Abstract
Legionella pneumophila, the etiological agent of Legionnaires disease, is known to trigger pore formation in bone marrow-derived macrophages (BMMs) by mechanisms dependent on the type IVB secretion system known as Dot/Icm. Here, we used several mutants of L. pneumophila in combination with knockout mice to assess the host and bacterial factors involved in pore formation in BMMs. We found that regardless of Dot/Icm activity, pore formation does not occur in BMMs deficient in caspase-1 and Nlrc4/Ipaf. Pore formation was temporally associated with interleukin-1β secretion and preceded host cell lysis and pyroptosis. Pore-forming ability was dependent on bacterial Dot/Icm but independent of several effector proteins, multiplication, and de novo protein synthesis. Flagellin, which is known to trigger the Nlrc4 inflammasome, was required for pore formation as flaA mutant bacteria failed to induce cell permeabilization. Accordingly, transfection of purified flagellin was sufficient to trigger pore formation independent of infection. By using 11 different Legionella species, we found robust pore formation in response to L. micdadei, L. bozemanii, L. gratiana, L. jordanis, and L. rubrilucens, and this trait correlated with flagellin expression by these species. Together, the results suggest that pore formation is neither L. pneumophila specific nor the result of membrane damage induced by Dot/Icm activity; instead, it is a highly coordinated host cell response dependent on host Nlrc4 and caspase-1 and on bacterial flagellin and type IV secretion system.
Legionella pneumophila is a Gram-negative, facultative intracellular bacterium that causes a severe form of pneumonia called Legionnaires disease in humans. Essential for L. pneumophila pathogenesis is its capacity to replicate within alveolar macrophages; thus, the bacteria utilize a type IVB secretion system called Dot/Icm to modulate phagosome biogenesis and create an intracellular niche that supports bacterial multiplication (reviewed in references 16, 26, and 53). The Dot/Icm is composed of about 26 genes that encode a diverse set of proteins: some are contained in the cytoplasm, such as IcmW (60), IcmQ, IcmR, IcmS (14), and the ATPase DotB (50); some are periplasmic, such as IcmX (36); and some are known to localize in the inner bacterial membrane, such as DotA (60), DotL (9), DotU, and IcmF (49, 54). Collectively, this system builds a complex that facilitates the secretion of a large repertoire of bacterial proteins directly into the host cell cytoplasm. The secreted proteins, so-called effectors, target different host cell processes and, whereas deletions of the Dot/Icm structural components abolish bacterial multiplication in bone marrow-derived macrophages (BMMs), deletions of single effector proteins have little impact on the capacity of L. pneumophila to multiply in BMMs (20, 44).
In contrast to human cells, macrophages obtained from most inbred mice strains are nonpermissive for L. pneumophila replication (57). We have previously demonstrated that caspase-1 activation effectively contributes to the restriction of L. pneumophila multiplication by mechanisms not yet identified (58). In addition to caspase-1, Naip5 and Nlrc4/Ipaf, which belong to the Nod-like receptor (NLR) family of proteins, are also required for the effective restriction of L. pneumophila multiplication in murine BMMs (5, 24, 41, 46, 58). This restriction is also dependent on bacterial flagellin as bacterial mutants deficient in flaA fail to trigger caspase-1 activation and freely multiply in restrictive BMMs (5, 41, 46). The demonstration that both Nlrc4−/− and Naip5−/− BMMs fail to trigger caspase-1 activation in response to L. pneumophila flagellin (35) has led to the speculation that both proteins form an inflammasome responsible for flagellin recognition. Whereas the Nlrc4/Naip5 inflammasome is required for flagellin recognition in a process that culminates with the restriction of L. pneumophila multiplication, another inflammasome seems to be engaged in response to L. pneumophila infection (11). This second inflammasome is dependent on ASC (apoptosis-associated speck-like protein with a caspase recruitment domain) but independent of Nlrp3/Nalp3 and apparently plays no role in the restriction of Legionella multiplication in BMMs (11). Interestingly, ASC−/− BMMs were severely impaired for caspase-1 activation and interleukin-1β (IL-1β) secretion in response to L. pneumophila; therefore, the ASC-dependent inflammasome may play a major role in the processing and secretion of IL-1β (11, 58). Although extensively investigated, the mechanism by which IL-1β is secreted from BMMs is still not clear; the protein lacks the signal for canonical endoplasmic reticulum (ER)-Golgi apparatus protein secretion. Possible mechanisms to explain IL-1β secretion include the following: (i) release upon host cell lysis, (ii) release via exocytosis of secretory lysosomes, (iii) release from shed plasma membrane microvesicles, (iv) release via fusion of multivesicular bodies with the plasma membrane and subsequent release of IL-1β-containing exosomes, or (v) release through a transporter or pores formed in host cell membranes (19).
In this context, Fink and Cookson have recently demonstrated that Salmonella enterica serovar Typhimurium (S. Typhimurium) induced pore formation in the membranes of J774 macrophage-like cells in a process temporally associated with secretion of active IL-1β and IL-18 (22). This process was inhibited by caspase-1 inhibitor Z-YVAD-FMK and was dependent on the S. Typhimurium type III secretion system (22). Interestingly, previous studies have demonstrated that L. pneumophila triggers pore formation in BMMs by mechanisms dependent on the Dot/Icm secretion system (31, 60). However, it was not clear whether this pore was a host cell response or the result of membrane damage induced directly by the Dot/Icm secretion system. Because recent reports demonstrated that the cytotoxic effects of L. pneumophila were independent of ASC and Nlrp3 but dependent on flagellin, Naip5, and Nlrc4 (11, 35, 46), we used L. pneumophila as a model to assess the host and bacterial factors involved in pore formation in BMMs and their relation to IL-1β secretion. By using several isogenic mutants of L. pneumophila in combination with primary BMMs from different knockout mice, we obtained results that account for the determination of host and bacterial factors responsible for pore formation in response to L. pneumophila infection. Our data support the hypothesis that pore formation is not the result of membrane damage induced by Dot/Icm activity. Instead, pore formation is a highly coordinated host response that requires host Nlrc4 and caspase-1, and it is triggered in response to pathogenic bacteria expressing virulence factors such as flagellin and type IV secretion systems.
MATERIALS AND METHODS
Bacteria.
L. pneumophila strains used included Lp01 (7) and the ΔdotA (60), ΔdotB (50), ΔdotI (6), ΔicmR, ΔicmQ, ΔicmS, ΔicmW and ΔicmW ΔicmS (14), ΔicmX (36), and ΔflaA (46) isogenic mutants. Bacteria used in the experiment shown in Fig. 6 were in the JR32 background (47). All bacteria were grown on buffered charcoal-yeast extract (BCYE) agar [1% yeast extract, 1% N-(2 acetoamido)-2-aminoethaneosulfonic (ACES), pH 6.9, 3.3 mM l-cysteine, 0.33 mM Fe(NO)3, 1.5% Bacto agar, and 0.2% activated charcoal) at 37°C (21). The thyA (7) mutant was grown on BCYE agar supplemented with 100 mg/ml thymidine (BCYET). Wild-type and L. pneumophila ΔdotA strains expressing green fluorescent protein (GFP) cloned into the pMMB207 plasmid (15) were grown on BCYET medium with chloramphenicol (10 mg/liter). GFP expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). F2111, a clinical isolate of L. pneumophila was previously described (18). The Legionella species L. gratiana (ATCC 49413), L. jordanis (ATCC 33623), L. longbeachae (ATCC 33462), L. micdadei (ATCC 33218), L. parisiensis (ATCC 35299), L. sainthelensi (ATCC 35248), L. wadsworthii (ATCC 33877), L. bozemanii (ATCC 33217), L. rubrilucens (ATCC 35304), L. dumoffii (ATCC 332799), and L. tucsonensis (ATCC 49180) were previously described (3, 13). Before infection, bacteria were resuspended in sterile water and diluted to an appropriate multiplicity of infection (MOI) according to the optical density (OD) at 600 nm.
FIG. 6.
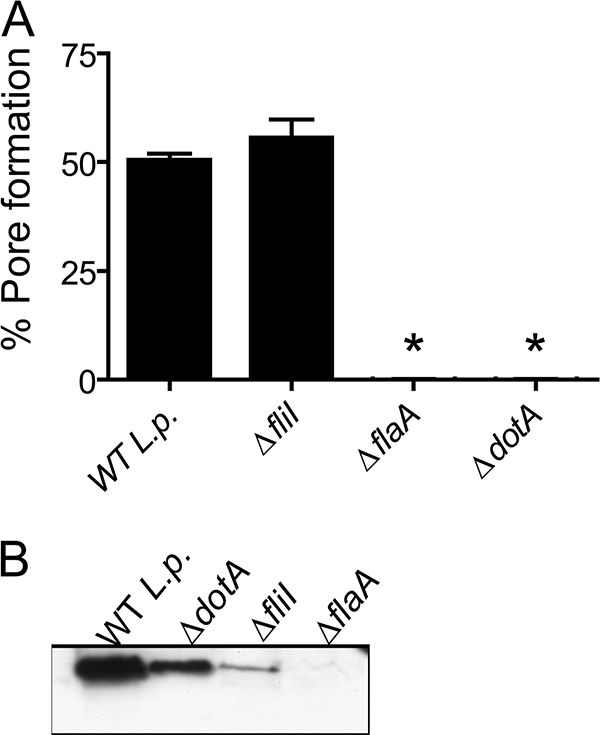
Flagellin expression is required for L. pneumophila induced pore formation. (A) BMMs from C57BL/6 mice were infected with wild-type L. pneumophila (WT L.p.) strain JR32 or ΔfliI, ΔflaA, or ΔdotA isogenic mutants at an MOI of 10 in the presence of anti-L. pneumophila antibody for 1 h and assessed for pore formation. (B) Bacteria grown on BCYE agar plates for 2 days were used to prepare extracts of total bacterial proteins. Extracts were separated by SDS-PAGE, blotted, and probed with antiflagellin antibody. Data are representative of three independent experiments. *, P < 0.05 in comparison to BMMs infected with WT L. pneumophila.
Preparation and treatment of macrophages.
BMMs were obtained from C57BL/6 (BL/6) mice or from animals deficient in caspase-1 or Nlrc4 (33, 34). Mice deficient in Nlrc4 or caspase-1 were backcrossed to BL/6 mice for eight generations. BMMs were isolated from femurs of BL/6 caspase-1−/− and Nlrc4−/− mice and differentiated in L929 cell-conditioned medium (LCCM) as previously described (59). BMMs were replated in RPMI medium supplemented with 10% fetal bovine serum and 5% LCCM. In the experiment for which the results are shown in Fig 3C, chloramphenicol (100 mg/ml) was added to the medium 30 min before infection and maintained throughout the experiment.
FIG. 3.
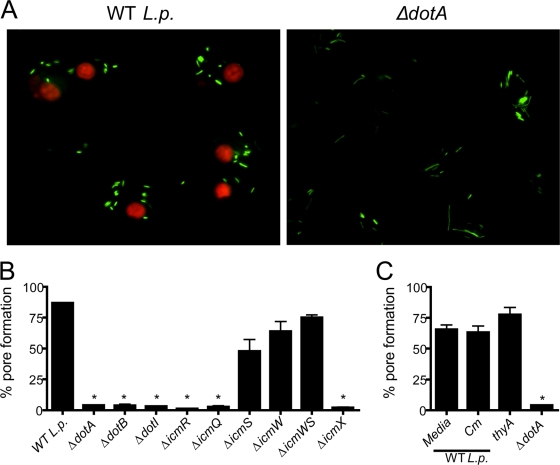
Pore formation is dependent on the type IV secretion system but independent of bacterial multiplication and de novo protein synthesis. BMMs from C57BL/6 mice were infected with wild-type L. pneumophila (WT L.p.) or isogenic mutants for 1 h and assessed for pore formation. (A) BMMs were infected with GFP-expressing WT L. pneumophila or ΔdotA mutants at an MOI of 10 in the presence of anti-L. pneumophila antibody. Shown are merged images of GFP-expressing bacteria (green) and EtBr-positive nuclei (red). (B) BMMs were infected at an MOI of 100 of the wild-type strain and indicated isogenic mutants. ΔicmWS, ΔicmW ΔicmS. (C) BMMs were infected at an MOI of 100 of WT L. pneumophila in the absence (media) or presence of 20 μg/ml of chloramphenicol (Cm), or BMMs were infected with thyA and ΔdotA mutants. Data are representative three (A and B) or two (C) independent experiments. *, P < 0.05 in comparison to BMMs infected with WT L. pneumophila.
Pore formation.
Pore formation was assayed by ethidium bromide (EtBr) staining as described previously (31). In this assay, bacteria were added at an MOI of 2, 10, 50, or 100 to 1.5 × 105 BMMs plated on 13-mm glass coverslips in 24-well tissue culture dishes. In all experiments, except for the one for which the results are shown in Fig. 8, rabbit anti-L. pneumophila polyclonal antibody (described below) was added to each well at a dilution of 1:2,000 before infection. For some experiments (those in which we also measured IL-1β secretion) the cultures were pretreated with 1 μg/ml lipopolysaccharide ([LPS] Escherichia coli 055:B5 LPS; Sigma, St. Louis, MO) for 4 h before infection. The plates were centrifuged at 200 × g for 5 min at room temperature and incubated for 1, 2, or 3 h at 37°C in 5% CO2. The coverslips were then inverted onto a 5-μl drop of phosphate-buffered saline (PBS) containing 25 μg/ml EtBr and 5 μg/ml acridine orange. All cells were stained with acridine orange, whereas only cells with membrane pores allowed diffusion of EtBr into the cell. Pore-forming activity was measured as the percentage of BMMs that stained positive with EtBr. Images were acquired using a Leica microscope (DMI4000B) with 10× and 40× objectives and analyzed using ImageJ software.
FIG. 8.
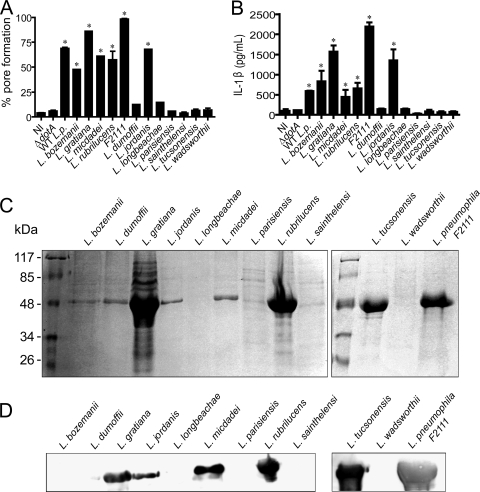
Pore formation is a host response triggered in response to different Legionella species. Different species of Legionella were used to assess pore formation (A), IL-1β release (B), direct protein staining of flagellin extractions (C), and Western blotting with antiflagellin (D). (A and B) BMMs obtained from C57BL/6 mice were pretreated with LPS for 4 h and were either left uninfected (NI) or infected for 1 h at an MOI of 50 with wild-type L. pneumophila (WT L.p.), the isogenic ΔdotA mutant, a highly virulent clinical isolate of L. pneumophila (F2111), or with the indicated species of Legionella. Infected cultures were assayed for pore formation, and the supernatants of the same experiment were assayed for secretion of IL-1β by ELISA. Bacterial species were grown on BCYE agar plates for 6 days and used to extract flagellin. The proteins were separated by SDS-PAGE and stained with Coomassie blue (C) or were blotted and probed with antiflagellin antibody raised in mouse (D). Data are representative of three independent experiments. *, P < 0.05 in comparison to noninfected BMMs.
IL-1β secretion and LDH release.
For quantification of IL-1β secretion by enzyme-linked immunosorbent assay (ELISA), BMMs in 24-well plates (2.0 × 105 cells/well) were pretreated with 1 μg/ml LPS for 4 h and then infected with the bacteria described above. The cytokine in the supernatant was measured with a mouse IL-1β ELISA kit (BD OptEIA; Franklin Lakes, NJ) according to the manufacturer's instructions. Cytotoxicity was quantified colorimetrically by evaluation of the activity of lactate dehydrogenase (LDH) released from infected BMMs. The culture supernatants were collected after 1, 2, and 3 h of infection and assayed for LDH with a CytoTox96 LDH-release kit (Promega, Madison, WI).
Flagellin purification and antibody generation.
Flagellin preparations were obtained from bacteria grown in BCYE medium for 4 or 6 days at 37°C. Bacteria were resuspended in 1 ml of PBS, and the suspensions were vigorously vortexed for 2 min to shear off the flagella. The cells were kept in an ice bath, and the procedure was repeated three times. The suspension was centrifuged at 20,000 × g at 4°C for 10 min to remove the bacterial cells. The flagella were precipitated with 3 ml of acetone followed by centrifugation 20,000 × g at 4°C for 45 min. The protein concentration was determined by Bradford assay (St. Louis, MO). Flagellin obtained from the JR32 strain of L. pneumophila was used to generate mouse antiflagellin. Animals were challenged with 5 μg of protein three times with a 15-day interval. The blood was collected 1 week after the last dose, and the serum was adsorbed to formalin-fixed ΔflaA mutant bacteria. Anti-L. pneumophila polyclonal antibodies were generated by injecting rabbits with 2 × 1010 heat-killed Lp01 cells. Animals were injected three times with a 15-day interval, and the serum of each rabbit was tested for the presence of anti-L. pneumophila and stocked at −80°C.
Transfection of bacterial flagellin.
Flagellum preparations were monomerized at 65°C for 1 h. For flagellin degradation, flagella were incubated with 1 μg/ml proteinase K (Fermentas Life Sciences, Burlington, ON, Canada) overnight at 37°C, followed by inactivation of the protease at 75°C for 15 min. The cationic lipid 1,2-dioleoyl-3-(trimethylammonium) propane (DOTAP) (Roche Diagnostics, Indianapolis, IN) was used according to the manufacturer's instructions. Briefly, 3 μl of DOTAP was incubated for 30 min with 0.6 μg of flagellin and used to stimulate (for 6 h) 2 × 105 macrophages seeded in 24-well plates.
Immunofluorescence.
BMMs were plated to confluence at 2 × 105 cells/well on 13-mm glass coverslips in 24-well tissue culture dishes and infected with L. pneumophila at an MOI of 10. The plates were centrifuged at 200 × g for 5 min at room temperature, incubated for 1 h at 37°C in 5% CO2, and then fixed and permeabilized for 10 min in ice-cold methanol. Coverslips were washed with PBS and blocked in PBS containing 5% bovine serum albumin (BSA) and 2% goat serum for 1 h. Bacteria were stained with a rabbit anti-L. pneumophila polyclonal antibody (1:2,000) followed by Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:300 dilution; Invitrogen-Molecular Probes, Carlsbad, CA). Ubiquitin was stained with anti-ubiquitinylated protein clone FK2 (1:1,000 dilution; Millipore, Billerica, MA) followed by Alexa Fluor 594-conjugated goat anti-mouse secondary antibody (1:300 dilution; Invitrogen). All antibody washes were in PBS, and all antibody dilutions were in PBS-BSA. The coverslips were inverted onto Prolong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) mounting medium (Invitrogen) on glass slides for fluorescence microscopy. Images were acquired with a Leica TCS SP2 SE inverted microscope using a 40×/1.25 oil objective (Leica HCX PL APO) and analyzed using Leica Advanced Fluorescence software. At least 100 cell-associated bacteria were counted, and standard errors were calculated from three coverslips.
Western blotting.
For detection of active caspase-1, 1 × 106 BMMs were added per well in 24-well plates and infected with L. pneumophila at an MOI of 10 in the presence of anti-L. pneumophila antibody (1:2,000). The plates were centrifuged at 200 × g for 5 min at room temperature and then incubated at 37°C in 5% CO2. Supernatants were collected after 1 h of infection and used for Western blotting using rat anti-caspase-1 p20 monoclonal antibody clone 4B4 (Genentech, San Francisco, CA). For detection of Legionella flagellin, bacteria were grown in BCYE medium for 6 days. Flagellin preparations were used for Western blotting using the antiflagellin described above. Proteins were resuspended in SDS-containing loading buffer, separated on a 15% SDS-PAGE gel, and transferred (Semidry Transfer Cell; Bio-Rad Laboratories, Inc., Hercules, CA) at 15 V for 45 min to a nitrocellulose membrane (GE Healthcare, Piscataway, NJ) in transfer buffer (50 mM Tris, 40 mM glycine, and 10% methanol). Membranes were blocked for 1 h at 25°C in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBS-T) and 5% nonfat dry milk and stained with primary antibodies for 1 h. Membranes were washed in TBS-T and were incubated for 1 h at 25°C with the appropriate horseradish peroxidase-conjugated secondary antibody (1:3,000 dilution; KPL, Gaithersburg, MD,). Western blotting Luminol reagent (Santa Cruz Biotechnology, CA) was used for antibody detection.
Statistical analysis.
Data are expressed as the mean ± standard error of the mean (SEM). In time course studies, data were analyzed by two-way analysis of variance (ANOVA), followed by Bonferroni posttest analysis. For all other data, one-way ANOVA was used followed by Bonferroni posttest analysis. Statistical analyses were performed using GraphPad Prism, version 5.0, software. Differences were considered statistically significant at a P value of <0.05.
RESULTS
Caspase-1 is required for pore formation and pyroptosis in macrophages infected with Legionella expressing a functional Dot/Icm system.
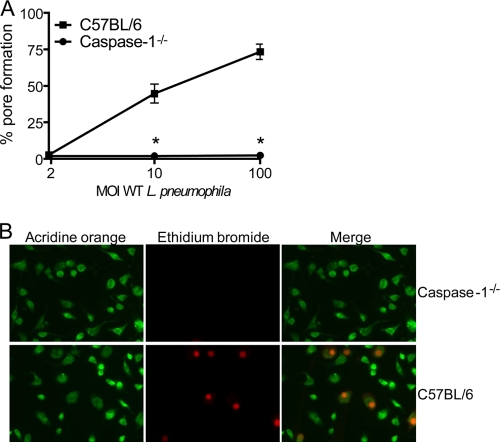
Murine BMMs undergo caspase-1-dependent cell death in response to infection with virulent L. pneumophila (5, 41, 46, 58). Because it has been previously reported that L. pneumophila inserts pores into the macrophage membranes (4, 31, 40, 60), we aimed to investigate if the pore-forming ability of L. pneumophila was dependent on host caspase-1. To test this hypothesis, we used L. pneumophila to infect BMMs obtained from BL/6 and caspase-1-deficient mice and assessed the uptake of the membrane-impermeant dye EtBr. BMMs with intact membranes fail to internalize EtBr while cells containing pores or a rupture in plasma membranes become permeable to this dye (37). The pore formation assay was performed by using EtBr in combination with acridine orange, a nonselective acidophilic green dye that stains both permeabilized and intact cells and therefore allows the determination of the percentage of pore formation (60). Because the assays described herein were performed within 1 h after infection, we opsonized the bacteria to guarantee uniform and efficient macrophage infection (60). At 1 h after infection, pore formation in BL/6 BMMs was evident at an MOI of 10 bacteria per cell, and the pore formation efficiency was dependent on the bacterial MOI (Fig. 1A). In contrast, BMMs from caspase-1−/− mice failed to trigger pore formation even at an MOI of 100 bacteria per cell (Fig. 1A). Representative images of BMMs infected for 1 h at an MOI of 10 bacteria per cell are shown in Fig. 1B. Next, we used an MOI of 10 bacteria per cell and infected BMMs for 1, 2, or 3 h to investigate the kinetics of the pore formation. Whereas BL/6 BMMs allowed the influx of EtBr in a time-dependent manner, caspase-1−/− BMMs were not permissive for EtBr influx even at 3 h after infection (Fig. 2A). Experiments carried out for longer times indicate that pore formation was strictly dependent on caspase-1 within the first 6 h of infection while we detected caspase-1-independent permeabilization at a later time (data not shown). Caspase-1-dependent pore formation in BMM membranes leads to host cell lysis in a process called pyroptosis (8, 22, 23). Thus, to further evaluate pore formation and pyroptosis in response to L. pneumophila, we measured the release of the cytosolic enzyme LDH. In BL/6 BMMs we found low levels of LDH released into the supernatants at 1 and 2 h after infection (Fig. 2B), yet at these time points we detected a high percentage of EtBr-positive cells (Fig. 2A). These data suggest that caspase-1-dependent pore formation precedes host cell lysis. Because caspase-1 activation in BMMs culminates with the processing and secretion of IL-1β, we measured secretion of this cytokine during this time course experiment to gain insight into the relation between pore formation, pyroptosis, and IL-1β secretion. We found that whereas IL-1β secretion was temporally coordinated with pore formation (Fig. 2C), BMMs lysis occurred later after infection (Fig. 2B). These data support the hypothesis that IL-1β secretion is an active and regulated process that precedes host cell lysis and pyroptosis.
FIG. 1.
Caspase-1 is required for pore formation in response to Legionella infection. BMMs adherent to glass coverslips were infected with L. pneumophila at different MOIs for 1 h in the presence of anti-L. pneumophila antibody, and the cultures were processed for pore formation assays by double staining with EtBr and acridine orange. (A) Caspase-1−/− and C57BL/6 BMMs were infected for 1 h with L. pneumophila at an MOI of 2, 10, or 100. (B) Representative images of pore formation in caspase-1−/− or C57BL/6 BMMs at 1 h after infection with L. pneumophila at an MOI of 10 per cell. Acridine orange-stained BMMs (green) show total cells in each field, and EtBr (red) indicates permeabilized BMMs (middle panels). Right panels show merged images. Data are representative of those found in four independent experiments. *, P < 0.05 in comparison to C57BL/6 BMMs. WT, wild type.
FIG. 2.
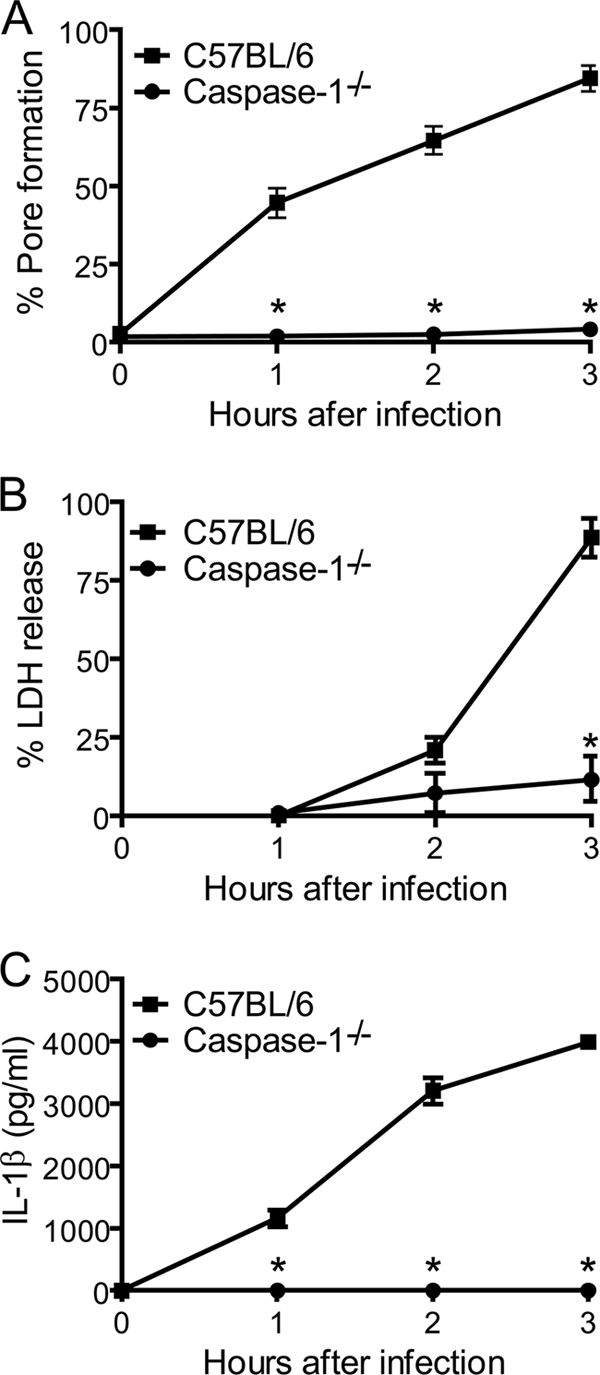
Pore formation is temporally coordinated with IL-1β secretion and precedes host cell lysis. BMMs from caspase-1−/− and C57BL/6 mice were treated with LPS for 4 h and infected with L. pneumophila at an MOI of 10 in the presence of anti-L. pneumophila antibody. Cultures were infected for 1, 2, and 3 h and processed for pore formation (A), release of LDH (B), or secretion of IL-1β (C). Data shown in all panels were obtained from the same cultures and are representative of three independent experiments. *, P < 0.05 in comparison to C57BL/6 BMMs.
Because previous work has demonstrated that the bacterial Dot/Icm secretion system is required for pore formation (1, 2, 11, 31, 41, 46, 58, 60), we investigated the requirement of Dot/Icm proteins to trigger caspase-1-dependent pore formation by using a single cell assay with GFP-expressing bacteria. Thus, pore formation assays were performed by infecting BMMs with either wild-type or ΔdotA mutant bacteria expressing GFP. BMMs infected with wild-type bacteria efficiently triggered pore formation (Fig. 3A, red nuclei); in contrast, BMMs infected with ΔdotA bacteria showed intact membranes and failed to allow EtBr influx (Fig. 3A). The experiments using GFP-expressing bacteria showed that nearly all cells were infected (Fig. 3A and data not shown); therefore, the lack of pore formation observed in cultures infected with the dotA mutants was not due to a lack of host cell infection.
To further investigate the components of the Dot/Icm system required to trigger pore formation, we infected BL/6 BMMs with isogenic mutants for the structural components of the L. pneumophila Dot/Icm. The expression of the Dot/Icm proteins DotA, DotI, DotB, IcmX, IcmR, and IcmQ, was required for pore formation (Fig. 3B). In contrast, the chaperones IcmW and IcmS, which facilitate the secretion of a subset of Dot/Icm effectors (10, 45), were not required for pore formation in BMMs (Fig. 3B). Previous studies have demonstrated that icmW and icmS mutants trigger pore formation in BMMs (60). Indeed, we have previously demonstrated that icmW and icmS mutants effectively trigger caspase-1 activation and IL-1β secretion in BL/6 BMMs (58). Therefore, caspase-1 activation and pore formation may not be affected by the Dot/Icm effectors secreted in an IcmS/IcmW-dependent manner. We have used bacterial mutants defective for Dot/Icm substrates such as the ankX, drrA, lidA, ralF, or ylfA ylfB mutant (reviewed in references 20 and 44) and found that these mutants effectively triggered pore formation in BL/6 BMMs at similar levels to the wild-type L. pneumophila (data not shown). To investigate the requirement of bacterial multiplication and bacterial de novo protein synthesis for pore formation in BMMs, we performed the pore assay either in the presence of the bacteriostatic antibiotic chloramphenicol or by using thymidine auxotroph mutants of L. pneumophila (thyA), which are able to modulate phagosome biogenesis but show limited multiplication because of the small amount of thymidine within the L. pneumophila-containing vacuole (LCV) (27). As shown in Fig. 3C, we found that neither bacterial multiplication nor de novo protein synthesis was required for pore formation in response to L. pneumophila infection.
Dot/Icm activity is not sufficient to trigger pore formation in infected macrophages.
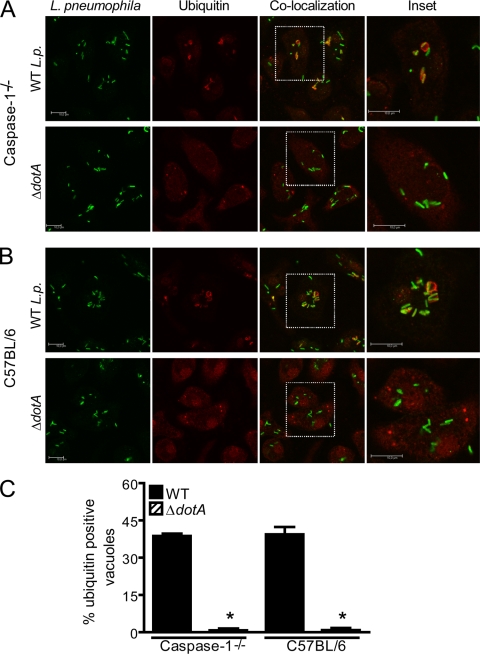
The demonstration that Dot/Icm mutant bacteria fail to trigger pore formation in BMMs has led to the speculation that pore formation could be a direct result of the insertion of Dot/Icm proteins into host cell membranes (31, 60). However, data shown in Fig. 1, 2, and 3 suggest that pore formation is, rather, a caspase-1-dependent host cell response against virulent Legionella bacteria that express the Dot/Icm secretion system. Thus, to determine if Dot/Icm activity was sufficient to trigger pore formation in BMMs, we used caspase-1−/− BMMs to perform the pore assay under conditions in which Dot/Icm was active. A known trait of Dot/Icm activity is the recruitment of polyubiquitinated proteins to the LCV (17, 28, 32). Thus, we infected BL/6 and caspase-1−/− BMMs with wild-type or ΔdotA mutant L. pneumophila for 1 h and immunostained the cells with antipolyubiquitin. At 1 h after infection, we found a similar percentage of polyubiquitination-positive LCVs in caspase-1−/− and BL/6 BMMs (Fig. 4A). In contrast, dotA mutant bacteria failed to recruit proteins to the LCV (Fig. 4A). Since recruitment of polyubiquitinated proteins is strictly dependent on Dot/Icm, these data demonstrate that despite Dot/Icm activity, pores were not formed in caspase-1−/− BMMs. Importantly, this was not due to a delay in pore formation kinetics because experiments carried out for longer times showed that caspase-1−/− BMMs still failed to trigger pore formation within the first 6 h of infection (data not shown). In fact, at 6 h after infection, we found that both BL/6 and caspase-1−/− BMMs supported the establishment of L. pneumophila replicative vacuoles that contained about three to five replicating bacteria (data not shown). Since Dot/Icm functions are absolutely required for bacterial multiplication (48, 55), these data provide independent confirmation that, despite the Dot/Icm activity, pore formation does not occur in the absence of caspase-1. Overall, these studies support the proposition that pore formation is, rather, a caspase-1-dependent host cell response and not the result of Dot/Icm activity itself.
FIG. 4.
Pore formation is not a result of membrane damage by the Dot/Icm apparatus. BMMs from caspase-1−/− and C57BL/6 mice were infected with wild-type (WT) L. pneumophila (WT L.p.) or isogenic ΔdotA mutants for 1 h at an MOI of 10 in the presence of anti-L. pneumophila antibody. Recruitment of ubiquitin (red) to L. pneumophila vacuoles (green) in caspase-1−/− (A) or C57BL/6 (B) infected BMMs was visualized by confocal microscopy. Confocal high-resolution detail corresponds to ubiquitin recruitment to the vacuoles in the white inset. Scale bar, 10 μm. (C) Percentage of infected cells showing ubiquitin-positive L. pneumophila vacuoles upon infection with WT L. pneumophila or ΔdotA mutants. Data are representative of three independent experiments. *, P < 0.05 in comparison to BMMs infected with WT L. pneumophila.
Transfection of purified flagellin is sufficient to trigger pore formation in a caspase-1-dependent pathway.
Because previous studies have demonstrated that flagellin mutants of L. pneumophila fail to trigger cytotoxicity to macrophages (2, 35, 41, 46), we tested whether delivery of purified flagellin to the host cytosol could trigger pore formation independently of infection. BL/6 BMMs were stimulated with purified Legionella flagellin in the presence or absence of DOTAP, a cationic lipid formulation that mediates delivery of negatively charged molecules into the host cell cytoplasm (51). Stimulation of BMMs with flagellin plus DOTAP, but not with flagellin or DOTAP alone, induced pore formation in BMMs (Fig. 5A). Experiments performed with BMMs from caspase-1−/− mice indicate that caspase-1 was required for pore formation in response to transfected flagellin (Fig. 5A). To further demonstrate that flagellin was required for the cytotoxic effects in BMMs, we transfected BMMs (i) with a nonrelated protein (p131, from the pathogenic parasite Schistosoma mansonii), (ii) with a mock flagellin preparation obtained from ΔflaA mutant L. pneumophila, or (iii) with a flagellin preparation that was treated with proteinase K. In all cases pore formation was significantly reduced compared to transfection of L. pneumophila flagellin (Fig. 5). These data further indicate that flagellin specifically triggers pore formation in macrophages in a process dependent on caspase-1. Therefore, pore formation in BMMs may be the mechanism underlying the flagellin-dependent cytotoxicity previously described (2, 35, 41, 46). To further evaluate the requirement of flagellin in triggering pore formation, we evaluated pore formation in BMMs infected with Legionella mutants for the flagellin gene (ΔflaA). We found no pore formation in response to ΔflaA mutant bacteria (Fig. 6A). In contrast, BMMs infected with ΔfliI, which encodes an ATPase necessary for flagellin secretion for flagellum assembly (38), showed pore formation at similar levels to wild-type bacteria (Fig. 6A). Thus, the pore formation in BMMs is dependent on Legionella flagellin itself but not on flagella, motility, or the bacterial type IV secretion system. To assess flagellin expression, we performed a Western blot analysis using an antibody generated against flagellin purified from L. pneumophila. We showed that whereas wild-type L. pneumophila, ΔdotA mutants, and, to a lesser extent, ΔfliI mutants do express flagellin, ΔflaA mutant bacteria were defective for flagellin expression (Fig. 6B).
FIG. 5.
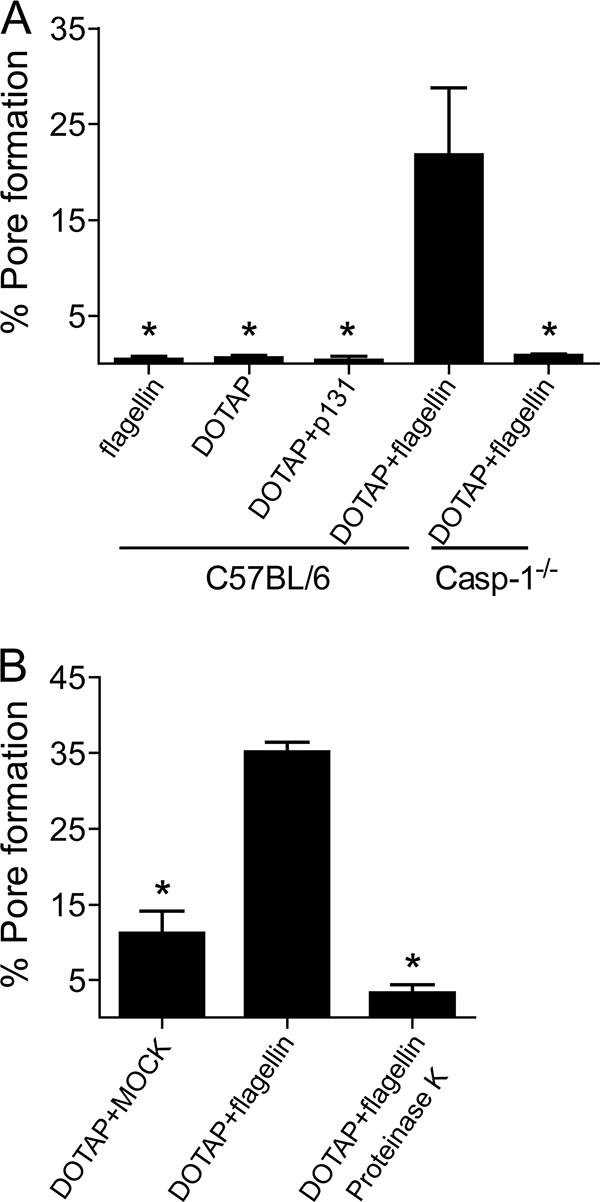
Bacterial flagellin is sufficient to trigger pore formation. DOTAP was used to transfect proteins into BMMs obtained from C57BL/6 and caspase-1−/− (casp-1−/−) mice. Cultures were stimulated for 6 h and assayed for pore formation. (A) Cultures were treated with flagellin alone, DOTAP alone, DOTAP and p131 from S. mansoni, or DOTAP and flagellin. (B) BMM cultures were treated with DOTAP and a mock-flagellin preparation (extracted from ΔflaA L. pneumophila), DOTAP and flagellin, or DOTAP and flagellin treated with proteinase K. Data are representative of three independent experiments. *, P < 0.05 in comparison to BMMs treated with DOTAP and flagellin.
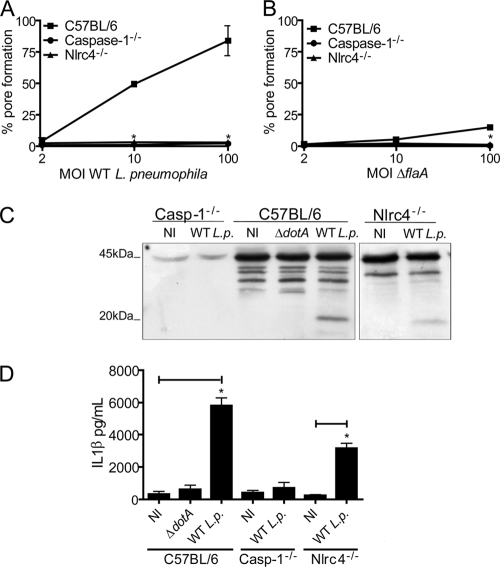
Pore formation is induced upon recognition of bacterial flagellin by the Nlrc4 inflammasome.
Within the macrophage cytosol, bacterial flagellin is detected by an inflammasome possibly composed of Nlrc4, Naip5, and caspase-1 (25, 35, 39, 41, 46, 52). Thus, we used BMMs from Nlrc4−/− mice to test whether the Nlrc4 inflammasome was required for pore formation in response to L. pneumophila. Whereas BL/6 BMMs showed pore formation at an MOI of 10 bacteria per cell, Nlrc4−/− and caspase-1−/− BMMs failed to trigger pore formation even at an MOI of 100 bacteria per cell (Fig. 7A). Indeed, flagellin appeared to be upstream of those signaling pathways as both BL/6 and Nlrc4−/− BMMs failed to trigger pore formation in response to infection with ΔflaA mutant bacteria (Fig. 7B). Since our data showed that Nlrc4 was required for pore formation, we used BMMs from Nlrc4−/− mice to further investigate the possible role of pore formation for IL-1β secretion, a process that is not yet clear. Thus, we performed infections using Nlrc4−/− cells under conditions in which we did not detect pore formation (MOI of 10:1 for 1 h) and found that despite the absence of pore formation, we still detected caspase-1 activation and IL-1β secretion by Nlrc4−/− cells (Fig. 7). These data indicate that mechanisms different from pore formation may account for IL-1β released by the infected BMM.
FIG. 7.
Nlrc4 is required for pore formation. BMMs obtained from C57BL/6, caspase-1−/− (Casp-1−/−), and Nlrc4−/− mice were infected at an MOI of 2, 10, or 100 of the wild-type L. pneumophila (A) or ΔflaA mutant (B) strain for 1 h and assayed for pore formation. (C) Proteins from uninfected cultures (NI) or BMMs infected with wild-type L. pneumophila (WT L.p.) or the ΔdotA mutant at an MOI of 10 for 1 h were separated by SDS-PAGE, blotted, and probed with a caspase-1 antibody. Indicated at left are procaspase-1 (45 kDa) and subunit p20 of active caspase-1 (20 kDa). (D) BMMs were pretreated with LPS for 4 h and either left uninfected (NI) or infected with wild-type L. pneumophila (WT L.p.) or the ΔdotA mutant for 1 h at an MOI of 10. IL-1β secretion was measured by ELISA. Data are representative of four independent experiments. *, P < 0.05 in comparison to C57BL/6 BMMs (A) or uninfected BMMs (D).
Next, we aimed to investigate if pore formation was a specific host response against L. pneumophila or a general response against intracellular bacteria. Thus, we investigated whether other species of the Legionella genus would trigger pore formation in BMMs. We infected BMMs with 11 species of nonpneumophila Legionella and found that 6 species fail to trigger pore formation, namely, L. dumoffii, L. longbeachae, L. parisiensis, L. sainthelensi, L. tucsonensis, and L. wadsworthii. In contrast, five other species did trigger pore formation at a variable efficiency: L. bozemanii, L. gratiana, L. jordanis, L. micdadei, and L. rubrilucens (Fig. 8A). In this experiment, we also tested a highly virulent and cytotoxic clinical isolate of L. pneumophila SG1 (F2111) and found that this isolate was very efficient in triggering pore formation (Fig. 8A). We found that secretion of IL-1β in response to the different Legionella species directly correlated with pore formation; i.e., species that formed pores also triggered IL-1β secretion (Fig. 8B). Because flagellin was required for caspase-1-dependent pore formation in response to L. pneumophila, we aimed to investigate if flagellin expression by the different Legionella species correlated with pore formation. Thus, we grew bacteria to late stationary phase and performed a direct protein staining of flagellin preparations obtained from these species. As shown in Fig. 8, the species that did trigger pore formation showed expression of a 48-kDa protein (Fig. 8C). To further assess if this 48 kDa protein is flagellin, we performed Western blot analysis using a polyclonal antibody raised against L. pneumophila flagellin (antiflagellin). As shown in Fig. 8D, the extractions obtained from L. pneumophila, L. gratiana, L. jordanis, L. micdadei, L. rubrilucens, and L. tucsonensis stained positive with antiflagellin. Of note, some species did express a 48-kDa protein that did not react with antiflagellin, possibly because they expressed different serotypes of flagellin and therefore were not recognized by the anti-L. pneumophila flagellin. Interestingly, some species, such as L. dumoffii and L. tucsonensis, expressed a 48-kDa protein but failed to trigger pore formation. Possibly, these bacteria do express flagellin (as shown here for L. tucsonensis) but are unable to deliver the flagellin to the macrophage cytoplasm and therefore fail to trigger pore formation. According to these speculations, it was recently demonstrated that although flagellins from L. parisiensis and L. tucsonensis were toxic for BMMs, these species were able to multiply in BL/6 BMMs (56). Regardless of the flagellin secretion through the Dot/Icm system of these other Legionella species, it is important that none of the flagellin-negative bacteria triggered pore formation; therefore, the studies performed herein support our assertions that pore formation is a host cell response dependent on bacterial flagellin and is not L. pneumophila specific.
DISCUSSION
Activation of caspase-1 has emerged as an important feature that accounts for different host cell processes including unconventional protein secretion and response against intracellular pathogens (23, 29, 58). Although little is known regarding host cell targets for active caspase-1, it is clear that BMMs expressing active caspase-1 release large amounts of active IL-1β and undergo an unconventional form of cell death called pyroptosis (23). In this context, our investigation of host and bacterial factors required for pore formation in response to L. pneumophila increase our understanding of the induction of pyroptosis in response to intracellular bacteria. It is possible that the pores formed early after infection of BMMs directly account for host cell swelling, culminating with the lysis of the infected cells (22). We here demonstrated that pore formation was temporally associated with IL-1β secretion and preceded host cell lysis and LDH release (Fig. 2). Therefore, the caspase-1-dependent pore formation investigated herein may be an important step required for pyroptosis.
The temporal coordination of pore formation and IL-1β secretion has led to the speculation that pore formation could be the mechanism by which IL-1β is secreted (22). Here, we took advantage of Nlrc4-deficient cells to address this question. This was possible because it was previously demonstrated that Legionella triggers an ASC-dependent caspase-1 activation (and IL-1β secretion) independently of Nlrc4 (11). In this context, we performed infections using Nlrc4−/− cells under conditions in which we did not detect pore formation (MOI of 10:1 for 1 h). Of note, we found that despite the absence of pore formation, we still detected caspase-1 activation and IL-1β secretion by Nlrc4−/− cells (Fig. 7). These results may indicate that pore formation is neither a prerequisite for caspase-1 activation nor absolutely required for IL-1β secretion. However, we cannot rule out the possibility that a few pores are formed in Nlrc4−/− cells that are not sufficient to allow dye uptake but are sufficient to allow cytokine secretion.
Although it has been previously reported that L. pneumophila cytotoxic activity is cell associated, it was not clear whether phagocytosis was required for full cytotoxicity in BMMs (30). Here, we demonstrated that transfection of purified flagellin was sufficient to trigger pore formation in macrophages. Lightfield and colleagues demonstrated that transduction of BMMs with a retroviral construct that encodes bacterial flagellin renders cytotoxicity to BMMs (35). Indeed, it was recently demonstrated that transfection of flagellin from L. parisiensis and L. tucsonensis triggers cytotoxicity to BMMs (56). Collectively, these data indicate that bacterial internalization and phagosome formation are not required for pyroptosis. These conclusions contrast with a previous publication reporting that actin activity was required for caspase-1-dependent pore formation in response to S. Typhimurium infection (22). However, the authors did not investigate the kinetics of pore formation in cytochalasin-treated J774 cells. In an attempt to address this issue, we performed kinetics assays of L. pneumophila-induced pore formation in cytochalasin-treated BMMs. We showed that pore formation does occur in the presence of cytochalasin with a delayed kinetics (unpublished data). Thus, we suggest that bacterial internalization and phagosome formation are not required for pore formation in BMMs. Given the recent demonstration that phagocytosis is required to trigger effector delivery unless intimate contact between the bacteria and the host is artificially generated (12, 43), it is possible that the bacterial internalization facilitates the secretion of L. pneumophila products into the host cell cytoplasm. Among these products may be flagellin, which may then trigger the Nlrc4 and caspase-1-dependent pore formation.
Although we have demonstrated that Nlrc4 is required for pore formation in response to L. pneumophila, the role of Naip5 in this process is yet unclear. BMMs from the A/J mice strain, which harbors a deficient Naip5 allele, effectively trigger pore formation (31, 42, 60). However, the Naip5−/− mouse was recently generated, and it was demonstrated that Naip5−/− BMMs failed to trigger caspase-1 activation and failed to release LDH in response to Legionella infection (35). Therefore, we speculate that Naip5 does play a role in the pore-forming ability of L. pneumophila. Regardless of the participation of Naip5, we have shown that pore formation in BMMs is a highly coordinated host cell response against virulent bacteria. This response involves host cell Nlrc4 and caspase-1 and is possibly triggered in response to bacterial flagellin that gains access to the macrophage cytoplasm. The future determination of the molecular composition of the pore and the physiological functions of pore formation and pyroptosis may contribute to our understanding of how the innate immune cells resist infection and contribute to shaping appropriate acquired immune responses.
Acknowledgments
We are grateful to Catarina V. Horta for excellent technical assistance and to Liliana M. Massis for help with flagellin preparations and antibody generation. Knockout mice were kindly provided by Richard Flavell and Anthony Coyle. Caspase-1 antibody was kindly provided by Vishva Dixit, and the polyubiquitin antibody was provided by Marcelo Damario. Legionella sp. strains used in this work were kindly provided by Craig Roy, Nicholas Cianciotto, Howard Shuman, and Paul Edelstein.
This work was supported by CNPq and FAPESP grant number 06/52867-4 (D.S.Z.). T.N.S. is the recipient of a doctoral fellowship from FAPESP (06/55084-0). D.S.Z. is a research fellow from CNPq.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 4 January 2010.
REFERENCES
- 1.Abu-Zant, A., R. Asare, J. E. Graham, and Y. Abu Kwaik. 2006. Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect. Immun. 74:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhter, A., M. A. Gavrilin, L. Frantz, S. Washington, C. Ditty, D. Limoli, C. Day, A. Sarkar, C. Newland, J. Butchar, C. B. Marsh, M. D. Wewers, S. Tridandapani, T. D. Kanneganti, and A. O. Amer. 2009. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog. 5:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alli, O. A., L. Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amer, A., L. Franchi, T. D. Kanneganti, M. Body-Malapel, N. Ozoren, G. Brady, S. Meshinchi, R. Jagirdar, A. Gewirtz, S. Akira, and G. Nunez. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281:35217-35223. [DOI] [PubMed] [Google Scholar]
- 6.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 8.Bergsbaken, T., S. L. Fink, and B. T. Cookson. 2009. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscher, B. A., G. M. Conover, J. L. Miller, S. A. Vogel, S. N. Meyers, R. R. Isberg, and J. P. Vogel. 2005. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol. 187:2927-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambronne, E. D., and C. R. Roy. 2007. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Case, C. L., S. Shin, and C. R. Roy. 2009. Asc and Ipaf inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect. Immun. 77:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charpentier, X., J. E. Gabay, M. Reyes, J. W. Zhu, A. Weiss, and H. A. Shuman. 2009. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 5:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cianciotto, N. P., J. M. Bangsborg, B. I. Eisenstein, and N. C. Engleberg. 1990. Identification of mip-like genes in the genus Legionella. Infect. Immun. 58:2912-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 15.Coers, J., C. Monahan, and C. R. Roy. 1999. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1:451-453. [DOI] [PubMed] [Google Scholar]
- 16.Cunha, B. A. 2008. Atypical pneumonias: current clinical concepts focusing on Legionnaires' disease. Curr. Opin. Pulm. Med. 14:183-194. [DOI] [PubMed] [Google Scholar]
- 17.Dorer, M. S., D. Kirton, J. S. Bader, and R. R. Isberg. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelstein, P. H., and M. A. Edelstein. 1989. WIN 57273 is bactericidal for Legionella pneumophila grown in alveolar macrophages. Antimicrob. Agents Chemother. 33:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eder, C. 26 February 2009. Mechanisms of interleukin-1β release. Immunobiology. [Epub ahead of print.] doi: 10.1016/j.imbio.2008.11.007. [DOI] [PubMed]
- 20.Ensminger, A. W., and R. R. Isberg. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12:67-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fink, S. L., and B. T. Cookson. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 8:1812-1825. [DOI] [PubMed] [Google Scholar]
- 23.Fink, S. L., and B. T. Cookson. 2007. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 9:2562-2570. [DOI] [PubMed] [Google Scholar]
- 24.Fortier, A., K. Doiron, M. Saleh, S. Grinstein, and P. Gros. 2009. Restriction of Legionella pneumophila replication in macrophages requires concerted action of the transcriptional regulators Irf1 and Irf8 and nod-like receptors Naip5 and Nlrc4. Infect. Immun. 77:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franchi, L., A. Amer, M. Body-Malapel, T. D. Kanneganti, N. Ozoren, R. Jagirdar, N. Inohara, P. Vandenabeele, J. Bertin, A. Coyle, E. P. Grant, and G. Nunez. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nat. Immunol. 7:576-582. [DOI] [PubMed] [Google Scholar]
- 26.Isberg, R. R., T. J. O'Connor, and M. Heidtman. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isberg, R. R., S. Rankin, C. R. Roy, M. S. Swanson, and K. H. Berger. 1993. Legionella pneumophila: factors involved in the route and response to an intracellular niche. Infect. Agents Dis. 2:220-223. [PubMed] [Google Scholar]
- 28.Ivanov, S. S., and C. R. Roy. 2009. Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell Microbiol. 11:261-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller, M., A. Ruegg, S. Werner, and H. D. Beer. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132:818-831. [DOI] [PubMed] [Google Scholar]
- 30.Kirby, J. E., and R. R. Isberg. 1998. Legionnaires' disease: the pore macrophage and the legion of terror within. Trends Microbiol. 6:256-258. [DOI] [PubMed] [Google Scholar]
- 31.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 32.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67:1307-1319. [DOI] [PubMed] [Google Scholar]
- 33.Kuida, K., J. A. Lippke, G. Ku, M. W. Harding, D. J. Livingston, M. S. Su, and R. A. Flavell. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1β converting enzyme. Science 267:2000-2003. [DOI] [PubMed] [Google Scholar]
- 34.Lara-Tejero, M., F. S. Sutterwala, Y. Ogura, E. P. Grant, J. Bertin, A. J. Coyle, R. A. Flavell, and J. E. Galan. 2006. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203:1407-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lightfield, K. L., J. Persson, S. W. Brubaker, C. E. Witte, J. von Moltke, E. A. Dunipace, T. Henry, Y. H. Sun, D. Cado, W. F. Dietrich, D. M. Monack, R. M. Tsolis, and R. E. Vance. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews, M., and C. R. Roy. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGahon, A. J., S. J. Martin, R. P. Bissonnette, A. Mahboubi, Y. Shi, R. J. Mogil, W. K. Nishioka, and D. R. Green. 1995. The end of the (cell) line: methods for the study of apoptosis in vitro. Methods Cell Biol. 46:153-185. [DOI] [PubMed] [Google Scholar]
- 38.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao, E. A., C. M. Alpuche-Aranda, M. Dors, A. E. Clark, M. W. Bader, S. I. Miller, and A. Aderem. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7:569-575. [DOI] [PubMed] [Google Scholar]
- 40.Molmeret, M., O. A. Alli, S. Zink, A. Flieger, N. P. Cianciotto, and Y. A. Kwaik. 2002. icmT is essential for pore formation-mediated egress of Legionella pneumophila from mammalian and protozoan cells. Infect. Immun. 70:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molofsky, A. B., B. G. Byrne, N. N. Whitfield, C. A. Madigan, E. T. Fuse, K. Tateda, and M. S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73:5720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15:372-380. [DOI] [PubMed] [Google Scholar]
- 45.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55:912-926. [DOI] [PubMed] [Google Scholar]
- 46.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72:5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simberg, D., S. Weisman, Y. Talmon, and Y. Barenholz. 2004. DOTAP (and other cationic lipids): chemistry, biophysics, and transfection. Crit. Rev. Ther. Drug Carrier Syst. 21:257-317. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian, N., and A. Qadri. 2006. Lysophospholipid sensing triggers secretion of flagellin from pathogenic Salmonella. Nat. Immunol. 7:583-589. [DOI] [PubMed] [Google Scholar]
- 53.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 54.VanRheenen, S. M., G. Dumenil, and R. R. Isberg. 2004. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect. Immun. 72:5972-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 56.Whitfield, N. N., B. G. Byrne, and M. S. Swanson. 2010. Mouse macrophages are permissive to motile Legionella species that fail to trigger pyroptosis. Infect. Immun. 78:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto, Y., T. W. Klein, C. A. Newton, R. Widen, and H. Friedman. 1988. Growth of Legionella pneumophila in thioglycolate-elicited peritoneal macrophages from A/J. mice. Infect. Immun. 56:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zamboni, D. S., K. S. Kobayashi, T. Kohlsdorf, Y. Ogura, E. M. Long, R. E. Vance, K. Kuida, S. Mariathasan, V. M. Dixit, R. A. Flavell, W. F. Dietrich, and C. R. Roy. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318-325. [DOI] [PubMed] [Google Scholar]
- 59.Zamboni, D. S., and M. Rabinovitch. 2003. Nitric oxide partially controls Coxiella burnetii phase II infection in mouse primary macrophages. Infect. Immun. 71:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuckman, D. M., J. B. Hung, and C. R. Roy. 1999. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32:990-1001. [DOI] [PubMed] [Google Scholar]