Abstract
Hepatocyte growth factor (HGF) is important for cell proliferation, differentiation, and related activities. HGF acts through its receptor c-Met, which activates downstream signaling pathways. HGF binds to c-Met at the plasma membrane, where it is generally believed that c-Met signaling is initiated. Here we report that c-Met rapidly translocates to the nucleus upon stimulation with HGF. Ca2+ signals that are induced by HGF result from phosphatidylinositol 4,5-bisphosphate hydrolysis and inositol 1,4,5-trisphosphate formation within the nucleus rather than within the cytoplasm. Translocation of c-Met to the nucleus depends upon the adaptor protein Gab1 and importin β1, and formation of Ca2+ signals in turn depends upon this translocation. HGF may exert its particular effects on cells because it bypasses signaling pathways in the cytoplasm to directly activate signaling pathways in the nucleus.
Hepatocyte growth factor (HGF)2 is secreted by stromal cells and binds to its receptor c-Met, which is a prototypic receptor tyrosine kinase (RTK) (1). Activation of c-Met is responsible for cell proliferation under normal conditions such as tissue regeneration (2), as well as under abnormal conditions such as neoplasia (3). For example, in the liver, HGF is secreted by stellate cells and then binds to c-Met on hepatocytes (1) to mediate processes such as liver regeneration following liver resection (2) and development of hepatocellular carcinoma (4). The mechanisms of action of c-Met are not entirely understood, but it is generally believed that this and other RTKs act at the plasma membrane (5-7). However, several RTKs have been found in the nucleus, including receptors for insulin, epidermal growth factor, and fibroblast growth factor (8-10). The function of these receptors in the nucleus is controversial (11, 12).
C-met and other RTKs act in part through Ca2+ signaling, which is initiated by the phospholipase C-γ (PLCγ)/inositol 1,4,5-trisphosphate (InsP3)/Ca2+ signaling cascade (13). A number of peptide hormones also increase Ca2+ via InsP3 but typically do so via G protein-coupled receptors that activate PLCβ (13). Ca2+ signals induced by growth factors typically have separate effects from hormone-induced Ca2+ signals, although both are mediated by PLC and InsP3 (13). In hepatocytes, for example, vasopressin activates the V1a receptor to modulate secretion (14), glucose release (15), and apical contractility (16), whereas HGF activates c-Met to regulate cell proliferation (17). The versatility of Ca2+ as a second messenger in part reflects that Ca2+ signals have distinct effects in different parts of the cell (13). Moreover, cell proliferation depends upon Ca2+ signals in the nucleus rather than in the cytoplasm (18). Therefore, we examined the subcellular distribution of c-Met and the mechanism by which HGF induces c-Met to form Ca2+ signals.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
SkHep1 cells were cultured at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum, 1 mm sodium pyruvate, 50 units/ml penicillin, and 50 g/ml streptomycin (Invitrogen).
Immunoblots
SkHep1 cell immunoblots were performed as described previously (19, 20). The NE-PER kit (Pierce) was used to prepare nuclear and cytosolic cell fractions (20). Briefly, cell membranes were disrupted to release cytoplasmic contents. Intact nuclei were recovered from the cytoplasmic extract by centrifugation, and then the nuclei were washed with PBS and lysed to yield the nuclear extract. Protease and phosphatase inhibitors (Sigma) were added to all buffers. Blots were visualized by enhanced chemiluminescence and quantitatively analyzed using a GS-700 imaging densitometer (Bio-Rad).
Detection of PIP2
The phosphatidylinositol 4,5-bisphosphate mass strip kit was used for isolation and phosphatidylinositol 4,5-bisphosphate (PIP2) detection (Echelon). SkHep1 cells were starved overnight in serum-free Dulbecco's modified Eagle's medium. Cells were used in aliquots of 5 × 106 cells/sample and incubated without or with HGF (100 ng/ml) for 4 min or arginine vasopressin (AVP) (100 nm) for 30 s, and then the medium was aspirated, and cellular material was precipitated by the immediate addition of 3 ml of ice-cold 0.5 m trichloroacetic acid. The NE-PER kit (Pierce) was used to prepare the nuclear and cell fraction, as described above (20). Briefly, cell membranes were disrupted to release cytoplasmic contents. Intact nuclei were recovered from the cytoplasmic extract by centrifugation, and then the nuclei were washed with PBS and precipitated with 3 ml of ice-cold 0.5 M trichloroacetic acid. Isolation of lipids was performed according to the manufacturer's instructions and as described previously (21). The organic phase was collected into a clean tube and dried in a SpeedVac centrifuge. The pellet at this stage was faintly visible. The lipids were then resuspended by sonication in a cold water bath in 10 μl of CHCl3:MeOH:H2O (1:2:0.8) and spotted onto nitrocellulose membrane strips prespotted with phosphatidylinositol 4,5-bisphosphate standards, PIP controls, and space for spotting unknown samples for probing with anti-PIP2 monoclonal antibody (Echelon) to specifically detect PIP2. Blots were visualized by enhanced chemiluminescence and quantitatively analyzed using a GS-700 imaging densitometer (Bio-Rad).
Assay for Cell Surface Protein by Biotinylation
Cells were washed three times with 10 ml of PBS and subsequently incubated for 30 min at room temperature in 5 ml of PBS containing 1 mm sulfo-NHS-biotin (Pierce). Reactions were quenched for 10 min with 10 ml of PBS containing 100 mm glycine. Cells were stimulated with 100 ng/ml HGF (R&D Systems) for 4 min and lysed on ice for 10 min in 500 μl of NE-PER to prepare nuclear and cytosolic cell fractions (20). Streptavidin-agarose beads (Pierce) (50 μl of a 50:50 slurry) or anti-c-Met antibody were added to 500 μl of adjusted cell lysate, containing 200–500 μg of protein, and the suspension was rocked overnight at 4 °C. The samples were incubated with anti-c-Met antibody and 50 μl of agarose G for 2 h at room temperature. The beads were washed five times with 1 ml of PBS and analyzed by immunoblotting. Mouse monoclonal anti-c-Met antibody (Upstate Biotechnology) or streptavidin conjugated to horseradish peroxidase (Pierce) was used.
Immunofluorescence
Confocal immunofluorescence was performed as described (19, 20). Cells were triple-labeled with monoclonal antibodies against c-Met (Upstate Biotechnology) and polyclonal antibodies against Lamin B1 (AbCam) plus the nuclear stain TO-PRO-3 (Invitrogen) and then incubated with secondary antibodies conjugated to Alexa Fluor 488 and 555 (Invitrogen). Images were obtained using a Zeiss LSM 510 confocal microscope using a ×63, 1.4 NA objective lens with excitation at 488 nm and observation at 505–550 nm to detect Alexa Fluor 488, excitation at 543 nm and observation at 560–610 nm to detect Alexa Fluor 555, and excitation at 633 nm and observation using LP650 nm to detect TO-PRO-3 (Invitrogen).
InsP3 Buffer Constructs
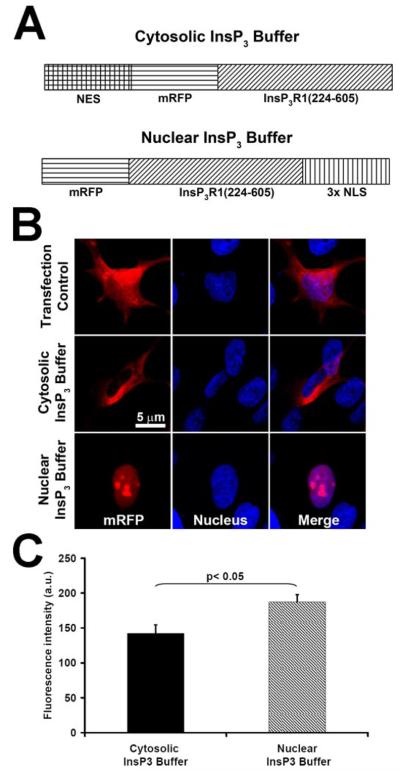
The InsP3 binding domain (residues 224–605) of the human type I InsP3 receptor was tagged with monomeric red fluorescent protein (mRFP), and then the nuclear localization signal was subcloned to generate the nuclear InsP3 buffer expression vector. The nuclear exclusion signal sequence derived from MKK1 was subcloned in the InsP3 binding domain tagged with the mRFP construct to generate the cytoplasmic InsP3 buffer expression vector, as described previously (22, 23).
Transfection of siRNA
SkHep1 cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen) with 50 nm siRNA for Gab1 (siRNA ID 1.927) (Ambion). The cells were incubated at 37 °C in an atmosphere of 5% CO2, 95% O2 for 48 h prior to use.
Detection of Ca2+ Signals
For Ca2+ imaging, cells were incubated with fluo-4/AM (6 μm) (Invitrogen) for 30 min at 37 °C, and then coverslips containing the cells were transferred to a custom-built perfusion chamber on the stage of a Zeiss LSM 510 confocal microscope, and fluo-4 fluorescence was monitored using a ×63, 1.4 NA objective lens at 1–5 frames/s. Changes in fluorescence were normalized by the initial fluorescence (F0) and were expressed as (F/F0) × 100% (19, 20). In selected experiments, cells were transfected 48 h prior to study with targeted mRFP-InsP3 buffer constructs using FuGENE 6 (Roche Applied Science).
Statistical Analysis
Significance of changes in treatment groups relative to controls were determined by Student's t test. Data are represented as mean ± S.E.
RESULTS
c-Met Translocates to the Nucleus
Studies were performed in the SkHep1 liver cell line because of the well known effects of HGF in liver and because this liver cell line contains Ca2+ signaling machinery in both the cytoplasm and the nucleus (20). As expected, c-Met was excluded from the nucleus of SkHep1 cells prior to stimulation with HGF (Fig. 1A). The phosphorylated, and hence active, form of c-Met accumulated in both the non-nuclear and the nuclear cell fraction of stimulated cells, reaching a peak in both compartments within 4 min (Fig. 1, A–C). The purity of nuclear preparations was confirmed using several nuclear and non-nuclear markers (Fig. 1D), although endoplasmic reticulum markers were not used because endoplasmic reticulum proteins can be found in the nucleus (20). Confocal immunofluorescence microscopy demonstrated that this receptor was at the plasma membrane or within the cytoplasm prior to stimulation and revealed that c-Met appeared both at the nuclear envelope and within the nuclear interior within 4 min of exposure to HGF (Fig. 2, A and B). In addition, c-Met that was biotinylated at the cell surface prior to stimulation with HGF could be recovered in the nucleus after cells were stimulated (Fig. 3). Together, these findings demonstrate that both total and activated c-Met rapidly translocates from the plasma membrane to the nucleus upon stimulation of cells with HGF.
FIGURE 1. c-Met rapidly translocates to the nucleus.
A, immunoblots of total and phosphorylated (activated) c-Met in nuclear and non-nuclear fractions of SkHep1 cells stimulated with HGF (100 ng/ml). Both total and phosphorylated receptor are absent from the nucleus at baseline but are detected in the nucleus within 1 min and reach peak intensity within 4 min of stimulation. Blots are representative of what was observed in n = 5 separate experiments. Na+/K+-ATPase and Lamin B were used as controls for the non-nuclear and nuclear fraction, respectively. C-met measurements were normalized by Na+/K+-ATPase and Lamin B levels in non-nuclear and nuclear fractions, respectively. B and C, densitometric measurement of cellular fractions of total and phosphorylated c-Met, respectively. Nuclear c-Met is maximal within 2–4 min of stimulation (n=5). D, the cytoplasmic protein α-tubulin and the nuclear protein histone 3 confirm the separation of non-nuclear (Non-Nuc.) from nuclear markers in subcellular fractions.
FIGURE 2. Confocal immunofluorescence images of c-Met before and 4 min after stimulation with HGF.

A, confocal immunofluorescence images of c-Met before and 4 min after stimulation with HGF, respectively. The images confirm that the receptor moves to the nucleus. In each image, c-Met labeling is in green, and the nucleus is stained with TO-PRO-3 (blue). Serial optical sections were collected for three-dimensional reconstruction; x-z sections are shown at the top, and y-z sections are shown at the right of each image. Note that c-Met redistributes to the region of the nuclear envelope as well as in a reticular pattern within the nuclear interior (arrows). B, additional confocal immunofluorescence images of c-Met before and after stimulation with HGF. Lamin B1 staining of the nuclear envelope (red) has been added to these triple-labeled images, which confirm that the receptor moves to the nucleus.
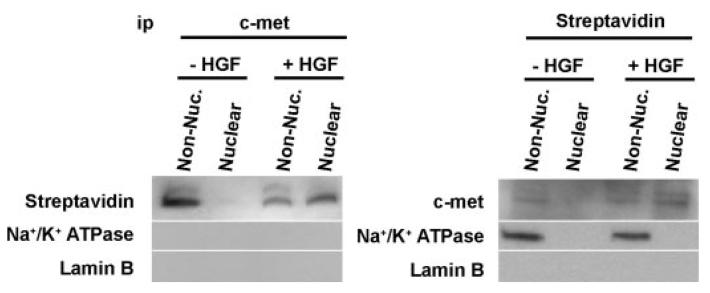
FIGURE 3. c-Met from the plasma membrane reaches the nucleus.
Biotinylated c-Met is recovered from the nucleus. c-Met at the plasma membrane was biotinylated prior to stimulation. After treatment with HGF, the non-nuclear (Non-Nuc.) and nuclear fractions were used for immunoprecipitation (ip) with either c-Met antibody or streptavidin. Both immunoprecipitations confirm that c-Met within the nucleus is labeled with streptavidin. The plasma membrane protein Na+/K+-ATPase co-precipitated with streptavidin but not c-Met, whereas the nuclear protein Lamin B did not precipitate with either streptavidin or c-Met.
c-Met Forms InsP3 in the Nucleus
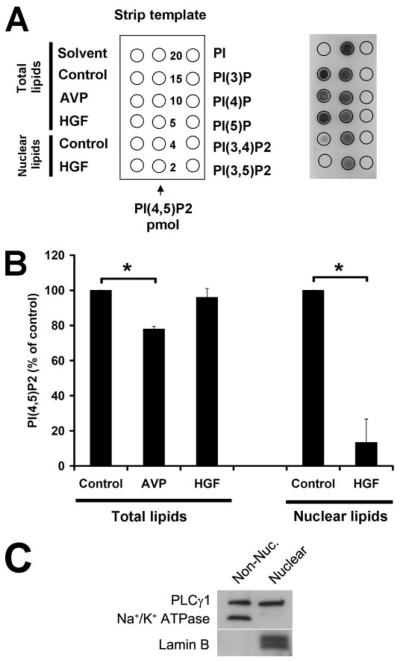
HGF stimulates cell proliferation, which has been linked to nuclear rather than cytoplasmic Ca2+ signals (18), so we examined whether c-Met forms InsP3 in the nucleus or cytoplasm. The nucleus is known to contain PLC and PIP2 (24), which are necessary for the formation of InsP3. The nucleus also contains InsP3-gated Ca2+ stores, within both the nuclear envelope (25-27) and the nucleoplasmic reticulum (20). To define whether c-Met forms InsP3 in the cytoplasm or nucleus, we targeted the ligand binding domain (residues 224–605) of the InsP3 receptor (InsP3R) type 1 (22) to one or the other of these two compartments using a nuclear exclusion signal (Fig. 4A, NES) or nuclear localization signal (NLS) sequence, respectively, plus mRFP to verify localization (Fig. 4, A and B). RFP fluorescence was slightly greater in the construct expressed in the nucleus (Fig. 4C), perhaps because the construct was distributed in a slightly smaller compartment. This segment of InsP3R1 specifically binds to InsP3 with sufficient affinity to compete for binding to the native receptor (22). These targeted InsP3 buffer constructs were expressed in the SkHep1 liver cell line, which express not only c-Met but also the G protein-coupled V1a vasopressin receptor (28). Stimulation of the V1a receptor was used as a cytoplasmic InsP3 control since G protein-coupled receptors activate PLC and form InsP3 at the plasma membrane (29). Vasopressin (100 nm) increased Ca2+ in both the cytoplasm and the nucleus, in control (non-transfected) cells as well as in cells transfected with the nuclear InsP3 buffer (Fig. 5, A, B, and E). However, Ca2+ signaling was nearly abolished in cells expressing the cytoplasmic InsP3 buffer (Fig. 5, A, B, and E). These results show that vasopressin increases InsP3 only in the cytoplasm and that Ca2+ signals in both the cytoplasm and the nucleus result from this. Like vasopressin, HGF (100 ng/ml) increased Ca2+ in both the cytoplasm and the nucleus (Fig. 5, C–E). However, Ca2+ signals induced by HGF were nearly abolished in cells expressing the nuclear InsP3 buffer rather than the cytoplasmic buffer (Fig. 5, C–E). These results show that although phosphorylated c-Met is in both the nucleus and the cytoplasm (Fig. 1A), HGF increases InsP3 only in the nucleus, and Ca2+ signals throughout the cell result from this. Consistent with this, Ca2+ signaling detected in cells in which images were collected more rapidly (Fig. 5F) occasionally identified cells in which vasopressin-induced signals began in the cytoplasm, whereas HGF-induced signals began in the nucleus, similar to what has been observed previously (20). In other cells, Ca2+ signals appeared to begin simultaneously in both compartments, but vasopressin-induced signals never began in the nucleus, whereas HGF signals never began in the cytoplasm. To further demonstrate that HGF directly forms InsP3 within the nucleus, total and nuclear PIP2 hydrolysis was examined. Total cellular PIP2 was 17.8 ± 1.9 pmol/5 × 106 cells under control conditions. PIP2 was detected in the nucleus as well, although this represented less than 15% of total cellular PIP2. Vasopressin was more effective than HGF in reducing total cellular PIP2, but HGF selectively and potently reduced PIP2 within the nucleus (Fig. 6, A and B). Vasopressin had minimal effect on nuclear PIP2 (n = 2, not shown). PLC-γ is thought to be responsible for HGF-induced hydrolysis of PIP2, and both immunoblot (Fig. 6C) and confocal immunofluorescence (not shown) demonstrated that this isoform of PLC is in the nucleus as well as in the cytoplasm. Together, these findings identify c-Met as an endogenous factor that selectively activates the nuclear machinery responsible for hydrolyzing PIP2 and thus generating InsP3 and Ca2+ signals within the nucleus.
FIGURE 4. Subcellular localization of targeted InsP3 buffer fusion proteins.
A, schematic representation of mRFP-targeted InsP3 buffer fusion proteins. NES, nuclear exclusion signal; NLS, nuclear localization signal. B, targeted InsP3 buffer constructs localize correctly to the cytoplasm or nucleus. SkHep1 cells were transiently transfected with either the cytosolic or the nuclear InsP3 buffer construct or else with mRFP alone as a transfection control. Cells were labeled with the nuclear marker TO-PRO-3 (blue) to confirm nuclear localization or exclusion. C, RFP fluorescence is slightly but significantly greater in cells expressing the nuclear InsP3 buffer. Values are mean ± S.E. of measurements made from 30 cells in each group. a.u., arbitrary units.
FIGURE 5. c-Met generates InsP3 in the nucleus rather than the cytoplasm.
A, vasopressin-induced Ca2+ signals are blocked by the cytosolic but not the nuclear InsP3 buffer. Cells were loaded with fluo-4 and then stimulated with 100 nm AVP while examined by confocal microscopy. The red box indicates the region of interest in the nucleus, and the white box represents the region of interest in the cytoplasm that was used to monitor Ca2+ signals. min., minimum; max., maximum. B, graphical representation of the nuclear and cytosolic Ca2+ signal detected in a representative cell from each experimental group stimulated with vasopressin. The tracings confirm that the cytosolic but not the nuclear InsP3 buffer blocks all Ca2+ signaling. C, HGF-induced Ca2+ signals are blocked by the nuclear but not the cytosolic InsP3 buffer. Cells were stimulated with HGF (100 ng/ml) while examined by confocal microscopy. The red box indicates the region of interest in the nucleus, and the white box represents the region of interest in the cytoplasm that was used to monitor Ca2+ signals. D, graphical representation of the nuclear and cytosolic Ca2+ signal detected in a representative cell from each experimental group stimulated with HGF. The tracings confirm that the nuclear but not the cytosolic InsP3 buffer blocks all Ca2+ signaling. E, summary of InsP3 buffer studies confirms that AVP Ca2+ signaling depends upon cytosolic InsP3, whereas HGF Ca2+ signaling depends upon nuclear InsP3. Values are mean ± S.E. of the peak fluo-4 fluorescence attained during the observation period (expressed as % of baseline) and include the response from 10–15 cells in each experimental group (*, p < 0.05). F, cytoplasmic and nuclear Ca2+ signals in representative cells examined at a rate of 83 ms/image. The vasopressin-induced Ca2+ signal begins in the cytoplasm, whereas the HGF-induced signal begins in the nucleus. Note the expanded time scales; each agonist was added at t = 0.
FIGURE 6. HGF hydrolyzes nuclear PIP2.
A, lipid samples were spotted onto nitrocellulose membranes (right), as illustrated by the strip template (left), and the PIP2 levels were detected using an anti-PIP2 monoclonal antibody that reacts with PIP2 with a high degree of specificity. Left column, experimental samples; middle column, PIP2 standards; right column, PIP controls (20 pmol each). PI(3)P, phosphatidylinositol-3-phosphate; PI(4)P, phosphatidylinositol 4-phosphate; PI(5)P, phosphatidylinositol 5-phosphate; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; and PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate. B, densitometric measurement shows that AVP hydrolyzes 22 ± 2% of PIP2 in whole cell preparations (n = 3, *, p < 0.05), whereas HGF stimulation does not hydrolyze significant amounts of PIP2 in such preparations. However, upon HGF stimulation, PIP2 levels in the nucleus are decreased by 87 ± 13% (n = 3, *, p < 0.05). Total lipids and nuclear lipids were isolated 4 min after stimulation with HGF (100 ng/ml) or 30 s after stimulation with AVP (100 nm). Values mean ± S.E. C, phospholipase C-γ1 is found in both the cytoplasm and the nucleus of SkHep1 cells. Non-Nuc., non-nuclear.
c-Met Must Translocate to the Nucleus to Induce Ca2+ Signals
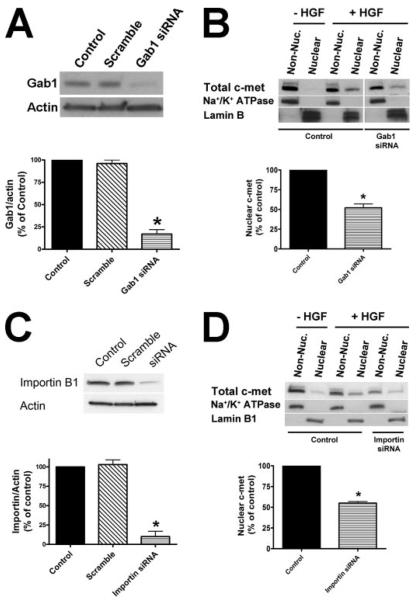
To determine whether c-Met must go to the nucleus to initiate nuclear Ca2+ signaling, the role of Gab1 and importin-β1 was examined. Gab1 is an adaptor protein that contains a nuclear localization sequence (30) and also binds to c-Met and to other RTKs (31). Transport of proteins through the nuclear pore complex typically involves importins α/β and exportins (32). Specifically, importin-α binds to the classical lysine-rich nuclear localization signal in the cargo, and importin-β interacts with the importin-α/cargo complex to guide it through the nuclear pore (32). Immunoblots of cell fractions isolated at serial time points following HGF stimulation demonstrated that HGF induced translocation of Gab1 to the nucleus (Fig. 7). To determine whether Gab1 or importin-β1 are necessary for the translocation of c-Met to the nucleus, siRNAs were developed that reduced Gab1 expression by 83 ± 6% (Fig. 8A) and importin expression by 90 ± 7% (Fig. 8C) in SkHep1 cells, relative to non-transfected and scrambled siRNA-transfected controls. Immunoblots of nuclear and non-nuclear cell fractions demonstrated that treatment of cells with Gab1 and importin siRNAs reduced the translocation of c-Met to the nucleus by 48 ± 6% (Fig. 8B) and 55% ± 2% (Fig. 8D), respectively. These results suggest that Gab1 and importin each play a role in c-Met translocation. Finally, HGF-induced Ca2+ signals in both the nucleus and the cytoplasm were eliminated in cells transfected with either Gab1 or importin siRNA (Fig. 9, A–C). This action of these siRNAs did not reflect a nonspecific inhibition of Ca2+ signaling pathways because vasopressin-induced Ca2+ signals were not inhibited (Fig. 9B). These findings demonstrate that Gab1- and importin-mediated translocation of c-Met to the nucleus is necessary to initiate nuclear Ca2+ signaling.
FIGURE 7. Gab1 translocates to the nucleus upon HGF stimulation.
A, immunoblots of Gab1 in nuclear and non-nuclear (Non-Nuc.) fractions of SkHep1 cells before and after stimulation with HGF (100 ng/ml) demonstrate that HGF induces Gab1 to shift to the nucleus. Na+/K+-ATPase and Lamin B were used as controls for the non-nuclear and nuclear fraction, respectively. B, the graphical representation confirms that Gab1 rapidly shifts to the nucleus after stimulation with HGF. Values are scaled relative to the initial amount in the non-nuclear fraction (mean ± S.E.; n = 3).
FIGURE 8. Gab1 and importin β1 are required for c-Met translocation.
A, an siRNA construct specifically reduces expression of Gab1 in SkHep1 cells. Cells were transfected with 50 nm siRNA for Gab or a scrambled siRNA control. Densitometric analysis confirms that the Gab1 siRNA reduces protein expression by 83 ± 6% (n = 4, *, p < 0.001). B, immunoblots of total c-Met in nuclear and non-nuclear (Non-Nuc.) fractions of SkHep1 cells stimulated with HGF show that Gab1 siRNA reduces translocation of c-Met to the nucleus. Densitometry confirms that the amount of c-Met in the nucleus in siRNA-treated cells is reduced by 48 ± 5% (n = 3; *, p < 0.05). C, an siRNA construct specifically reduces expression of importin β1 in SkHep1 cells. Cells were transfected with 50 nm siRNA for importin β1 or a scrambled siRNA control. Densitometric analysis confirms that the importin siRNA reduces protein expression by 90 ± 7% (n = 3, *, p < 0.001). D, immunoblots of total c-Met in nuclear and non-nuclear fractions of SkHep1 cells stimulated with HGF show that importin β1 siRNAs reduces translocation of c-Met to the nucleus. Densitometry confirms that the amount of c-Met in the nucleus in siRNA-treated cells is reduced by 55% ± 2% (n = 3; *, p < 0.05).
FIGURE 9. c-Met must translocate to the nucleus to generate Ca2+ signals.
A, HGF-induced Ca2+ signals are blocked by reduced expression of Gab1 or importinβ1. Cells were stimulated with HGF (100 ng/ml) while examined by confocal microscopy. The red box indicates the region of interest in the nucleus, and the white box represents the region of interest in the cytoplasm that was used to monitor Ca2+ signals. min., minimum; max., maximum. B, graphical representation of the nuclear and cytosolic Ca2+ signal detected in a representative non-transfected cell and in cells transfected with Gab-1 or importin siRNA. Each cell was stimulated with HGF and then with AVP. Note that reduced expression of Gab-1 or importin β1 blocks the response to HGF but does not impair the response to vasopressin. C, a summary of Gab1 and importin studies confirms that HGF Ca2+ signaling is eliminated in cells treated with siRNA for either Gab1 or importinβ1. Values are mean ± S.E. of the peak fluo-4 fluorescence (expressed as % of baseline) and include the response from 15–35 cells in each experimental group (*, p < 0.05).
DISCUSSION
Our results suggest that Ca2+ signaling induced by RTKs such as c-Met occurs through direct formation of InsP3 in the nucleus, which differs fundamentally from Ca2+ signaling induced by G protein-coupled receptors such as the V1a receptor (Fig. 10). The cell nucleus contains the machinery necessary for locally generating InsP3 and InsP3-dependent Ca2+ signals, including PIP2 (24), PLC (33), and InsP3-dependent Ca2+ stores (20, 27). Several lines of evidence suggest that growth factors (33) and integrins (34) may activate nuclear isoforms of PLC, and the current work demonstrates the functional significance of this by showing that HGF, via c-Met, hydrolyzes PIP2 to form InsP3 and increase free Ca2+ within the nucleus. It is now appreciated that Ca2+ signals within the nucleus exert a range of cellular effects that cannot be induced by cytoplasmic Ca2+ signals, including stimulation of cell proliferation (18), activation of transcription factors such as cAMP-response element-binding protein (35) and Elk-1 (23), and activation of kinases such as protein kinase C (20) and calmodulin kinase IV (36). The observation that c-Met induces InsP3 formation directly within the nucleus, and must do so to generate Ca2+ signals, provides insight as to how Ca2+-dependent events within the nucleus are controlled. The current findings also provide evidence that activation of c-Met generates little or no InsP3 in the cytoplasm, in contrast to what is widely believed (5-7). It is not yet clear why phosphorylated c-Met activates PLC only in the nucleus, and not at the plasma membrane, before the receptor translocates to the nucleus. One possible explanation is that the proper isoform of PLC does not associate with c-Met until the receptor reaches the nucleus, perhaps because the internalization process of c-Met at the plasma membrane makes it inaccessible to cytoplasmic PLC or because adaptor proteins in the nucleus but not the cytosol are required for this association. In any case, the versatility of Ca2+ as a second messenger is thought to result from tight spatial control of Ca2+ signals in cells (13). Therefore, local intranuclear generation of InsP3 by c-Met or other RTKs would provide a mechanism to selectively activate Ca2+-dependent events that occur in the nucleus and not elsewhere.
FIGURE 10. c-Met Ca2+ signaling within the nucleus.
Stimulation with HGF induces c-Met to translocate to the nucleus, and this depends upon importin β and the adaptor protein Gab1. In the nucleus, activated c-Met forms InsP3 (although the topology of c-Met in the nucleus, and whether HGF remains bound to nuclear c-Met, are unknown). This acts on nuclear InsP3 receptors to increase free nucleoplasmic Ca2+. In contrast, vasopressin induces the V1a receptor to form InsP3 in the region of the plasma membrane, which acts on InsP3 receptors in the endoplasmic reticulum to increase free Ca2+ in the cytoplasm.
A number of RTKs have been found in the nucleus, including receptors for insulin, epidermal growth factor receptor, and fibro-blast growth factor (8-10). These nuclear RTKs have been shown to act as transcription factors (9, 38). However, the mechanism by which RTKs travel from the cell surface to the nucleus is unknown. RTKs generally are internalized by clathrin-dependent endocytosis (39). Desensitization of both the epidermal growth factor receptor and the HGF receptor occurs by a common mechanism in which internalization is dependent upon clathrin but also depends upon association with a complex consisting of Cbl, CIN85, and endophilin. This complex enables Cbl to induce ubiquitination and then degradation of these receptors (40, 41). It is unclear whether transport of RTKs to the nucleus begins with this same mechanism of internalization because disruption of this tripartite protein complex prevents internalization of the HGF receptor yet enhances rather than impairs the biological response to HGF (40). Additional questions include how c-Met enters the nucleus, the topology of c-Met within the nucleus, and whether HGF remains bound to nuclear c-Met.
Ca2+ signals can consist of complex spatial patterns, including gradients and waves, and the subcellular distribution of InsP3Rs helps shape these patterns. For example, presynaptic increases in Ca2+ regulate neurotransmitter release (39), and local increases in Ca2+ also regulate extension of neural growth cones (42). Accessory proteins such as Homer and protein 4.1N are thought to localize InsP3Rs to these regions in neurons (43, 44), which may in turn permit these highly localized Ca2+-mediated events to occur. Similarly, apical increases in Ca2+ regulate fluid and electrolyte secretion (45) and exocytosis (46) in polarized epithelia, and these localized Ca2+ signals result from the local accumulation of InsP3Rs in lipid rafts (47, 48). Furthermore, apoptosis can be induced by transmission of Ca2+ signals from InsP3Rs to mitochondria (49), and physical connections between these two structures may facilitate this (37). Finally, gene transcription (23) and cell growth (18) are regulated by increases in Ca2+ in the nucleus, which in turn can be induced by release of Ca2+ from intranuclear InsP3Rs (20). Moreover, there are differences in the types of InsP3R isoforms found in the nucleus in different cell types, and this variability can affect the sensitivity of the nucleus to InsP3 (19). By demonstrating that growth factors can selectively increase InsP3 within the nucleus, the current findings reveal an even more sophisticated level of regulation in which nuclear InsP3Rs can be activated not by virtue of their subcellular distribution but rather as a result of local, subcellular production of InsP3.
Footnotes
This work was supported by National Institutes of Health Grants DK57751, DK34989, and DK45710, and by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico. Fundaçao de Amparo à Pesquisa do Estado de Minas Gerais, and Howard Hughes Medical Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
The abbreviations used are: HGF, hepatocyte growth factor; PIP2, phosphatidylinositol 4,5-bisphosphate; InsP3, inositol 1,4,5-trisphosphate; InsP3R, InsP3 receptor; RTK, receptor tyrosine kinase; PLC, phospholipase C; PBS, phosphate-buffered saline; AVP, arginine vasopressin; mRFP, red fluorescent protein; siRNA, small interfering RNA.
REFERENCES
- 1.Maher JJ. J. Clin. Investig. 1993;91:2244–2252. doi: 10.1172/JCI116451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalopoulos GK, DeFrances MC. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 3.Furge KA, Zhang YW, Vande Woude GF. Oncogene. 2000;19:5582–5589. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- 4.Shiota G, Rhoads DB, Wang TC, Nakamura T, Schmidt EV. Proc. Natl. Acad. Sci. U. S. A. 1992;89:373–377. doi: 10.1073/pnas.89.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okano Y, Mizuno K, Osada S, Nakamura T, Nozawa Y. Biochem. Biophys. Res. Commun. 1993;190:842–848. doi: 10.1006/bbrc.1993.1125. [DOI] [PubMed] [Google Scholar]
- 6.Anderson D, Koch CA, Grey L, Ellis C, Moran MF, Pawson T. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- 7.Wahl MI, Olashaw NE, Nishibe S, Rhee SG, Pledger WJ, Carpenter G. Mol. Cell. Biol. 1989;9:2934–2943. doi: 10.1128/mcb.9.7.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SJ, Kahn CR. J. Cell. Physiol. 1993;157:217–228. doi: 10.1002/jcp.1041570203. [DOI] [PubMed] [Google Scholar]
- 9.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 10.Reilly JF, Maher PA. J. Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massie C, Mills IG. Nat. Rev. Cancer. 2006;6:403–409. doi: 10.1038/nrc1882. [DOI] [PubMed] [Google Scholar]
- 12.Wells A, Marti U. Nat. Rev. Mol. Cell. Biol. 2002;3:697–702. doi: 10.1038/nrm905. [DOI] [PubMed] [Google Scholar]
- 13.Berridge MJ, Lipp P, Bootman MD. Nat. Rev. Mol. Cell. Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 14.Nathanson MH, Gautam A, Bruck R, Isales CM, Boyer JL. Hepatology. 1992;15:107–116. doi: 10.1002/hep.1840150119. [DOI] [PubMed] [Google Scholar]
- 15.Nathanson MH, Rios-Velez L, Burgstahler AD, Mennone A. Gastroenterology. 1999;116:1176–1183. doi: 10.1016/s0016-5085(99)70021-1. [DOI] [PubMed] [Google Scholar]
- 16.Nathanson MH, Gautam A, Ng OC, Bruck R, Boyer JL. Am. J. Physiol. 1992;262:G1079–G1086. doi: 10.1152/ajpgi.1992.262.6.G1079. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues MA, Gomes DA, Leite MF, Grant W, Zhang L, Lam W, Cheng YC, Bennett AM, Nathanson MH. J. Biol. Chem. 2007;282:17061–17068. doi: 10.1074/jbc.M700490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, Ehrlich BE, Nathanson MH. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2975–2980. doi: 10.1073/pnas.0536590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Nat. Cell Biol. 2003;5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray A, Olsson H, Batty IH, Priganica L, Peter DC. Anal. Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 22.Varnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spat A, Rhee SG, Hajnoczky G, Balla T. J. Biol. Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- 23.Pusl T, Wu JJ, Zimmerman TL, Zhang L, Ehrlich BE, Berchtold MW, Hoek JB, Karpen SJ, Nathanson MH, Bennett AM. J. Biol. Chem. 2002;277:27517–27527. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- 24.Irvine RF. Nat. Rev. Mol. Cell. Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 25.Stehno-Bittel L, Luckhoff A, Clapham DE. Neuron. 1995;14:163–167. doi: 10.1016/0896-6273(95)90250-3. [DOI] [PubMed] [Google Scholar]
- 26.Malviya AN, Rogue P, Vincendon G. Proc. Natl. Acad. Sci. U. S. A. 1990;87:9270–9274. doi: 10.1073/pnas.87.23.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerasimenko OV, Gerasimenko JV, Tepikin AV, Petersen OH. Cell. 1995;80:439–444. doi: 10.1016/0092-8674(95)90494-8. [DOI] [PubMed] [Google Scholar]
- 28.Leite MF, Hirata K, Pusl T, Burgstahler AD, Okazaki K, Ortega JM, Goes AM, Prado MA, Spray DC, Nathanson MH. J. Biol. Chem. 2002;277:16313–16323. doi: 10.1074/jbc.M109207200. [DOI] [PubMed] [Google Scholar]
- 29.Birnbaumer M. Trends Endocrinol. Metab. 2000;11:406–410. doi: 10.1016/s1043-2760(00)00304-0. [DOI] [PubMed] [Google Scholar]
- 30.Osawa M, Itoh S, Ohta S, Huang Q, Berk BC, Marmarosh NL, Che W, Ding B, Yan C, Abe J. J. Biol. Chem. 2004;279:29691–29699. doi: 10.1074/jbc.M309371200. [DOI] [PubMed] [Google Scholar]
- 31.Gu H, Neel BG. Trends Cell Biol. 2003;13:122–130. doi: 10.1016/s0962-8924(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 32.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martelli AM, Billi AM, Manzoli L, Faenza I, Aluigi M, Falconi M, De PA, Gilmour RS, Cocco L. FEBS Lett. 2000;486:230–236. doi: 10.1016/s0014-5793(00)02313-9. [DOI] [PubMed] [Google Scholar]
- 34.Shankar G, Davison I, Helfrich MH, Mason WT, Horton MA. J. Cell Sci. 1993;105:61–68. doi: 10.1242/jcs.105.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Hardingham GE, Chawla S, Johnson CM, Bading H. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 36.Deisseroth K, Heist EK, Tsien RW. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 37.Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni CY, Murphy MP, Golde TE, Carpenter G. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 39.Di Fiore PP, De Camilli P. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 40.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 41.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 42.Lohmann C, Myhr KL, Wong RO. Nature. 2002;418:177–181. doi: 10.1038/nature00850. [DOI] [PubMed] [Google Scholar]
- 43.Fukatsu K, Bannai H, Zhang S, Nakamura H, Inoue T, Mikoshiba K. J. Biol. Chem. 2004;279:48976–48982. doi: 10.1074/jbc.M408364200. [DOI] [PubMed] [Google Scholar]
- 44.Xiao B, Tu JC, Worley PF. Curr. Opin. Neurobiol. 2000;10:370–374. doi: 10.1016/s0959-4388(00)00087-8. [DOI] [PubMed] [Google Scholar]
- 45.Kasai H, Augustine GJ. Nature. 1990;348:735–738. doi: 10.1038/348735a0. [DOI] [PubMed] [Google Scholar]
- 46.Ito K, Miyashita Y, Kasai H. EMBO J. 1997;16:242–251. doi: 10.1093/emboj/16.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagata J, Guerra MT, Shugrue CA, Gomes DA, Nagata N, Nathanson MH. Gastroenterology. 2007;133:256–267. doi: 10.1053/j.gastro.2007.03.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez E, Leite MF, Guerra MT, Kruglov EA, Bruna-Romero O, Rodrigues MA, Gomes DA, Giordano FJ, Dranoff JA, Nathanson MH. J. Biol. Chem. 2007;282:10057–10067. doi: 10.1074/jbc.M700746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendes CC, Gomes DA, Thompson M, Souto NC, Goes TS, Goes AM, Rodrigues MA, Gomez MV, Nathanson MH, Leite MF. J. Biol. Chem. 2005;280:40892–40900. doi: 10.1074/jbc.M506623200. [DOI] [PubMed] [Google Scholar]