Abstract
Yersinia pestis, the causative agent of plague, autoaggregates within a few minutes of cessation of shaking when grown at 28°C. To identify the autoaggregation factor of Y. pestis, we performed mariner-based transposon mutagenesis. Autoaggregation-defective mutants from three different pools were identified, each with a transposon insertion at a different position within the gene encoding phosphoglucomutase (pgmA; y1258). Targeted deletion of pgmA in Y. pestis KIM5 also resulted in loss of autoaggregation. Given the previously defined role for phosphoglucomutase in antimicrobial peptide resistance in other organisms, we tested the KIM5 ΔpgmA mutant for antimicrobial peptide sensitivity. The ΔpgmA mutant displayed >1,000-fold increased sensitivity to polymyxin B compared to the parental Y. pestis strain, KIM5. This sensitivity is not due to changes in lipopolysaccharide (LPS) since the LPSs from both Y. pestis KIM5 and the ΔpgmA mutant are identical based on a comparison of their structures by mass spectrometry (MS), tandem MS, and nuclear magnetic resonance analyses. Furthermore, the ability of polymyxin B to neutralize LPS toxicity was identical for LPS purified from both KIM5 and the ΔpgmA mutant. Our results indicate that increased polymyxin B sensitivity of the ΔpgmA mutant is due to changes in surface structures other than LPS. Experiments with mice via the intravenous and intranasal routes did not demonstrate any virulence defect for the ΔpgmA mutant, nor was flea colonization or blockage affected. Our findings suggest that the activity of PgmA results in modification and/or elaboration of a surface component of Y. pestis responsible for autoaggregation and polymyxin B resistance.
The etiologic agent of plague is the Gram-negative bacterium Yersinia pestis. Plague is a deadly infectious disease that has traumatized civilizations throughout history (10, 12). Plague outbreaks still occur, and from 1989 to 2003, 876 to 5,419 cases of plague with 103 to 232 deaths were reported to the World Health Organization each year (71). Plague is usually transmitted from rats to humans by fleas. Occasionally, human-to-human transmission occurs by inhalation of infectious droplets spread by pneumonic plague cases (53, 54).
A correlation between autoaggregation and virulence has been shown for many Gram-negative bacteria such as Yersinia enterocolitica (19, 38, 44), Escherichia coli (67), Campylobacter jejuni (32), and Vibrio cholerae (14). In one study, autoaggregation was observed in about 70% of human and animal clinical isolates of Y. enterocolitica, whereas all environmental isolates tested were negative for autoaggregation (38). Thus, the autoaggregation phenotype has been used as a virulence marker for enteropathogenic Yersinia (19, 38). Enteropathogenic Yersiniae species, Y. enterocolitica and Y. pseudotuberculosis, autoaggregate via a YadA-mediated hydrophobic interaction in tissue culture medium at 37°C (60). YadA is encoded by virulence plasmid pYV (pCD1 in Y. pestis). However, YadA is disrupted by a frameshift mutation and rendered nonfunctional in Y. pestis (57, 61). Even though Y. pestis does not have a functional YadA, it autoaggregates quickly, following cessation of shaking, when grown at 28°C. Y. pestis does not autoaggregate when grown at 37°C due to expression of capsule (Caf1) at this temperature (S. Felek and E. S. Krukonis, unpublished observations).
Since autoaggregation is reported to be important for virulence in many pathogenic organisms, we hypothesized that the autoaggregation factor of Y. pestis may be an important virulence factor. In this study we demonstrated that phosphoglucomutase (PgmA in Y. pestis) is required for efficient autoaggregation in Y. pestis and plays an important role in antimicrobial peptide resistance. Phosphoglucomutase (PGM) is an enzyme that catalyzes the interconversion between glucose-6-phosphate and glucose-1-phosphate. The conversion from glucose-6-phosphate to glucose-1-phosphate initiates the pathway leading to formation of nucleotide sugars such as UDP-glucose and UDP-galactose. These nucleotide sugars can then be used to elaborate sugar modifications on cellular components such as lipopolysaccharide ([LPS] particularly the core region), teichoic acid, and secreted matrix components. Loss of PGM activity in several organisms has been reported to increase their sensitivity to antimicrobial peptides (9, 48, 66, 69), and PGM-dependent modifications of bacterial surfaces have been shown to be critical for virulence in a number of pathogenic organisms (3, 9, 39, 48, 66, 69), usually due to alterations in LPS structure. Accordingly, the studies presented here assess the role of Y. pestis KIM5 PgmA in LPS structure, antimicrobial peptide resistance, and virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Y. pestis strains were grown in heart infusion broth (HIB) or on heart infusion agar (HIA). Strains were grown at 28°C to mimic flea temperature or at 37°C to mimic mammalian temperature. E. coli strains were grown in Luria-Bertani (LB) medium or LB agar at 37°C.
The bacterial strains used in this study are listed in Table 1. The following antibiotic concentrations were used: streptomycin (Sm), 100 μg/ml; kanamycin (Km), 30 μg/ml; chloramphenicol (Cm), 10 μg/ml; ampicillin (Amp), 100 μg/ml. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a concentration of 100 μM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| AAEC185 | supE44 hsdR17 mcrA mcrB endA1 thi-1 ΔfimB-fimH ΔrecA | 4 |
| DH5α | supE44 ΔlacU169(F80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| Y. pestis | ||
| KIM5-3001 | KIM5-3001 Pgm− Strr pCD1+ pCP1+ pMT1+; referred to as KIM5 in the text | 46 |
| KIM5-3001 ΔpsaA ΔyapC Δcaf1 | ΔpsaA ΔyapC Δcaf1 | 20 |
| KIM5-3001 ΔpgmA | ΔpgmA | This study |
| KIM5-3001 ΔyapC | ΔyapC | 21 |
| KIM5-3001Δail | Δail | 20 |
| KIM5-3001Δail ΔyapC | Δail ΔyapC | This study |
| KIM5-3001 ΔpgmA ΔyapC | ΔpgmA ΔyapC Kanr | This study |
| KIM5-3001 ΔpgmA Δail | ΔpgmA Δail | This study |
| KIM5 ΔpgmA ΔyapC Δail | ΔpgmA ΔyapC Δail Kanr Cmr | This study |
| KIM5-3001 pCD1− | pCD1− | 21 |
| KIM6+ | pPCP1+ pMT1+ pCD1− | 22 |
| KIM6+ ΔpgmA | ΔpgmA | This study |
| Plasmidsa | ||
| pFD1 | Kmr transposon and Himar1 transposase on an Ampr R6K-based suicide plasmid | 58 |
| pMMB207 | IPTG-inducible expression vector; Cmr | 50 |
| pMMB207-pgmAKIM5 | pMMB207 encoding the KIM5 pgmA gene; Cmr | This study |
| pMMB207-pgmE. coli | pMMB207 encoding the E. coli DH5α pgm gene; Cmr | This study |
| pMMB208 | IPTG-inducible expression vector; inverted polylinker relative to pMMB207; Cmr | 50 |
| pMMB208-pgmA-HA | pMMB208 encoding the HA-tagged KIM5 pgmA gene; Cmr | This study |
| pMMB208-pgmA-S160A-HA | pMMB208 encoding the HA-tagged KIM5 pgmA gene with mutation S160A; Cmr | This study |
| pMMB208-pgmA-R521S-HA | pMMB208 encoding the HA-tagged KIM5 pgmA gene with mutation R521S; Cmr | This study |
| pcDNA3-HA | HA-tagging vector; Ampr | 36 |
| pKD46 | Ampr; λ-RED recombinase expression plasmid | 17 |
| pKD4 | Kmr; template plasmid | 17 |
| pCP20 | Flp recombinase expression plasmid; Ampr, Cmr | 17 |
The origin of the pgm or pgmA gene is indicated by the subscript.
Identification of autoaggregation-defective strains.
In previous mutagenesis experiments with Y. pestis KIM5, we observed spontaneous mutants expressing Caf1 (capsule) at 28°C, which can mask surface components and abolish autoaggregation of Y. pestis (Felek and Krukonis, unpublished). To exclude this possibility, we used a Δcaf1 mutant for transposon mutagenesis. It was previously shown that YapC of Y. pestis promotes autoaggregation when it is expressed in E. coli (21, 73). But deletion of yapC does not abolish autoaggregation in Y. pestis strain KIM5, suggesting that there are other factors involved in autoaggregation (21). To determine the autoaggregation factor(s) of Y. pestis, we used previously described mariner transposon-mutagenized (58) pools of the Y. pestis starting strain KIM5-3001 Δcaf1 ΔyapC ΔpsaA (20). psaA encodes the structural subunit of pH 6 antigen (Psa), a surface structure known to bind host cells but not mediate autoaggregation (46, 72). To enrich for mutants deficient in autoaggregation, mutant pools were grown in HIB overnight (ON) at 28°C with shaking and left for 2 to 3 h at room temperature to settle for autoaggregation. Five microliters of supernatant from the top of the tube was transferred to a new tube and shaken ON in HIB and again left to settle for autoaggregation. The enrichment was repeated for five consecutive days, and the enriched mutant population was plated on HIA. A total of 100 single colonies from enrichment steps were tested for autoaggregation.
DNA sequencing and analysis.
Three strains defective for autoaggregation from three different pools were sequenced to identify the transposon insertion site. Total genomic DNA was purified using a genomic DNA purification kit (Qiagen, Valencia, CA). A nested PCR using genomic DNA template and sequencing of PCR products were performed as described previously (20).
Deletion of pgmA in Y. pestis.
Genes were deleted in Y. pestis KIM5 and KIM6+ using the λ-RED system (17, 75). Briefly, the kanamycin resistance cassettes were amplified from pKD4 by PCR with primers containing 5′ extension sequences from regions flanking the pgmA (y1258) gene (pgmdf, 5′-AGTGTGCGGAACTATCTGATTTCATTGAAACCATCCTCTTTCTCAGCCATG TGTAGGCTGGAGCTGCTTC-3′; pgmdr, 5′-CGAGTGCCAAGATTACAGCGTTGGTCTTTAAGGACAGGAATATACCCGCCATATGAATATCCTCCTTAGT-3′). PCR products were purified by using a genomic DNA purification kit (Qiagen), digested with DpnI, and then transformed into Y. pestis KIM5 or KIM6+ that was previously transformed with pKD46 and induced with arabinose. PCR was performed on kanamycin-resistant colonies to confirm deletion of the pgmA gene with primers from about 200 bp upstream and downstream of the gene (pgmf, 5′-ACTTTGCCGGTGATCAACAG-3′; pgmr, 5′-TTTCCAGGCAGACTTAAACTC-3′). Finally, the strains were transformed with pCP20 to resolve kanamycin resistance cassettes. The deletion mutant was then cured of all plasmids required for λ-RED mutagenesis.
Cloning of pgmA.
The pgmA gene (including the ribosomal binding site of pgmA) was amplified from Y. pestis KIM5 by PCR (pgmcf, 5′-GCGCGAATTCTGTGCGGAACTATCTGATTTC-3′; pgmcr, 5′-GCGCCTGCAGCAGCGTTGGTCTTTAAGGAC-3′). PCR products were purified, digested with EcoRI and PstI, ligated into pMMB207 (50), digested with the same enzymes, and introduced into the pgmA mutant for complementation experiments. Recombinant pgmA clones were sequenced at the University of Michigan DNA Sequencing Core.
Cloning of E. coli pgm.
To find the pgm homologue of E. coli, we performed a PubMed BLAST search. Our search showed that the EcDH1_2949 locus of E. coli DH1 strain (GenBank accession number CP001637) is the homologue of KIM5 pgm. The phosphoglucomutase activity of this locus was demonstrated previously (47). Protein sequences of KIM5 and DH1 Pgm proteins were 82.6% identical. To clone E. coli pgm, DH5α pgm was amplified with primers pgmeccf1 (5′-GCGCGAATTCCTAAAACGTTGCAGACAAAGG) and pgmeccr1 (3′-GCGCCTGCAGAAAAAGGGCGATCTTGCGAC), cut with EcoRI and PstI, and ligated into pMMB207. The upstream primer (pgmeccf1) was designed to begin 29 bp upstream of E. coli pgm to include the ribosomal binding site. The DNA sequences of DH5α pgm and DH1 pgm were determined to be 100% identical.
Pgm-HA construction and point mutations.
To determine whether point mutants of PgmA were expressed at the same level as wild-type PgmA in KIM5, we constructed a C-terminally epitope-tagged version of each PgmA derivative in the vector pMMB208-pgmA-HA. These alleles encode PgmA from KIM5 with a hemagglutinin (HA) tag derived from the influenza virus hemagglutinin protein. First, we amplified the KIM5 pgmA gene using primers pgmcf2 (5′-GCGCAAGCTTTGTGCGGAACTATCTGATTTC-3′) and pgmcr2 (5′-GCGCCTCGAGTTTCGCCTCAGCCAGCACT-3′), cut with HindIII and XhoI, and ligated into similarly digested pcDNA3-HA (36). The pgmA-HA fragment was then liberated from pcDNA3-HA using HindIII and EcoRI and ligated into similarly digested pMMB208 (50). pgmA derivatives encoding the mutants S160A and R521S were generated by PCR mutagenesis. pMMB208-pgmA-HA was amplified by PCR using the primer pair pgmS160Af (5′-GATGGTATTGTCATTACGCCAGCTCACAACCCACCGGAAGACG-3′) and pgmS160Ar (5′-CGTCTTCCGGTGGGTTGTGAGCTGGCGTAATGACAATACCATC-3′) and the pair pgmR521Sf (5′-GAACGGCTGGTTTGCCGCTAGCCCATCAGGCACTGAAGAAGC-3′) pgmR521Sr (5′-GCTTCTTCAGTGCCTGATGGGCTAGCGGCAAACCAGCCGTTC-3′), respectively. PCR products were DpnI digested and transformed into E. coli DH5α. Purified plasmids were sequenced to confirm mutations and transformed into the KIM5 ΔpgmA strain.
Surface charge and hydrophobicity assays.
For the surface charge assay, overnight 28°C cultures in HIB were pelleted and washed two times with 20 mM (pH 7) morpholinepropanesulfonic (MOPS) acid (Fisher Scientific) buffer. Pellets were resuspended in 0.5 ml of MOPS buffer to an optical density at 620 nm (OD620) of 7.5, and cytochrome c (Sigma) was added to a final concentration of 0.1 mg/ml. Samples were incubated for 15 min at room temperature and centrifuged for 5 min at 13,000 rpm, and the OD530 values were determined in supernatants. To determine hydrophobicity, bacteria were cultured as above and washed two times with phosphate-buffered saline (PBS), and the final OD620 was adjusted to 1.0. Two milliliters of bacterial suspension and 600 μl of n-hexadecane (Sigma) were mixed, vortexed for 60 s, and allowed to stand for 30 min for phase separation at room temperature. n-Hexadecane was removed, and the OD620 values were recorded for the aqueous phase.
Phosphoglucomutase and PMM assays.
Phosphoglucomutase and phosphomannomutase (PMM) activities were determined as described previously (9). Briefly, overnight cultures of bacteria were diluted to an OD620 of 0.15 in HIB, and 100 μM IPTG was added to strains containing expression plasmids. Strains were cultured for 4 h at 28°C with shaking, and 3 ml of cultures at an OD620 of 0.6 was washed two times with 1 ml of cold PBS, resuspended in 400 μl of CelLytic B 2× cell lysis solution (Sigma), and allowed to stand for 10 min at room temperature for lysis. Supernatants were collected after 5 min of centrifugation at 13,000 rpm and frozen at −80°C. For phosphoglucomutase assays, 5 mM MgCl2, 0.4 mM NADP+, 2 U/ml glucose-6-phosphate dehydrogenase, and 50 μM α-glucose-1,6-bisphosphate were combined in 160 μl of double-distilled H2O (ddH2O) in 96-well flat-bottom microtiter plates (Costar), and 20 μl of bacterial lysate was added. Twenty microliters of lysis solution was added to blank wells. Plates were incubated at 30°C for 3 to 4 min, and 20 μl of α-glucose-1-phosphate was added to a final concentration of 1.4 mM to start the reaction, and the OD340 was recorded after 5 min at 30°C. For phosphomannomutase assays, in addition to the above reagents (except α-glucose-1-phosphate), 2 U/ml phosphomannose isomerase and 2 U/ml phosphoglucose isomerase were added, and reactions were initiated with the addition of 1.4 mM α-mannose-1-phosphate. OD340 values were recorded after 1 h at 30°C. All reagents were obtained from Sigma.
Autoaggregation assay.
Y. pestis KIM5 and derivative strains were shaken overnight in HIB at 28°C, and 2 ml of each culture was transferred into test tubes at an OD620 of ∼1.5 and left to stand at room temperature for autoaggregation. The OD620 was recorded at different time points. Chloramphenicol and 100 μM IPTG were added to pMMB207-containing strains to induce pgmA expression. In this experiment, if the bacteria autoaggregate, they settle, and the supernatant clears, leading to an OD620 decrease. To visualize autoaggregation under the microscope, 50 μl of an overnight culture was transferred to a glass slide with coverslip and left at room temperature in a humidified chamber for 45 min. Then, photographs were obtained.
Antimicrobial peptide susceptibility test.
Strains were cultured overnight in HIB. Serial dilutions of polymyxin B (Sigma) were prepared in HIB in 96-well flat-bottom culture plates, and 105 bacteria pregrown at 28°C or 37°C were added to each well. Chloramphenicol and 100 μM IPTG were added to pMMB207-containing strains in all steps. Plates were incubated at 28°C or 37°C ON with shaking. CaCl2 (2.5 mM) was added to HIB for the culture at 37°C. The MIC was determined by assessing growth inhibition in a microtiter plate. After overnight culture in HIB, the Y. pestis minimal medium PMH2 (27, 62) was used for assessment of susceptibility to human antimicrobial peptides. Other steps were similar to those described above. Human α-defensin-1 and -3 and β-defensin-1 and -2 were obtained from Peptides International Louisville, KY. LL-37 was obtained from Panatecs GmbH, Tübingen, Germany.
Intracellular survival assay.
RAW264.7 macrophages were grown in 24-well plates in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) to ∼60% confluence. Bacteria were grown overnight at 28°C in HIB, pelleted, and resuspended in DMEM at an OD620 of 0.006 (0.6 followed by a 1:100 dilution). RAW cells were washed twice with DMEM (no serum), and 400 μl of DMEM (no serum)/well was added. Bacteria (100 μl) were added to each well. Infections were incubated at 37°C in 5% CO2 for 1 h. Gentamicin was added to each well to a final concentration of 7.5 μg/ml for Y. pestis or 20 μg/ml for E. coli, and cells were incubated for an additional hour at 37°C in 5% CO2. At this 2-h time point, triplicate wells were washed twice with PBS and lysed with 100 μl of 0.1% Triton X-100 for 10 min at room temperature with shaking. Lysates were resuspended in additional 500 μl of PBS, and serial dilutions were plated to enumerate surviving bacteria by CFU analysis. For cultures allowed to be infected for 4, 6, or 24 h, triplicate wells were washed at hour 2, DMEM supplemented with 5% FBS and 2 μg/ml gentamicin for Y. pestis or 5 μg/ml for E. coli was added, and bacteria were allowed to grow or be killed within macrophages for an additional 2, 4, or 22 h.
LPS isolation and structural analysis.
Phenol-killed cells from both Y. pestis KIM5 and the ΔpgmA mutant grown at 28°C were washed first with PBS buffer, then three times with 95% (vol/vol) ethanol, and finally two times with acetone prior to LPS extraction. LPS was extracted using the phenol-chloroform-petroleum ether (PCP) procedure (25). Crude LPS was further purified using RNase, DNase, and proteinase K (Sigma); dialyzed against deionized water (1,000-molecular-weight-cutoff [MWCO] dialysis tubing); and ultracentrifuged at 100,000 × g for 6 h. An electrophoretic profile of LPS was performed using deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) with 18% resolving and 4% stacking gels, respectively. LPS samples were developed with silver using a Bio-Rad Silver Stain kit (Bio-Rad, Hercules, CA) (43).
To identify the glycosyl residues as well as the fatty acid profile of the purified intact LPS, composition analysis was performed by the preparation of trimethylsilyl methyl esters (TMS) as described by York et al. (74). The TMS derivatives were analyzed on a Hewlett-Packard HP5890 gas chromatograph equipped with mass selective detector 5970 MSD using an Alltech AT-1 fused silica capillary column (30 m with a 0.25-mm internal diameter). Helium was used as the carrier gas. The initial oven temperature was 80°C for 2 min, which was then ramped to 160°C at 20°C/min with a 2-min hold and then to 200°C at 2°C/min and to 250°C at 10°C/min with an 11-min hold.
To determine whether there is any difference in the noncarbohydrate components of LPS from KIM5 and the ΔpgmA mutant, lipid A was released from LPS by mild hydrolysis (10 mM sodium, pH 4.5; 100°C in the presence of 1% SDS) (13). The resulting lipid A samples were dissolved in chloroform-methanol (3:1, vol/vol), mixed 1:1 by volume with 0.5 M 2,4,6-trihydroxyacetophenone monohydrate (THAP) matrix in methanol, spotted onto a matrix-assisted laser desorption ionization (MALDI) plate, and analyzed by MALDI-time of flight mass spectrometry (MALDI-TOF MS) (Applied Biosystems 4700 Proteomics System Spectrometer). Spectra were obtained in the negative reflector ion mode.
Core oligosaccharide (OS) was released from LPS with 1% acetic acid (HOAc) at 100°C for 1.5 h and purified from lipid A precipitate by extensive centrifugation and further by Biogel P4 gel permeation chromatography. The eluting fractions were monitored with a refractive index detector. Core OSs were chemically characterized by the alditol acetate method (74) with Xyl used as an internal standard. Structural comparison of OSs was performed using electrospray ionization (ESI)-MS and proton nuclear magnetic resonance (NMR) spectroscopy experiments. ESI-MS was performed on an ion-trap LCQ-MS instrument (Thermo-Finnigan), using helium as the buffering and target gas. Spectra were acquired using collision energy in the negative positive ion polarity mode. NMR analysis was done by exchanging the sample two times with 99.9% deuterium oxide and finally dissolving it in 100% D2O (Cambridge Isotope Laboratories, Inc.). Samples were then transferred to 3-mm Shigemi tubes, and one-dimensional (1-D) proton spectra were recorded at 25°C using a Varian Inova 600 MHz NMR spectrometer (Varian, Palo Alto, CA).
NO assay.
To analyze whether there is any difference in binding and neutralization of polymyxin B to LPS from KIM5 and the ΔpgmA mutant, we mixed each LPS with 10-fold increasing concentrations of polymyxin B and incubated the mixture for 30 min at 37°C. Approximately 1 × 105 RAW264.7 murine macrophage cells were mixed with LPS and LPS-polymyxin B mixtures in 24-well tissue culture plates and incubated for 20 h at 37°C in 5% CO2. To analyze nitric oxide (NO) production by murine macrophages, NO2− was measured in tissue culture medium supernatants by a Griess reaction assay (40). Briefly, 100 μl of culture supernatant was mixed with 100 μl of Griess reagent in a 96-well flat-bottom plate and incubated for 10 min at room temperature. The OD540 was read by a plate reader.
Mouse experiments.
Female Swiss Webster mice, 6 to 8 weeks old, were obtained from Harlan Sprague-Dawley, Indianapolis, IN. Mice were infected intravenously (i.v.) through the tail vein using 5-fold increasing concentrations of Y. pestis KIM5 and ΔpgmA mutant to determine the median lethal dose (LD50). Three different doses were tested with 10 mice for each dose. Survival was noted daily for 16 days. As a control group, 5 mice were infected with 1 × 106 CFU of Y. pestis KIM5 pCD1−, a strain that lacks the Yop-encoding virulence plasmid. Our preliminary studies indicated that the LD50 for intranasal (i.n.) inoculation for Y. pestis KIM5 is 350,000. Twenty mice for KIM5 and the ΔpgmA mutant and 5 mice for KIM5 pCD1− were inoculated i.n. with one LD50 dose to determine any attenuation for the i.n. route of infection. Mice were observed for 16 days for survival.
Flea infections.
Xenopsylla cheopis fleas were fed with blood containing about 108 KIM6+ or the KIM6+ ΔpgmA mutant bacteria per milliliter, using a previously described artificial feeding system (34, 35). Equal numbers of male and female fleas that took a blood meal (n = 106 to 112) were maintained at 21°C and 75% relative humidity, fed twice weekly on uninfected mice for 4 weeks, and monitored for proventricular blockage, as previously described (34, 35, 37). The infection rate was determined by CFU count of the bacterial load in samples of 20 individual females collected immediately after the infectious blood meal and at 28 days postinfection (34, 35).
RESULTS
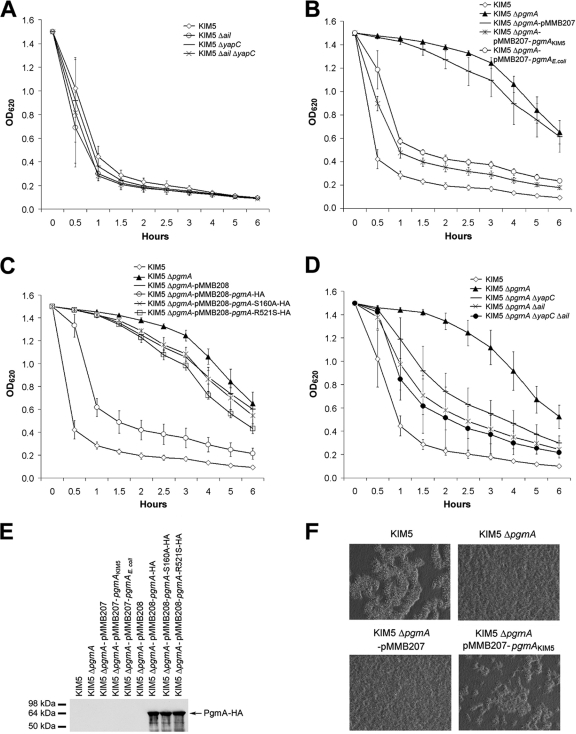
Previous studies demonstrated that two outer membrane proteins of Y. pestis, YapC (21) and Ail (42), can mediate autoaggregation. We first tested single and double deletion mutants of yapC and ail for autoaggregation. None of these strains showed a defect in autoaggregation (Fig. 1A). To determine the gene required for autoaggregation at 28°C in Y. pestis, we used previously described mariner transposon mutant pools of a Y. pestis KIM5-3001 Δcaf1 ΔyapC ΔpsaA strain (20). Six strains from three different pools that aggregate at a significantly slower rate than the parent strain were identified and analyzed. Sequencing and diagnostic PCR results showed that all transposon insertions were in the gene encoding phosphoglucomutase (pgm, or y1258) at different positions. To avoid confusion with the well-studied pigmentation locus of Y. pestis (also called pgm), we propose to name the Y. pestis phosphoglucomutase gene pgmA. A reconstructed pgmA mutant in Y. pestis KIM5 exhibited delayed autoaggregation like that observed in the transposon insertion mutants. After a 1-h static incubation at room temperature (following overnight shaking at 28°C), 70% of KIM5 cells autoaggregated (settled out of solution), whereas only 4% of the ΔpgmA mutant cells autoaggregated (Fig. 1B). Delayed autoaggregation was complemented by expression of Y. pestis pgmA or the E. coli pgm homolog (locus EcDH1_2949 annotated in strain DH1) from plasmid pMMB207 (Fig. 1B and 2A). Complementation was dependent upon the activity of the protein encoded by Y. pestis pgmA since two different mutations, shown previously to disrupt the active site of Pgm enzymes (51, 56, 79), eliminated the ability of PgmA to restore autoaggregation (Fig. 1C). Both HA-tagged mutant proteins were expressed to levels similar to wild-type HA-tagged PgmA (Fig. 1E). To test whether double or triple deletions of pgmA, ail, and yapC may lead to a complete inhibition of autoaggregation, we constructed KIM5 ΔpgmA ΔyapC, KIM5 ΔpgmA Δail, and KIM5 ΔpgmA ΔyapC Δail mutants. None of these strains showed any further delayed autoaggregation compared to Y. pestis KIM5 ΔpgmA (Fig. 1D). In fact, deletion of yapC and/or ail appears to restore some level of autoaggregation to the KIM5 ΔpgmA derivative by an unknown mechanism (Fig. 1D).
FIG. 1.
PgmA is required for Y. pestis autoaggregation. (A to D) Overnight cultures grown at 28°C were transferred to a test tube and allowed to sit, without shaking, for autoaggregation at room temperature. The OD620 values of the tubes were recorded at the indicated time points. The results are from duplicate assays of three independent experiments (n = 6). (E) Expression levels of PgmA-HA or PgmA-HA catalytic mutants (S160A and R521S) were determined by Western blotting using an anti-HA tag antibody directed against the C terminus of the hybrid protein. (F) Autoaggregation of identical numbers of Y. pestis KIM5 and the ΔpgmA mutant after a 45-min incubation at room temperature. Defects in autoaggregation were restored upon complementation with plasmid-encoded PgmA.
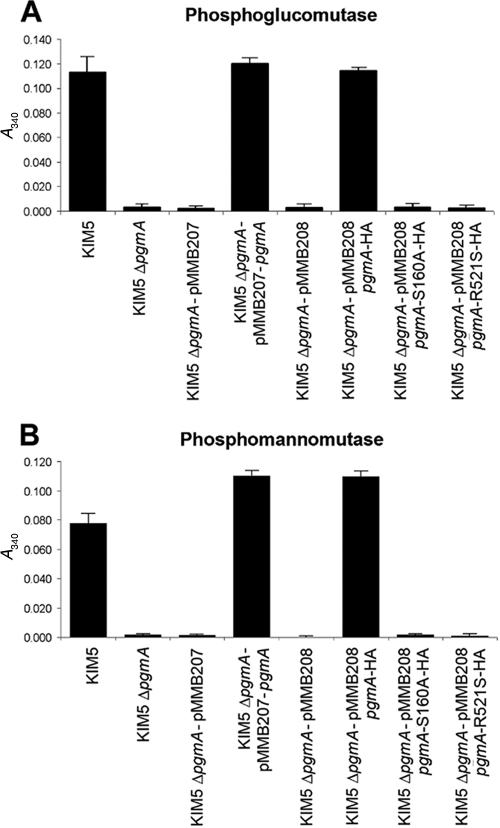
FIG. 2.
PGM and PMM activities of the KIM5 ΔpgmA mutant. PGM assays were recorded 5 min after addition of α-glucose-1-phosphate as the assay reached saturation. PMM assays were recorded at 60 min after the addition of α-mannose-1-phosphate when the reaction had reached saturation. The ΔpgmA mutant was complemented with PgmA expressed from plasmid pMMB207 or HA-tagged wild-type PgmA or catalytic mutants of PgmA expressed from the plasmid pMMB208.
To observe the architecture of Y. pestis autoaggregation, we examined bacteria under the light microscope. Y. pestis KIM5 showed large aggregates after 45 min of incubation at room temperature. The pgmA mutant showed very few aggregates at this time point, and the defect for autoaggregation could be restored when pgmA was expressed by an inducible plasmid (Fig. 1F).
To confirm the role of PgmA in phosphoglucomutase activity, the KIM5 ΔpgmA mutant and complemented strains were tested in a standard phosphoglucomutase assay. PgmA was required for phosphoglucomutase activity in Y. pestis, and the active-site residues, S160 and R521, were required for this activity (Fig. 2A). PgmA of Y. pestis was not anticipated to have strong phosphomannomutase (PMM) activity based on the lack of a highly conserved GEMS(G/A) motif in enzymes containing both PGM and PMM activities (56). Analysis of the KIM5 ΔpgmA strain for PMM activity showed that under the conditions tested, it was the only PMM enzyme detected (Fig. 2B). However, completion of the PMM assay took 60 min, whereas the PGM assay was complete within 3 to 5 min. These data demonstrate that PgmA of Y. pestis is a phosphoglucomutase with some phosphomannomutase activity as well. E. coli phosphoglucomutase, having strong PGM activity and weak PMM activity (18; also data not shown) is also able to restore Y. pestis autoaggregation (Fig. 1B). Thus, we feel that it is likely that the PGM activity of Y. pestis PgmA plays a role in autoaggregation.
Effects of the ΔpgmA mutation on cell surface characteristics.
pgm mutations in a number of bacteria have been shown to affect surface characteristics (9, 47). The effects of the KIM5 ΔpgmA mutation on surface charge and hydrophobicity, two aspects that may contribute to autoaggregation, were determined. Binding to the positively charged protein, cytochrome c, was assessed to monitor surface charge. Whole KIM5 cells bound 69% ± 3% of the cytochrome c added, while the ΔpgmA mutant bound 67% ± 3%, indicating that there was no obvious charge difference on the cell surface. Partitioning of cells into the n-hexadecane phase of a PBS-n-hexadecane solution is an indication of cell surface hydrophobicity. A total of 28% ± 5% of KIM5 cells partitioned to the n-hexadecane phase after mixing, while 30% ± 6% of the ΔpgmA mutant partitioned to the n-hexadecane phase. Thus, cell surface hydrophobicity was also not affected by the ΔpgmA mutation.
Antimicrobial peptide resistance.
Deletion of the gene encoding phosphoglucomutase has been shown to result in modestly increased susceptibility of various bacteria to antimicrobial peptides (9, 66). To determine whether PgmA plays a role in Y. pestis resistance to antimicrobial compounds, we tested our KIM5 ΔpgmA mutant in the presence of various antimicrobial peptides. The MIC for polymyxin B was 128 μg/ml for Y. pestis KIM5 and 0.031 μg/ml for the ΔpgmA mutant when bacteria were grown at 28°C (Table 2). Thus, the ΔpgmA mutant has a >1,000-fold decrease in MIC of polymyxin B. Resistance to polymyxin B could be restored by expression of pgmA from pMMB207-pgmA or pMMB208-pgmA-HA but not by PgmA-HA derivatives harboring point mutations in their active sites (PgmA-S160A and PgmA-R521S) (Table 2). When Y. pestis was grown at 37°C, the MIC of polymyxin B for KIM5 was 1 μg/ml while for the ΔpgmA mutant it was 0.5 μg/ml. Thus, the increased resistance of KIM5 to polymyxin B relative to the ΔpgmA mutant is temperature dependent, and bacteria grown at 28°C are much more resistant to polymyxin B than those grown at 37°C. We also tested the Δail ΔpgmA and ΔyapC ΔpgmA double mutants and a Δail ΔyapC ΔpgmA triple mutant for polymyxin B sensitivity. While deletion of ail and/or yapC in combination with the ΔpgmA mutation resulted in partial restoration of autoaggregation (Fig. 1D), this restoration did not provide increased polymyxin B resistance (Table 2).
TABLE 2.
Polymyxin B resistance of Y. pestis KIM5 and various deletion mutants
| Straina | Polymyxin B MIC (μg/ml) at:b |
|
|---|---|---|
| 28°C | 37°C | |
| KIM5 | 128 | 1 |
| KIM5 ΔpgmA | 0.031 | 0.5 |
| KIM5 ΔpgmA/pMMB207 | 0.031 | 0.5 |
| KIM5 ΔpgmA/pMMB207-pgmAKIM5 | 64 | 1 |
| KIM5 ΔpgmA/pMMB207-pgmE. coli | 64 | |
| KIM5 ΔpgmA/pMMB208 | 0.031 | |
| KIM5 ΔpgmA/pMMB208-pgmA-HA | 64 | |
| KIM5 ΔpgmA/pMMB208-pgmA-S160A-HA | 0.12 | |
| KIM5 ΔpgmA/pMMB208-pgmA-R521S-HA | 0.063 | |
| KIM5 Δail | 64 | |
| KIM5 ΔyapC | 128 | |
| KIM5 ΔpgmA ΔyapC | 0.031 | |
| KIM5 ΔpgmA Δail | 0.0039 | |
| KIM5 Δail ΔyapC | 64 | |
| KIM5 ΔpgmA ΔyapC Δail | 0.0039 | |
The origin of the pgm or pgmA gene is indicated by the subscript.
Results are based on three independent experiments that gave identical MICs.
We also tested our strains for sensitivity to the human antimicrobial peptides α-defensin-1, α-defensin-3, β-defensin-1, β-defensin-2, and LL-37 (a derivative of cathelicidin [30]). The MICs of all five antimicrobial peptides were the same for KIM5 and the ΔpgmA mutant at both 28°C and 37°C (Table 3). Kocuria rhizophila (ATCC 9341), an organism commonly used as a reference strain for antimicrobial susceptibility tests (65), was sensitive to antimicrobial peptides (Table 3), confirming the activity of the compounds used.
TABLE 3.
Resistance of Y. pestis KIM5 and KIM5 ΔpgmA to various human antimicrobial peptides
| Strain (temp) | MIC (μg/ml) of:a |
||||
|---|---|---|---|---|---|
| α-Defensin-1 | α-Defensin-3 | β-Defensin-1 | β-Defensin-2 | LL-37 | |
| KIM5 (28°C) | >64 | >64 | >64 | >64 | 32 |
| KIM5 ΔpgmA (28°C) | >64 | >64 | >64 | >64 | 32 |
| K. rhizophila ATCC 9341 (28°C) | 32 | 64 | >64 | 32 | 4 |
| KIM5 (37°C) | >64 | >64 | >64 | >64 | >64 |
| KIM5 ΔpgmA (37°C) | >64 | >64 | >64 | >64 | >64 |
Results are based on two independent experiments that gave identical MICs.
LPS structural analysis.
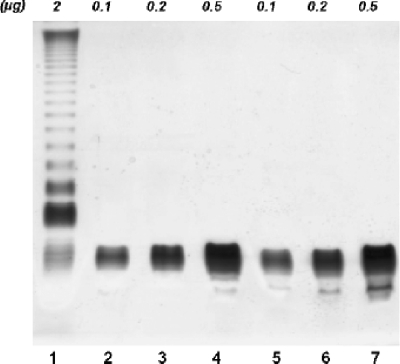
The phosphoglucomutase pathway of other bacterial species has been shown to affect LPS core structure (15, 69), and polymyxin B is known to bind the LPS of Gram-negative bacteria (2, 76). Given the increased polymyxin B sensitivity of the KIM5 ΔpgmA mutant, we hypothesized that the LPS structure might be altered in this strain. LPS compositional analysis was performed on samples extracted from cultures of Y. pestis KIM5 and the KIM5 ΔpgmA mutant grown at 28°C. DOC-PAGE analysis of both LPS preparations showed only low-molecular-weight LPS, indicating an oligosaccharide, rather than a polysaccharide, attached to lipid A (Fig. 3). Y. pestis is known to lack O antigen on its LPS due to inactivation of O-antigen gene clusters (59). LPSs from both strains contained a minor species that migrated more rapidly on the gel (Fig. 3), likely representing cleaved lipid A.
FIG. 3.
DOC-PAGE analysis of purified Y. pestis LPS samples by the phenol-chloroform extraction method. For each strain 0.1, 0.2, and 0.5 μg of LPS were loaded. Lanes 2 to 4, KIM5; lanes 5 to 7; KIM5 ΔpgmA. In lane 1, 2 μg of LPS from Salmonella enterica serovar Minnesota served as a control for an O-antigen-containing LPS molecule.
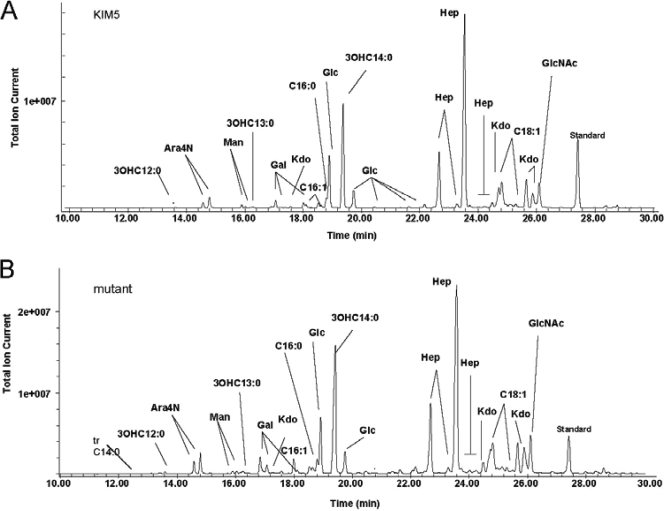
Composition analysis of the intact LPS samples from each strain showed that the glycosyl and fatty acyl components of KIM5 and the KIM5 ΔpgmA mutant LPSs were very similar to one another (Table 4 and Fig. 4). Both LPS molecules contained ld-Hep, Glc, and Kdo as major glycosyl residues, with smaller amounts of GlcNAc, Gal, and dd-Hep. This indicates that structural differences between these two LPS molecules may reside in noncarbohydrate components not detected by the methods used.
TABLE 4.
The glycosyl residue and fatty acid composition of the LPS isolated from Y. pestis KIM5 and its ΔpgmA mutant
| LPS composition | Relative peak area percentage for the indicated strain |
|
|---|---|---|
| KIM5 | KIM5 ΔpgmA | |
| Glycosyl residues | ||
| Ara4N | 3 | 4 |
| Man | 1 | 0 |
| Gal | 3 | 4 |
| Glc | 15 | 12 |
| GlcNAc | 5 | 7 |
| Kdo | 16 | 19 |
| Hepa | 57 | 53 |
| Fatty acidsb | ||
| 3OHC12:0 | 1 | 1 |
| 3OHC13:0 | 1 | 1 |
| C16:1 | 3 | 6 |
| C16:0 | 5 | 7 |
| 3OHC14:0 | 68 | 71 |
| C18:1c | 23 | 14 |
Separate analysis of the glycosyl components by alditol acetates revealed that Hep was present as l,d-Hep and dd-Hep in a 3:1 ratio.
The lipid A samples contained trace amounts of C12:0, a known component of Y. pestis lipid A.
The molecular ions obtained by MALDI-TOF MS analysis are consistent with the published Y. pestis lipid A structures which do not contain C18:1, and, therefore, it is likely that the presence of C18:1 is due to contaminating phospholipids.
FIG. 4.
Composition analysis of the LPS from Y. pestis KIM5 (A) and a Y. pestis ΔpgmA mutant (B). The analysis was performed by preparation and gas chromatography (GC)-MS analysis of trimethylsilyl methyl glycosides. The analysis reveals the glycosyl residues as well as the fatty acids as fatty acid methyl esters. The quantification values for each residue are shown in Table 4.
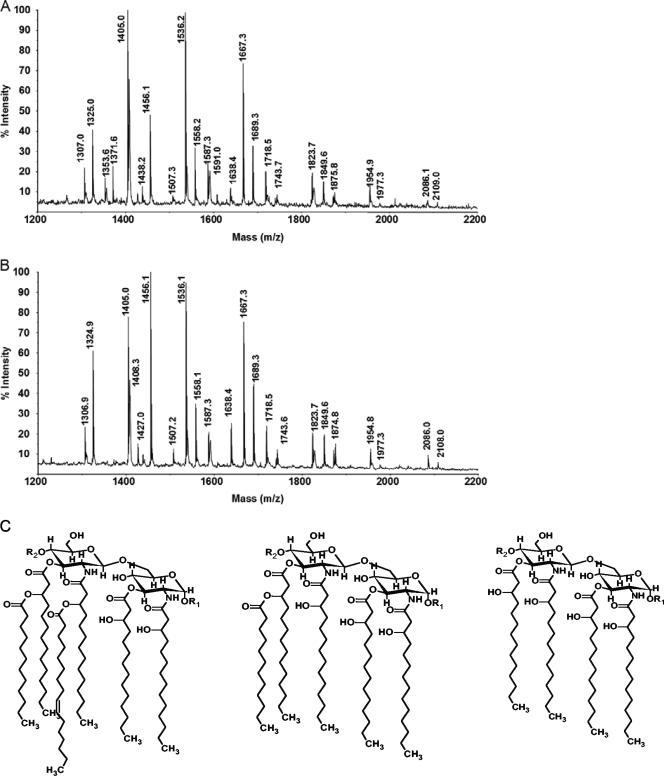
To determine whether there is any difference in the noncarbohydrate components of the KIM5 and KIM5 ΔpgmA LPS molecules, lipid A was released from each LPS by very mild acid hydrolysis in the presence of SDS (13), and each lipid A was analyzed by MALDI-TOF MS analysis. The mass spectra for these lipid A preparations are almost identical to one another, indicating that the structures of these two lipid A moieties are the same (Fig. 5 and Table 5). Each lipid A contains hexa-acylated, penta-acylated, and tetra-acylated species which contain a bis-phosphorylated glucosamine disaccharide that has zero, one, or two 4-aminoarabinosyl residues (Fig. 5 and Table 5). The tetra-acylated species give the most intense ions, consistent with the composition analysis showing that the acyloxylacyl fatty acids, C12:0 and C16:1, are present in trace or small amounts, respectively (Fig. 5 and Table 5).
FIG. 5.
The MALDI-TOF mass spectra of the lipid A from Y. pestis KIM5 (A) and its ΔpgmA mutant (B). Both spectra were acquired in the negative reflector mode, and observed ions are (M-H)− deprotonated and monosodiated forms of lipid A (Table 5 gives details). Representative structures of the three identified lipid A molecules are provided: hexa-acylated, penta-acylated, and tetra-acylated (C).
TABLE 5.
Observed and calculated ions, (M-H)−, for the lipid A of Y. pestis KIM5a
| Observed ion (m/z) | Calculated ion (m/z) | Proposed compositionc |
|---|---|---|
| 1324.9 | 1,324.7 | P1GlcN2βOHC14:0(4) |
| 1405.1 | 1,404.7 | P2GlcN2βOHC14:0(4) |
| 1456.2 | 1,455.9 | P1AraN1GlcN2βOHC14:0(4) |
| 1536.3 (1,558.3)b | 1,535.8 | P2AraN1GlcN2βOHC14:0(4) (Na) |
| 1667.5 (1,669.6)b | 1,667.0 | P2AraN2GlcN2βOHC14:0(4) (Na) |
| 1718.8 | 1,718.1 | P2AraN1GlcN2βOHC14:0(4)C12:0(1) |
| 1743.9 | 1,743.4 | P1GlcN2βOHC14:0(4)C12:0(1)C16:1(1) |
| 1824.0 | 1,823.4 | P2GlcN2βOHC14:0(4)C12:0(1)C16:1(1) |
| 1850.0 | 1,849.3 | P2AraN2GlcN2βOHC14:0(4)C12:0(1) |
| 1876.3 | 1,874.6 | P1AraN1GlcN2βOHC14:0(4)C12:0(1)C16:1(1) |
| 1955.5 | 1,954.3 | P2AraN1GlcN2βOHC14:0(4)C12:0(1)C16:1(1) |
| 2086.6 (2,109.6)b | 2,085.7 | P2AraN2GlcN2βOHC14:0(4)C12:0(1)C16:1(1) (Na) |
The lipid A from the pgmA mutant of Y. pestis KIM5 gave the same ions. The most intense ions are indicated in bold.
Occasionally the sodiated form of the lipid A was observed for some ions (shown in parentheses).
Subscript numbers in parentheses indicate the number of carbon chains with the indicated composition.
Since no difference was detected between the KIM5 and KIM5 ΔpgmA LPS molecules with the first two methods, each intact LPS molecule was further analyzed by MALDI-TOF MS analysis in the linear negative mode. The spectra were very similar to one another. Each LPS shows two higher-mass ion clusters. One ion cluster is in the m/z 3,200 to 3,350 range, and the second is in the m/z 3,100 to 3,200 range (data not shown). These ion ranges are similar to those reported for Y. pestis LPS by Knirel et al. (41). The ion cluster at m/z 3,100 to 3,200 likely differs from the cluster at m/z 3,200 to 3,350 due to molecules that lack a 4-aminoarabinose (Ara4N) residue (a difference of 131 mass units). In addition, the ions for the major lipid A forms at m/z 1,540 and 1,406 are also observed.
Each LPS was de-O-acylated by treatment with anhydrous hydrazine at 37°C for 50 min, and the de-O-acylated LPS was then subjected to MALDI-TOF MS analysis. The spectra for the two de-O-acylated LPS species are very similar to one another, and each spectrum shows two clusters of ions; e.g., m/z 2,690/2,705 and m/z 2,821/2,837. The approximately 131-mass unit difference between these ion clusters is, again, due to an Ara4N residue. De-O-acylation likely removed two ester-linked β-hydroxymyristic acid residues and any acyloxylacyl fatty acid residues from each intact LPS (data not shown).
The oligosaccharide from each LPS was released by mild acid hydrolysis in the presence of SDS (13). Each oligosaccharide preparation was then analyzed by MALDI-TOF. The spectra are very similar to each other. Each spectrum shows (M-H)− ions at m/z 1,371.7, 1,342.7, and 1,168.3 (data not shown). The ion at m/z 1,353.7 is due to the loss of water from 1,371.7. Oligosaccharides from Y. pestis with these masses have been previously reported by Knirel et al. (41). Proton NMR analysis demonstrated that the KIM5 and KIM5 Δpgm mutant oligosaccharides had identical spectra (data not shown).
Binding of LPS to polymyxin B.
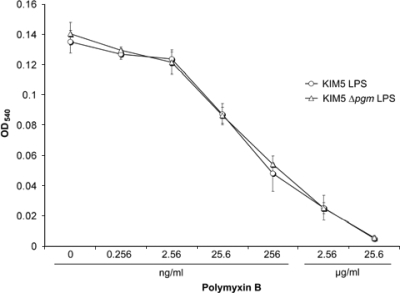
Based on tandem MS (MS/MS) and NMR structural analyses, there appears to be little or no difference between the LPS from Y. pestis KIM5 and the LPS from the KIM5 ΔpgmA mutant. To determine whether binding affinities of the two LPS molecules to polymyxin B differ, we assessed the ability of polymyxin B to neutralize LPS-mediated toxicity using the murine macrophage cell line RAW264.7. Upon interaction with LPS, macrophages release NO− as an indication of LPS stimulation. This stimulation is known to be blocked by polymyxin B (64). Purified LPS from KIM5 and the ΔpgmA mutant was mixed with 10-fold increasing concentrations of polymyxin B and incubated for 30 min at 37°C, and NO2− production (a by-product of nitric oxide) was determined after 20 h of incubation with RAW264.7 cells. Our results demonstrate that neutralizing concentrations of polymyxin B for the two LPS molecules were the same. The average OD540 for control wells (PBS and no LPS) was 0.005 ± 0.002. Endotoxic activity of 200 ng/ml LPS was completely neutralized by 25.6 μg/ml polymyxin B, and 50% neutralization was achieved with ∼25.6 ng/ml polymyxin B for both LPS preparations (Fig. 6). These results indicate that sensitivity to polymyxin B is not due to differential binding to LPS of KIM5 compared to KIM5 ΔpgmA.
FIG. 6.
Neutralization of LPS toxicity by polymyxin B. RAW264.7 murine macrophages were incubated with 200 ng/ml LPS and 10-fold increasing concentrations of polymyxin B. NO2− was detected in culture supernatants by a Griess reaction assay after 20 h of incubation. Each point represents averages and standard deviations of two independent experiments, with triplicate assays in each experiment.
Mouse virulence experiments.
To determine whether the ΔpgmA mutant is attenuated for virulence, we tested our strains with i.v. and i.n. infection models in mice. The LD50 was 4 bacteria for i.v. infection of both Y. pestis KIM5 and KIM5 ΔpgmA (10 mice/dose group). All five mice infected with 106 organisms of a KIM5 pCD1-negative (pCD1−) strain (negative control, lacking the Yop encoding plasmid) survived. For the i.n. infection model, we inoculated 20 mice each with KIM5 and the ΔpgmA mutant and 5 mice with KIM5 pCD1− at one LD50 dose (5 × 105 bacteria). While 17 out of 20 mice survived in the KIM5 group, 13 out of 20 mice survived in the ΔpgmA mutant group. All 5 mice survived in the KIM5 pCD1− group. Our mouse experiments demonstrate that pgmA is not required for i.v. or i.n. infection in mice in the KIM5 strain background.
Flea blockage.
Previous studies demonstrated that biofilms block the proventriculus of the flea, and this is an important factor in the transmission of bubonic plague (35). Since we determined an autoaggregation defect and increased polymyxin B sensitivity for the KIM5 ΔpgmA mutant when cells were grown at 28°C, we speculated that PgmA might play a role in Y. pestis survival in fleas or in flea blockage. Since Y. pestis KIM5 lacks the large pgm (pigmentation) locus, which is required for biofilm formation (16, 35, 63), we used the Y. pestis KIM6+ strain for flea experiments. This strain has the large pgm locus but lacks the Yop-encoding virulence plasmid pCD1 (this plasmid is not required for flea colonization or blockage of the proventriculus). A KIM6+ ΔpgmA mutant has a similar autoaggregation defect as the KIM5 ΔpgmA mutant (data not shown). For flea blockage experiments, equal numbers of male and female fleas were infected and monitored for proventricular blockage. The blockage rate for Y. pestis KIM6+ was 21%, and for KIM6+ ΔpgmA it was 18% (Table 6). These rates were within the normal expected range. Flea samples were also collected for each group at 0 and 4 weeks after an infectious blood meal to determine bacterial loads by counting CFU. Groups infected with either KIM6+ or the KIM6+ ΔpgmA mutant showed similar average CFU counts per flea at 0 and 4 weeks (Table 6). Our results indicate that pgmA is not required for flea infection or long-term colonization.
TABLE 6.
Flea colonization and persistence assay
| Strain | Postinfection blockage rate (%) | Flea colonization ata: |
|||
|---|---|---|---|---|---|
|
t = 0 |
t = 4 |
||||
| % Infected | Avg no. of CFU/flea | % Infected | Avg no. of CFU/flea | ||
| KIM6+ | 21 | 100 | 6.0 × 103 | 75 | 5.6 × 105 |
| KIM6 ΔpgmA | 18 | 100 | 4.1 × 104 | 80 | 5.2 × 105 |
t, time of infection.
DISCUSSION
The aim of this study was to identify Y. pestis factors required for autoaggregation and assess the effect of autoaggregation on virulence. Previous studies have demonstrated that, when expressed in E. coli, the autotransporter YapC of Y. pestis causes autoaggregation (21, 73). However, deletion of yapC in Y. pestis strain KIM5 does not lead to loss of autoaggregation, suggesting that other factor(s) play a role in autoaggregation (21). Ail in Y. pestis strain KIM6+ has also been reported to facilitate autoaggregation (42). However, we did not observe an autoaggregation defect upon deletion of ail in Y. pestis KIM5 (20). This difference may be due to the use of different KIM strains in these studies. In this report we demonstrated that phosphoglucomutase (encoded by pgmA) is required for autoaggregation of Y. pestis KIM5 (Fig. 1). To assess the possibility that, in addition to phosphoglucomutase (PgmA), YapC and Ail may also contribute to autoaggregation, we constructed KIM5 ΔpgmA Δail, KIM5 ΔpgmA ΔyapC, and KIM5 ΔpgmA Δail ΔyapC mutants. None of these mutant strains demonstrated a further delay in autoaggregation compared to Y. pestis KIM5 ΔpgmA. On the contrary, deletion of yapC and ail in the ΔpgmA background led to partial restoration of autoaggregation. This may indicate that deletion of pgmA leads to a general defect in assembly of envelope components, and deletion of secreted surface proteins like YapC and Ail may partially relieve the secretion defects of the functioning autoaggregation factor (potential mechanisms for PgmA effects on protein secretion are discussed further below).
PGM facilitates the interconversion of glucose-6-phosphate to glucose-1-phosphate. Glucose-1-phosphate is a precursor of UDP-glucose and UDP-galactose (78), which are precursors of carbohydrate-containing anionic envelope polymers such as LPS, teichoic acid, lipoteichoic acid, cyclic β-1,2-glucans, and exopolysaccharides (5, 45, 66, 78). Deletion of the gene encoding PGM activity modestly increases antimicrobial peptide susceptibility of Streptococcus iniae (9), Brucella abortus (66), Bordetella bronchiseptica (69), and Stenotrophomonas maltophilia (48). In some cases, mutants lacking PGM activity are also defective for intracellular survival (9, 66, 69), serum resistance (9, 15, 39, 48), and in vivo colonization and/or virulence (3, 9, 39, 48, 66, 69).
In the absence of PGM, UDP-glucose and UDP-galactose cannot be incorporated into LPS (78). As a result, lack of PGM activity often results in an incomplete core and the inability to attach O antigen to the LPS core. In many cases this is believed to be the cause of serum sensitivity and antimicrobial peptide sensitivity of PGM-negative strains (15, 39, 48, 66, 69). LPS from the KIM5 Δpgm strain had no alterations in LPS chemistry compared to the parental strain (Fig. 4 and 5; Tables 4 and 5). Thus, the effects of the Δpgm mutation on polymyxin B resistance are not due to changes in LPS chemistry, as might have been anticipated since the fatty acid side chain of polymyxin B interacts with LPS, facilitating insertion into the outer membrane (2, 76).
Purified LPS is known to stimulate immune cells, and this LPS-mediated activation can be neutralized by polymyxin B (24, 64). Nitric oxide production can be used as an indicator of LPS toxicity on macrophages (40, 64). Our nitric oxide experiments demonstrated that binding and neutralization of both purified LPS species (KIM5 and KIM5 ΔpgmA) by polymyxin B are the same, indicating that increased polymyxin B susceptibility of the ΔpgmA mutant is not due to differential LPS binding compared to parent strain LPS. This result is consistent with our LPS compositional analysis findings that showed that the two LPS molecules are identical and that polymyxin B sensitivity is not due to an LPS structure change. Further studies are required to explain why the Y. pestis ΔpgmA mutant is susceptible to polymyxin B and causes delayed autoaggregation at 28°C.
It is possible that other outer membrane and/or inner membrane structures might be altered in the absence of PgmA, and this may explain the susceptibility of the ΔpgmA mutant to polymyxin B. One such candidate is cyclic β-1,2-glucan, a periplasmic sugar polymer that is known to be dependent upon the UDP-glucose-forming activity of Pgm in B. abortus (8). This periplasmic polymer has been shown to affect antibiotic sensitivity as well as proper assembly of structures at cell membranes (6, 11) and is known to play a role in B. abortus virulence (1, 7). Cyclic β-1,2-glucan has not been studied in Y. pestis. Given that the biologic effect of polymyxin B appears to depend on lipid exchange between that outer and inner membrane, the presence of extensive cyclic β-1,2-glucan in the periplasm may interfere with polymyxin B-mediated contacts between the two lipid bilayers. We note that Y. pestis contains a locus with homology to β-1,2-glucan synthetase.
The PhoP/PhoQ and PmrA/PmrB systems of several bacterial pathogens have been shown to play a role in LPS modifications (23, 28, 29, 33, 49, 52, 55, 70), and mutations in these loci have been shown to result in increased susceptibility to polymyxin B (23, 49, 55). PhoP/PhoQ- and PmrA/PmrB-dependent resistance to antimicrobial peptides has been proposed to be due to a reduction in negative charge via addition of 4-aminoarabinose to the lipid A portion of LPS, resulting in decreased binding to cationic peptides (28, 33, 68). These modifications are predicted to reduce the affinity of cationic peptides for LPS. In addition to LPS modification, a number of other pathways are affected in a phoP mutant. In Y. pestis, microarray analysis has shown a large number of genes up- or downregulated in a phoP mutant compared to the starting strain (77). Two genes upregulated by PhoP in Y. pestis are pgi (encoding glucose-6-phosphate isomerase) and gpm (encoding phosphoglycerate mutase 1). Both enzymes are in the glycolysis pathway and affect the cellular levels of glucose-6-phosphate, the substrate for PgmA. In a phoP mutant, higher levels of glucose-6-phosphate would be expected. While this would initially be anticipated to increase the amount of substrate for PgmA and lead to increased polymyxin B resistance, a phoP mutant displays decreased polymyxin B resistance. Thus, while much of the effect of PhoP on polymyxin B resistance is likely due to effects on LPS modifications, some of the effects of a phoP mutation on Y. pestis polymyxin B sensitivity may be due to disruption of the metabolic pathway involved in balancing glucose-6-phosphate and glucose-1-phosphate. Such effects would be consistent with the effect on polymyxin B sensitivity that we identified in the ΔpgmA mutant. It is unclear why we did not identify other genes in this pathway in our transposon mutagenesis screen; however, it should be emphasized that our original screen was for genes affecting autoaggregation, and the pgmA mutation may have a stronger effect on this phenotype than other genes in the pathway.
Since the ΔpgmA mutant showed increased polymyxin B susceptibility, we tested Y. pestis KIM5 and the ΔpgmA mutant for sensitivity to five different human antimicrobial peptides. The ΔpgmA mutant does not show increased sensitivity to any of the human antimicrobial peptides tested (Table 3). Phagocytic cells can also be a source of antimicrobial peptides during infection. We tested the ability of the KIM5 Δpgm mutant to survive within cultured macrophage-like cells from mouse (RAW264.7) or human (THP-1) origin. Monitoring survival at 2-, 4-, 6-, and 24 h time points revealed no defect in intracellular survival of the KIM5 Δpgm strain compared to parental KIM5. E. coli was readily killed by 24 h (data not shown).
Autoaggregation is a first step in the development of microcolonies and biofilm formation for many pathogens (31), and phosphoglucomutase plays a role in biofilm formation of Bacillus subtilis (45). Since biofilm formation is known to play a role in flea colonization by Y. pestis, we tested Y. pestis KIM6+ (a strain containing the large pigmentation locus involved in biofilm formation) and a KIM6+ ΔpgmA mutant strain for flea blockage. There was no difference in flea blockage, suggesting that PgmA does not play a role in Y. pestis biofilm formation and blockage in fleas (Table 6).
Phosphoglucomutase has been reported to be an important virulence factor for many Gram-negative and Gram-positive bacteria such as S. iniae (9), B. abortus (66), Pseudomonas aeruginosa (26), Bordetella bronchiseptica (69), and Stenotrophomonas maltophilia (48). We did not detect any virulence defect for the KIM5 ΔpgmA mutant following i.v. or i.n. infection of mice. These represent two natural infection routes of Y. pestis. It should be noted that our parental strain, KIM5, lacks the ability to acquire iron in peripheral tissues. Thus, we were unable to assess the role of PgmA in a bubonic plague mode. Furthermore, lethality following lung infection by KIM5 appears to be due to eventual septicemia and not classic plague pneumonia (Felek and Krukonis, unpublished). Thus, in a fully virulent plague model, PgmA may also play a role in establishment of true pneumonic plague.
Antibiotic resistance is a growing problem worldwide. This increasing resistance limits the availability of effective antimicrobials. Our results suggest that in the absence of PgmA, altered carbohydrate metabolism causes some change in the membrane structure(s) of Y. pestis, and these changes make the bacterium susceptible to polymyxin B and unable to mediate autoaggregation. Our results also demonstrate that absence of PgmA does not change Y. pestis LPS structure. Polymyxin B sensitivity of the ΔpgmA mutant does not depend on differential binding to LPS of the ΔpgmA mutant compared to KIM5 parent strain. Differences in other membrane structures likely lead to the susceptibility of Y. pestis KIM5 ΔpgmA to polymyxin B.
ADDENDUM IN PROOF
A recent publication by H. Lee-Lewis and D. M. Anderson (Infect. Immun. 78:220-230, 2010) confirms our observation that KIM5 does not cause plague pneumonia.
Acknowledgments
We thank Stephen Trent for helpful discussions concerning this work, Clayton Jarrett for assistance with the flea experiments, and J. Chris Fenno for critical reading of the manuscript.
This study was supported by a grant from the Biomedical Research Council at the University of Michigan School of Medicine and by grants from the University of Michigan's Office of the Vice President for Research and the Rackham School of Graduate Studies. This work was also partially supported by Department of Energy Grant FG02-93ER20097 to the Complex Carbohydrate Research Center.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 22 December 2009.
REFERENCES
- 1.Arellano-Reynoso, B., N. Lapaque, S. Salcedo, G. Briones, A. E. Ciocchini, R. Ugalde, E. Moreno, I. Moriyon, and J. P. Gorvel. 2005. Cyclic beta-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat. Immunol. 6:618-625. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, T. M., G. N. Forrest, and K. J. Messmer. 2007. Polymyxin antibiotics for gram-negative infections. Am. J. Health Syst. Pharm. 64:819-826. [DOI] [PubMed] [Google Scholar]
- 3.Bizzini, A., P. Majcherczyk, S. Beggah-Moller, B. Soldo, J. M. Entenza, M. Gaillard, P. Moreillon, and V. Lazarevic. 2007. Effects of alpha-phosphoglucomutase deficiency on cell wall properties and fitness in Streptococcus gordonii. Microbiology 153:490-498. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., M. S. McClain, J. A. Princ, P. J. Calie, and B. I. Eisenstein. 1991. Type 1 fimbriation and fimE mutants in Escherichia coli K-12. J. Bacteriol. 173:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breedveld, M. W., and K. J. Miller. 1994. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58:145-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briones, G., N. Inon de Iannino, M. Roset, A. Vigliocco, P. S. Paulo, and R. A. Ugalde. 2001. Brucella abortus cyclic β-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69:4528-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briones, G., N. Inon de Iannino, M. Steinberg, and R. A. Ugalde. 1997. Periplasmic cyclic 1,2-β-glucan in Brucella spp. is not osmoregulated. Microbiology 143:1115-1124. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan, J. T., J. A. Stannard, X. Lauth, V. E. Ostland, H. C. Powell, M. E. Westerman, and V. Nizet. 2005. Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development. Infect. Immun. 73:6935-6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler, T., and D. T. Dennis. 2005. Yersinia species, including plague, p. 2691-2700. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 6th ed. Churchill Livingstone, Philadelphia, PA.
- 11.Cangelosi, G. A., G. Martinetti, and E. W. Nester. 1990. Osmosensitivity phenotypes of Agrobacterium tumefaciens mutants that lack periplasmic beta-1,2-glucan. J. Bacteriol. 172:2172-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor, N. 2001. In the wake of the plague. Perennial, New York, NY.
- 13.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 14.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 17:1133-1142. [DOI] [PubMed] [Google Scholar]
- 15.Coyne, M. J., Jr., K. S. Russell, C. L. Coyle, and J. B. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwivedi, K., A. F. Post, and G. S. Bullerjahn. 1996. Cloning and functional analysis of the pmmA gene encoding phosphomannomutase from the photosynthetic prokaryote Prochlorothrix hollandica. Biochim. Biophys. Acta 1290:210-214. [DOI] [PubMed] [Google Scholar]
- 19.Falcão, J., D. Falcão, A. Pitondo-Silva, A. Malaspina, and M. Brocchi. 2006. Molecular typing and virulence markers of Yersinia enterocolitica strains from human, animal and food origins isolated between 1968 and 2000 in Brazil. J. Med. Microbiol. 55:1539-1548. [DOI] [PubMed] [Google Scholar]
- 20.Felek, S., and E. S. Krukonis. 2009. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 77:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felek, S., M. B. Lawrenz, and E. S. Krukonis. 2008. The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 154:1802-1812. [DOI] [PubMed] [Google Scholar]
- 22.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 23.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 24.Fuhrmann, O., M. Arvand, A. Gohler, M. Schmid, M. Krull, S. Hippenstiel, J. Seybold, C. Dehio, and N. Suttorp. 2001. Bartonella henselae induces NF-κB-dependent upregulation of adhesion molecules in cultured human endothelial cells: possible role of outer membrane proteins as pathogenic factors. Infect. Immun. 69:5088-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg, J. B., M. J. J. Coyne, A. N. Neely, and I. A. Holder. 1995. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect. Immun. 63:4166-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect. Immun. 69:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groisman, E., J. Kayser, and F. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudmundsson, G. H., B. Agerberth, J. Odeberg, T. Bergman, B. Olsson, and R. Salcedo. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238:325-332. [DOI] [PubMed] [Google Scholar]
- 31.Guerry, P. 2007. Campylobacter flagella: not just for motility. Trends Microbiol. 15:456-461. [DOI] [PubMed] [Google Scholar]
- 32.Guerry, P., C. P. Ewing, M. Schirm, M. Lorenzo, J. Kelly, D. Pattarini, G. Majam, P. Thibault, and S. Logan. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 34.Hinnebusch, B. J., E. R. Fischer, and T. G. Schwan. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406-1415. [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 36.Inohara, N., L. del Peso, T. Koseki, S. Chen, and G. Nunez. 1998. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 273:12296-12300. [DOI] [PubMed] [Google Scholar]
- 37.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 38.Kapperud, G., and J. Lasse. 1983. Relationship of virulence-associated autoagglutination to hemagglutinin production in Yersinia enterocolitica and Yersinia enterocolitica-like bacteria. Infect. Immun. 42:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, S. H., S. H. Ahn, J. H. Lee, E. M. Lee, N. H. Kim, K. J. Park, and I. S. Kong. 2003. Genetic analysis of phosphomannomutase/phosphoglucomutase from Vibrio furnissii and characterization of its role in virulence. Arch. Microbiol. 180:240-250. [DOI] [PubMed] [Google Scholar]
- 40.Kirikae, T., F. U. Schade, F. Kirikae, E. T. Rietschel, and D. C. Morrison. 1993. Isolation of a macrophage-like cell line defective in binding of lipopolysaccharide. Influence of serum and lipopolysaccharide chain length on macrophage activation. J. Immunol. 151:2742-2752. [PubMed] [Google Scholar]
- 41.Knirel, Y. A., B. Lindner, E. V. Vinogradov, N. A. Kocharova, S. N. Senchenkova, R. Z. Shaikhutdinova, S. V. Dentovskaya, N. K. Fursova, I. V. Bakhteeva, G. M. Titareva, S. V. Balakhonov, O. Holst, T. A. Gremyakova, G. B. Pier, and A. P. Anisimov. 2005. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry 44:1731-1743. [DOI] [PubMed] [Google Scholar]
- 42.Kolodziejek, A. M., D. J. Sinclair, K. S. Seo, D. R. Schnider, C. F. Deobald, H. N. Rohde, A. K. Viall, S. S. Minnich, C. J. Hovde, S. A. Minnich, and G. A. Bohach. 2007. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 153:2941-2951. [DOI] [PubMed] [Google Scholar]
- 43.Krauss, J. H., J. Weckesser, and H. Mayer. 1988. Electrophoretic analysis of lipopolysaccharides of purple non-sulphur bacteria. Int. J. Syst. Bacteriol. 38:157-163. [Google Scholar]
- 44.Laird, W. J., and D. C. Cavanaugh. 1980. Correlation of autoagglutination and virulence of Yersiniae. J. Clin. Microbiol. 11:430-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazarevic, V., B. Soldo, N. Médico, H. Pooley, S. Bron, and D. Karamata. 2005. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 71:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindler, L., M. Klempner, and S. Straley. 1990. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect. Immun. 58:2569-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu, M., and N. Kleckner. 1994. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 176:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKay, G. A., D. E. Woods, K. L. MacDonald, and K. Poole. 2003. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 71:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, S. I., W. S. Pulkkinen, M. E. Selsted, and J. J. Mekalanos. 1990. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect. Immun. 58:3706-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 51.Naught, L. E., and P. A. Tipton. 2001. Kinetic mechanism and pH dependence of the kinetic parameters of Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Arch. Biochem. Biophys. 396:111-118. [DOI] [PubMed] [Google Scholar]
- 52.Oyston, P. C. F., N. Dorrell, K. Williams, S.-R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pollitzer, R. 1953. Plague studies. IX. Epidemiology. Bull. World Health Organ. 9:131-170. [PMC free article] [PubMed] [Google Scholar]
- 55.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 56.Regni, C., P. A. Tipton, and L. J. Beamer. 2002. Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10:269-279. [DOI] [PubMed] [Google Scholar]
- 57.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-525. [DOI] [PubMed] [Google Scholar]
- 58.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skurnik, M., and J. A. Bengoechea. 2003. The biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic yersiniae. Carbohydr. Res. 338:2521-2529. [DOI] [PubMed] [Google Scholar]
- 60.Skurnik, M., I. Bolin, H. Heikkinen, S. Piha, and H. Wolf-Watz. 1984. Virulence plasmid-associated autoagglutination in Yersinia spp. J. Bacteriol. 158:1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 62.Staggs, T. M., and R. D. Perry. 1991. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 173:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Straley, S., and M. Cibull. 1989. Differential clearance and host-pathogen interactions of YopE− and YopK− YopL− Yersinia pestis in BALB/c mice. Infect. Immun. 57:1200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.St Swierzko, A., T. Kirikae, F. Kirikae, M. Hirata, M. Cedzynski, A. Ziolkowski, Y. Hirai, S. Kusumoto, T. Yokochi, and M. Nakano. 2000. Biological activities of lipopolysaccharides of Proteus spp. and their interactions with polymyxin B and an 18-kDa cationic antimicrobial protein (CAP18)-derived peptide. J. Med. Microbiol. 49:127-138. [DOI] [PubMed] [Google Scholar]
- 65.Tang, J. S., and P. M. Gillevet. 2003. Reclassification of ATCC 9341 from Micrococcus luteus to Kocuria rhizophila. Int. J. Syst. Evol. Microbiol. 53:995-997. [DOI] [PubMed] [Google Scholar]
- 66.Ugalde, J. E., C. Czibener, M. F. Feldman, and R. A. Ugalde. 2000. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68:5716-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ulett, G. C., J. Valle, C. Beloin, O. Sherlock, J. M. Ghigo, and M. A. Schembri. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75:3233-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaara, M., T. Vaara, and M. Sarvas. 1979. Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J. Bacteriol. 139:664-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.West, N. P., H. Jungnitz, J. T. Fitter, J. D. McArthur, C. A. Guzmán, and M. J. Walker. 2000. Role of phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence. Infect. Immun. 68:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winfield, M. D., T. Latifi, and E. A. Groisman. 2005. Transcriptional regulation of the 4-amino-4-deoxy-l-arabinose biosynthetic genes in Yersinia pestis. J. Biol. Chem. 280:14765-14772. [DOI] [PubMed] [Google Scholar]
- 71.World Health Organization. 2004. Human plague in 2002 and 2003. Wkly. Epidemiol. Rec. 79:301-306. [PubMed] [Google Scholar]
- 72.Yang, Y., and R. R. Isberg. 1997. Transcriptional regulation of the Yersinia pseudotuberculosis pH 6 antigen adhesin by two envelope-associated components. Mol. Microbiol. 24:499-510. [DOI] [PubMed] [Google Scholar]
- 73.Yen, Y. T., A. Karkal, M. Bhattacharya, R. C. Fernandez, and C. Stathopoulos. 2007. Identification and characterization of autotransporter proteins of Yersinia pestis KIM. Mol. Membr. Biol. 24:28-40. [DOI] [PubMed] [Google Scholar]
- 74.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1985. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 75.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zavascki, A. P., L. Z. Goldani, J. Li, and R. L. Nation. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206-1215. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, D., Y. Han, L. Qin, Z. Chen, J. Qiu, Y. Song, B. Li, J. Wang, Z. Guo, Z. Du, X. Wang, and R. Yang. 2005. Transcriptome analysis of the Mg2+-responsive PhoP regulator in Yersinia pestis. FEMS Microbiol. Lett. 250:85-95. [DOI] [PubMed] [Google Scholar]
- 78.Zhou, D., D. S. Stephens, B. W. Gibson, J. J. Engstrom, C. F. McAllister, F. K. Lee, and M. A. Apicella. 1994. Lipooligosaccharide biosynthesis in pathogenic Neisseria. Cloning, identification, and characterization of the phosphoglucomutase gene. J. Biol. Chem. 269:11162-11169. [PubMed] [Google Scholar]
- 79.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]