Abstract
Colonization of the gastric mucosa by Helicobacter pylori can lead to serious clinical outcomes, including gastric cancer. Toll-like receptors (TLRs) play an important role in the host response to H. pylori through the recognition of pathogen-associated molecular patterns. TLR9, in particular, is partly responsible for initiating bacterial induced immunity by binding unmethylated CpG-DNA, which is abundant in bacteria. A well-documented single nucleotide polymorphism (SNP) within the TLR9 promoter (TLR9 −1237T/C), is associated with a variety of inflammatory disorders, including allergic asthma, inflammatory bowel disease, and atopy. Analysis of the TLR9 promoter gene sequence has shown that carriage of the variant “C” allele at position −1237 creates a potential NF-κB binding site that would theoretically increase the transcriptional activity of the gene. In this study, we report that the TLR9 −1237 C allele was significantly associated with the development of H. pylori-induced premalignant gastric changes. Functional analysis of the SNP, supporting the data generated from the genetic association study, showed that carriage of the C allele increased TLR9 transcriptional activity driven mainly by activation of NF-κB. Collectively, these findings confirm that the TLR9 −1237T/C polymorphism is a risk factor for the development of H. pylori-induced premalignant gastric changes and provide a plausible mechanistic explanation.
Helicobacter pylori infection is associated with a variety of clinical outcomes including gastric cancer and duodenal ulcer disease (24, 30, 35). These differing outcomes are defined in part by changes in gastric acid secretion, which is influenced by the severity and distribution of H. pylori-induced gastritis. Severe inflammation of the gastric mucosa in the corpus region can inhibit parietal cells from secreting acid and may eventually cause gastric atrophy and hypochlorhydria (HC), both of which are precursors of gastric cancer. On the other hand, duodenal ulcers are associated with an antral predominant pattern of gastritis, high acid secretion, and a decreased risk of gastric cancer (9, 14). The host immune response has a strong role in defining the outcome of H. pylori infection, and polymorphisms in genes that control this immune response have been shown to influence risk of gastric cancer (6, 7, 23, 25). More recently, polymorphisms in genes involved in H. pylori recognition have also been shown to be important (11, 18).
Toll-like receptors (TLRs) are important innate immunity regulators that can be activated upon recognition of bacterial and viral ligands known as pathogen-associated molecular patterns (PAMPs). TLR-mediated signaling pathways are primarily NF-κB dependent, NF-κB being a key transcription factor in the release and production of proinflammatory mediators, which have important functions with regard to antigen presentation and controlling the release of antibodies (36). In humans, 10 different TLRs have been identified; in particular, TLR9 recognizes unmethylated CpG oligonucleotides that are abundant in bacterial DNA, with TLR9 expression mainly restricted to plasmacytoid dendritic cells (pDC), monocytes, and B cells (15, 39). CpG-DNA binds to TLR9 and initiates MyD88 recruitment leading to the phosphorylation of IRAK and TRAF6 and ultimately NF-κB activation (19, 21). PAMP recognition by TLR9 is distinctive from the other TLRs that recognize bacterial PAMPs in that recognition takes place on the surface of the endosomal compartment as opposed to the cell surface (15). The merit of interlocalization is to allow TLR9 to interact with pathogens that have previously been phagocytosed and minimize the chance of the recognition of self-antigen. Although pDC levels are low in peripheral blood, their presence is responsible for interleukin-12 (IL-12) and type I interferon (IFN) production that has implications in the activation of NK cells and monocytes and in Th-1 cell differentiation (20).
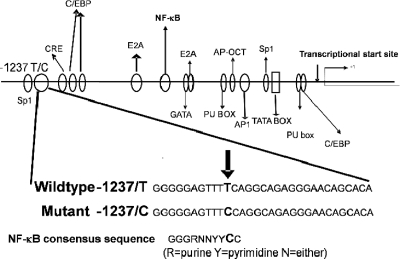
Several single nucleotide polymorphisms (SNPs) have been identified within the TLR9 gene. The −1237T/C SNP (rs5743836) is the most reported and has been shown to be associated with various inflammatory related diseases, including allergic asthma and Crohn's disease (22, 37). In silico analysis of rs5743836 shows that carriage of the variant “C” allele creates a putative NF-κB binding site (Fig. 1). This extra binding site was postulated to enhance the transcriptional activation of TLR9 and potentially affect CpG-DNA-induced activation of proinflammatory chemokines, cytokines, and the adaptive immune response (13). However, a recent report examining the association of rs5743836 with atopic eczema showed that the risk allele was the “T” allele. This finding was published, along with an assessment of promoter activity using a reporter assay, which indicated that basal promoter activity was higher in the TT allelic variant sequence than in the CC allelic variant (29).
FIG. 1.
Schematic diagram showing transcriptional factors and activator binding sites in the TLR9 promoter region. In silico analysis shows that the C allele at position −1237 creates an extra putative NF-κB binding site.
Since both H. pylori infection and TLR9-mediated immune responses are mainly Th-1 phenotype, an increased TLR9 activation through carriage of the risk “C” allele could exaggerate the subsequent inflammatory response. The aim of the present study was to determine whether the TLR9 −1237T/C polymorphism is important with respect to the different clinical outcomes of H. pylori infection and to define the molecular mechanism involved. In order to assess the changes in transcriptional activity, we designed and used a luciferase reporter system containing each of the promoter regions from the TLR9 allelic variants. Changes in binding affinity of NF-κB to the TLR9 promoter region were also assessed using Noshift transcriptional factor (DNA-protein interaction) analysis.
MATERIALS AND METHODS
Study populations.
To determine whether the TLR9 −1237T/C polymorphism is associated with differing outcomes of H. pylori infection, we studied a cohort of 168 healthy Caucasian first-degree relatives of gastric cancer patients from the West of Scotland. These subjects had been extensively investigated in relation to their H. pylori status (assessed by [14C]urea breath test, serology, rapid slide urease test, culture, and histology). Their gastric phenotype was defined histologically by assessment of antral and corpus biopsies for H. pylori density, combined inflammatory scores (active and chronic giving a maximum score of 6, with a range from 0 to 6) and the presence of mucosal atrophy (score of 0 to 3) (31). In addition, these subjects had their peak acid output measured in response to pentagastrin stimulation (PAOpg), and the subjects were designated as having HC if their PAOpg was <15 mmol/h. Corpus atrophy was absent in all subjects with a PAOpg of ≥15 mmol/h. The 168 subjects were then classified into three distinct groups: (i) 51 subjects had H. pylori infection, HC, and gastric atrophy; (ii) 66 subjects had H. pylori infection but no HC or corpus atrophy; and (iii) 51 subjects had no evidence of H. pylori infection and had normal acid secretion and gastric morphology. One hundred randomly selected umbilical cord blood DNA samples from the West of Scotland were used as population controls for the genetic studies. The institutional review boards of the participating centers approved the study, and written informed consent was obtained from all subjects.
TLR9 genotyping.
All genotyping was performed on genomic DNA extracted from leukocytes. The TLR9 −1237T/C polymorphism (rs5743836) was genotyped by using a predesigned Applied Biosystems 5′ nuclease SNP genotyping assay, using minor groove binding probes 5′-labeled with VIC or FAM (6-carboxyfluoresceine) fluorochromes to detect the T or C allele, respectively. For TLR9 −1237T/C, the forward primer 5′-CAGAGACATAATGGAGGCAAAGGA-3′ and the reverse primer 5′-GCCTTGGGATGTGCTGTTC-3′ were used, along with the wild-type probe VIC (CTGCCTGAAAACT) and the variant allele probe FAM (TCTGCCTGGAAACT). Allelic discrimination analyses were prepared by using standard reactions conditions. Real-time and endpoint analyses were performed by using an ABI Prism 7700 sequence detection system (PE Applied Biosystems). The results were confirmed by direct sequencing of selected samples of each genotype. Representative samples from these confirmed genotypes were then taken and used for functional assessment of the polymorphism.
Luciferase reporter assays.
TLR9 promoter sequences (−1471 to +29) from subjects homozygous for wild-type (TLR9 −1237T/T) or variant (TLR9 −1237C/C) genotypes were generated by PCR using the primers TLR9f (5′-CTAGTGGTACCAGCAGGGGAATAAGACGAT-3′) and TLR9r (5′-CAGGGGACTGAGAGCTGTTG-3′). PCR cycling conditions to generate TLR9 promoter sequences were as follows: 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 62.4°C for 30 s, and extension at 68°C for 90 s. The last cycle was followed by an extension step at 68°C for 10 min. The PCR fragments were inserted into the pGL3-Basic vector (Promega, Madison, MI) using the KpnI and XhoI restriction sites, generating TLR9-T-Luc and TLR9-C-Luc. Once the promoter sequences had been successfully incorporated into the reporter vectors, sequence analysis was used to verify that no PCR or cloning errors had been introduced.
HEK293 cells (American Type Culture Collection, Middlesex, United Kingdom) were maintained in α-MEM growth medium supplemented with 1% l-glutamine and 10% (vol/vol) fetal calf serum (Sigma Aldrich, Dorset, United Kingdom), and TLR9-luciferase constructs and an internal control vector —thymidine kinase promoter-driven Renilla plasmid (pRL-TK-Renilla)—were transiently transfected by using Fugene-6 (Roche Diagnostics, East Sussex, United Kingdom) at a ratio of 3:1 (volume [μl] Fugene to mass [μg] DNA constructs). After 8 h of incubation at 37°C in 5% CO2, various stimulants (tumor necrosis factor alpha [TNF-α; 0.25 to 0.75 ng/ml]; lipopolysaccharide [LPS] from Salmonella enterica serotype Typhimurium [Sigma commercial preparation L6143], and H. pylori [ATCC 26695 prepared in-house according to the Westphal method and subsequent lipoprotein purification by ultracentrifugation] [16, 40] at 100 to 500 ng/ml) and E. coli CpG-DNA (5 μg/ml) were added, and the cells were incubated for a further 16 h. The cell lysates were collected and transferred to 96-well black wall plates. Dual Luciferase (Stop and Glow; Promega) measurements were performed by using a luminometric plate reader (Victor3; Wallac, Finland; BioTek Instruments, Winooski, VT). The effect of each stimulant was assessed in six independent experiments with quadruplicate samples for each stimulant concentration. The results are reported as the fold increase in relative luminescence in arbitrary units (RLA) of TLR9 compared to the promoterless control vector pGl3-Basic.
Noshift NF-κB transcriptional factor assays.
Initial Noshift transcriptional factor assays were performed using activated HeLa nuclear extracts as positive control experiments, with subsequent experiments performed using freshly extracted nuclear proteins from HEK293 cells that had undergone stimulation as described previously in the reporter assays. Nuclear protein extraction from HEK293 cells was performed by using a NucBuster nuclear extraction kit according to the manufacturer's protocol (Merck Bioscience, Nottingham, United Kingdom). Protein concentrations of the nuclear protein extracts were determined by using the bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL).
Biotinylated oligonucleotide probes (Sigma Aldrich, Dorset, United Kingdom) for the Noshift assay were designed for the region surrounding the TLR9 −1237T/C polymorphism: wild-type sequence 46T, 5′-CAAAGGAGGGGTCATATGAGACTTGGGGGAGTTTTCAGGCAGAGG-3′; and mutant sequence 46C, 5′-CAAAGGAGGGGTCATATGAGACTTGGGGGAGTTTCCAGGCAGAGG-3′. An unlabeled copy of the probes was synthesized for competitive binding studies.
Noshift transcription factor analysis was performed according to the manufacturer's protocol using a mouse anti-human NF-κB (p65) monoclonal antibody (Merck Bioscience, Nottingham, United Kingdom). Competition studies were performed with either unlabeled competitor DNA supplied in the kit as a standard, unlabeled 46C, and unlabeled 46T used at a concentration 10 times in excess of 46C. The absorbance was measured at 450 nm by using a Labsystem Multiskan photometric plate reader (VWR international, Leicestershire, United Kingdom).
Statistical analysis.
The effect of the TLR9 −1237 T/C polymorphism on acid secretory status and histological parameters (inflammation and atrophy) was assessed by using a Mann-Whitney U test with significance taken at the 5% level. Hardy-Weinberg equilibrium of alleles at individual loci was assessed by χ2 statistics. Odds ratios (OR) with Cornfield 95% confidence intervals (CIs) were computed by logistical regression using STATA software (version 7.0; STATA Press, College Station, TX). ORs for HC were age adjusted (categorized as ≤35, 36 to 45, 46 to 55, and >55 years in age) because of its age dependence, and their CIs were based on robust variance estimates, adjusted for within-family correlation, to account for sampling of several members of a given family.
Luciferase reporter data were calculated as means ± the standard errors of the mean (SEM) unless otherwise stated. Statistical analysis was performed by using either Tukey's post hoc test or an unpaired two-sample t test for equal means using SPSS software (LEAD Technologies, Chicago, IL). DNA-NF-κB interaction results were expressed as means ± the standard deviations (SD) and analyzed using t test statistics with SPSS (LEAD Technologies). P values of ≤0.05 were considered significant.
RESULTS
In the control population, the alleles at the TLR9 −1237 locus were in Hardy-Weinberg equilibrium, with nonsignificant χ2 values. The frequency of the variant allele in the control population was 14%, which is similar to those reported from other Caucasian studies.
Association of TLR9 −1237T/C polymorphism with risk of H. pylori infection and clinical outcome.
The TLR9 polymorphism was not associated with risk of H. pylori infection. Comparing all infected subjects (with or without precancerous abnormalities) to uninfected subjects, the adjusted OR for infection was 1.0 (95% CI = 0.43 to 2.2). However, there was a significantly higher frequency of the variant C allele in H. pylori-infected subjects with HC and gastric atrophy compared to infected subjects with neither abnormality. A total of 26 (51%) of the 51 subjects with HC and atrophy were variant carriers compared to only 14 (21%) of 66 infected subjects without these precancerous changes. The OR of HC/atrophy for carriers of the C allele was 3.9 (95% CI = 1.7 to 8.6), adjusted for age and multiple sampling from the same family (Table 1).
TABLE 1.
Genotype frequencies and adjusted odds ratios for the TLR9 −1237T/C polymorphism
| Subject genotype |
H. pylori-positive subjects |
H. pylori-negative subjects |
Population controls (n = 100) | |||
|---|---|---|---|---|---|---|
| Low acid/atrophy (n = 51) | Normal/high acid (n = 66) | OR (95% CI)a | No. of subjects (n = 51) | OR (95% CI)b | ||
| T/T | 25 | 52 | 1.0 | 35 | 1.0 | 75 |
| T/C | 26 | 14 | 3.9 (1.7-8.6) | 15 | 0.9 (0.2-3.8) | 22 |
| C/C | 0 | 0 | 1 | 3 | ||
The OR (and Cornfield 95% confidence interval) for low-acid versus normal/high-acid conditions, adjusted for age and within-family sampling, is given. ORs were calculated for the carriage of “C” (TT versus T/C + C/C).
The OR for H. pylori-positive versus H. pylori-negative subjects is given.
Effect of TLR9 −1237T/C polymorphism on TLR9 transcriptional activity.
The results from genotyping analyses indicated that carriage of the variant “C” allele is a risk factor for development of premalignant gastric changes in H. pylori-infected subjects. In silico sequence analysis also indicated that the presence of the variant “C” allele created an extra putative NF-κB transcriptional binding site (Fig. 1). In order to define the effect on promoter activity caused by the TLR9 −1237T/C polymorphism, luciferase reporter constructs were generated that contained either the wild-type (TLR9-T-Luc) or the variant (TLR9-C-Luc). TLR9 promoter regions were transfected into HEK293 cells together with a control TK-Renilla reporter plasmid, and the cells were stimulated with TNF-α, LPS, and CpG-DNA at various concentrations.
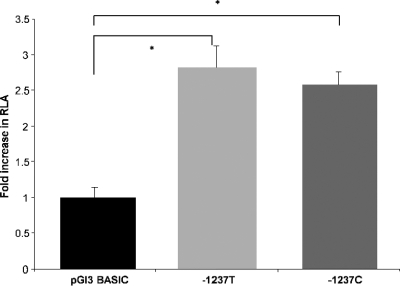
The constructs containing the TLR9 promoter region showed an ∼3-fold increase (P ≤ 0.001) in luciferase activity compared to the promoter-less pGl3-Basic vector (Fig. 2). However, the difference in transcriptional activation between the wild-type and variant constructs under basal conditions was not statistically significant.
FIG. 2.
Basal transcriptional activity of pGl3-Basic and TLR9-T-Luc or TLR9-C-Luc in HEK293. Control plasmid pRL-TK (Renilla) was used for transfection efficiency normalization of luciferase activity. The results are reported as the fold increase in RLA of TLR9 compared to the promoterless control vector pGl3-Basic. The figure shows the mean ± the SEM of results obtained from six experiments, each performed in quadruplicate. The statistical significance of differences in luciferase activity between TLR9-T-Luc and TLR9-C-Luc was assessed by using Tukey's post hoc test (*, P < 0.001).
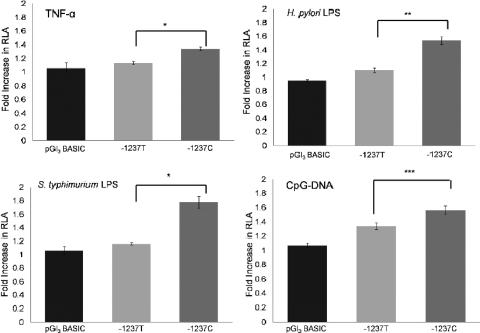
Since the “C” variant of the promoter is predicted to introduce an additional NF-κB consensus site, we next studied promoter activity in response to activators of the NF-κB pathway. A range of concentrations of TNF-α were examined for effectiveness at inducing luciferase expression. After 16 h of stimulation, 0.5 ng of TNF-α/ml was the most effective concentration and gave maximal transcriptional activation for both wild-type and variant TLR9 promoter constructs (data not shown). TNF-α induced a ∼20% increase in signal for the wild type (TLR9-T-Luc)-containing vector compared to a 40% increase seen in the variant (TLR9-C-Luc)-containing vector (P ≤ 0.001) (Fig. 3).
FIG. 3.
Reporter analysis of TLR9 promoter variants in response to various stimulants. HEK293 cells were transiently transfected with promoterless pGl3-Basic, TLR9-T-Luc, or TLR9-C-Luc. The transfection efficiency was normalized by cotransfection of a pRL-TK Renilla control plasmid. Cultures were stimulated with, TNF-α (0.5 ng/ml), serovar Typhimurium and H. pylori LPS (200 ng/ml) or CpG-DNA (5 μg/ml) for 16 h before the cells were lysed for luciferase measurements. The results are reported as the fold increase in RLA of the TLR9 constructs compared to the promoterless pGl3-Basic vector. The data represent the mean ± the SEM of six experiments each performed in quadruplicate. The statistical significance of differences in luciferase activity between TLR9-T-Luc and TLR9-C-Luc was assessed by using the Student t test (unpaired) (*, P < 0.001, **, P < 0.025, ***, P < 0.01).
The effect of the “C” variant on transcriptional activation was also assessed after stimulation with microbial ligands, including LPS derived from S. Typhimurium and H. pylori (both assessed at between 100 and 500 ng/ml) and CpG-DNA (5 μg). An increase in transcriptional activity was observed with both S. Typhimurium and H. pylori LPS over the range of concentrations (data not shown), with the 200-ng/ml concentration giving a maximal increase for both LPS formulations. The pattern of increase in transcriptional activity was similar, with S. Typhimurium LPS and H. pylori LPS showing 16 and 9% elevations in transcriptional activity of the wild type (TLR9-T-Luc)-containing vector compared to a 77 and 53% increase when the cells were transfected with the variant (TLR9-C-Luc; P ≤ 0.01 for S. Typhimurium LPS and P ≤ 0.025 for H. pylori LPS). CpG-DNA-induced promoter transcriptional activity was also more significant in cells transfected with the C allelic variant containing vector (TLR9 −1237C) than the wild type (TLR9-T-Luc) (Fig. 3), with a 31% increase in transcriptional activity observed (P ≤ 0.025). Taken together, these data show that the presence of the extra putative NF-κB binding site within the C allelic variant promotes TLR9 transcription in response to various stimuli more effectively than the wild-type TLR9 −1237T sequence.
Comparison of NF-κB binding affinity between wild-type and variant TLR9 −1237 promoter sequences.
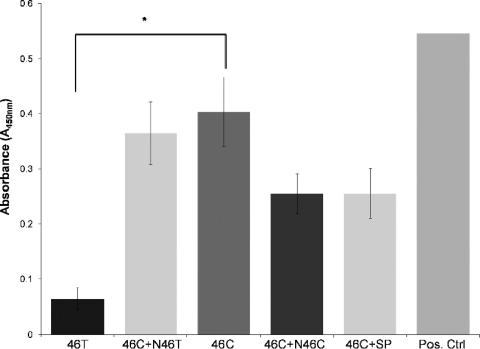
In order to confirm that NF-κB binding was responsible for the increase in transcriptional activity seen in the variant (TLR9-C-Luc) containing vector, the sequence encompassing the polymorphism was analyzed for the binding capacity of potential NF-κB transcriptional binding sites by using Noshift transcriptional factor analysis. The interaction between NF-κB and both TLR9 −1237T and TLR9 −1237C allelic variants was assessed initially using 46-bp biotin-labeled oligonucleotides and HeLa cell nuclear protein extracts (positive control). The results showed that NF-κB could bind to both wild-type 46T (TLR9 −1237T) and variant 46C (TLR9 −1237C), but the binding affinity of variant 46C was significantly higher compared to the wild-type 46T (unpaired t test; *, P < 0.01) (Fig. 4). These findings were further substantiated through competitor studies using unlabeled wild-type 46T probe (N46T), unlabeled 46C probe (N46C), and unlabeled positive control standard NF-κB probe (SP), with the inclusion of the unlabeled wild-type 46T probe showing a nonsignificant level of change compared to the use of 46C alone.
FIG. 4.
Noshift assays using HeLa nuclear extracts were performed to assess the difference in binding affinity to NF-κB between the two allelic variant promoters, 46C and 46T, using HeLa nuclear extracts. Competition studies with unlabeled 46C (N46C), 46T (N46T), and standard unlabeled NF-κB competitor probes (SP) were performed. Unlabeled probes were added 10 times in excess compared to 46C by using the Student t test (unpaired) (*, P < 0.01). The figure shows the means ± the SD of results obtained in three separate experiments.
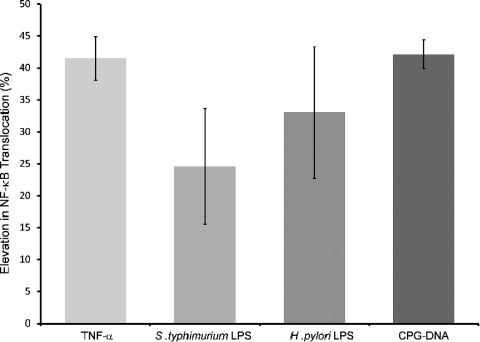
In order to correlate the results obtained from our reporter assays with our DNA-protein interaction analysis, it was essential to assess the binding affinity of the variant promoter region sequence to nuclear protein extracts obtained from HEK293 cells after stimulation with the same stimulants as for the previously described luciferase reporter assays. A twofold increase in NF-κB/DNA interactions was observed when the 46C probe was incubated with nuclear extracts of nonstimulated HEK293 cells compared to the 46T probe. The pattern of NF-κB-DNA interaction was similar to that detected previously using HeLa nuclear protein extracts (data not shown). This experiment was then repeated with HEK293 nuclear extracts that had been subjected to stimulation with the various stimulants used in the reporter assays. The increase in NF-κB after stimulation was calculated relative to the increase seen with nuclear protein extracts of unstimulated HEK293 cells. An increase in nuclear NF-κB was detected with TNF-α, LPS from S. Typhimurium and H. pylori, and CpG-DNA (Fig. 5). The maximal increase in nuclear NF-κB varied according to the stimulant and also occurred in a time-dependent manner (data not shown), with TNF-α- and LPS-stimulated extracts showing maximal increases within 30 min compared to CpG-DNA stimulation, whose maximal increase was detected at 8.5 h (Fig. 5).
FIG. 5.
46C NF-κB interaction analysis utilizing nuclear protein extracts obtained from HEK293 cells stimulated with TNF-α (0.5 ng/ml for 20 min), S. Typhimurium and H. pylori LPS (both at 200 ng/ml for 30 min), and CpG-DNA (0.5 μg/ml for 8.5 h). The percent increase in NF-κB of individual stimulation was calculated by normalizing to the negative, i.e., unstimulated, control. Binding activity is clearly observed between 46C and HEK293 NF-κB, and the amount of increase in NF-κB nuclear translocation correlates with the increase in luciferase activity, i.e., the TLR9 promoter activity, shown earlier. The figure shows the means ± the SD of results obtained in three separate experiments.
DISCUSSION
Toll receptors play a crucial role within the innate immune system. In the case of TLR9, its role is primarily associated with the maturation of dendritic cells and the release of proinflammatory cytokines via activation of NF-κB (19). The TLR9 gene, although highly conserved across species, has distinct sequence variations between hosts that in turn elicit different responses to CpG motifs. These variations may partly explain how genetic polymorphisms in TLR9 play a role in disease risk. Interestingly, however, few studies to date have shown a positive correlation between SNPs in TLR9 and disease susceptibility (8).
Of the four common TLR9 SNPs, the −1237T/C promoter polymorphism has been the most evaluated for association with various diseases (13, 22, 28, 29, 37). Lazarus et al. performed a case control study reporting that the C allele was a risk factor for asthma but with only marginal statistical significance (22). In contrast, Novak et al. suggested that the T allele was associated with increased risk of atopic eczema (29). Since evidence already existed to indicate that carriage of the C allele created a putative NF-κB binding site, we speculated that the TLR9 −1237 C allele would be associated with an increased inflammatory state, which in our disease model meant increased gastric damage characterized by the presence of HC and gastric atrophy, the premalignant states of gastric cancer seen in H. pylori-positive subjects.
Our findings showed that the C allele at TLR9 −1237 was associated with a significantly increased risk of HC and gastric atrophy with an OR of 3.9 (95% CI = 1.7 to 8.6). This finding represents the second TLR polymorphism that our group has shown to increase an individual's risk of developing gastric cancer. Previously TLR4 +896G carriers were shown to have an 11-fold (95% CI = 2.5 to 48) increased OR of developing gastric cancer (18). The association of the TLR4 +896G polymorphism and also the TLR4 +1196T polymorphism with increased risk of gastric cancer have subsequently been assessed in several studies, although their prevalence is known to vary dramatically depending on the ethnic background (1, 32, 38, 41).
We subsequently assessed the functionality of the TLR9 −1237 polymorphism for transcriptional activity and binding affinity to NF-κB. Our data suggest that TLR9 transcriptional activity of the variant C allele is consistently higher than the wild-type T allele when innate immunity pathways are activated. We also demonstrated that this increase in TLR9 transcriptional activity is due to increased NF-κB activation and binding to the TLR9 promoter region when the C allele is present.
Recently, data have been published examining the functional activity of the TLR9 −1237T/C SNP under basal conditions, which showed that the T allele is transcribed more effectively than the C allele (29). Under similar experimental conditions, i.e., basal, our findings are in concordance with Novak et al. in that the wild-type construct elicits higher transcriptional activity compared to the variant C allele. However, unlike Novak et al., we did not observe a statistically significant difference between constructs. Assuming that nuclear NF-κB concentrations are low prior to stimulation, assessing TLR9 basal transcriptional activity is a suboptimal approach to validating the functionality of this polymorphism in the context of an infectious disease model such as our study. However, taking the major differences of the etiology between allergic and infectious disease models into the account, assessing basal transcriptional activity of the allelic variants of the −1237T/C polymorphism may not be an inappropriate method in the atopic eczema study. This may also help to explain the apparent opposite association with risk alleles between atopic eczema and infectious inflammatory diseases. Nevertheless, further clarification is required to identify not only the competitiveness of NF-κB binding caused by the creation of an extra NF-κB binding site but also how the extra NF-κB binding on the TLR9 promoter region of the C allele carriers leads to the formation of a more effective transcriptional machinery. Previous studies have suggested that H. pylori LPS is less potent than other LPS preparations at inducing NF-κB-dependent proinflammatory cytokine production (26, 34, 42). In the present study, all LPS preparations were ultrapurified to remove contaminating lipoproteins to ensure activation was purely due to LPS. In the luciferase studies, NF-κB activation seen after H. pylori LPS stimulation was between 30 and 50% lower than the Salmonella LPS levels, suggesting the H. pylori LPS was capable but not as effective at activating NF-κB as Salmonella LPS, a finding which is entirely consistent with the previous studies. Our findings from the NF-κB binding affinity study showed a nonsignificant difference between Salmonella and H. pylori LPS.
One of the main questions relating to H. pylori gastric colonization is how the organism, which primarily resides within the gastric lumen, is recognized by the immune system. It is known that H. pylori can induce both humoral and cellular immune responses, with several studies demonstrating that H. pylori can invade gastric epithelial cells both in vitro (2) and in vivo in the stomachs of humans and monkeys (33). H. pylori was also shown to be in direct contact with immune cells of the lamina propria in the majority of gastritis and gastric cancer cases in a study by Necchi et al. (27). Dendritic cells, which express a variety of TLRs, including TLR9, are an important group of antigen-presenting cells within the gastric lamina propria. Once stimulated, dendritic cells influence the direction of the immune response, and the stimulation of human dendritic cells has been shown to respond directly to H. pylori bacteria (12). H. pylori bacteria have also been shown to bind to the dendritic cell receptor DC-specific ICAM-3-grabbing nonintegrin (DC-SIGN) (3). Cytokine profiling of the gastric mucosa suggests that both H. pylori infection and TLR9 activation induce a predominantly Th-1 immune response (4, 10). Therefore, it is assumed that elevated TLR9 expression increases host sensitivity to H. pylori CpG-DNA, which at least in part enhances the Th-1 phenotype inflammatory effect seen during the infection. TLR9 activation is known to stimulate IL-12/IFN-γ production, which inhibits the expression of IgE receptors on pDCs (20). This is partially responsible for the Th-1 phenotype suppressing Th-2 phenotype inflammation. Interestingly, the TLR9 −1237 “C” allelic variant, although shown to increase TLR9 expression, has been shown to have no effect on total serum IgE levels (5, 28). On this basis, we would have anticipated that the TLR9 −1237 “C” allele would continue to be a risk factor for as long as the chronic inflammatory state persists, i.e., at all stages of the H. pylori-induced gastric cancer process. However, we have examined this possibility in two large case control studies of gastric cancer based in Poland and the United States, but we found no increased risk associated with either study (17). It is clear, therefore, that this genetic risk factor applies at the early stages of the neoplastic process. Other factors assume more significance in the latter stages which culminate in malignant transformation. The TLR9 −1237 polymorphism may be relevant in setting the scene with induction of severe inflammation, and this may allow other factors to assume more significance later on. Having also provided evidence for the potential mechanisms by which this effect occurs, it is now appropriate to assess the effect of the polymorphism on other multistage infectious inflammatory disease processes.
Acknowledgments
This study was supported in part by Cancer Research UK (A6657) and NHS Grampian endowment funds.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Achyut, B. R., U. C. Ghoshal, N. Moorchung, and B. Mittal. 2007. Association of Toll-like receptor-4 (Asp299Gly and Thr399Ileu) gene polymorphisms with gastritis and precancerous lesions. Hum. Immunol. 68:901-907. [DOI] [PubMed] [Google Scholar]
- 2.Amieva, M. R., N. R. Salama, L. S. Tompkins, and S. Falkow. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 4:677-690. [DOI] [PubMed] [Google Scholar]
- 3.Appelmelk, B. J., I. van Die, S. J. van Vliet, C. M. J. E. Vandenbroucke-Grauls, T. B. H. Geijtenbeek, and Y. van Kooyk. 2003. Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 170:1635-1639. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1:63-96. [DOI] [PubMed] [Google Scholar]
- 5.Berghofer, B., T. Frommer, I. R. Konig, A. Ziegler, T. Chakraborty, G. Bein, and H. Hackstein. 2005. Common human Toll-like receptor 9 polymorphisms and haplotypes: association with atopy and functional relevance. Clin. Exp. Allergy 35:1147-1154. [DOI] [PubMed] [Google Scholar]
- 6.El-Omar, E. M., C. S. Rabkin, M. D. Gammon, T. L. Vaughan, H. A. Risch, J. B. Schoenberg, J. L. Stanford, S. T. Mayne, J. Goedert, W. J. Blot, J. F. Fraumeni, and W. Chow. 2003. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124:1193-1201. [DOI] [PubMed] [Google Scholar]
- 7.El-Omar, E. M., M. Carrington, W. H. Chow, K. E. McColl, J. H. Bream, H. A. Young, J. Herrera, J. Lissowska, C. C. Yuan, N. Rothman, G. Lanyon, M. Martin, J. F. Fraumeni, Jr., and C. S. Rabkin. 2000. The role of interleukin-1 polymorphisms in the pathogenesis of gastric cancer. Nature 404:398-402. [DOI] [PubMed] [Google Scholar]
- 8.El-Omar, E. M., M. T. Ng, and G. L. Hold. 2008. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene 27:244-252. [DOI] [PubMed] [Google Scholar]
- 9.El-Omar, E. M., K. Oien, L. S. Murray, A. El-Nujumi, A. Wirz, D. Gillen, C. Williams, G. Fullarton, and K. E. McColl. 2000. Increased prevalence of precancerous changes in relatives of gastric cancer patients: critical role of Helicobacter pylori. Gastroenterology 118:22-30. [DOI] [PubMed] [Google Scholar]
- 10.Gangloff, S. C., and M. Guenounou. 2004. Toll-like receptors and immune response in allergic disease. Clin. Rev. Allergy Immunol. 26:115-125. [DOI] [PubMed] [Google Scholar]
- 11.Goto, Y., T. Ando, K. Yamamoto, A. Tamakoshi, E. El-Omar, H. Goto, and N. Hamajima. 2006. Association between serum pepsinogens and polymorphism of PTPN11 encoding SHP-2 among Helicobacter pylori-seropositive Japanese. Int. J. Cancer 118:203-208. [DOI] [PubMed] [Google Scholar]
- 12.Guiney, D. G., P. Hasegawa, and S. P. Cole. 2003. Helicobacter pylori preferentially induces interleukin 12 (IL-12) rather than IL-6 or IL-10 in human dendritic cells. Infect. Immun. 71:4163-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamann, L., C. Glaeser, A. Hamprecht, M. Gross, A. Gomma, and R. R. Schumann. 2006. Toll-like receptor (TLR)-9 promoter polymorphisms and atherosclerosis. Clin. Chim. Acta 364:303-307. [DOI] [PubMed] [Google Scholar]
- 14.Hansson, L. E., O. Nyren, A. W. Hsing, R. Bergstrom, S. Josefsson, W. H. Chow, J. F. Fraumeni, Jr., and H. O. Adami. 1996. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N. Engl. J. Med. 335:242-249. [DOI] [PubMed] [Google Scholar]
- 15.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 16.Hirschfeld, M., Y. Ma, J. H. Weis, S. N. Vogel, and J. J. Weis. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165:618-622. [DOI] [PubMed] [Google Scholar]
- 17.Hold, G. L., C. S. Rabkin, M. D. Gammon, S. H. Berry, M. G. Smith, J. Lissowska, H. A. Risch, W. H. Chow, N. A. G. Mowat, T. L. Vaughan, and E. M. El-Omar. 2009. CD14−159C/T and TLR9−1237T/C polymorphisms are not associated with gastric cancer risk in Caucasian populations. Eur. J. Cancer Prev. 18:117-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hold, G. L., C. S. Rabkin, W. H. Chow, M. G. Smith, M. D. Gammon, H. A. Risch, T. L. Vaughan, K. E. McColl, J. Lissowska, W. Zatonski, J. B. Schoenberg, W. J. Blot, N. A. Mowat, J. F. Fraumeni, Jr., and E. M. El-Omar. 2007. A functional polymorphism of Toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology 132:905-912. [DOI] [PubMed] [Google Scholar]
- 19.Krieg, A. M. 2002. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20:709-760. [DOI] [PubMed] [Google Scholar]
- 20.Krug, A., A. Towarowski, S. Britsch, S. Rothenfusser, V. Hornung, R. Bals, T. Giese, H. Engelmann, S. Endres, A. M. Krieg, and G. Hartmann. 2001. Toll-like receptor expression reveals CpG DNA as a unique microbial stimulus for plasmacytoid dendritic cells which synergizes with CD40 ligand to induce high amounts of IL-12. Eur. J. Immunol. 31:3026-3037. [DOI] [PubMed] [Google Scholar]
- 21.Latz, E., A. Schoenemeyer, A. Visintin, K. A. Fitzgerald, B. G. Monks, C. F. Knetter, E. Lien, N. J. Nilsen, T. Espevik, and D. T. Golenbock. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190-198. [DOI] [PubMed] [Google Scholar]
- 22.Lazarus, R., W. T. Klimecki, B. A. Raby, D. Vercelli, L. J. Palmer, D. J. Kwiatkowski, E. K. Silverman, F. Martinez, and S. T. Weiss. 2003. Single-nucleotide polymorphisms in the Toll-like receptor 9 gene (TLR9): frequencies, pairwise linkage disequilibrium, and haplotypes in three U.S. ethnic groups and exploratory case-control disease association studies. Genomics 81:85-91. [DOI] [PubMed] [Google Scholar]
- 23.Lochhead, P., and E. M. El-Omar. 2007. Helicobacter pylori infection and gastric cancer. Best Pract. Res. Clin. Gastroenterol. 21:281-297. [DOI] [PubMed] [Google Scholar]
- 24.Macarthur, M., G. L. Hold, and E. M. El-Omar. 2004. Inflammation and cancer. II. Role of chronic inflammation and cytokine gene polymorphisms in the pathogenesis of gastrointestinal malignancy. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G515-G520. [DOI] [PubMed] [Google Scholar]
- 25.Machado, J. C., C. Figueiredo, P. Canedo, P. Pharoah, R. Carvalho, S. Nabais, C. Castro Alves, M. L. Campos, L. J. Van Doorn, C. Caldas, R. Seruca, F. Carneiro, and M. Sobrinho-Simoes. 2003. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 125:364-371. [DOI] [PubMed] [Google Scholar]
- 26.Mandell, L., A. P. Moran, A. Cocchiarella, J. M. Houghton, N. Taylor, J. G. Fox, T. C. Wang, and E. A. Kurt-Jones. 2004. Intact gram-negative Helicobacter pylori, Helicobacter felis, and Helicobacter hepaticus bacteria activate innate immunity via Toll-like receptor 2 but not Toll-like receptor 4. Infect. Immun. 72:6446-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Necchi, V., M. E. Candusso, F. Tava, O. Luinetti, U. Ventura, R. Fiocca, V. Ricci, and E. Solcia. 2007. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by helicobacter pylori. Gastroenterology 132:1009-1023. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi, E., F. Nishimura, H. Fukai, J. Kim, K. Ichikawa, M. Shibasaki, and T. Arinami. 2004. An association study of asthma and total serum immunoglobulin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin. Exp. Allergy 34:177-183. [DOI] [PubMed] [Google Scholar]
- 29.Novak, N., C. F. Yu, C. Bussmann, L. Maintz, W. M. Peng, J. Hart, T. Hagemann, A. Diaz-Lacava, H. J. Baurecht, N. Klopp, S. Wagenpfeil, H. Behrendt, T. Bieber, J. Ring, T. Illig, and S. Weidinger. 2007. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy 62:766-772. [DOI] [PubMed] [Google Scholar]
- 30.Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 31.Price, A. B. 1991. The Sydney system: histological division. J. Gastroenterol. Hepatol. 6:209-222. [DOI] [PubMed] [Google Scholar]
- 32.Santini, D., S. Angeletti, A. Ruzzo, G. Dicuonzo, S. Galluzzo, B. Vincenzi, A. Calvieri, F. Pizzagalli, N. Graziano, and E. Ferraro. 2008. Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in gastric cancer of intestinal and diffuse histotypes. Clin. Exp. Immunol. 154:360-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semino-Mora, C., S. Q. Doi, A. Marty, V. Simko, I. Carlstedt, and A. Dubois. 2003. Intracellular and interstitial expression of helicobacter pylori virulence genes in gastric precancerous intestinal metaplasia and adenocarcinoma. J. Infect. Dis. 187:1165-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, M. F., A. Mitchell, G. Li, S. Ding, A. M. Fitzmaurice, K. Ryan, S. Crowe, and J. B. Goldberg. 2003. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-κB activation and chemokine expression by epithelial cells. J. Biol. Chem. 278:32552-32560. [DOI] [PubMed] [Google Scholar]
- 35.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 36.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 37.Torok, H. P., J. Glas, L. Tonenchi, G. Bruennler, M. Folwaczny, and C. Folwaczny. 2004. Crohn's disease is associated with a Toll-like receptor-9 polymorphism. Gastroenterology 127:365-366. [DOI] [PubMed] [Google Scholar]
- 38.Trejo-de la O, A., J. Torres, M. Pérez-Rodríguez, M. Camorlinga-Ponce, L. F. Luna, J. M. Abdo-Francis, E. Lazcano, and C. Maldonado-Bernal. 2008. TLR4 single-nucleotide polymorphisms alter mucosal cytokine and chemokine patterns in Mexican patients with Helicobacter pylori-associated gastroduodenal diseases. Clin. Immunol. 129:333-340. [DOI] [PubMed] [Google Scholar]
- 39.Vollmer, J. 2006. TLR9 in health and disease. Int. Rev. Immunol. 25:155-181. [DOI] [PubMed] [Google Scholar]
- 40.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-91. [Google Scholar]
- 41.Wu, M. S., T. Y. Cheng, C. T. Shun, M. T. Lin, L. C. Chen, and J. T. Lin. 2006. Functional polymorphisms of CD14 and Toll-like receptor 4 in Taiwanese Chinese with Helicobacter pylori-related gastric malignancies. Hepatogastroenterology 53:807-810. [PubMed] [Google Scholar]
- 42.Yokota, S., T. Okabayashi, M. Rehli, N. Fujii, and K. Amano. 2010. Helicobacter pylori lipopolysaccharides upregulate Toll-like receptor 4 expression and proliferation of gastric epithelial cells via MEK1/2-ERK1/2 mitogen-activated protein kinase pathway. Infect. Immun. 78:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]