Summary
ATP (energy production) production is not the only function of the mitochondria. Mitochondria perform multiple cellular functions. Among others, these functions include control of cell death, growth, development, integration of signals from mitochondria to nucleus and nucleus to mitochondria, and various metabolic pathways. Although defects in mitochondrial function are most commonly associated with bioenergetic deficiencies, our studies demonstrate that mitochondrial defects lead to genome instability in the nuclear DNA, resistance to apoptosis and induction of NADPH oxidase, a designated producer of reactive oxygen species. These transformations in cellular phenotype are known contributors to the development of tumors in humans. Consistent with the role of mitochondria in carcinogenesis, studies in the past few years have described an increased risk of cancers associated with specific mitochondrial DNA (mtDNA) polymorphism among various different haplogroups in human population. However, molecular mechanisms underlying increased risk of cancer due to specific mtDNA polymorphisms is currently lacking. It is likely that mtDNA polymorphisms in mitochondrial genes involved in electron transport chain and oxidative phosphorylation result in increased oxidative stress and hypermutagenesis of mitochondrial as well as nuclear DNA. We suggest that in studies relating to cancer epidemiology, the significance of a particular mtDNA polymorphism(s) should be analyzed together with other polymorphisms in mtDNA and in nuclear DNA.
Keywords: Apoptosis, mitochondria, oxidative phosphorylation, polymorphism, reactive oxygen species
1. Introduction
Mitochondria perform multiple cellular functions. Mitochondria are the predominant source of energy (ATP) production and reactive oxygen species (ROS) in the cell. ROS are generated in the mitochondria as a by-product of oxidative phosphorylation (OXPHOS) involved in ATP production. Mitochondria contain their own genetic material (mitochondrial DNA, mtDNA) which encodes proteins that are an integral part of various OXPHOS complexes (1,2). It is estimated that 90% of cellular oxygen is consumed in the mitochondria and about 2–4% of that oxygen is converted to ROS. This amounts to approximately 107 ROS molecules/mitochondrion every day (1,2). Consistent with these observations, studies have indicated somatic mtDNA mutations in almost all cancers examined to date. The mutations found in tumors cells reinforce the suggestion made by Warburg in 1930 (3). Warburg hypothesized that mitochondrial abnormalities were implicated in development of cancers. Since then, many mitochondrial abnormalities in cancers, both at genetic and metabolic levels, have been reported. In this chapter, we intend to discuss abnormalities at the mtDNA level. With regard to mitochondrial metabolic and other related abnormalities in cancers, see recent reviews (1,2).
2. Mitochondrial Genome
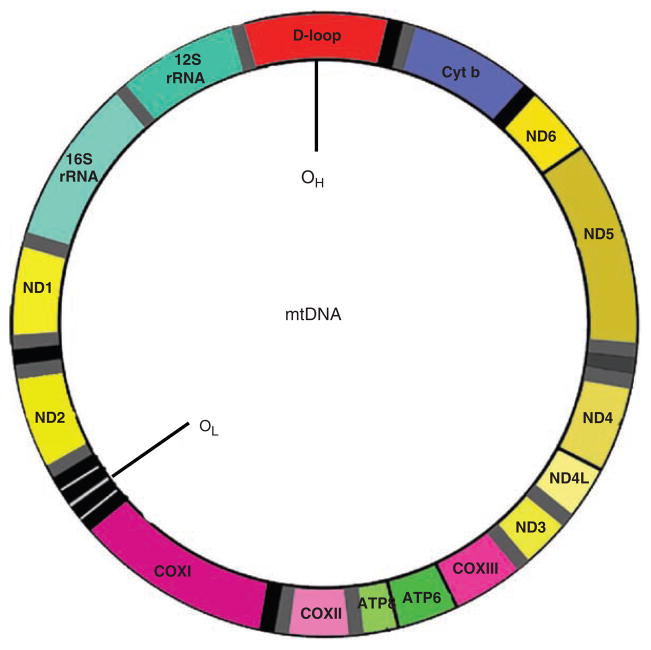
The mitochondrial genome is a 16.6-kb closed circular, double-helical molecule. The mitochondrial genome is inherited only through the mother. The mtDNA encodes two rRNAs, 22 tRNAs, and 13 proteins (Fig. 15.1; [4]). Each of these proteins is a subunit of one of four respiratory enzyme complexes localized into the mitochondria. They include seven subunits of respiratory enzyme complex I, one subunit of complex III, three subunits of complex IV, and two subunits of complex V. All other (~1,500) mitochondrial proteins, including those involved in the replication, transcription, and translation of mtDNA, are encoded by nuclear genes, and they are targeted to the mitochondrion by a specific transport system (5). Although mtDNA represents <1% of total cellular DNA, its gene products are essential for normal cell function. Unlike nuclear DNA, mammalian mtDNA contains no introns, has no protective histones, and is exposed to deleterious reactive oxygen species generated by OXPHOS. In addition, replication of mtDNA is error prone (4). The accumulation of mutations in mtDNA is approximately 10-fold greater than that in nuclear DNA (6,7). This feature of mtDNA makes it a useful tool for studying human migration around the globe and identifying specific human populations.
Fig. 15.1.
mtDNA map.
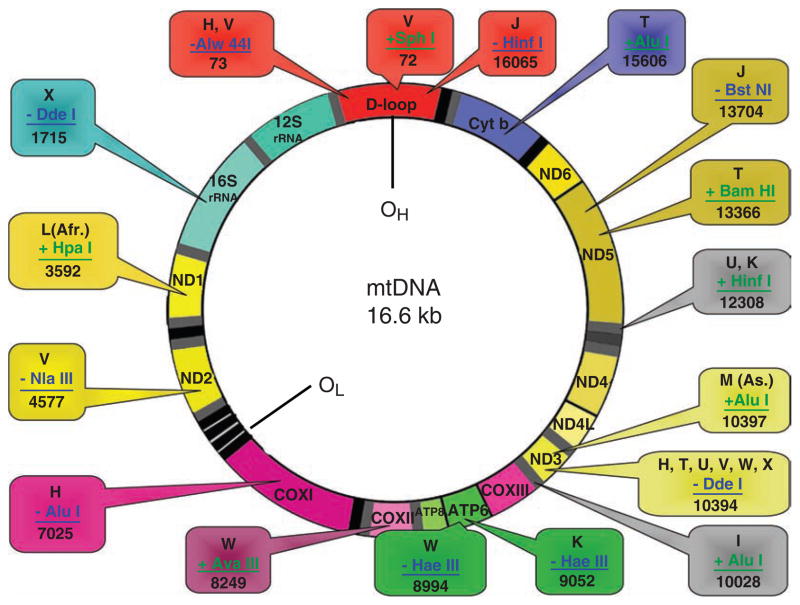
A haplogroup is defined on the basis of a particular mutation that is well established and widely distributed among individual of the populations. These haplogroups can be further divided into haplotypes, generally based on restriction fragment length polymorphism (RFLP). RFLP are a result of restriction enzymes that cut DNA sequences at a particular point, priming them for electrophoresis and allowing a comparison to be made showing a pattern among individuals based on the length of the DNA fragments (8). More than 2 dozen mtDNA haplogroups are known among human populations around the world. Figure 15.2 describes some of the major mitochondrial haplogroups identified throughout the world. Figure 15.2 also reveals the RFLPs used for haplotype analyses. However, haplogroups and subgroups are best defined by mtDNA sequencing.
Fig. 15.2.
Haplotype analysis by RFLP: summary of various restriction enzymes used to define haplotype.
3. Mitochondrial Polymorphism and Risk of Cancer
mtDNA mutations causing defects in mitochondrial respiratory enzyme complexes are thought to increase risk of cancer. In support of this hypothesis, we have demonstrated that defects in mitochondrial respiratory chain activity lead to 1) overexpression of superoxide producing NADPH oxidase (9); 2) increased damage and hypermutagenesis in the nuclear DNA (10–13); and 3) resistance to apoptosis (14). These transformations in cellular phenotype are known to contribute to the development of cancer.
Studies in the past few years have described the risk of cancers associated with various mtDNA haplogroups in the human population. Tanaka’s group analyzed a population with 1,503 autopsied cases (696 male, 807 female) at Tokyo Metropolitan Geriatric Hospital registered. The genotypes for 149 polymorphisms in the coding region of the mitochondrial genome were determined. The haplogroups, were classified into 30 haplotypes, i.e., F, B5, B4a, B4b, B4c, A, N9a, N9b, Y, M10+M11+M12, M7a, M7b2, M7c, M8+Z+C, G1, G2, M9, D5, D4a, D4b, D4d, D4e, D4g, D4h, D4j, D4k, D4k, D4l, D4m, and D4n. Multivariate logistic regression analysis was performed with adjustment for age, the prevalence of drinking, and that of smoking. Among 1,363 subjects whose data were available for presence or absence of cancer, age, gender, history of alcohol intake, history of smoking, and mitochondrial haplogroups, 819 subjects carried a pathologically confirmed cancer(s) at the time of autopsy. Subjects with the haplogroup M7b2 tended to have an increased risk for hemopoietic cancer (P = 0.037), having an odds ratio of 2.46 (95% confidence interval [CI] = 1.06–5.73). Results also indicated that haplogroup M7b2 is a risk factor for leukemia (15).
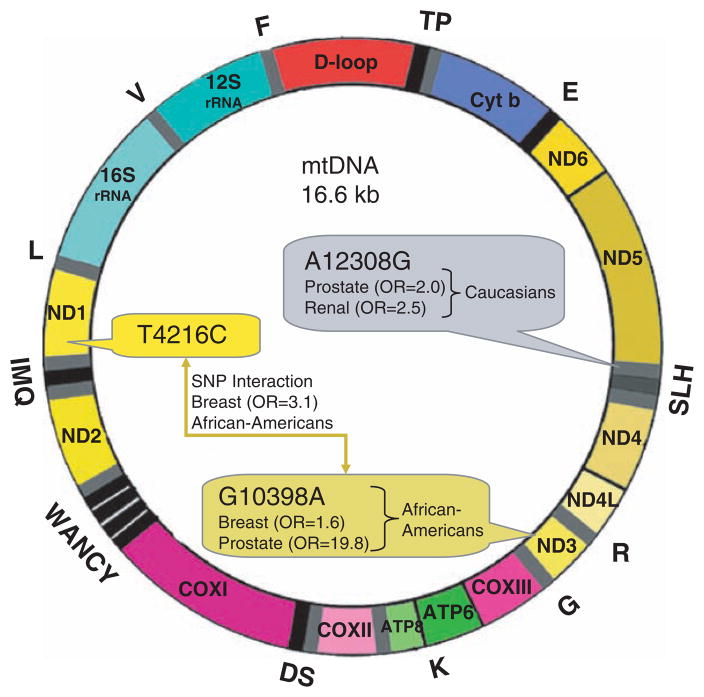
Nine main European haplotypes (H, I, J, K, T, U, V, W, and X) were analyzed in a series of patients with prostate and renal cancers studied by Booker et al. (16). Using the A12308G substitution in tRNALeu2 (Fig. 15.3) as a marker of the mtDNA haplogroup U, it was found that patients carrying this haplogroup had an increased risk of renal (odds ratio [OR] = 2.52) and prostate cancer (OR = 1.95) (16).
Fig. 15.3.
mtDNA oncomap: summary of mtDNA mutations found in various tumors examined.
Canter’s epidemiologic studies in breast cancer showed the G10398A substitution in ND3 was associated with an increased risk (OR = 1.6) in African-American women (Fig. 15.3; (17)). His group also reported one of the first cases of synergy among single-nucleotide polymorphisms. The T4216C substitution alone in ND1 conferred no increased risk. However, when the T4216C substitution was present with G10398A, the risk of breast cancer was increased (OR = 3.1; Fig 15.3). It has been hypothesized also that the prevalence of germline cyclooxygenase (COX) I mutations may explain the increased incidence of prostate cancer in African American men (18).
Darvishi et al. (19) analyzed mtG10398A (Ala→Thr) polymorphism in a haplotype constituting mtDNA haplogroup N and its sublineages for its contribution to higher risk for breast cancer. They analyzed approximately 1,000 complete human mtDNA sequences worldwide and collated information on 2,334 individuals belonging to 18 regions in India. They confirmed that mt10398A allele provides a risk toward breast cancer by using a case-control comparison study of 124 sporadic breast cancer patients and 273 controls. They also included 55 squamous cell carcinoma of esophagus and 163 controls, matched for age, ethnicity, and sex from north India. Their study also found that 10398A mtDNA polymorphism confers a higher risk for esophageal cancer. In a recent study, Li et al. (62) showed an association between mtDNA haplogroup D4a and D5a and increased risk of esophageal cancer in China.
Bai et al. (20) analyzed mtDNA polymorphism in European-American females and reported that A10398G (OR = 1.79) and T16519C (OR = 1.98) increase breast cancer risk. In contrast, T3197C (OR = 0.31) and G13708A (OR = 0.47) were found to decrease breast cancer risk.
Wang et al. (21) evaluated polymorphisms in mtDNA associated with increased risk of pancreatic cancer. They screened Caucasian cases and found no significant associations with pancreatic cancer. It is likely that polymorphism in other mitochondrial genes or in conjunction with nuclear genes involved in OXPHOS may contribute to pancreatic cancer.
In some of the above-mentioned studies, it is noteworthy that the potential influence of population stratification was not taken in to account. A population stratification of these studies, independent positive replication of these studies, or both are needed to provide convincing evidence for the association of mtDNA polymorphism and risk for cancer.
4. Mitochondrial Polymorphism and Risk of Other Diseases
Recent studies in Japanese centenarians and super-centenarians (those ≥105 years) have demonstrated that specific mtDNA haplogroup that are associated with these age groups. Centenarians seem to be resistant against lifestyle-related diseases such as type 2 diabetes, myocardial infarction, and cerebrovascular infarction, as well as geriatric diseases such as Alzheimer’s disease (AD) and Parkinson disease (22).
Tanaka’s group described that mtDNA mutations in these age groups provide protection from disease and confer longevity. Data from this study suggest that longevity is associated with the D4a haplogroup, whereas noncentenarians mitochondrial haplogroups F and A are risk factors for diabetes. Furthermore, haplogroup N9a was found to be a protective against diabetes, especially in women. A large-scale association studies will have a major impact on the functional studies of mitochondrial haplogroups and on the elucidation of their contributions to longevity or pathogenesis of lifestyle-related diseases (23).
Van der Walt (24) analyzed risk of AD associated with European mitochondrial haplogroups. Interestingly, they found that males in haplogroup U showed an increase in risk (OR = 2.30; 95% CI = 1.03–5.11; P = 0.04) of AD relative to the most common haplogroup H, whereas females demonstrated a significant decrease in risk with haplogroup U (OR = 0.44; 95% CI = 0.24–0.80; P = 0.007).
Van der Walt (24) also demonstrated a significant decrease in risk of PD among European haplogroup J versus individuals carrying the most common haplogroup, H. Furthermore, a specific mutation 10398G, that defines these two haplogroups, is strongly associated with this protective effect (OR = 0.53; 95% CI = 0.39–0.73; P = 0.0001). Interestingly, these studies suggest that a single mtDNA substitution may exert different effects in different disease states.
5. Mitochondrial DNA Mutations in Human Tumors
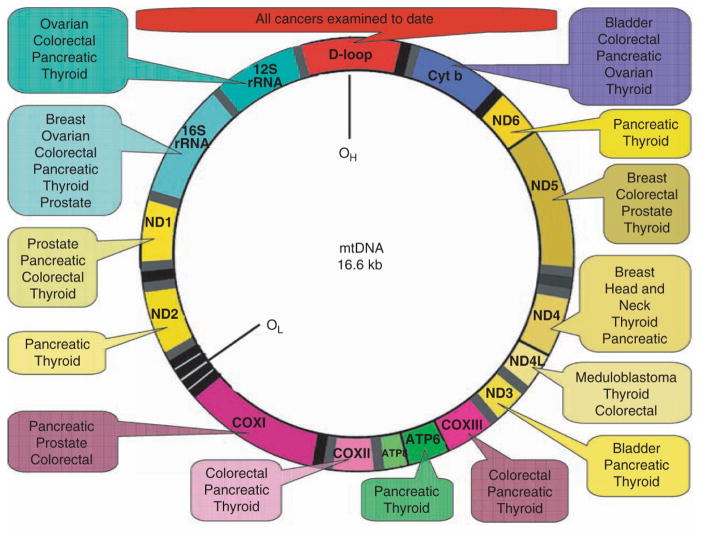
Mutations in mtDNA have been reported in all cancers examined to date (1,2). A summary of mtDNA mutations in various tumors is presented in Fig. 15.4. Although these mutations are known to occur throughout the mitochondrial genome, the displacement loop (or D-loop) region has been shown to be a mutational “hot spot” in human cancer. The D-loop is a noncoding control region of mtDNA (np 16024–516) that houses cis-regulatory elements required for replication and transcription of the molecule. Thus, mtDNA mutations in this region might be expected to affect copy number and gene expression of the mitochondrial genome. Somatic mutations in the D-loop region have been observed in all tumors examined to date (1,2). In a recent comprehensive study involving 54 hepatocellular carcinomas, 31 gastric, 31 lung, and 25 colorectal cancers, the incidence of somatic D-loop mutations in cancers of later stage was found to be higher that that of early stage cancers (25). These findings suggest that instability in the D-loop region of mtDNA may be involved in carcinogenesis of human cancers.
Fig. 15.4.
mtDNA polymorphism associated with risk of cancers: G10398A in ND3 is associated with increased risk of breast (OR = 1.6) and prostate cancer (OR = 19.8) in African-Americans (37). A12308G in tRNALeu2 is a marker of the mtDNA haplogroup U. This haplogroup is associated with increased risk of prostate (OR = 1.95) and renal cancer (OR = 2.52) in Caucasians. *, the T4216C substitution in ND1 conferred no increased risk in isolation. However, when the T4216C substitution was present with G10398A, synergy was observed, and the risk of breast cancer was increased (OR = 3.1) in African-Americans.
In addition to the mtDNA mutations found in the D-loop region, deletions, point mutations, insertions, and duplications in other parts of the mitochondrial genome have been noted in a variety of human cancers, including ovarian, thyroid, salivary, kidney, liver lung, colon, gastric brain, bladder, head and neck, prostate, and breast cancer, and leukemia (26,18). For example, a 40-bp insertion localized in the COX I gene seems to be specific for renal cell oncocytoma (27), and a deletion mutation resulting in the loss of mtDNA within NADH dehydrogenase subunit III is associated with renal carcinoma (28). In a recent population based study involving 260 prostate cancer patients of European and African-American descent and 54 benign controls without cancer, the frequency of COX I missense mutations was found to be significantly higher in prostate cancer patients compared with the no-cancer controls (12 vs. 1.9%, respectively). At least some of these COX I sequence variants were thought to represent germ line mutations) (18). The co-occurrence of somatic and germline mtDNA mutations in renal cell carcinoma also has been reported (29).
Tumor-specific changes in mtDNA copy number also have been noted in human cancers. For example, the mtDNA content was found to be elevated in primary tumors of head and neck squamous cell carcinoma (30), and to increase with histopathologic grade in premalignant and malignant head and neck lesions (31). In addition, mtDNA copy number was shown to increase in papillary thyroid carcinomas (32) and during endometrial cancer development (33). Conversely, it was reported that mtDNA content is reduced in 80% of breast tumors relative to normal controls. mtDNA depletion also occurs in gastric cancers (32,33) and during endometrial cancer development (34). Conversely, it was reported that mtDNA content is reduced in 80% of breast tumors relative to normal controls. mtDNA depletion also occurs in breast cancers (35). The actual mtDNA copy number in cancers might depend upon the specific site of mutation associated with that cancer. For example, mutations in the D-loop region, which controls mtDNA replication, would be expected to result in a decrease in copy number. Conversely, an increase in mtDNA copy number might occur as a compensatory response to mitochondrial dysfunction or due to mutations in nuclear genes involved in controlling mtDNA copy number indirectly.
Mitochondrial abnormalities resulting from mutation in mtDNA or copy number changes invokes mitochondria-to-nucleus retrograde response in human cells (36). To identify proteins involved in retrograde response and their potential role in tumorigenesis, Kulawiec et al. carried out a comparative proteomic analysis using a parental cell line, a rho0 cell line in which the mitochondrial genome was completely depleted (and the cells were therefore lacking all mtDNA-encoded protein subunits), and a cybrid cell line in which mtDNA was restored (10,36). This comparative proteomic approach revealed marked changes in the cellular proteome, including quantitative changes in expression of several proteins in breast and ovarian tumors, which suggest that retrograde responsive genes may potentially function as tumor suppressor or oncogenes.
Transmitochondrial hybrid (cybrid) studies also suggest that mtDNA plays a key role in establishing the tumorigenic phenotype, maintaining the tumorigenic phenotype, or both. In a recent study, Singh et al. demonstrated that a rho0 derivative of human osteosarcoma cells display increased tumorigenic phenotype as evidenced by increased anchorage independent growth compared with the parental cells (10). Interestingly, the tumorigenic phenotype was restored to parental level by transfer of wild-type mitochondrial DNA to rho0 cells. These studies suggest that intergenomic cross-talk between mitochondria and cancer plays an important role in tumorigenesis and that mitochondria-to-nucleus retrograde signaling may be an important factor in restoration of the nontumorigenic phenotype. Indeed, additional studies by Singh’s group demonstrate that retrograde mitochondria-to-nucleus signaling involves regulation of NADPH oxidase (Nox1) and that this enzyme is overexpressed in a majority of breast and ovarian tumors (9). The family of Nox enzymes comprises seven structurally related homologs, Nox 1–5 and dual oxidase 1 and 2 (37,38), and nuclear DNA encoded Nox 1 is a major source of endogenous ROS in the cell (39). Until recently, these enzymes were known only to function as phagocytic respiratory burst oxidases that catalyze the NADPH-dependent reduction of molecular oxygen to superoxide, hydrogen peroxide, and other ROS, and to participate in host defense by killing or damaging invading microbes (39).
Transmitochondrial cybrid studies also suggest the functional significance of mtDNA mutations. Cybrids harboring the ATP6T8993G mtDNA mutation in prostate cancer (PC3) cells were found to generate tumors that were 7 times larger than wild-type cybrids, which barely grew in mice (8). Additionally, cybrids constructed using a common HeLa nucleus and mitochondria containing a point mutation in ATP synthase subunit 6 were conferred a growth advantage in early tumor stages after transplantation into nude mice. It was suggested that this growth advantage might possibly occur via prevention of apoptosis (40). These studies support the hypothesis that mtDNA mutations might directly promote tumor growth in vivo.
6. Possible Mechanism of mtDNA-Mediated Carcinogenesis
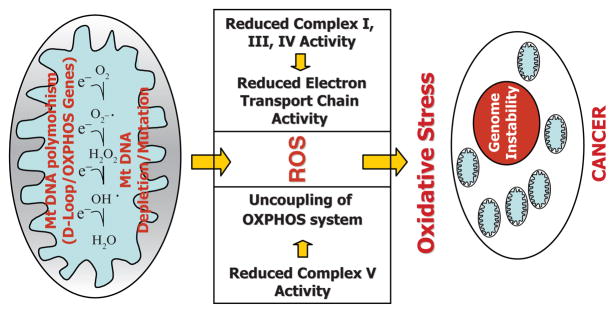
Polymorphisms in human mtDNA may affect the efficiency of ETC and ROS production. This argument is supported by the fact that common mtDNA variants are indeed involved in ROS production (41). Mitochondrial respiratory activity is associated with the generation of ROS. Under physiological conditions, a small fraction of reducing equivalents from complex I, complex III, or both of the mitochondrial electron transport chain may be transferred directly to molecular oxygen, generating the superoxide anion O2−. This ROS is converted to H2O2 by mitochondrial manganese superoxide dismutase. H2O2 can be converted to water by glutathione peroxidase or catalase, or it can acquire an additional electron from a reduced transition metal to generate the highly reactive hydroxyl radical ·OH. ROS generation has been shown to increase in mitochondria under conditions of excess electrons or as a result of respiratory enzyme complex inhibition. High levels of ROS, or oxidative stress, can not only mutagenize mtDNA but also induce hypermutagenesis and instability of nuclear (chromosomal) DNA (Fig. 15.5). These factors can contribute to development of cancer.
Fig. 15.5.
Mechanisms of mitochondria-mediated mutagenesis and development of cancer (see text for details).
7. Future Prospectives
In past year, studies have been conducted that identify the role of mtDNA polymorphism of cancer. However, to fully understand the importance of mtDNA polymorphism and risk of cancer, it is necessary to 1) identify genetic and epigenetic changes in the nucleus associated with mtDNA polymorphisms; 2) identify D-loop region polymorphisms resulting in changes in mtDNA copy number; 3) identify nuclear genes controlling mtDNA copy number; 4) identify functional changes associated with mtDNA copy number; 5) identify genetic alteration in the nuclear DNA (nDNA) associated with mtDNA copy number changes; 5) identify epigenetic alteration in the nDNA associated with mtDNA copy number changes; and understand the role of synergy between different polymorphisms in mtDNA as well in nDNA. This is particularly important in view of constant cross-talk between the mitochondria and mitochondria and nucleus.
Acknowledgments
Studies in our laboratory were supported by National Institutes of Health grants R01 CA121904, 113655 and 116430 (to K.K.S.) and the New York State Department of Health (to M.K. and K.K.S.).
References
- 1.Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–762. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Modica-Napolitano JS, Singh K. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med. 2002;4:1–19. doi: 10.1017/S1462399402004453. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O. Metabolism of Tumors. Arnold Constable; London, UK: 1930. [Google Scholar]
- 4.Singh KK, editor. Mitochondrial DNA Mutations in Aging, Disease, and Cancer. Springer; New York: 1998. [Google Scholar]
- 5.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 6.Grossman LI, Shoubridge EA. Mitochondrial genetics and human disease. Bioessays. 1996;18:983–991. doi: 10.1002/bies.950181208. [DOI] [PubMed] [Google Scholar]
- 7.Johns DR. Seminars in medicine of the Beth Israel Hospital, Boston: mitochondrial DNA and disease. N Engl J Med. 1995;333:638–644. doi: 10.1056/NEJM199509073331007. [DOI] [PubMed] [Google Scholar]
- 8.Malhi RS, Mortensen HM, Eshleman JA, Kemp BM, Lorenz JG, Kaestle FA, Johnson JR, Gorodezky C, Smith DG. Native American mtDNA prehistory in the American Southwest. Am J Phys Anthropol. 2003;120(2):108–24. doi: 10.1002/ajpa.10138. [DOI] [PubMed] [Google Scholar]
- 9.Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Crosstalk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol Ther. 2005;4:1367–1373. doi: 10.4161/cbt.4.12.2233. [DOI] [PubMed] [Google Scholar]
- 10.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, Matsui S. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–146. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Singh KK. Mitochondrial damage checkpoint in apoptosis and genome stability. FEMS Yeast Res. 2004;2:127–132. doi: 10.1016/j.femsyr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–3917. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delsite RL, Rasmussen LJ, Rasmussen AK, Kalen A, Goswami PC, Singh KK. Mitochondrial impairment is accompanied by impaired oxidative DNA repair in the nucleus. Mutagenesis. 2003;18:497–503. doi: 10.1093/mutage/geg027. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA depleted cells against cell death: Role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279:7512–7520. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- 15.Verma M, Naviaux RK, Tanaka M, Kumar D, Franceschi C, Singh KK. Mitochondrial DNA and cancer epidemiology. Cancer Res. 2007;67(2):437–9. doi: 10.1158/0008-5472.CAN-06-4119. [DOI] [PubMed] [Google Scholar]
- 16.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, Lim SD, Fernandez-Golarz C, Lyles RH, Brown MD, Marshall FF, Petros JA. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol. 2006;175:468–472. doi: 10.1016/S0022-5347(05)00163-1. discussion 472–463. [DOI] [PubMed] [Google Scholar]
- 17.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 18.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249(2):249–55. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67(10):4687–94. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol. Biomarkers Prev. 2007;16(7):1455–9. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 22.Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, Segawa T, Watanabe S, Kato K, Yokoi K, Nozawa Y, Lee HK, Tanaka M. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007;80(3):407–15. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexe G, Fuku N, Bilal E, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Bhanot G, Tanaka M. Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum Genet. 2007;121(3–4):347–56. doi: 10.1007/s00439-007-0330-6. [DOI] [PubMed] [Google Scholar]
- 24.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72(4):804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CF, Liu CY, Chen SM, Sikorska M, Lin CY, Chen TL, Wei YH. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann NY Acad Sci. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 26.Clayton DA, Vinograd J. Circular dimer and catenate forms of mitochondrial DNA in human leukaemic leucocytes. J Pers. 1967;35:652–657. doi: 10.1038/216652a0. [DOI] [PubMed] [Google Scholar]
- 27.Welter C, Kovacs G, Seitz G, Blin N. Alteration of mitochondrial DNA in human oncocytomas Genes Chromosome. Cancer. 1989;1:79–82. doi: 10.1002/gcc.2870010112. [DOI] [PubMed] [Google Scholar]
- 28.Selvanayagam P, Rajaraman S. Detection of mitochondrial genome depletion by a novel cDNA in renal cell carcinoma. Lab Invest. 1996;74:592–599. [PubMed] [Google Scholar]
- 29.Sangkhathat S, Chiengkriwate P, Kusafuka T, Patrapinyokul S, Fukuzawa M. Renal cell carcinoma in a pediatric patient with an inherited mitochondrial mutation. Pediatr Surg Int. 2005;21:745–748. doi: 10.1007/s00383-005-1471-0. [DOI] [PubMed] [Google Scholar]
- 30.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Koch W, Califano J. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–2491. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]
- 31.Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA. Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res. 2004;10:8512–8515. doi: 10.1158/1078-0432.CCR-04-0734. [DOI] [PubMed] [Google Scholar]
- 32.Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D. Tumor-specific changes in mtDNA content in human cancer. Int J Cancer. 2005;116:920–924. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- 33.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee C. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY. The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured microdissected tissues. Gynecol Oncol. 2005;98:104–110. doi: 10.1016/j.ygyno.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 36.Kulawiec M, Arnouk H, Desouki MM, Kazim L, Still I, Singh KK. Proteomic anlysis of proteins involved in mitochondria-to-nucleus retrograde response in human cancer cells. Cancer Biol Ther. 2006;8:967–975. doi: 10.4161/cbt.5.8.2880. [DOI] [PubMed] [Google Scholar]
- 37.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci. 2002;59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambeth JD. Oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 39.Cheng G, Cao Z, Xu X, van Meir EG, Lambeth JD. Homologs of gp91phox: cloning and tissue expression of Nox3, Nox4, and Nox5. Gene. 2001;269:131–140. doi: 10.1016/s0378-1119(01)00449-8. [DOI] [PubMed] [Google Scholar]
- 40.Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- 41.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes. Nat Genet. 2007;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]