Abstract
Computer simulations have been performed to determine how accurately and precisely structures of DNA oligomers can be generated from distance data obtained from two-dimensional NMR experiments. A hexamer fragment d(CGAATT) of the Dickerson dodecamer [Drew, H.R., Wing, R.M., Takano, T., Broka, C., Tanaha, S., Itakura, K. & Dickerson, R.E. (1981) Proc. Natl. Acad. Sci. USA 78, 2179-2183] was used as the model structure in these simulations. Protons were added to the coordinates of the original x-ray structure, which was then subjected to a regularization procedure to minimize deviations from standard bond lengths and bond angles. The proton-proton distances normally observed in NMR experiments were measured from this regularized target structure and used as input for a distance geometry algorithm. Distance geometry structures were generated from two distance sets, one with essentially exact distances (+/- 0.005 A) and one set with a precision (+/- 0.2 A) that simulates an optimal NMR experiment. The results of these calculations were used to judge how accurately and precisely the following helical parameters could be reproduced from this simulated NMR distance data: helical twist, helical rise, dislocation, roll, tilt, glycosidic angle, delta torsion angle, and pseudorotation angle. These data provide a basis from which to judge the quality of DNA structures produced from real NMR experiments.
Full text
PDF

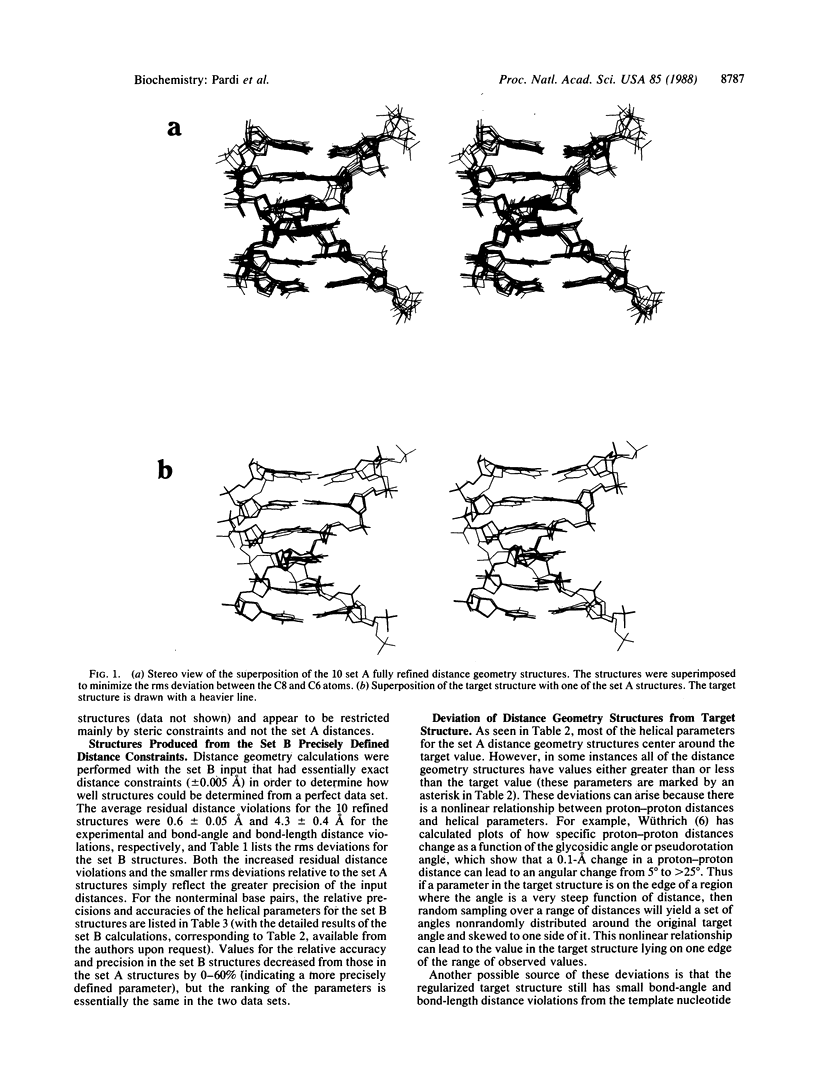
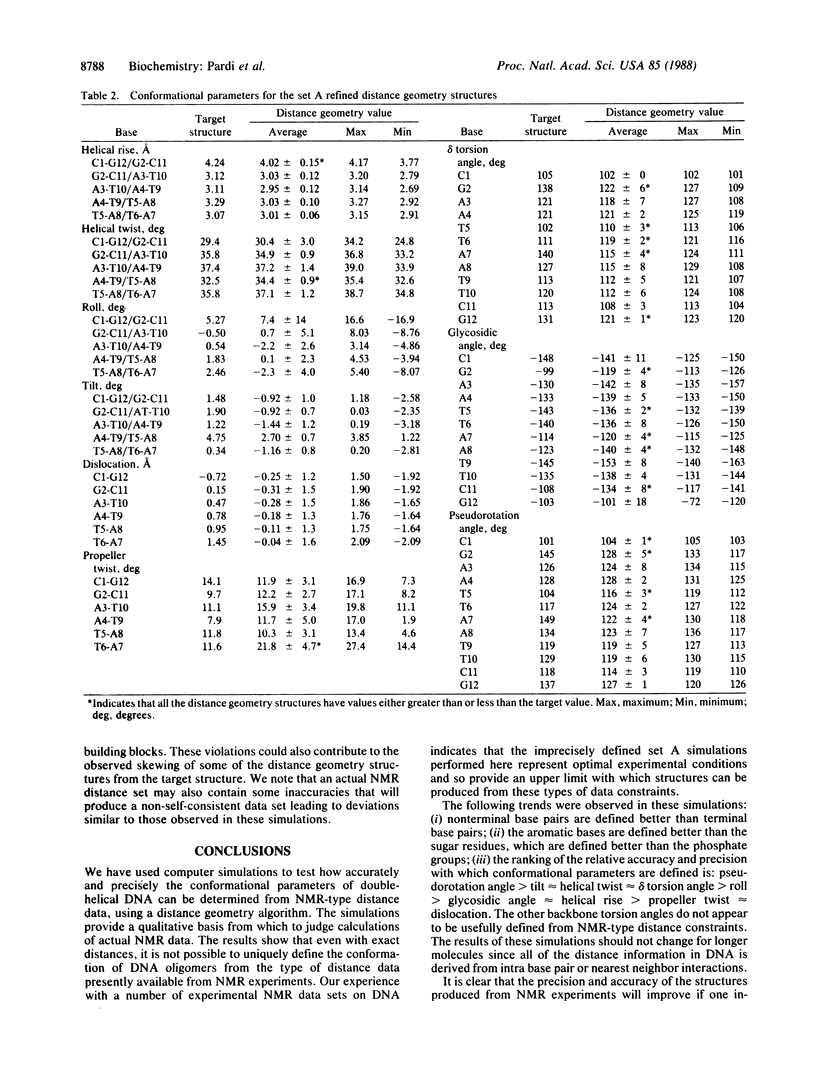
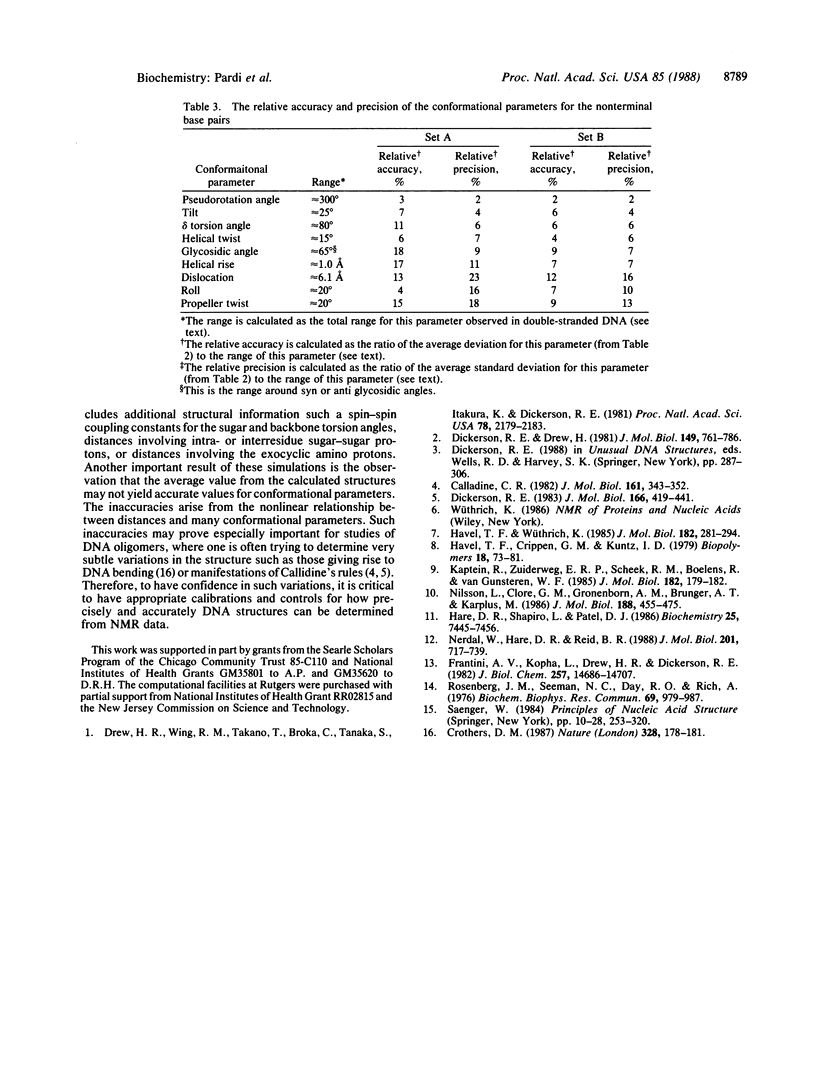
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Wobble dG X dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- Havel T. F., Wüthrich K. An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol. 1985 Mar 20;182(2):281–294. doi: 10.1016/0022-2836(85)90346-8. [DOI] [PubMed] [Google Scholar]
- Kaptein R., Zuiderweg E. R., Scheek R. M., Boelens R., van Gunsteren W. F. A protein structure from nuclear magnetic resonance data. lac repressor headpiece. J Mol Biol. 1985 Mar 5;182(1):179–182. doi: 10.1016/0022-2836(85)90036-1. [DOI] [PubMed] [Google Scholar]
- Nerdal W., Hare D. R., Reid B. R. Three-dimensional structure of the wild-type lac Pribnow promoter DNA in solution. Two-dimensional nuclear magnetic resonance studies and distance geometry calculations. J Mol Biol. 1988 Jun 20;201(4):717–739. doi: 10.1016/0022-2836(88)90469-x. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Structure refinement of oligonucleotides by molecular dynamics with nuclear Overhauser effect interproton distance restraints: application to 5' d(C-G-T-A-C-G)2. J Mol Biol. 1986 Apr 5;188(3):455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Day R. O., Rich A. RNA double helices generated from crystal structures of double helical dinucleoside phosphates. Biochem Biophys Res Commun. 1976 Apr 19;69(4):979–987. doi: 10.1016/0006-291x(76)90469-1. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]



