Abstract
Copy number variation (CNV) is increasingly recognized as a source of phenotypic variation among humans. We hypothesized that a CNV in the human arginine vasopressin receptor-2 gene (AVPR2) would be associated with serum sodium concentration based on the following lines of evidence: 1) the protein product of the AVPR2 gene is essential for renal water conservation; 2) mutations in the AVPR2 gene are associated with aberrant water balance in humans; 3) heritability of serum sodium concentration may be greater in females than in males; 4) the AVPR2 gene is X-linked; and 5) a common CNV spanning the AVPR2 gene was recently described in a non-Hispanic Caucasian population. We developed a highly reproducible assay for AVPR2 CNV. Among 279 subjects with measured serum sodium concentration in the Offspring Cohort of the Framingham Heart Study, no subjects exhibited CNV at the AVPR2 locus. Among 517 subjects in the Osteoporotic Fractures in Men Study (MrOS)—including 152 with hyponatremia and 183 with hypernatremia—no subjects with CNV at the AVPR2 locus were identified. CNV at the AVPR2 locus could not be independently confirmed, and CNV at the AVPR2 gene is unlikely to influence systemic water balance on a population-wide basis in non-Hispanic Caucasian subjects. A novel AVPR2 single nucleotide polymorphism affecting the reporter hybridization site gave rise to an artifactually low copy number signal (i.e., less than unity) in one male African American subject. Reanalysis of the original comparative genomic hybridization data revealed bona fide CNVs flanking—but not incorporating—the AVPR2 gene, consistent with our new genotyping data.
Keywords: osmoregulation, arginine vasopressin, hyponatremia, hypernatremia
systemic tonicity is among the most tightly regulated of physiological parameters. In humans, dysregulated water balance results in neurological dysfunction and death. Even relatively subtle changes in systemic tonicity cause reversible defects in coordination and cognition (26, 27). Clinically, water balance is reflected in the serum (or plasma) sodium concentration. Water excess relative to total body sodium content gives rise to hyponatremia, the most prevalent of the electrolyte abnormalities. Hyponatremia complicates a wide range of common medical conditions including congestive heart failure and liver failure and occurs in an unpredictable fashion in response to a number of frequently prescribed medications (reviewed in Refs. 13, 32).
In mammals, systemic water balance is regulated via the renal water-conserving role of the hormone arginine vasopressin (AVP). Release of AVP from the posterior pituitary into the circulation is governed by the hypothalamic sensor(s) of systemic osmolality. On reaching the kidney, AVP interacts with arginine vasopressin receptor-2 (the product of the AVPR2 gene) on the basolateral aspect of principal cells lining the collecting duct. Ligand-dependent activation of this receptor causes sequestered aquaporin-2 water channel proteins to enter the apical plasma membrane such that transcellular water uptake from the urinary space can be achieved (reviewed in Ref. 7).
Recent data have underscored genetic influences on systemic water balance. For example, a number of single-gene (i.e., Mendelian) defects in the AVP/AVPR2 axis have been identified. These give rise to diabetes insipidus (DI, renal water wasting) and a tendency toward hypernatremia via mutations in AVP (4, 18) or AVPR2 (31) or to the nephrogenic syndrome of inappropriate antidiuresis (NSIAD) and a tendency toward hyponatremia via mutation in AVPR2 (15). Mutation of a single amino acid residue in the AVPR2 protein—Arg137—can impact serum sodium concentration bidirectionally: the Arg → His mutation causes nephrogenic DI, whereas the Arg → Cys mutation gives rise to NSIAD (15). Therefore, the AVPR2 gene is a pivotal locus for genetic impact on systemic water balance.
In contrast to these studies involving rare familial clustering of grossly aberrant water balance, little is known about the genetic contribution to water balance on a population-wide basis. A single twin study addressing genetic influences on a panel of biochemical laboratory values suggested heritability of serum sodium concentration (5), and the effect was more pronounced in females than males. Because the AVPR2 gene is X-linked, we hypothesized that a copy number variation (CNV) at this locus might account for this potential discrepancy and impact systemic water balance in humans. In support of this hypothesis, a curated public database of human CNV (Database of Genomic Variants, http://projects.tcag.ca/variation/; Ref. 17) included a common CNV spanning the AVPR2 gene at Xq28. This putative CNV, designated Variation_7770, was identified through a genomewide comparative genomic hybridization-based approach (11). Among 50 healthy white males from Northern France, there were a total of 5 copy number gains and 62 copy number losses across a region spanning ∼1.5 Mb (11). These individuals exhibited overlapping, but not necessarily identical, CNVs; in addition, some individuals had more than one discrete region of copy number change within the overall CNV region.
CNV is increasingly recognized as a source of phenotypic variation in human populations. Specific CNVs have been linked to a number of clinically relevant conditions including autism (8, 28, 30), schizophrenia (1, 8, 29, 34), susceptibility to human immunodeficiency virus (HIV) infection (16), and the glomerulonephritis associated with systemic lupus erythematosis (2). Therefore, we tested for the presence of this AVPR2 CNV in normonatremic and dysnatremic subjects from two large populations.
METHODS
Genotyping—Offspring Cohort of Framingham Heart Study.
The Offspring Cohort of the Framingham Heart Study included men and women who were either offspring of the Original Cohort of the study or spouses of these offspring; 5,135 of the 6,838 eligible individuals participated (14, 19). The Offspring Cohort has undergone repeat examinations approximately every 3–4 yr. Genomic DNA was obtained and banked by the parent study. The Original Cohort included 5,209 respondents to a random sample of two-thirds of the adult population of Framingham, Massachusetts, 30–62 yr of age (by household), in 1948 (10). Offspring Cohort genomic DNA from plates 1 through 5 of the Gen2A/unr genomic DNA plate set was genotyped for AVPR2 copy number (see below). Samples were excluded from analysis if they were duplicates of included samples (n = 46) or if phenotypic data (i.e., serum sodium concentration) were absent (n = 154). A total of 286 unique genomic DNA samples were tested; 7 were excluded from the analysis for absence of signal in both target and internal control (n = 2), presence of only a single rather than replicated successful determination (n = 1), or SD > 0.25 between technical replicates (n = 4). Demographic data reflecting the 279 successful genotypes are shown in Table 1. In this and the following sections, all genotyping studies using human DNA were approved by the Institutional Review Board of the Portland Department of Veterans Affairs (VA) Medical Center, or were deemed exempt by this body under the Code of Federal Regulations, Title 45-Public Welfare, Department of Health and Human Services; Part 46-Protection of Human Subjects; Paragraph 46.101(b)(4), i.e., Exemption 4.
Table 1.
Phenotypic data for successfully genotyped subjects in Framingham Heart Study Offspring Cohort and MrOS cohort
| Ethnicity |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Sex | Age, yr | Na+, meq/l | Creatinine, mg/dl | 1 | 2 | 3 | 4 | 5 | |
| FHS | 279 | M 50.2% | 43.3 ± 9.3 | 139.3 ± 2.7 | 1.13 ± 0.24 | * | ||||
| MrOS High | 183 | M 100% | 72.7 ± 5.8 | 145.6 ± 0.8 | 0.98 ± 0.14 | 172 | 10 | 0 | 0 | 1 |
| MrOS Mean | 182 | M 100% | 72.5 ± 5.4 | 141.5 ± 0.5 | 0.96 ± 0.14 | 175 | 5 | 0 | 0 | 2 |
| MrOS Low | 152 | M 100% | 73.1 ± 5.9 | 136.6 ± 2.2 | 0.95 ± 0.18 | 143 | 7 | 1 | 1 | 0 |
Phenotypic data for the successfully genotyped subjects from the Framingham Heart Study (FHS) Offspring Cohort and the Osteoporotic Fractures in Men Study (MrOS) cohort. All data are shown as means ± SD for n subjects. The MrOS High sodium (Na+ ≥; 145 meq/l) and Low sodium (Na+ ≤; 138 meq/l) groups consisted of subjects with serum sodium concentration in the highest and lowest deciles, respectively (i.e., more than ∼1.5 SD units above and below the population mean, respectively). Subjects in the MrOS Mean sodium group had serum sodium concentration of 141 or 142 meq/l; mean for the entire MrOS population was 141.4 meq/l. Ethnicities: 1, non-Hispanic Caucasian; 2, African American; 3, Asian; 4, Hispanic; 5, other.
Ethnicity for FHS Offspring cohort was inferred to be non-Hispanic Caucasian.
Genotyping—Osteoporotic Fractures in Men Study.
Data from the Osteoporotic Fractures in Men Study (MrOS) were used to corroborate our findings with the Framingham Offspring Cohort, above, and to extend these observations to include more subjects with aberrant water balance. MrOS was designed to assess the determinants of fracture in 5,995 healthy community-dwelling U.S. male subjects over 65 yr of age (22). Subjects were recruited from six geographically diverse centers (Birmingham, Minneapolis, Palo Alto, Pittsburgh, Portland, and San Diego); details were published previously (6). Banked serum and genomic DNA were obtained by the parent study from 5,840 subjects. Serum sodium and creatinine were measured on a single instrument with thawed previously frozen serum (Clinical Laboratory, Portland VA Medical Center), and values were obtained for 5,528 and 5,534 subjects, respectively. Subjects with serum creatinine >1.3 mg/dl were excluded from further analysis because abnormal renal function may lead to impaired water excretion (see, e.g., Ref. 35). The Low sodium concentration group included subjects with serum sodium concentration ≤138 meq/l, and the High sodium concentration group included subjects with sodium ≥145 meq/l. These groups approximately corresponded to the lowest and highest deciles (i.e., more than ∼1.5 SD units below or above the population mean, respectively) for the MrOS population, in terms of serum sodium concentration. The MrOS population mean for sodium concentration was 141.4 meq/l; for the Mean sodium group, we selected every third subject when subjects with serum sodium concentration of 141 and 142 meq/l were ordered by serum sodium concentration and then by coded alphanumeric identifier (sodium concentrations were “binned” as integers at the time of reporting by the clinical laboratory). Subjects with glucose ≥150 mg/dl were excluded, as were subjects with estimated glomerular filtration rate >2 SD below the population mean; both conditions are associated with reduced serum sodium concentration and/or aberrant water balance independent of central osmoregulation (20, 35). Genomic DNA from the first 524 MrOS subjects, when ordered by coded subject identifier, was genotyped for AVPR2 copy number. All MrOS subjects were male. As was done for the analysis of the Framingham Offspring Cohort, subjects with SD between replicates of >0.25 were excluded (n = 2), as were subjects from whom insufficient DNA was available for replication (n = 5). Demographic data for the 517 successfully genotyped subjects are shown in Table 1.
Probe selection and genotyping.
For CNV determination, unamplified banked genomic DNA was subjected to a real-time PCR-based quantitative assay using a commercially prepared probe set designed to our specifications and directed against exon 3 of the human AVPR2 gene (Applied Biosystems). Crossing threshold (Ct) was determined for the VIC-labeled AVPR2 probe set and for the FAM-labeled RPPH1 internal control probe set in a single reaction in multiplexed fashion with NFQ quencher and a StepOnePlus instrument (Applied Biosystems). The exon 3 forward primer sequence was GAAGCTCAGCTGCCTTCCT, the reverse primer sequence was CCAGGGCCACACAGTGT, and the reporter (probe) sequence was CTGGCCAATTCTC. These encompass a 109-bp region within exon 3, beginning ∼35 bp downstream of the AVPR2 coding sequence in the reference cDNA NM_000054. There are no known “rs” SNPs in this region; there are four (rs5201 through rs5204) in the coding region of exon 3, and all are upstream of the interrogated region. This AVPR2 cDNA sequence was aligned with the corresponding genomic sequence (nucleotides 152823564-152825834 of NC_000023.9) in Spidey (www.ncbi.nlm.nih.gov/spidey/; Ref. 33) to assess exon/intron boundaries. All reactions were performed in duplicate except where indicated (i.e., sample availability was limiting). Although the manufacturer recommends performing CNV assays in quadruplicate (Applied Biosystems part no. 4380021 Rev A), the very small amount of genomic DNA made available to us, coupled with the high reproducibility among replicates, permitted duplicate determinations to suffice. Within each 96-well assay plate (n = 48 samples, in duplicate), copy number was estimated as ΔCt with the equation 2CtVIC−CtFAM. Copy number was normalized to the whole plate average copy number (for the all-male MrOS samples) or to the average of the male samples (for the Framingham Offspring Cohort samples) to maintain consistency between assays. Whole plate average ΔCt for AVPR2 copy number in male subjects (for MrOS) was generally ∼0.4 (range: 0.3–0.6; n = 11 experiments). For confirmation, an additional probe set was generated against exon 2 of the AVPR2 gene and produced similar results in pilot experiments (data not shown). Given that AVPR2 spans only ∼2 kb and that CNV were reportedly much larger—spanning the entire gene—it was not expected that probe location would affect our ability to detect CNV.
To amplify the genomic context of the exon 3 CNV probe binding site, PCR primers AVPR2-1706-5′ (CGAAGCTTGCTCTGCTGTG) and AVPR2-2005-3′ (CCTCATACAGCTGGGGATGT) were used; the 300-bp amplicon inclusive of the 109-bp exon 3 primer/reporter target region was then sequenced with primer AVPR2-1748-5′ (CTGGGTCCCCAAGATGAGT).
Modeling impact of an X-linked polymorphism on heritability.
We modeled in preliminary fashion the effect of a hypothetical polymorphism in the X-linked AVPR2 gene, considering two alleles with dissimilar effects on serum sodium concentration. We hypothesized the presence of a minor allele (i.e., copy number >1) with an arbitrarily selected frequency of 0.2 that conferred a decreased serum sodium concentration of 130 meq/l and a corresponding major allele (i.e., copy number = 1) with frequency 0.8 that conferred a (normal) serum sodium concentration of 140 meq/l. An additive model was presumed, in which each allele contributed equally to the serum sodium concentration in females and the single allele dictated serum sodium concentration in males. We then generated 1,000 hypothetical male sib pairs and 1,000 hypothetical female sib pairs, based on expected Hardy-Weinberg distribution. Among male sib pairs sib-sib regression of serum sodium concentration showed a slope of 0.5 with an r2 of 0.25, whereas for female sib pairs the slope was 0.75 with an r2 of 0.56 (Prism 4.0c, GraphPad). Although we did not model a universe of possible minor allele frequencies and did not consider a range of genetic models, we inferred that a polymorphism in an X-linked gene could result in an increased estimate of heritability (i.e., sib-sib correlation) for sodium concentration in female sib pairs relative to their male counterparts. Skewed X-inactivation, in which preferential expression of either the normal or the variant allele modulates disease severity in a heterozygous female (3, 12, 21), was not modeled.
RESULTS
In a study of mono- and dizygotic twins, the estimated heritability of serum sodium concentration was greater among females (5). Our preliminary modeling indicated that an X-linked polymorphism associated with hyponatremia could give rise to an increased correlation of serum sodium concentration among female sib pairs, relative to male sib pairs (see methods). In part because one had recently been reported (11), we hypothesized that a polymorphism in the form of a CNV in the X-linked water-regulatory AVPR2 gene would impact systemic water balance on a population-wide basis. The AVPR2 exon 3 probe set was designed to ascertain copy number. We screened deidentified genomic DNA obtained from a panel of male (n = 6) and female (n = 6) subjects to establish the discriminatory ability of this probe. In all instances, estimated copy number was approximately twice as high in female subjects as in male subjects, consistent with AVPR2 being an X-linked gene (data not shown). A second probe set recognizing exon 2 of this gene gave equivalent results (data not shown).
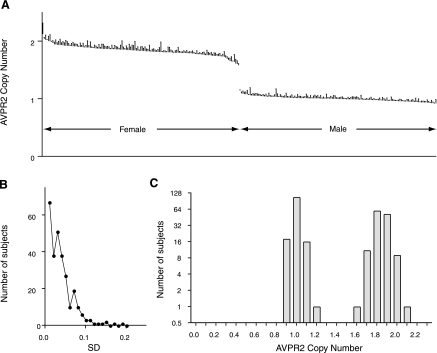
We assessed AVPR2 gene copy number in banked genomic DNA from subjects enrolled in the Offspring Cohort of the Framingham Heart Study. This population was chosen in part because, in terms of ethnicity, it was consistent with the non-Hispanic Caucasian population in which the putative AVPR2 CNV was identified. In addition, phenotypic data reflective of systemic water balance—including serum sodium concentration—were available from this population; these demographic data are shown in Table 1. We tested 286 unique unamplified genomic DNA samples for AVPR2 copy number (see methods). All copy number assessments were performed in duplicate (i.e., 2 technical replicates). For each sample, AVPR2 copy number was normalized to an internal copy number control for the RPPH1 gene (encoding the RNA component of the RNase P complex); there is no reported CNV for this gene, and deviation from expected copy number for this autosomal gene is expected to occur in <0.1% of amplifications (Technical Support, Applied Biosystems). Of the 286 genotyped samples, 7 were excluded from final analysis (see methods). AVPR2 CNV is shown in Fig. 1A. Data are ordered by descending copy number; for clarity, only the error bars (indicating SD of the 2 determinations) are shown. Calculated mean copy number was 1.00 ± 0.09 for males (n = 137) and 1.84 ± 0.09 for females (n = 142). Importantly, the mean copy number of <2 in female subjects was not a consequence of reduced copy number in a subset of female subjects (Fig. 1A); rather, it was a function of nonlinearity of the assay. Duplicates were very tight, with data for nearly all subjects exhibiting SD <0.1 (Fig. 1B). Of note, as these were duplicates, SD of 0.1 equates to an intersample difference between duplicates of 0.14 copy number units because SD equals the intersample difference times 2−0.5. The lowest estimated copy number among female subjects exceeded the highest estimated copy number in male subjects, and a threshold of 1.5 discriminated between male and female subjects with 100% accuracy (Fig. 1A). When data were viewed as a histogram (Fig. 1C), there was a clear bimodal distribution; no subjects had three copies of the gene, no males had two copies, and no females had only a single copy. Because copy number must be an integer, it was inferred that all male subjects in this genotyped cohort had one copy and all female subjects had the expected two copies of the AVPR2 gene; hence, no evidence of CNV was detected. Of note, few subjects in this relatively healthy population were dysnatremic. Among male subjects (n = 137), 14 (10.2%) were hyponatremic, 15 (10.9%) were hypernatremic, and the balance (n = 108) were normonatremic. Among female subjects (n = 142), 12 (8.5%) were hyponatremic, 16 (11.3%) were hypernatremic, and the balance (n = 114) were normonatremic.
Fig. 1.
Arginine vasopressin receptor 2 (AVPR2) copy number in successfully genotyped subjects from the Offspring Cohort of the Framingham Heart Study. A: AVPR2 calculated copy number (see methods), ordered by copy number, in 279 successfully genotyped subjects. Shown is SD of 2 technical replicates for AVPR2 copy number, normalized to RPPH1 copy number in a multiplexed assay as an internal control. For clarity, only the error bars are shown. B: distribution of SD (rounded to the nearest 0.01) for technical replicates in determining AVPR2 copy number; for nearly all samples, SD < 0.1, which equates to an interassay difference in estimated copy number of <0.14 for a given sample of genomic DNA (see methods). C: distribution of estimated AVPR2 copy number, where data from A are “binned” in increments of 0.1 copy number “units.” (For example, “1.0” is 0.95 < copy number < 1.05). Note that the y-axis is logarithmic (base 2) to more clearly demonstrate the individual subjects at the extremes of the distribution. No subjects exhibited copy number deviating from the expected single copy for male subjects or 2 copies for female subjects.
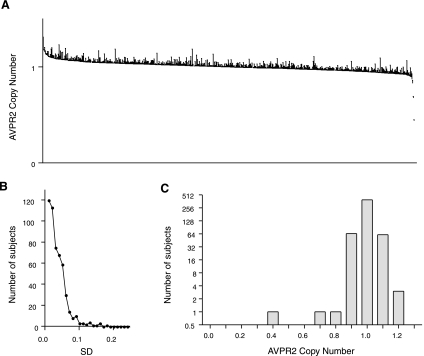
We sought a second population to confirm these findings. In addition, reasoning that AVPR2 CNVs might be more prevalent in human subjects exhibiting aberrant water balance, we wished to investigate a larger sample of subjects with hypo- and hypernatremia. The MrOS was designed to assess the determinants of fracture in 5,995 healthy community-dwelling U.S. male subjects over 65 yr of age (22). Banked serum was obtained from the parent study and assayed for sodium, glucose, and creatinine concentration, among other analytes. Subjects with glucose ≥150 mg/dl were excluded, as were subjects with estimated glomerular filtration rate >2 SD below the population mean; both conditions are associated with reduced serum sodium concentration and/or aberrant water balance independent of central osmoregulation (20, 35). Banked genomic DNA was requested from the parent study for three subgroups of subjects: 1) Low sodium—subjects with serum sodium concentration in the lowest decile (i.e., hyponatremic); 2) High sodium—subjects with serum sodium concentration in the highest decile (i.e., hypernatremic); and 3) Mean sodium—subjects with serum sodium concentration approximating the population mean (i.e., 141 or 142 meq/l). With this approach, roughly a third of subjects were relatively hyponatremic, a third were hypernatremic, and a third were eunatremic. DNA from the first 524 subjects, when ordered by coded subject identifier, was genotyped to assess AVPR2 copy number; 7 samples were excluded (see methods). Demographic data for the 517 successfully genotyped included subjects are shown in Table 1; all MrOS subjects were male. With two exceptions (see below), subjects exhibited the expected AVPR2 copy number of unity (Fig. 2A). Technical replicates for the CNV assay were again very tight: nearly all (i.e., 97%) showed SD <0.1 (Fig. 2B), and the highest SD among all duplicated samples was 0.3 (not shown; 1 of 1 excluded for SD >0.25). Estimated AVPR2 gene copy number is shown in Fig. 2C; a tight distribution around unity is seen, with the exception of a single sample at ∼0.4 (0.44 ± 0.01) and a sample at ∼0.7 (0.68 ± 0.01). Readings from these two subjects were confirmed with additional replicates using the same primer/reporter set. In aggregate, these data do not support the presence of CNV in the AVPR2 gene.
Fig. 2.
AVPR2 copy number in successfully genotyped subjects from the Osteoporotic Fractures in Men Study (MrOS). Included are subjects with serum sodium concentration approximating the mean for the MrOS population (n = 182) and subjects with serum sodium concentration in the highest and lowest deciles of the population distribution (n = 183 and 152, respectively; see methods). A: AVPR2 calculated copy number (see methods), ordered by copy number, in 517 successfully genotyped subjects. Shown is SD of 2 technical replicates for AVPR2 copy number, normalized to RPPH1 copy number in a multiplexed assay as an internal control. For clarity, only the error bars are shown. B: distribution of SD (rounded to the nearest 0.01) for technical replicates in determining AVPR2 copy number; for nearly all samples, SD < 0.1. C: distribution of estimated AVPR2 copy number, where data from A were “binned” in increments of 0.1 copy number “units.” No subjects exhibited copy number exceeding the expected single copy for male subjects (range: 0.83–1.18), although 2 subjects exhibited lower-than-expected copy number (A and C, 0.44 and 0.68; see text). The subject with estimated copy number of 0.44 was hemizygous for a single nucleotide polymorphism in the hybridization site for the AVPR2 reporter oligonucleotide (see text).
Whereas African Americans comprised only 7% of the genotyped MrOS population, the two subjects exhibiting copy number substantially less than unity were both self-reported African American; this suggested a polymorphism in either a primer or a probe binding site within the third exon of the AVPR2 gene. Follow-up studies in these two subjects using a primer/reporter set directed against exon 2 (rather than exon 3) showed copy number of unity (data not shown). After resequencing of this region in both subjects, we noted that the lower of the two subjects, in terms of normalized copy number (Fig. 2A), was hemizygous for a polymorphism spanned by the reporter of the exon 3-directed assay (CTGGCCAATTCTC → TTGGCCAATTCTC), potentially giving rise to weaker hybridization and accounting for the lower-than-anticipated signal amplitude; however, in the other subject, no polymorphism was detected within the 109-bp interval over which reporter and primers were expected to hybridize. We sequenced this same region in 19 additional African American subjects in the MrOS cohort, all of whom exhibited copy number of unity in our initial analysis. None showed this polymorphism; all were hemizygous for the wild-type allele. We infer in preliminary fashion that the minor allele frequency for this single nucleotide polymorphism (SNP) in African American subjects is ≤0.05. There are no reported SNPs in this genomic region.
DISCUSSION
We hypothesized that a CNV in the AVPR2 gene may be associated with aberrant water balance for the following reasons: 1) the AVPR2 gene product is integral to systemic osmoregulation; 2) heritability of serum sodium concentration—an index of systemic water balance—is likely greater among females than males; 3) AVPR2 is X-linked; and 4) a common CNV spanning the AVPR2 gene was recently reported in non-Hispanic Caucasians. The present data derived from two distinct populations of primarily non-Hispanic Caucasian subjects fail to establish the presence of a CNV at this locus. Although negative, we believe these findings are important because of the plausibility of the initial hypothesis and because the presence of this putative CNV was previously inferred based on comparative genomic hybridization. In addition, of the genes known to impact systemic water balance, CNV has been reported only for AVPR2.
These data strongly support the absence of a CNV. In the mixed-sex Framingham Offspring Cohort, validity of the CNV assay was demonstrated by complete separation of male and female subjects (i.e., no female subject exhibited estimated CNV less than any male subject). In addition, technical replicates using a single subject's genomic DNA were highly reproducible. The copy number assay was not perfectly linear. Mean calculated copy number among females was not twice that among males; however, when calculated copy number was interpreted in terms of biological plausibility (i.e., copy number must be an integer), all female subjects had two copies of the AVPR2 gene and all males had the expected single copy for an X-linked gene. We reasoned that a CNV in the AVPR2 gene may be more prevalent among subjects with abnormal water balance. Because the Framingham subjects had not been selected by serum sodium concentration, only a limited number had serum sodium concentrations in the highest and lowest deciles (e.g., ∼10%, respectively). We had previously assayed serum sodium concentration in nearly all of the 5,995 MrOS enrollees and had grouped subjects into low sodium, high sodium, or mean sodium concentration. We reasoned that testing a subset of these subjects for AVPR2 copy number would increase sensitivity at the extremes of the serum sodium concentration distribution. Despite this enrichment for subjects with aberrant water balance, no subjects with AVPR2 CNV were identified.
It cannot be concluded that a CNV in this region does not exist; however, our data in nearly 800 non-Hispanic Caucasian subjects do not demonstrate a CNV with the frequency predicted by the previously published comparative genomic hybridization data set. The tested subjects largely mirrored the racial makeup of this earlier cohort. An AVPR2 CNV unrelated to the putative one reported in the Database of Genomic Variants data set (http://projects.tcag.ca/variation/) may yet exist in other populations.
Although no instances of increased AVPR2 copy number were detected, two instances of measured copy number below the expected level were identified. However, because these were male subjects and AVPR2 is an X-linked gene, a copy number <1 is not biologically plausible. In addition, assessment with a probe set directed against an adjacent exon was consistent with a copy number of unity. Possible explanations include: 1) one or more polymorphisms in the region of primer or probe hybridization resulting in diminished AVPR2 signal or 2) CNV in the internal control RPPH1 gene. We excluded the latter possibility because copy number estimations of other genes—when normalized to RPPH1 copy number—were not affected in these two individuals (data not shown). Because both affected subjects were African American, and because African Americans constituted only a small percentage of the investigated subjects, we hypothesized that a polymorphism affected amplification or binding of the fluorescent copy number probe. Accordingly, one individual was found to be hemizygous for a novel polymorphism within the AVPR2 reporter hybridization site.
Reexamination of the raw data from the original array comparative genomic hybridization study (11) indicated the presence of bona fide CNVs flanking—but not incorporating—the AVPR2 gene. These regions are ∼0.24 Mb downstream (e.g., Variation_37014) and 0.64 Mb upstream (e.g., Variation_3259) of the AVPR2 gene and have been confirmed by other investigators (9, 23–25). Because comparative genomic hybridization data from this region were noisy, the computational algorithm “smoothed” the CNVs together into one larger, artifactual signal. This is a recognized analytical problem in reported CNVs, and, as a result of the present study, this particular CNV entry has been removed from the Database of Genomic Variants at the authors' request. We urge researchers to exercise caution in interpreting such information and recommend checking the original data carefully where possible before embarking on large-scale follow-up studies. The original array comparative genomic hybridization data from the de Smith et al. study (11) are publicly available (//www.ncbi.nlm.nih.gov/sites/entrez?db=gds&cmd=search&term=GSE8691), and new ultra-high-resolution array comparative genomic hybridization data on 40 HapMap individuals have also been placed in the public domain by the Sanger Centre (http://www.sanger.ac.uk/humgen/cnv/42mio/).
GRANTS
This work is supported by grants from the National Institutes of Health (NIH), the Department of Veterans Affairs, and the American Heart Association. The Osteoporotic Fractures in Men Study (MrOS) is supported by NIH funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Institute on Aging, the National Center for Research Resources, and NIH Roadmap for Medical Research under the following grant numbers: U01-AR-45580, U01-AR-45614, U01-AR-45632, U01-AR-45647, U01-AR-45654, U01-AR-45583, U01-AG-18197, U01-AG-027810, UL1-RR-024140, and R01-AR-051124.
A portion of this work was supported by the National Heart, Lung, and Blood Institute's (NHLBI) Framingham Heart Study (Contract No. N01-HC-25195). The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University. This article was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI.
DISCLOSURES
The authors are not aware of financial conflict(s) with the subject matter or materials discussed in this manuscript with any of the authors, or any of the authors' academic institutions or employers.
ACKNOWLEDGMENTS
The authors thank Joanne Murabito (Framingham Heart Study and Boston University School of Medicine) for guidance.
REFERENCES
- 1.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455: 237–241, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851–855, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Arthus MF, Lonergan M, Crumley MJ, Naumova AK, Morin D, De Marco LA, Kaplan BS, Robertson GL, Sasaki S, Morgan K, Bichet DG, Fujiwara TM. Report of 33 novel AVPR2 mutations and analysis of 117 families with X-linked nephrogenic diabetes insipidus. J Am Soc Nephrol 11: 1044–1054, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Bahnsen U, Oosting P, Swaab DF, Nahke P, Richter D, Schmale H. A missense mutation in the vasopressin-neurophysin precursor gene cosegregates with human autosomal dominant neurohypophyseal diabetes insipidus. EMBO J 11: 19–23, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bathum L, Fagnani C, Christiansen L, Christensen K. Heritability of biochemical kidney markers and relation to survival in the elderly—results from a Danish population-based twin study. Clin Chim Acta 349: 143–150, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Blank JB, Cawthon PM, Carrion-Petersen ML, Harper L, Johnson JP, Mitson E, Delay RR. Overview of recruitment for the Osteoporotic Fractures in Men Study (MrOS). Contemp Clin Trials 26: 557–568, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem 68: 425–458, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature 455: 919–923, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 40: 1199–1203, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawber TR, Moore FE, Mann GV. Coronary heart disease in the Framingham study. Am J Public Health 47: 4–24, 1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Smith AJ, Tsalenko A, Sampas N, Scheffer A, Yamada NA, Tsang P, Ben-Dor A, Yakhini Z, Ellis RJ, Bruhn L, Laderman S, Froguel P, Blakemore AI. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet 16: 2783–2794, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Decaux G, Vandergheynst F, Bouko Y, Parma J, Vassart G, Vilain C. Nephrogenic syndrome of inappropriate antidiuresis in adults: high phenotypic variability in men and women from a large pedigree. J Am Soc Nephrol 18: 606–612, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Ellison DH, Berl T. Clinical practice. The syndrome of inappropriate antidiuresis. N Engl J Med 356: 2064–2072, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med 4: 518–525, 1975 [DOI] [PubMed] [Google Scholar]
- 15.Feldman BJ, Rosenthal SM, Vargas GA, Fenwick RG, Huang EA, Matsuda-Abedini M, Lustig RH, Mathias RS, Portale AA, Miller WL, Gitelman SE. Nephrogenic syndrome of inappropriate antidiuresis. N Engl J Med 352: 1884–1890, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O'Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307: 1434–1440, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Mori Y, Oiso Y, Saito H. A single base substitution in the coding region for neurophysin II associated with familial central diabetes insipidus. J Clin Invest 87: 725–728, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol 110: 281–290, 1979 [DOI] [PubMed] [Google Scholar]
- 20.Katz MA. Hyperglycemia-induced hyponatremia—calculation of expected serum sodium depression. N Engl J Med 289: 843–844, 1973 [DOI] [PubMed] [Google Scholar]
- 21.Nomura Y, Onigata K, Nagashima T, Yutani S, Mochizuki H, Nagashima K, Morikawa A. Detection of skewed X-inactivation in two female carriers of vasopressin type 2 receptor gene mutation. J Clin Endocrinol Metab 82: 3434–3437, 1997 [DOI] [PubMed] [Google Scholar]
- 22.Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, Lewis C, Cawthon PM, Marcus R, Marshall LM, McGowan J, Phipps K, Sherman S, Stefanick ML, Stone K. Design and baseline characteristics of the Osteoporotic Fractures in Men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials 26: 569–585, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, Park HS, Kim JI, Seo JS, Yakhini Z, Laderman S, Bruhn L, Lee C. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet 82: 685–695, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet 16: R168–R173, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature 444: 444–454, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renneboog B, Musch W, Vandemergel X, Manto MU, Decaux G. Mild chronic hyponatremia is associated with falls, unsteadiness, and attention deficits. Am J Med 119: 71e71–78, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Schrier RW, Gross P, Gheorghiade M, Berl T, Verbalis JG, Czerwiec FS, Orlandi C. Tolvaptan, a selective oral vasopressin V2-receptor antagonist, for hyponatremia. N Engl J Med 355: 2099–2112, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science 316: 445–449, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature 455: 232–236, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Autism Genome Project Consortium Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39: 319–328, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Ouweland AM, Dreesen JC, Verdijk M, Knoers NV, Monnens LA, Rocchi M, van Oost BA. Mutations in the vasopressin type 2 receptor gene (AVPR2) associated with nephrogenic diabetes insipidus. Nat Genet 2: 99–102, 1992 [DOI] [PubMed] [Google Scholar]
- 32.Verbalis JG, Berl T. Disorders of water balance. In: Brenner and Rector's The Kidney, edited by Brenner BM. Philadelphia, PA: Saunders, 2007 [Google Scholar]
- 33.Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res 11: 1952–1957, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 40: 880–885, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Yee J, Parasuraman R, Narins RG. Selective review of key perioperative renal-electrolyte disturbances in chronic renal failure patients. Chest 115: 149S–157S, 1999 [DOI] [PubMed] [Google Scholar]