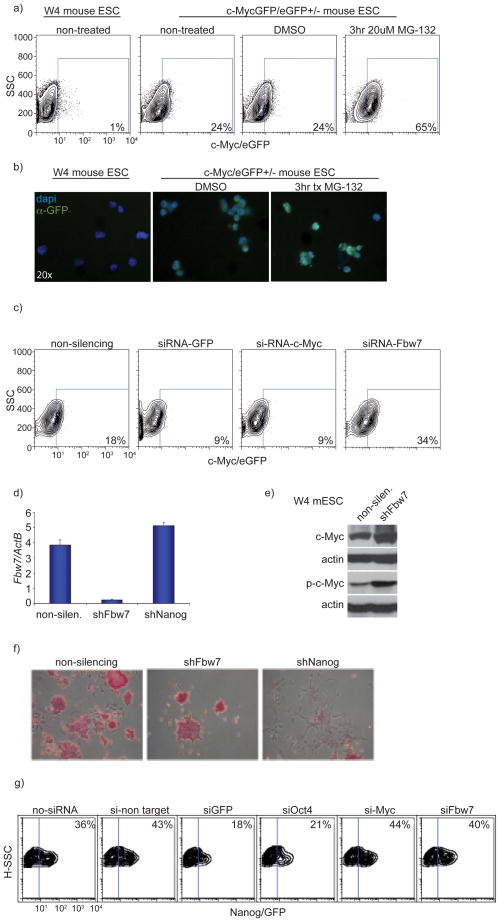
To address whether c-Myc stability could also be regulated by the UPS in ESCs, we treated self-renewing and differentiating ESCs with the proteasome inhibitor MG-132. We found that both total-and phospho-c-Myc protein accumulated in both self-renewing and differentiating culture conditions (Fig. 7d). Complimentary to this finding, we also treated MyceGFP/+ ESCs with MG-132 and observed stabilization of c-Myc–eGFP by flow cytometry as well as immunofluorescence using an antibody which detects eGFP (Fig. 8a,b). To begin assessing the role of Fbw7 in ESCs self-renewal and its regulation of c-Myc, we used siRNA-mediated gene silencing. ESC cell specific Fbw7 knockdown resulted in accumulation of c-Myc eGFP, suggesting that Fbw7 regulates c-Myc protein stability in self-renewing ESCs (Fig. 8c). Knockdown of Fbw7 was very efficient as measured by qRT-PCR expression assays (Figure 8d). We observed similar results in another mouse ESC line (W4) using shRNA directed against Fbw7 where knockdown resulted in accumulation of both total- and phospho-c-Myc protein (Figure 8e). However, there were no phenotypic changes (Fig. 8f) or changes in expression of germline markers demonstrating that ESCs retain characteristics associated with molecular (data not shown) effects of ESC self-renewal. In contrast, silencing of the pluripotency regulator Nanog, led to rapid cell differentiation and loss of self-renewal (Fig. 8f). To further test the dispensability of Fbw7 in ESCs – at least in this in vitro system – we used a transgenic ES cell line that reports for self-renewal due to the expression of GFP-driven by Nanog regulatory elements39. Fbw7 silencing was unable to affect Nanog–GFP expression and ES cell self-renewal in this system (Fig. 8g). Overall, these observations show that Fbw7 expression is upregulated during ESC differentiation and that Fbw7 silencing is dispensable for the in vitro self-renewal of ES cells. They also identify a significant molecular distinction between ESCs and quiescent adult stem cells (HSCs) (Supplemental Figure 9).
Figure 8. Fbw7 is dispensable for the self-renewal of Murine ESCs.
a) Inhibition of proteasomal degradation via MG-132 treatment resulted in accumulation of c-Myc protein in MyceGFP/− Murine ESCs. b) Visualization of c-Myc-eGFP in W4 and MyceGFP/− Murine ESCs with and without treatment with MG-132. GFP (green), nuclear staining (DAPI) c) Knock-down of Fbw7 using siRNA in c-MyceGFP/− ESCs resulted in accumulation of c-Myc protein (increase in eGFP levels) while siRNA targeting both Myc and GFP showed a reduction in c-Myc-eGFP levels as assessed by FACs analysis. d) qRT-PCR of Fbw7 mRNA expression in ESCs expressing shRNAs against Fbw7 and Nanog. A non silencing shRNA is used as a control. e) Western blot depicting accumulation of both phospho (T58)-and total-c-Myc protein upon shRNA mediated knock down of Fbw7 when compared to a non-silencing shRNA control. f) Alkaline phosphatase staining in Murine ESCs showed no difference in differentiation capacity upon shRNA mediated knock-down of Fbw7 when compared to non-silencing control while silencing of Nanog resulted in complete differentiation. g) siRNA knock-down of Fbw7 in a Nanog-eGFP reporter ESC line resulted in no difference in Nanog-eGFP expression levels when compared to non-silencing control. Knock-down of Oct4 resulted in a reduction of Nanog-eGFP expression levels (marking differentiation). Plots are a representation of at least 3 independent experiments
DISCUSSION
Our observations are the first to identify a specific ubiquitin ligase: substrate pair as a central regulator of stem cell differentiation. They demonstrate in vivo that slight changes in the stability and abundance of c-Myc protein expression could play the role of a rheostat controlling adult stem cell quiescence and self-renewal. Our work also explains the mechanism by which c-Myc carries out its function within the HSC population. It identifies, using whole-genome transcriptional analysis and ChIP2 approaches, a constellation of genes that are directly regulated by relative abundance of this transcription factor during these early stages of hematopoiesis. Most importantly, our studies suggest a differential need for c-Myc stability between adult and embryonic stem cells suggesting that different stem cell types sense and interpret c-Myc-regulated gene expression in distinct ways. All these studies prove the hypothesis that the ubiquitin–proteasome system can control stem cell differentiation by directly controlling important regulators of cell cycle entry, self-renewal and/or lineage commitment. Further studies are needed to define the extent of the involvement of the ubiquotome in different aspects of stem and progenitor cell biology.
Our findings explain the discrepancy between Myc mRNA expression and function that has been previously demonstrated. Indeed, c-Myc deletion studies suggested that c-Myc expression is essential for the exit of HSCs from the osteoblastic niche through regulating the expression of essential adhesion molecules, including N-Cadherin17. However, the same investigators have shown that Myc mRNA stability remains stable from the LT-HSC stage to the common myeloid progenitor (CMP) stage questioning the HSC-specific function of this transcription factor16. These findings were verified by our own independent analysis. We demonstrate here that Fbw7 expression is high in the LT-HSC stage and virtually undetectable at subsequent MPP and MP stages of differentiation. Similarly, c-Myc protein abundance increased as HSCs differentiated and reached their peak at the MP and CMP stages. These observations explain the previously reported discrepancy and underline the importance of ubiquitin-mediated protein stability in these early stages of hematopoiesis.
One of the most exciting aspects of this study is the differential dependence of embryonic (ESC) and adult HSCs on c-Myc stability and Fbw7 activity. Indeed, adult HSCs are characterized by high Fbw7 expression and a rapid c-Myc turnover while self-renewing ESC demonstrate only a basal Fbw7 activity and a very stable c-Myc protein. Few such factors that have distinct functions in the two stem cell types have been identified thus far, most of them members of the machinery that initiates cell cycle, including cyclins D and E, cell cycle inhibitors (CDKI) and the retinoblastoma protein (pRb). It is very likely that the differential Fbw7 expression is tightly linked to the cell cycle entry machinery. Indeed, studies in embryonic fibroblasts reveal that Fbw7 expression is cell cycle-dependent, with the highest levels recorded at the G0/G1 stages (B.A-O and I.A., unpublished). The Fbw7 behavior during ES cell differentiation is consistent with c-Myc availability and its essential role in the regulation of pluripotency, however at this point we cannot exclude a role for additional ESC Fbw7 substrates that could control cell cycle progression, including cyclin E. Finally, as c-Myc was recently proved to be one of the factors controlling adult cell reprogramming38, it is intriguing to propose that the ubiquitin system and more specifically Fbw7 could be a key regulator of this process. Our data also fit perfectly with other studies that showed that the kinase GSK3β, which phosporylates c-Myc on Thr58 and triggers Fbw7-mediated degradation in other cell systems40, is largely excluded from the nucleus in self-renewing ESCs. GSK3β enters the nucleus upon ESC differentiation due to the absence of LIF-induced Akt activation41. We demonstrate here that both proteasome inhibition and Fbw7 knockdown leads to stabilization and overexpression of c-Myc phosphorylated on Thr58 connecting ESC differentiation, kinase activation, SCFFbw7 complex recruitment and c-Myc degradation.
Our work also suggests that the ubiquitin complex is a novel regulator of stem cell differentiation and function. Indeed, activity of ubiquitin ligases (like Fbw7) or opposing deubiquitinases (DUB) could control both the abundance and the activation of central stem cell regulators. This close connection appears to be conserved between species as a recent report has demonstrated the requirement for the DUB Scrawny in the epigenetic control of gene expression in Drosophila stem cells10. These observations propose an additional layer of stem cell regulation and also suggest that genetic or chemical targeting of specific ubiquitin enzyme activity could affect somatic cell reprogramming, adult stem cell expansion and putatively cancer stem cell differentiation and oncogenicity.
METHODS
Animals
The Mycflox 42 mice were a kind gift from F. Alt (Harvard University). All mice were housed in a pathogen-free animal facility at New York University School of Medicine. All animal procedures were carried in compliance with Institutional Animal Care and Use Committee of the New York University School of Medicine.
Genotyping and in vivo animal studies
Tails were clipped from mice 1–2 weeks after birth and incubated in tail-lysis buffer containing Proteinase-K for 24 h at 55 °C. NaCl was added and cellular debris pelleted by ultra-centrifugation. DNA was precipitated upon addition of isoproanol, pelleted and washed using 70% ethanol. DNA pellet was dissolved in water and used for PCR analysis. Primers were designed to amplify both WT and floxed Myc and Fbw7 alleles. Furthermore, primers were designed to detect both WT and Myc-eGFP knockin alleles (Supplemental Table 2). Mice received 5 i.p. polyI:polyC injections of 20ug/gram of body weight 2 weeks after birth and analyzed 2 weeks post injection. Deletion of targeted alleles and transcript levels were measured by genomic PCR and qRT-PCR.
Quantitative real-time PCR
RNA was isolated using RNeasy-Plus Mini Kit (Qiagen). cDNA synthesis was carried out by using Super-Script III First Strand Synthesis System Reverse Transription Kit (Invitrogen) and used for qRT-PCR with iQ SYBR green supermix on an iCycler (BioRad). Relative expression was determined from cycle-threshold (Ct) and normalized to internal control, Actb. For primer sequences, see Supplemental table 2.
Flow cytometry and antibodies
Flow cytometric analysis was carried out as previously described 12. In experiments utilizing the Myc-eGFP knock in mouse we analyzed a fluorescence minus one (FMO) littermate control in parallel to set eGFP gates (Supplementary Fig. 9). Antibodies used for FACS analysis were procured from e-Bioscience. Ki-67 antibody used for cell cycle analysis was bought from BD Biosciences. Specifically, the antibodies we used were as follows: c-kit (2B8), Sca-1 (D7), CD150 (TC-15-12F2.2), CD48 (HM48-1), Mac-1 (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), TER-119, CD3 (145-2C11), CD19 (1D3), IL7R (RAM34), CD4 (RM4–5), CD4 (H129.19), CD8 (53 – 6.7), CD25 (PC61), and CD44 (IM7). BM lineage antibody cocktail includes: Mac-1, Gr-1, NK1.1, TER-119, CD3, IL7R and CD19.
Immunoblotting
Immunoblotting was done as previously described12. Briefly, c-Kit+ and c-Kit− bone marrow cells were isolated using a magnetic-bead based positive selection kit with a biotynylated c-Kit primary antibody (EasySep Positive Biotin Selection kit; Stem Cell Technologies). Upon selection, cells were lysed using RIPA lysis buffer, incubated at 0 °C for 20 min, and cellular debris was pelleted using ultra-centrifugation. Total cell lysates were separated on a 4–20% Tris-HCl gradient gel by SDS-PAGE electrophoresis and transferred to nitrocellulose membrane. Membrane was probed with the following antibodies: c-Myc (SC-764; Santa Cruz Biotechnology) and tubulin (Santa Cruz Biotechnology).
Immunofluorescence
Experiments were carried out as previously described43. c-Kit+ bone marrow cells were purified as described above (immunoblotting). Cells were aliquoted onto charged glass slides, fixed using 4% para-formalydehyde, cell membrane permeablized by 0.1% Triton-X, and incubated with GFP-specific rabbit polyclonal antibodies at 4 °C overnight (ab290; Abcam). Subsequently, washed slides were incubated with anti-Rabbit IgG alexa-488 conjugated secondary antibody (Invitrogen) and counterstained with Dapi nuclear stain (Sigma).
Gene Set Enrichment Analysis
GSEA was performed on normalized expression data as described44. Gene expression profiles of c-Myc-eGFPlo versus c-Myc-eGFPhi cell populations were used as background dataset and two different gene sets were used as foreground: “C2 curated gene sets” from the Broad Institute (http://www.broad.mit.edu/gsea/msigdb/index.jsp) and a homemade gene set constructed by considering c-Myc Chip on chip data, P-value < 0.005 (ref. 33). The differentially expressed genes are ranked using “signal to noise” metric.
Microarray Analysis
Five nanograms (5 ng) of total RNA from target cell populations was amplified and labeled using the Ovation Amplification v2 and Biotin cDNA Biotin systems (Nugen, Inc.). The resulting cDNA was hybridizated to GeneChip MG 430 2.0 arrays following recommendations of the array manufacturer (Affymetrix). The raw data were processed in GeneSpring 7.2 (Agilent). The Affymetrix CEL files were normalized using Robust Multi-Array Average expression measure (RMA) and baseline scaling44. To identify differentially abundant mRNAs between replicate experimental conditions, the abundance levels had to be identified as significantly different by at least one of three statistical algorithms available from TIGR Microarray Suite TM4: T-test (P < 0.05, α correction), Significance Analysis of Micro-array (SAM, false discovery rate set 5% and Pavlidis Template Matching (PTM, P < 0.05)45. KEGG pathway and DAVID tools were accessed through http://david.abcc.ncifcrf.gov/ for pathway enrichment analyses. The c-Myc-eGFP gene expression datasets are available through the Gene Expression Omnibus (GEO) repository (GSE19502).
Chromatin Immunoprecipitation Assay
Chip2 and ChIP assays were performed as previously described33 using T-ALL cell lines, Lin−c-Kit+ selected bone marrow cells and mouse embryonic stem cells (W4, 129/sj). Briefly, qRT-PCR was completed on chromatin immunoprecipitates and corresponding whole cell extracts to validate c-Myc promoter occupancy of 10 “hand-picked” genes using an ABI 7300 Real-Time PCR system (Applied Biosystems). β-actin was used as input reference.
In vitro colony forming assays
Total LSK from control, Fbw7−/−Myc+/− MxCre+ and Fbw7−/−Myc−/− MxCre+ mice or upper 30% c-Myc-eGFP+/− and lower 30% of c-Myc-eGFP+/− LSK cells were sorted from MyceGFP/eGFP knock-in adult mice or E14.5 embryos. 300 cells per subset were seeded in duplicate and cultured in cytokine supplemented methylcellulose medium (MethoCult 3434; Stem Cell Technologies). Subsequently, colonies were counted on day 7, isolated, re-plated and cultured for another 7 days (total 14 days).
ES cell culture, lenitviral transduction and siRNA transfection
All embryonic stem cell lines were maintained on MEFs cultured on gelatin-coated plates in the presence of DMEM (Cellgro) supplemented with 15% FBS. The Fbw7 ESC LacZ trapped line was obtained by the Texas Institute for Genomic Medicine. MyceGFP/− ESCs and Nanog-GFP ESCs were reverse transfected by mixing 30 nM siRNA (commercially available, siGENOME SMARTpool, Dharmacon) with Lipofectamine 2000 (Invitrogen) in supplemented DMEM media (Cellgro). siRNA mix was added to a gelatin coated 96-well plate, and ES cells were plated at 1,500 cells/well in DMEM and LIF (Chemicon). For differentiation experiments, ESC cells were pre-plated for 1 h (to remove MEFs), seeded at low density, and at 24 h post initial plating (Day 0) either only LIF was removed or LIF was removed and Retinoic Acid was added. eGFP fluorescence was measured using an LSRII (BD Biosciences) 48 h post-transfection. Nanog-GFP ESC cells (kind gift from I. Lemischka) were transduced by pLKO.1-PIG lentiviruses (a gift from Ihor Lemischka see reference 35 and Supplementary Table 3) containing respective shRNAs 1 h after preplating. Two days post-transduction, transduced cells were selected by puromycin (2 μg/ml) and subsequently used for self-renewal and differentiation experiments.
Bone marrow transplantation
Freshly dissected femur and tibia were flushed using a 1cc insulin syringe filled with PBS supplemented with 3% FBS. Cells were spun for 2.5 min at 200g and red blood cells lysed using ACK lysis buffer for 3 min. at 25 °C. The cells were then washed in PBS/3%FBS and spun. Subsequently, the cells were passed through a cell strainer, counted, and stained using antibodies directed against lineage markers (PE), c-Kit (APC), and Sca-1 (Pe-Cy7). Finally, bone marrow derived CD45.2+ c-Myc-eGFPHi or c-Myc-eGFPLo LSK subsets (from adult mice and E14 fetal livers) were FACS purified and 1000 cells from each subset were mixed with 1.0 × 106 wild-type CD45.1+ bone marrow cells and injected into the ocular cavity of lethally irradiated (2x 600rad) CD45.1 host mice. Chimerism was analyzed by peripheral blood analysis at 7,12, 17 and 25 weeks post transplantation. Chimerism in bone marrow, thymus and spleen was measured at 17 weeks.
5-FU treatment
Experiments using 5-FU were performed as previously described16. Briefly, MyceGFP/− mice were injected one time with 75 μg/gram of body weight of fluorouracil and LSK population within the bone marrow was analyzed 2 days post treatment using flow cytometry.
Statistical analysis
All the statistical analyses were performed using un-paired two-tailed Student’s t-test assuming experimental samples of equal variance, unless otherwise specified.
Supplementary Material
Acknowledgments
We would like to thank the members of the Aifantis Lab and specifically B. King for advice as well as time and effort spent towards this manuscript. P. Lopez and the NYU Flow Facility for expert cell sorting. The NYU Cancer Institute Genomics Facility for help with micro-array processing. M.Gostissa and F. Alt for the c-Mycflox mice. I. Lemishka and his lab for the Nanog-GFP line and valuable technical advice. Supported by the National Institutes of Health (RO1CA133379, RO1CA105129, R21CA141399, R56AI070310, P30CA016087 to I.A., RO1AI41428, RO1AI072039 to B.P.S. and R01CA120196 to A.F.), the American Cancer Society (RSG0806801 to I.A.), the Mallinckrodt Jr. Foundation, the Irma T. Hirschl Trust and the Alex’s Lemonade Stand Foundation (to I.A.). B.A-O. was supported by the Alexander von Humboldt Foundation. S.M.B. was supported by the NYU Hematology/Oncology Program and K.C. by the NYU Molecular Oncology and Immunology Training Grant (5T32CA009161). I.A and A.F. are Leukemia & Lymphoma Society Scholars. I.A. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Author Contributions
L.R. performed most of the experiments and participated in the preparation of the manuscript. G.D.G, T.P., A.F. and B.A-O have designed and performed the ChIP2 and ChIP experiments. K.C., E.L. and B.T. performed the ESC experiments. B.P.S, B.T., A.L.B, and B.A.H. generated and performed initial studies using the c-Myc/eGFP mice. J.Z. analyzed micro-array data. I.A. designed the study and prepared the manuscript.
References
- 1.Adams GB, Scadden DT. The hematopoietic stem cell in its place. Nat Immunol. 2006;7:333–337. doi: 10.1038/ni1331. [DOI] [PubMed] [Google Scholar]
- 2.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8:290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- 3.Kondo M, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida T, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 6.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 8.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki L, Pagano M. Cell cycle, proteolysis and cancer. Curr Opin Cell Biol. 2004;16:623–628. doi: 10.1016/j.ceb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuoka S, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008 doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson BJ, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008 doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whetton AD, et al. The time is right: proteome biology of stem cells. Cell Stem Cell. 2008;2:215–217. doi: 10.1016/j.stem.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 15.O’Neil J, Look AT. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene. 2007;26:6838–6849. doi: 10.1038/sj.onc.1210766. [DOI] [PubMed] [Google Scholar]
- 16.Laurenti E, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson A, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 20.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 22.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 23.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 25.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgantas RW, 3rd, et al. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova NB, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 30.Goldrath AW, Luckey CJ, Park R, Benoist C, Mathis D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc Natl Acad Sci U S A. 2004;101:16885–16890. doi: 10.1073/pnas.0407417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reya T, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson S. Is TGF-beta a stemness regulator? Blood. 2009;113:1208. doi: 10.1182/blood-2008-11-189738. [DOI] [PubMed] [Google Scholar]
- 33.Margolin AA, et al. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106:244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 36.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 37.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Schaniel C, et al. Delivery of short hairpin RNAs--triggers of gene silencing--into mouse embryonic stem cells. Nat Methods. 2006;3:397–400. doi: 10.1038/nmeth0506-397. [DOI] [PubMed] [Google Scholar]
- 40.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 41.Bechard M, Dalton S. Subcellular localization of glycogen synthase kinase 3beta controls embryonic stem cell self-renewal. Mol Cell Biol. 2009;29:2092–2104. doi: 10.1128/MCB.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 43.Demuth T, et al. MAP-ing glioma invasion: mitogen-activated protein kinase kinase 3 and p38 drive glioma invasion and progression and predict patient survival. Mol Cancer Ther. 2007;6:1212–1222. doi: 10.1158/1535-7163.MCT-06-0711. [DOI] [PubMed] [Google Scholar]
- 44.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 45.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.