Abstract
Outer membrane iron receptors are some of the major surface entities that are critical for meningococcal pathogenesis. The gene encoding the meningococcal hemoglobin receptor, HmbR, is both independently transcribed and transcriptionally linked to the upstream gene hemO, which encodes a heme oxygenase. The MisR/S two-component system was previously determined to regulate hmbR transcription, and its hemO and hmbR regulatory mechanisms were characterized further here. The expression of hemO and hmbR was downregulated in misR/S mutants under both iron-replete and iron-restricted conditions, and the downregulation could be reversed by complementation. No significant changes in expression of other iron receptors were detected, suggesting that the MisR/S system specifically regulates hmbR. When hemoglobin was the sole iron source, growth defects were detected in the mutants. Primer extension analysis identified a promoter upstream of the hemO-associated Correia element (CE) and another promoter at the proximal end of CE, and processed transcripts previously identified for other cotranscribed CEs were also detected, suggesting that there may be posttranscriptional regulation. MisR directly interacts with sequences upstream of the CE and upstream of the hmbR Fur binding site and thus independently regulates hemO and hmbR. Analysis of transcriptional reporters of hemO and hmbR further demonstrated the positive role of the MisR/S system and showed that the transcription of hmbR initiated from hemO was significantly reduced. A comparison of the effects of the misS mutation under iron-replete and iron-depleted conditions suggested that activation by the MisR/S system and iron-mediated repression by Fur act independently. Thus, the expression of hemO and hmbR is coordinately controlled by multiple independent regulatory mechanisms, including the MisR/S two-component system.
Neisseria meningitidis, an obligate human pathogen, is a leading cause of meningitis and rapidly fatal sepsis, usually in otherwise healthy individuals. Normally a commensal inhabitant of the human nasopharyngeal mucosal surface, meningococcus can occasionally cross the epithelial layer and gain access to the bloodstream, and replication in the bloodstream can result in septicemia. Subsequently, meningococci may also cross the blood-brain barrier to cause meningitis. Iron is essential for almost all microorganisms, and thus the survival of bacterial pathogens in their hosts depends on their ability to scavenge iron (55). The free iron concentration in the human body is extremely low, and most of the iron is associated with various proteins. Heme and hemoglobin (Hb) are the most abundant sources of iron in the body, and the majority of iron in the blood is sequestered in hemoglobin (35, 43). Meningococci are equipped with several TonB-dependent receptors for acquiring iron from various sources available at different sites (37). Two outer membrane binary receptor complexes, TbpA/B and LbpA/B, are involved in human transferrin and lactoferrin acquisition, respectively (13). A two-component transport system, HpuA/B, enables meningococci to use iron from hemoglobin and the hemoglobin-haptoglobin complex (28), and another hemoglobin-binding outer membrane receptor, HmbR, is believed to extract heme from hemoglobin and subsequently transport it into the periplasm (47).
While HpuBA-dependent hemoglobin utilization was not able to distinguish between different sources of hemoglobin (48), the efficiency of utilization of different hemoglobins by HmbR-expressing N. meningitidis was shown to be species specific, and human hemoglobin was the best source of iron. An hmbR mutant was cleared from the bloodstream much more rapidly than the wild-type parent strain in an infant rat infection model, indicating the importance of HmbR-dependent hemoglobin utilization in survival in the bloodstream (47). hmbR is located downstream of hemO, encodes a heme oxygenase responsible for heme degradation (57), and is in an exchangeable meningococcal genomic island (23), indicating that this virulence factor was recently acquired. Interestingly, of the best-characterized meningococcal TonB-dependent iron receptors, only receptors targeting hemoglobin undergo phase variation, which is mediated through slip-strand mispairing of the poly(G) tract in the coding sequences of hmbR and hpuA (27, 38). It is believed that meningococci employ “on-off” phase variation to evade the host immune response. In addition, most outer membrane receptors for various iron sources are controlled by Fur-dependent iron regulation (11, 37). Fur interacts with ferrous iron and binds to the Fur box, which in most cases overlaps the promoters of iron-regulated genes, resulting in inhibition of transcription. Regulation of hmbR by iron availability (48) and by direct binding of the Fur protein to the hmbR promoter has been demonstrated (10, 44).
Two-component regulatory systems are some of the most common signal transduction mechanisms governing bacterial responses and adaptation to environmental changes (19). Many such systems act globally to coordinate expression of virulence determinants. These signal transduction systems generally consist of a sensor histidine kinase and a response regulator protein. When a signal is sensed, the histidine kinase undergoes autophosphorylation, and the phosphoryl group is subsequently transferred to the response regulator, modifying its activity. Mutation in the meningococcal MisR/S system has been shown to increase sensitivity to cationic antimicrobial peptides and polymyxin B (22), which correlates with the loss of all phosphoethanolamine decorations of the lipooligosaccharide inner core heptose II residue (50). In addition, a misR mutation was shown to cause attenuation of virulence in a murine model of meningococcal infection (34). Previous microarray studies of the MisR/S regulon have shown that inactivation of misR results in decreased expression of hmbR (51). Control of hmbR and hemO expression by the MisR/S system was further characterized in this study. The role of the MisR/S two-component system in regulating hemoglobin-dependent growth was demonstrated, and MisR was shown to specifically interact with the hmbR promoter sequence and the sequence upstream of the hemO-associated Correia element (CE). Thus, the expression of hemO and hmbR, in addition to responding to iron availability via Fur repression, is coordinately regulated by the MisR/S two-component system when as-yet-unidentified host signals are sensed.
MATERIALS AND METHODS
Plasmids, strains, and media.
The strains and plasmids used in this study are listed in Table 1. N. meningitidis strain NMB (= CDC 8201085) is a serogroup B meningococcal strain that was originally isolated in 1982 from the cerebrospinal fluid of a patient with meningococcal meningitis in Pennsylvania. Meningococcal strains were grown with 5% CO2 at 37°C on GC base (GCB) (Difco) agar containing 0.4% glucose and 0.68 mM Fe(NO3)3 or in GC broth supplemented with 0.4% glucose, 0.68 mM Fe(NO3)3, and 0.043% NaHCO3. Brain heart infusion (BHI) medium (37 g/liter) with 1.25% fetal bovine serum was used when kanamycin selection was required. The Escherichia coli DH5α and Top10 strains were routinely grown in Luria-Bertani broth for cloning and propagation of plasmids. N. meningitidis was transformed by using the procedure of Janik et al. (20). E. coli strains were transformed by using chemical competence (Top10) or by electroporation (DH5α) with a GenePulser (Bio-Rad) used according to the manufacturer's protocol. The antibiotic concentrations used were as follows: for N. meningitidis, 80 μg/ml kanamycin and 3 μg/ml erythromycin; and for E. coli strains, 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 300 μg/ml erythromycin.
TABLE 1.
Strains and plasmids used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| N. meningitidis strains | ||
| NMB | B:2b:P1.2,5:L2 (CDC8201085) | 45 |
| IR3261 | NMB hpuB::erm hmbRoff | 38 |
| IR3287 | NMB hpuB::erm hmbRon | 38 |
| NMB310 | NMB misS::aphA-3 | 52 |
| YT391 | NMB310 complemented with Plac::misS at the lctP-aspC locus | 42 |
| SZT1001/YT0336 | NMB misR::aphA-3 | 51 |
| C1001 | NMB misR::aphA-3 complemented with Ptrc::misR in the iga locus | 51 |
| SZT1003 | NMB misRS::aphA-3 | 51 |
| SZT1004 | IR3261 misR::aphA-3 | This study |
| SZT1005 | IR3261 misS::aphA-3 | This study |
| SZT1006 | IR3261 misRS::aphA-3 | This study |
| SZT1009 | IR3287 misR::aphA-3 | This study |
| SZT1010 | IR3287 misS::aphA-3 | This study |
| GMT101 | PhmbR::lacZ | This study |
| GMT107 | PhmbR::lacZ ΔmisS | This study |
| O101 | lacZ-erm cassette inserted into BamHI site of hemO | This study |
| O101s | hemO::lacZ ΔmisS | This study |
| R103 | lacZ-erm cassette inserted into HincII site of hmbR | This study |
| R103s | hmbR::lacZ ΔmisS | This study |
| O1101 | PhemO1::lacZ | This study |
| O1101s | PhemO1::lacZ ΔmisS | This study |
| O2002 | PhemO2::lacZ | This study |
| O2002s | PhemO2::lacZ ΔmisS | This study |
| O305 | PhemO::lacZ with 918-bp promoter fragment | This study |
| O305s | PhemO::lacZ with 918-bp promoter fragment, ΔmisS | This study |
| O501 | PhemO::lacZ with 1,104-bp promoter fragment | This study |
| O501s | PhemO::lacZ with 1,104-bp promoter fragment, ΔmisS | This study |
| R1203 | PhemO::lacZ with 1,220-bp promoter fragment | This study |
| R1203s | PhmbR::lacZ with 1,220-bp promoter fragment, ΔmisS | This study |
| E. coli strains | ||
| DH5α | Cloning strain | New England Biolabs |
| TOP10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pUC18k | Source of aphA-3 cassette | 32 |
| pCR2.1 | TOPO cloning vector | Invitrogen |
| pSmartLCKan | Blunt-end cloning vector | Lucigen |
| pYT328 | Cloning vector for chromosomal lacZ fusion | 52 |
| pYT338 | 5′ sequence (YT174-YT185) and 3′ sequence (YT186-YT141) of misR inserted into HindIII-KpnI site and EcoRI site of pCR2.1 | This study |
| pSZ001 | aphA-3 cassette inserted into KpnI-BamHI site of pYT338 | This study |
| pGM1 | PCR product obtained with hmbR-pF2 and hmbR-PR in pCR2.1 | This study |
| pGM2 | EcoRI fragment of pGM1 inserted into EcoRI site of pYT328 | This study |
| pPK-hemO | PCR fragment obtained with hemO-pF1-ERI and hmbR-pR-ERI cloned into pSmartLCKan | This study |
| phemO101 | lacZ-erm cassette inserted into BamHI site of pPK-hemO | This study |
| pPK-hmbR | PCR fragment obtained with hmbR-pF2 and hmbR-F3R cloned into pCR2.1 | This study |
| phmbR103 | lacZ-erm cassette inserted into HincII site of pPK-hmbR | This study |
| pPK-hemO1101 | PCR fragment obtained with hemO-pF1-ER and hemO-pR1-ER inserted into pYT328 | This study |
| pPK-hemO2002 | PCR fragment obtained with hemO-pF2-ER and hemO-pR1-ER inserted into pYT328 | This study |
| pPK-hemO305 | PCR fragment obtained with hemO-pF1-ER and hemO-pR3-ER inserted into pYT328 | This study |
| pPK-hemO501 | PCR fragment obtained with hemO-pF1-ER and hemO-pR5-ER inserted into pYT328 | This study |
| pPK-hmbR1203 | PCR fragment obtained with hemO-pF1-ER and hmbR-pR-ER inserted into pYT328 | This study |
Construction of the misR::aphA-3 mutant.
A nonpolar aphA-3 insertional mutation with deletion of a 510-bp fragment of misR (678 bp) was generated. A 760-bp PCR product containing the misR promoter region and 5′ coding sequence was generated using primers YT174-HindIII and YT185-KpnI, and the resulting DNA fragment was cloned into the pCR2.1 vector, yielding pTA174-185. Another 500-bp PCR product containing the 3′ sequence of misR and a 450-bp 5′ sequence of misS was amplified using primers YT186-BamHI and YT141, and the PCR product was cloned into the pCR2.1 vector, yielding pTA186-141. A 760-bp fragment of plasmid pYT174-185 was obtained by HindIII and KpnI digestion and then subcloned into the corresponding sites in the polylinker of pYT186-141, yielding pYT338. The aphA-3 cassette released from pUC18K (32) by KpnI-BamHI digestion was inserted into the KpnI-BamHI sites of pYT338, yielding pSZ001. Removal of the misR internal sequence and the presence of a correctly oriented aphA-3 cassette in the resulting pSZ001 plasmid were confirmed by performing colony PCRs and a sequence analysis. The construction procedure removed most of misR region and replaced it with a kanamycin resistance gene. To generate the meningococcal misR mutant, pSZ001 was linearized with ScaI to disrupt the bla gene, and the digestion mixture was used to transform meningococcal strains. Colonies were selected on BHI agar plates with kanamycin. Mutants were examined using colony PCR linking the aphA-3 cassette to a chromosome-specific primer, DNA sequencing, and Southern blotting to confirm correct allelic exchange at the chromosomal misRS locus. In addition, Western blotting was performed to confirm the absence of the MisR protein in all misR and misRS mutants (data not shown).
Construction of the hmbR::lacZ transcriptional fusion.
A 536-bp fragment of the hmbR promoter region was obtained by PCR amplification using primers hmbR-p-F2 and hmbR-p-R with chromosomal DNA of N. meningitidis serogroup B strain NMB as the template. The PCR product was cloned into pCR2.1 by TOPO cloning (Invitrogen) to construct pGM1. pGM1 was digested with EcoRI, and the released insert was purified and cloned into the EcoRI site of pYT328 (52) to generate a transcriptional fusion to the lacZ gene that was flanked by the meningococcal NMB0428 and NMB0430 coding sequences. The ligation reaction mixtures were transformed into E. coli, and erythromycin-resistant colonies were selected. Correct orientation of the promoter relative to the lacZ gene was confirmed by colony PCR using an outward primer (YT168) at the 5′ end of lacZ and a forward primer in the cloned promoter fragment. The resulting plasmid, pGM2, was digested with NcoI for linearization and transformed into N. meningitidis NMB and NMB310 (misS::kan). Transformants (strains GMT101 and GMT107, respectively) were selected on plates containing GC medium supplemented with erythromycin or BHI medium supplemented with erythromycin and kanamycin, and integration of the hmbR::lacZ fusion via homologous recombination into an irrelevant intergenic region was verified by PCR amplification using YT168 and chromosome-specific primer YT02.
Similar strategies were used to generate hemO::lacZ reporters and hmbR::lacZ reporters originating from the 5′ region of hemO. Primers hemO-pF1-ERI and hemO-pR1-ERI (418 bp), primers hemO-PF2-ERI and hemO-pR1-ERI (267 bp), primers hemO-pF1-ERI and hemO-pR3-ERI (918 bp), primers hemO-pF1-ERI and hemO-pR5-ERI (1,104 bp), and primers hemO-pF1-ERI and hmbR-pR-ERI (1,220 bp) were used to generate promoter fragments with flanking EcoRI sites using Phusion polymerase (New England Biolab) and the chromosomal DNA of strain NMB as the template. After EcoRI digestion, the fragments were purified and cloned into the EcoRI site of pYT328. The resulting plasmids were linearized with NcoI and used to transform meningococcal strain NMB. Correct transformants were identified by colony PCR as described above and designated O1101, O2002, O305, O501, and R1203, respectively. Reporters in the misS mutant background were generated by transformation of strain NMB310 and designated like the reporters of the wild-type strain but with the suffix s.
The lacZ fusion integrated into the native hmbR locus was constructed as follows. A ∼1.4-kb PCR fragment obtained with primers hmbR-pF2 and hmbR-F3R was cloned into the pCR2.1 vector by TOPO cloning. The resulting plasmid, pPK-hmbR, was digested with HincII, which was followed by dephosphorylation with calf intestine alkaline phosphatase (CIP), and then it was ligated with the lacZ-erm cassette that was released from pAErmC′G (56) by BamHI digestion and filled in with Klenow DNA polymerase. The desired plasmid (designated phmbR103) with correct orientation of the lacZ cassette with respect to the hmbR coding sequence was identified by colony PCR using primers hmbR-pF2 and YT168. The plasmid was linearized with ScaI and then used to transform wild-type strain NMB and misS mutant strain NMB310 to obtain R103 and R103s, respectively. Integration into the hmbR locus via allelic exchange was confirmed by PCR using primers YT168 and hmbR-pg-1b, which annealed to sequences not included in the cloned fragment. A lacZ fusion at the native hemO locus was constructed in an analogous manner. A PCR product generated with primers hemO-pF1-ERI and hmbR-pR-ERI was cloned into the pSmartLCKan vector (Lucigen) by following the manufacturer's protocol. The resulting plasmid, pPK-hemO, was linearized at the unique BamHI site in the insert and ligated with the BamHI-digested lacZ-erm cassette. The orientation of the lacZ cassette was verified by PCR using primers YT168 and hemO-pF2-ERI. Plasmid phemO101 was used to obtain the reporter fusions O101 and O101s in the wild-type strain and the misS mutant, respectively.
β-Galactosidase assays.
β-Galactosidase activity was assayed by the method of Miller (33), and experiments were performed in triplicate. The reporter strains were grown in GC broth with supplements at 37°C with aeration at 200 rpm to mid-log phase (optical density at 600 nm [OD600], ∼0.5). The cultures were then split into two 2-ml cultures and treated with and without 100 μM 2,2′-dipyridyl (Sigma) for the iron-depleted sample and the iron-replete control, respectively.
Hb utilization assay.
Bacteria grown overnight on appropriate selection plates were collected and resuspended in GC broth without supplements at an OD600 of 0.1. Aliquots (100 μl) were plated onto GCB agar plates containing 100 μM deferoxamine mesylate (Desferal; Sigma). Filter disks (diameter, 0.25 in.) impregnated with 10 μl of human hemoglobin (Hb) (5, 3, and 1 mg/ml) or ferric nitrate (5 mg/ml) as a control were placed on the plates. The zones of growth around the disks were recorded after 48 h of incubation at 37°C in the presence of 5% CO2. Six independent experiments were performed.
Determination of the switching frequencies.
Meningococcal strains that were hmbR phase off were isolated on nonselective GCB agar plates and then resuspended in GC broth. Aliquots (100 μl) of 10−1 to 10−3 serial dilutions were plated on GCB agar plates containing 100 μM deferoxamine mesylate (DFS) and 100 μg/ml of Hb (GCB-DFS-Hb plates), while aliquots of 10−5 to 10−8 serial dilutions were plated on nonselective GCB agar plates. The hmbR “off-to-on” switching frequencies were determined by dividing the number of colonies on GCB-DFS-Hb plates by the number of colonies on the nonselective GCB agar plates. Four independent measurements were obtained, and an analysis with Student's t test indicated that there was no statistically significant difference between the parent strain and the mutants.
RNA isolation and real-time qRT-PCR.
Cultures of the wild-type strain and the mutants were grown and treated with dipyridyl as described above. Two volumes of RNAprotect reagent was added to cultures, and RNA was isolated using an RNeasy mini kit (Qiagen) by following the manufacturer's recommendations with on-column DNase digestion for 1 h at room temperature. PCR amplification of the RNA samples using primers hmbR-p-f and hmbR-p-r confirmed that there was no genomic DNA contamination. cDNA was obtained by reverse transcription (RT) of total RNA (1 μg) using a GeneAmp RNA PCR core kit (Applied Biosystems), and reactions without the reverse transcriptase were used as a negative control. The transcription of genes of interest was measured by real-time quantitative RT (qRT)-PCR using the SYBR green detection method (SYBR green Supermix; Bio-Rad); the reaction conditions used have been described previously (50). The constitutive and highly expressed meningococcal ribosomal gene rpsE, which was not affected by the growth conditions, was used as an internal control in each experiment for normalization. The primers designed with Primer 3 software (http://fokker.wi.mit.edu/primer3/) (41) are listed in Table 2, and they were confirmed to yield similar amplification efficiencies and thus were suitable for the 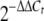 method for determining the relative transcriptional differences between the mutant strains and the wild-type strain. The values for the wild-type strain grown in iron-replete conditions were used for calibration (30). RT negative control reactions were also analyzed to measure whether there was contaminating chromosomal DNA, and melting curve analyses were performed following each RT-PCR experiment to ensure that each reaction mixture contained only one specific product. Each qRT-PCR was performed in triplicate. Student's t test with a two-tailed hypothesis was used to determine the significance of differences (P < 0.01) between two variables in this study.
method for determining the relative transcriptional differences between the mutant strains and the wild-type strain. The values for the wild-type strain grown in iron-replete conditions were used for calibration (30). RT negative control reactions were also analyzed to measure whether there was contaminating chromosomal DNA, and melting curve analyses were performed following each RT-PCR experiment to ensure that each reaction mixture contained only one specific product. Each qRT-PCR was performed in triplicate. Student's t test with a two-tailed hypothesis was used to determine the significance of differences (P < 0.01) between two variables in this study.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| hmbR-p-F2 | GCTTTATTGCGCCGAAGGATCCAATTTG |
| hmbR-p-R | ACCAGCGCGGCAATAGGGAGCAT |
| hmbR-pL-F | CAATCGGCTGGCTTTATTGCGCCGAAGGATCCAATTTG |
| hmbR-pL-R | AAAATACTGCCGACCAGCGCGGCAATAGGGAGCATTT |
| hmbR-pR-ER | CGGAATTCACCAGCGCGGCAATAGGGAGCATT |
| hemO-pF1-ERI | CGGAATTCCCGACTGCCGAACCCATTTGA |
| hemO-pF2-ERI | CGGAATTCAAATCAGGACAAGGCGGCGAAG |
| hemO-pR1-ERI | CGGAATTCCGCCGTGGTATCCGCCTTCA |
| hemO-pR3-ERI | CGGAATTCAACACGACTTTGTAGAATGCAAAG |
| hemO-pR5-ERI | CGGAATTCTAAGACAGTAATCCATGCAAACAAAGCCG |
| hemO-PE1 | TGCTTGATTTTCGGTTTCACTCATATTTTTTCC |
| hemO-PE3 | GTTCCGGTACTATTTGTACTGTCTGC |
| hmbR-F1 | CCCGTTATGGCAACTTCAAC |
| hmbR-R1 | GAAAGTCCGAGCGAAAAATG |
| hemO-F2 | CTTGAAAGATTTGGGCGAAG |
| hemO-R2 | CGCCGTTGTAATCGAGTTTT |
| NMB1436-F1 | TTTACCCATTTCCTGCTTGC |
| NMB1436-R1 | CGTTCGACGTTTTCCATACC |
| YT02 | GGATACCGCAAGAACAGG |
| YT45 | CGTAGATGACGATGCCCTGCTAACCG |
| YT46 | GGCGGATGCTGGAGATGTGTACGTCG |
| YT141 | GCAGGCGGCGGGCTTGGAGTTTG |
| YT168 | GCGGATCAGTCCGGCGTAGAGG |
| YT173 | CTTTGCATTCTACAAAGTCGTGTTG |
| YT174-HindIII | ACCCGGAAGCTTAACGCCTTTTCCC |
| YT185-KpnI | CCCGCTCAGGTACCCCTGTACGCCTGCTT |
| YT186-BamHI | AGTTGGGCGGATCCTCTCTGATTCAAA |
| rpsE F1/YT129 | TGCTAAAGAGGGTAGTGGCG |
| rpsE R1/YT130 | CAAACCATCTAATGTTGCACGT |
| lbpB-F1 | GGAACATCGACCTTTTCCTG |
| lbpB-R1 | TGCTTTTGCCGCTTCTTTAT |
| tbpB-F4 | GCTGCCTGTGTTTTTGTTGA |
| tbpB-R4 | TATCCGCCTTGGTCTTTTTG |
| YT150 | GCGAAATCCTGACGGAATGG |
EMSA and DNase I protection assay.
Both electrophoretic mobility shift assays (EMSA) and DNase I protection assays were performed using previously reported procedures (52). The hmbR promoter fragment (326 bp) was obtained by PCR amplification using primers YT173 and hmbR-p-r, while the probe for DNase I protection assays was prepared by PCR using 32P-labeled primer hmbR-p-r. The hemO promoter fragment (418 bp) was obtained by PCR amplification using primers hemO-pF1-ERI and hemO-pR1-ERI, while the probe for the DNase I protection assays was prepared using 32P-labeled primer hemO-pF1-ERI. The binding reactions were examined with phosphorylated (50 mM acetyl phosphate for 30 min at 37°C) or nonphosphorylated MisR. Competition with excess specific competitors (unlabeled probes) and nonspecific competitors (the 237-bp internal coding sequence of misS obtained by PCR amplification using primers YT150 and YT141) was performed to assess the specificity of the interaction.
Primer extension.
Total RNA (30 μg) was used in primer extension reactions with 32P-labeled hemO-specific primers (hemO-PE1, hemO-PE3, and hemO-pR1-ERI) using a previously described procedure (42). The corresponding DNA sequencing reactions were carried out using the same labeled oligonucleotides and PCR fragments of the promoter regions with a Thermo Sequenase dye primer manual cycle sequencing kit (USB). The extension products and the sequencing ladders were resolved on an 8% sequencing gel.
RESULTS
Expression of hmbR and hemO was reduced in the misR/S mutants.
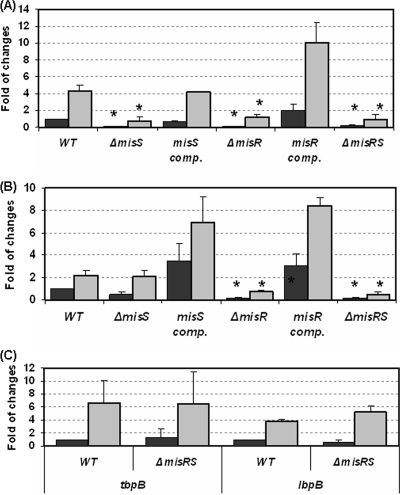
The HmbR hemoglobin receptor was identified as a member of the MisR/S regulon in a microarray study that compared the transcriptional profiles of the misR::erm mutant and the wild-type parent strain grown under iron-replete conditions (51). To better characterize the role of the MisR/S system in hmbR regulation under both iron-rich and iron-limited growth conditions, we measured hmbR-specific mRNA levels in wild-type strain NMB and compared these levels to the levels obtained for the corresponding ΔmisR, ΔmisS, and ΔmisRS mutants by quantitative qRT-PCR. In addition, complemented ΔmisR and ΔmisS mutants were also included in the analysis. RNAs were isolated from mid-log-phase cultures grown in iron-replete medium and iron-depleted medium (treated with 100 μM dipyridyl for 45 min). Dipyridyl is a ferrous iron chelator that rapidly produced an iron-limiting environment by sequestering ferrous iron, a cofactor of the Fur protein that mediates iron regulation. The transcripts of the wild-type strain and the mutants were induced in dipyridyl-treated cultures, confirming the iron-restricted growth conditions and that the iron-dependent transcriptional control occurred in all of the mutants. When cultures were grown under iron-replete conditions, the levels of hmbR transcription in all of the misRS mutants were significantly reduced (Fig. 1A). Similarly, downregulation of hmbR was detected in all mutants under iron-restricted conditions (Fig. 1A). These results were consistent with previous data for an independent misR mutant (51) and suggested that the MisR/S system, either directly or indirectly, plays an activator role in hmbR expression regardless of the Fur repressor activity. The data showing that the misS mutant behaved like the misR mutant suggest that MisR activation via phosphoryl transfer from the MisS kinase is important for hmbR regulation. Complementation of the misR and misS strains, which placed the misR and misS genes under isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter control, rescued the hmbR expression under both growth conditions, confirming that the effect on hmbR expression was due to a defect in the two-component system.
FIG. 1.
qRT-PCR determination of relative transcriptional changes for hmbR (A), hemO (B), and tbpB and lbpB (C) in the misR/S mutants. Total RNAs were isolated from mid-log-phase cultures that were treated (gray bars) or not treated (black bars) with 100 μM dipyridyl for 45 min. The relative transcriptional differences between the mutants and the wild-type strain were calculated by the 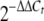 method (30), using the transcriptional level of the wild-type strain under iron-rich conditions as the calibrator. Each qRT-PCR was performed in triplicate and was repeated with at least two independent RNA preparations. The asterisks indicate statistically significant differences between the wild-type strain and the mutant as determined by the Student t test with a two-tailed distribution (P < 0.01). WT, wild type; comp., complemented.
method (30), using the transcriptional level of the wild-type strain under iron-rich conditions as the calibrator. Each qRT-PCR was performed in triplicate and was repeated with at least two independent RNA preparations. The asterisks indicate statistically significant differences between the wild-type strain and the mutant as determined by the Student t test with a two-tailed distribution (P < 0.01). WT, wild type; comp., complemented.
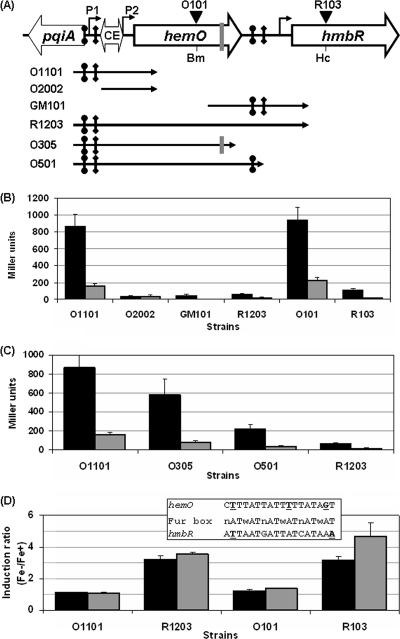
hmbR is downstream of hemO (Fig. 2A) (57). Studies of hemO mutants of a serogroup C strain suggested that hmbR and hemO may be transcriptionally linked (57). Using cDNA generated by a reverse transcriptase reaction performed with a reverse primer complementary to the hmbR coding sequence, we obtained PCR products spanning the hemO-hmbR intergenic region, as well as positive PCR amplification of the hemO gene, confirming that there is also transcriptional coupling of hmbR and hemO in the serogroup B strain NMB (data not shown). Because the qRT-PCR primers for hmbR measured total hmbR transcripts, the significantly decreased hmbR expression suggested that hemO transcription would likely be affected by the misR/S mutations. Thus, we examined hemO expression by using qRT-PCR, and, as shown in Fig. 1B, the transcription of hemO was indeed significantly reduced in the misR and misRS mutants, while the misS mutation resulted in a modest reduction in transcription. Again, the reduced hemO expression could be complemented by a second copy of the corresponding mutated gene, and the levels were higher than those of the wild-type strain, probably due to IPTG-induced overexpression of the two-component proteins. The fact that overexpressing either MisS or MisR appeared to enhance hemO expression in the complemented strains is consistent with a positive role for the two-component system.
FIG. 2.
(A) Organization of the hemO-hmbR locus in meningococcal strain NMB. The hemO gene is transcribed divergently from the pqiA (hmp) gene with a 286-bp intergenic space that includes a full-length Correia element (CE) (6), while hemO is separated from hmbR by a 191-bp intergenic region. The bent arrows represent the promoters of hemO and hmbR. The schematic diagram is not to scale. (B) Sequence of the 5′ region of hemO. The ATG start codons are in bold type and underlined, and the large arrows show the direction of transcription. The sequence of the CE is in gray type, the terminal inverted repeats (TIR) are indicated by dotted arrows, and the IHF binding sequence in the CE is underlined, with the nucleotides matching the consensus sequence nucleotides (7) in bold type. The putative transcript processing sites in the TIRs are indicated by bold type and labeled d and p. The putative Fur binding motif matching the (NATWAT)3 motif is indicated by an arrow. The sequence protected by MisR in the DNase I protection assay is in bold type and shaded, and the sequences matching the MisR binding motif are indicated by double overlining. The 5′ ends of two hemO::lacZ fusions are indicated by bent arrows, and the hemO-pR1-ERI primer is the corresponding 3′ end. The hemO-pF1-ERI primer used to generate probes for DNase I protection assay and the hemO-PE3 primer used in primer extension are indicated by dashed arrows. The two transcriptional start sites and the two corresponding promoter elements are enclosed in boxes. (C) Nucleotide sequence of the hmbR promoter region. The stop codon of hemO and the start codon of hmbR are in bold type and underlined. The putative −10 and −35 hexamers are in bold type and enclosed in boxes. The sequence protected by MisR is shaded, and the two matching MisR consensus motifs are indicated by double underlining. The Fur-protected sequence reported by Delany et al. (10) is indicated by a dashed line. The primers used for the EMSA and DNase I protection assay, YT173 and hmbR-pR, are indicated by dashed arrows. The dotted bent arrows indicate the 3′ ends of promoter fragments cloned in reporter strains O305, O501, and R1203.
To examine whether the MisR/S system specifically affected hmbR and hemO expression, mediating hemoglobin utilization in particular, the levels of transcription of tbpB and lbpB were also measured. tbpB and lbpB are the first genes in the tbpB/A and lbpB/A operons encoding the outer membrane receptors responsible for transferrin and lactoferrin acquisition, respectively. As shown in Fig. 1C, the misRS mutation resulted in no significant changes in expression of these genes. Several studies have demonstrated that these different iron acquisition receptors are all controlled by Fur-dependent repression, which is alleviated upon iron starvation (44, 48). The observation that mutations in the MisR/S system significantly affected only hmbR expression suggested that the regulatory effect of the MisR/S system does not broadly interfere with Fur-dependent regulation. Another Fur-dependent and iron-induced gene, NMB1436, which codes for a putative protein involved in oxidative stress protection (14), was also analyzed. As expected, the levels of the NMB1436 transcript were reduced in the absence of iron, and no significant changes were detected in the misS or misRS mutants (0.98- ± 0.38-fold and 0.69- ± 0.24-fold, respectively). Thus, inactivating the MisR/S two-component system significantly and specifically downregulated the transcription of hmbR and hemO.
Mutation in the misR/S system affected hemoglobin utilization.
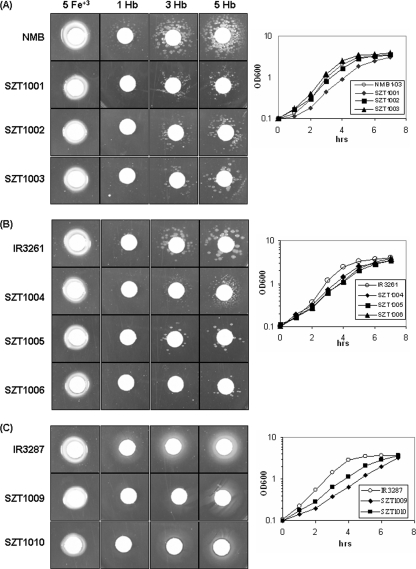
Two TonB-dependent outer membrane receptors have been shown to mediate Hb utilization in N. meningitidis; HmbR enables uptake of hemoglobin, and the HpuA/B binary protein complex allows utilization of both hemoglobin and haptoglobin-Hb complexes (37). Disk diffusion assays were performed to examine whether the decreased hmbR and hemO expression in the two-component mutants affected hemoglobin utilization and consequently altered hemoglobin-dependent growth. The strains were plated onto GCB agar plates containing 100 μM Desferal to remove free iron, and specific iron sources were provided on disks. All strains utilized free iron equally well; however, when Hb was the sole iron source, all mutants showed reduced growth (Fig. 3A). To remove the contribution from the HpuB/A receptor, the misR, misS, and misRS mutations were introduced into a derivative of strain NMB (IR3261) that carries an hpuB::erm mutation to create strains SZT1004, SZT1005 and SZT1006, respectively. As shown in Fig. 3B, all mutants again showed an apparent defect in hemoglobin-dependent growth but not in free-iron-dependent growth.
FIG. 3.
Hemoglobin utilization assay. Bacteria grown overnight on appropriate selection plates were collected and resuspended at an OD600 of 0.1 in GC broth. Aliquots (100 μl) were plated onto GCB agar plates containing 100 μM Desferal. Filter paper disks were placed on the plates and then soaked with 10 μl of a human hemoglobin solution (1, 3, and 5 mg/ml [1 Hb, 3 Hb, and 5 Hb respectively]) or a ferric nitrate solution (5 mg/ml [5 Fe+3]) as a control. The zones of growth around disks were measured after 48 h of incubation at 37°C. The results for one representative of six independent experiments are shown. The growth curves for all of the strains tested in standard GC broth are shown on the right. (A) hmbRoff hpuAoff background; (B) hmbRoff hpuB::erm background; (C) hmbRon hpuB::erm background. NMB, IR3261, and IR3287 are wild-type parental strains. SZT1001, SZT1004, and SZT1009 are misR::aphA-3 mutants. SZT1002, SZT1005, and SZT1010 are misS::aphA-3 mutants. SZT1003 and SZT1006 are misRS::aphA-3 double mutants.
Both meningococcal hmbR and hpuB are phase variable (27, 38) and are “phase off” in the serogroup B parental strain. When Hb was the sole iron source, all strains grew as single colonies due to the off-to-on switching of hmbR and/or hpuB. The off-to-on switching frequencies of the two wild-type strains, NMB and IR3261, and the six mutants were examined by plating either plate-grown cultures or broth cultures on standard GCB agar plates and GCB agar plates containing 100 μM Desferal and 100 μg/ml Hb, followed by a comparison of the CFU. No significant differences were detected between the parental wild-type strains and the corresponding mutants (data not shown), indicating that the observed growth phenotypes were not due to altered switching frequencies of the hemoglobin receptor genes. Finally, misS and misR mutations were created in an hmbR “on” derivative of IR3261 (IR3287) to generate SZT1009 and SZT1010, respectively, and both mutants displayed reduced Hb-dependent growth (Fig. 3C). The growth curves of all of the strains in standard GC broth were examined together with the plate growth assay results, which were recorded after 48 h of growth. Although slightly reduced growth rates were seen for most mutants, all mutants reached similar densities after 8 h of growth. Thus, the decreased hemO and hmbR expression in the meningococcal misRS mutants correlated with a diminished capacity to utilize Hb as the sole source of iron for growth.
To correlate the hmbR transcription with the results of the hemoglobin-dependent growth assay, which required use of the Fe3+ scavenger Desferal, we also examined the regulatory effect of the MisRS system under the iron-restricted conditions generated with Desferal, which may correspond to adaptation to iron starvation. Meningococcal strain IR3287, an hmbRon derivative of serogroup B strain NMB, and the corresponding misR and misS mutants, SZT1009 and SZT1010, respectively, were grown in the presence of 50 μM Desferal for 3 h, and qRT-PCR analyses of hmbR expression were performed with purified total RNAs. The overall changes in the hmbR expression patterns of these independent wild-type and mutant strains (see Fig. S1 in the supplemental material) were similar to those obtained with the dipyridyl treatment (Fig. 1A).
Multiple potential regulatory mechanisms are present in the hemO-hmbR locus.
hemO is separated from hmbR by 191 bp in strain NMB and is transcribed divergently with a 286-bp intergenic region from a putative paraquat-inducible protein homologue gene, pqiA (Fig. 2A). Examination of the upstream sequences of hemO and hmbR revealed potential binding motifs of several global regulators. Searches of the upstream sequence of hemO using the (NATWAT)3 meningococcal Fur binding motif (15) with a maximum of three mismatches identified a single putative Fur box sequence ∼250 bp upstream of the hemO start codon (Fig. 2B), while four potential Fur boxes clustered within 100 bp of the ATG start site of hmbR coincided with the Fur binding site mapped by Delany et al. (10) (Fig. 2C). The more distal location of the Fur box with respect to the hemO coding sequence is due to the presence of a 156-bp full-length insertion element termed the Correia element (CE) (6) or Correia repeat-enclosed element (29). CEs are transposon-like elements prominent in neisserial genomes that introduce TA dinucleotide duplications upon insertion, thus potentially creating divergent promoters at the ends of elements (5, 36). In addition, in the full-length element there is a functional binding site for integration host factor (IHF) that has been shown to interact with IHF proteins in vitro (5, 40). Finally, Mazzone et al. (31) and De Gregorio et al. (9) have reported that many CEs cotranscribed with the adjacent open reading frame (ORF) are posttranscriptionally processed at the terminal inverted repeats (TIRs) by RNase III. Either one or both TIRs that form a double-stranded RNA hairpin structure can be processed, and the stability of the processed products varies significantly (9).
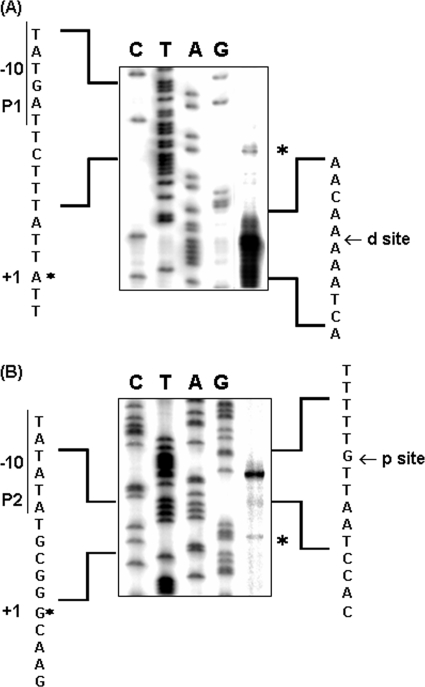
qRT-PCR detected increased expression of hemO and hmbR after dipyridyl treatment (Fig. 1), indicating that both of these genes are repressed by iron. Because putative Fur boxes typically overlap the promoter elements of iron-repressed genes, we predicted that the hemO promoter maps upstream of the CE. However, transcriptional initiation originating from the ends of a CE has also been reported (4, 5, 36). Thus, we performed primer extension experiments using three primers that anneal either within or downstream of the CE to map the transcriptional start site of hemO. Using a primer annealed inside the CE (hemO-PE3), a transcriptional start site (PhemO1) (Fig. 4A) was mapped to a location in the single predicted Fur box sequence, and an AATATG-17 bp-TATGAT promoter element could be derived (Fig. 2B). A shorter extended product was detected (d site in Fig. 4A) that corresponded to the RNase III-cleaved distal site reported by Mazzone et al. (31). Another transcriptional start site (PhemO2) (Fig. 4B) was detected using a primer that annealed within the hemO coding sequence (hemO-pR1-ER) at a location immediately downstream of the CE, and it may use an GTACTG-16 bp-TATATA promoter sequence, where the −10 hexamer contains the TA duplication of the proximal TIR (Fig. 2B). Similarly, we detected an additional major extended product (p site in Fig. 4B) that mapped to the G nucleotide of the TIR, which has been reported by Mazzone et al. (31) and De Gregorio et al. (9) to be an RNase III-processing site. Although the signals for both transcriptional start sites were relatively weak compared to those for the presumed RNase III-processed products, a third primer (hemO-PE1) located at the ATG start codon of hemO detected both start sites (data not shown). Thus, the transcription of hemO was driven by promoters located both upstream and downstream of the CE, and thus the Correia element was cotranscribed with hemO. In addition, the hemO mRNA was likely processed in a similar fashion, as described previously for other transcripts containing CE (9, 31, 40).
FIG. 4.
(A) Primer extension analysis of hemO using the hemO-PE3 primer (A) and hemO-pR1-ERI primer (B). Lanes G, A, T, and C contained the dideoxy sequencing reaction mixtures. The asterisks indicate the transcriptional start sites, while the mRNA processing sites (d site and p site) as described by Mazzone et al. are indicated by arrows.
Despite the fact that a reasonable −10 and −35 promoter element (TTATTA-17 bp-TAAAAT) overlapping the potential Fur boxes was predicted, attempts to map the transcriptional start site of hmbR were unsuccessful (data not shown). A 4-bp substitution that changed the predicted −10 sequence from TAAAAT to TAAGCA was generated in reporter strain GM101 (see below) to probe its involvement in promoter activity. This mutation decreased the hmbR promoter activity (the activity was ∼20% of the activity of the wild-type strain) (data not shown) under both iron-replete and iron-depleted conditions, confirming the importance of this sequence in the promoter function. In addition, the mutated promoter remained responsive to iron limitation, suggesting that the mutation did not interfere with iron regulation.
MisR response regulator directly interacts with both hemO and hmbR promoters.
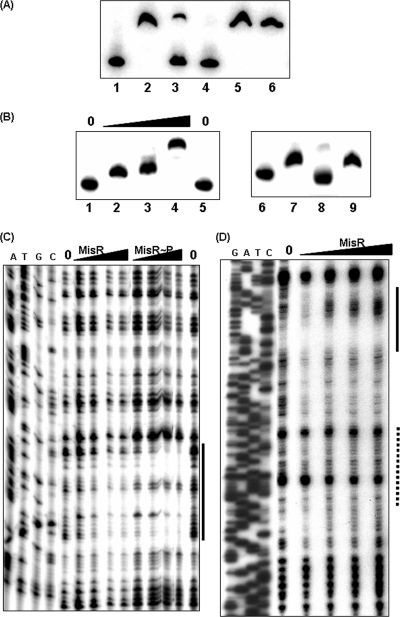
As the MisR/S two-component system was demonstrated to act as a positive regulator in the overall hmbR expression by qRT-PCR, we performed EMSA to determine whether MisR directly interacts with both the hemO and hmbR promoters. A direct interaction between MisR and the hmbR promoter has been found previously using electrophoretic mobility shift assays (EMSA) performed with purified His-tagged MisR proteins (52) to display a dose-dependent interaction (51), and the specificity of this interaction is further demonstrated in Fig. 5A. Unlabeled hmbR DNA specifically competed with the labeled probe for MisR binding, while excess nonspecific DNA did not interfere. Similarly, dose-dependent shifts of the hemO promoter with MisR were detected, and again, a competition EMSA demonstrated the specificity of this interaction (Fig. 5B).
FIG. 5.
(A) Competition EMSA experiments with the hmbR promoter and MisR. A 326-bp hmbR fragment (YT173-hmbR-pLR) was end labeled with [γ-32P]ATP using T4 kinase and incubated with the MisR protein as described previously (52). Lane 1, DNA probe; lanes 2 to 6, DNA probe with MisR protein (77 pmol, 5 μM) (lane 2, no DNA competitor; lanes 3 and 4, probe with 1.5 and 2 μg specific DNA competitor, respectively; lanes 5 and 6, probe with 1.5 and 2 μg nonspecific DNA competitor, respectively). (B) (Left panel) Dose-dependent shift of the hemO promoter with MisR. Lanes 1 and 5, free probe; lanes 2 to 4, 0.6, 2.6, and 3.8 μM MisR protein, respectively. (Right panel) Competition EMSA with the hemO promoter. Lane 6, free probe; lanes 7 to 9, DNA probe with MisR protein (38 pmol, 2.5 μM). One microgram of cold specific DNA was added to lane 8, while 1 μg of nonspecific DNA was added to lane 9. (C and D) DNase I protection assays with the hemO promoter (C) and the hmbR promoter (D). The noncoding strand of the promoter fragment was end labeled with 32P as described in Materials and Methods, incubated with increasing amounts of MisR for 20 min at 30°C, and then subjected to DNase I digestion in a 20-μl (total volume) reaction mixture. MisR∼P was prepared by incubation with acetyl phosphate (50 mM) at 37°C for 30 min. For hemO, the amounts of MisR tested were 0, 93, 186, 232, and 278 pmol and the total amounts of MisR∼P tested were 0, 93, 139, 232, and 371 pmol. For hmbR, the amounts of MisR tested were 0, 170, 340, 510, and 850 pmol. The results for dideoxy chain termination sequencing ladders corresponding to the probes are shown on the left (lanes A, T, G, and C). The solid line indicates the sequence protected by MisR, while the Fur protected region determined by Delany et al. (10) is indicated by a dotted line.
The MisR-interacting sequences in the hemO and hmbR promoters were further defined by DNase I protection assays. MisR interacted with a ∼30-bp sequence located upstream of the hemO promoter elements (Fig. 5C), consistent with the general location of a transcriptional activator (Fig. 2B). Analogously, MisR interacted with a ∼30-bp sequence located upstream of the predicted −35 element of the hmbR promoter, and a hypersensitive DNase cleavage site was detected at higher MisR protein concentrations (Fig. 5D). Putative MisR binding motifs, (A/T)(A/T)TGTAA(A/G/C)G (51), were detected in the protected sequences of both promoters (Fig. 2B and 2C). Thus, in addition to the Fur-dependent iron regulation and the possible CE-mediated transcriptional and posttranscriptional regulation, the MisR/S two-component system also directly and positively regulates the expression of hemO and hmbR.
Transcriptional regulation of hemO and hmbR expression.
As transcriptional coupling between hemO and hmbR was detected, we determined the contribution of each promoter's activity using lacZ transcriptional reporter fusions. Reporter fusions O1101, O2002, and GM101 were constructed as single copies at an irrelevant chromosomal location to determine the relative strengths of the PhemO1 and PhemO2 promoters and the proximal PhmbR promoter, respectively (Fig. 6A). A reporter that comprised all three promoters, R1203, was also constructed to assess the overall hmbR expression (Fig. 6A). As shown in Fig. 6B, the PhemO1 promoter was ∼20-fold stronger than the proximal PhmbR promoter (O1101 compared with GM101), indicating that the PhemO1 promoter likely contributed the majority of transcription. The PhemO2 promoter created by insertion of the CE (O2002) was relatively weak and thus did not contribute significantly to the overall expression of hemO and hmbR. Surprisingly, despite the presence of the PhemO1 promoter, the R1203 fusion exhibited low transcriptional activity similar to that of the weak promoters (∼6% of the strain O1101 activity), suggesting that the transcriptional read-through from hemO to hmbR was repressed or prematurely terminated. To ensure that the difference was not due to the location of the reporter fusions at a second site, we generated reporters at the native hemO-hmbR locus. The lacZ-erm cassette was inserted into the unique BamHI site in the hemO coding sequence to generate strain O101 and into the unique HincII site in hmbR to generate strain R103 (Fig. 6A). The significant difference in transcriptional activity between strains O1101 and R1203 was also observed for the corresponding strains O101 and R103 with insertions at the native locus (Fig. 6B), confirming that hemO-directed expression of hmbR was limited. Further, the transcription of hmbR detected in the R103 strain was not growth phase dependent (see Fig. S2 in the supplemental material), which ruled out the possibility that the change was caused by activities measured at different growth phases.
FIG. 6.
(A) Schematic diagram of the hemO-hmbR locus. The MisR binding sites are indicated by vertical lines with circles at the ends, while the Fur binding sites are indicated by vertical lines with diamonds at the ends. The location of clustered repeats is indicated by a gray vertical line. The BamHI (Bm) and HincII (Hc) sites used for inserting the lacZ-erm cassette to generate the hemO::lacZ (O101) and hmbR::lacZ (R103) fusions, respectively, at the native locus are also indicated. The DNA fragments used to generate transcriptional lacZ reporter fusions at a permissive ectopic genomic location are indicated by black arrows. The schematic diagram is not drawn to scale. (B) Promoter activities of hemO::lacZ (O1101 and O2002) and hmbR::lacZ (GM101 and R1203) reporter strains integrated into a permissive chromosomal locus or the native locus (O101 and R103). Black bars, wild-type background; gray bars, misS background. (C) Comparison of reporter activities with various promoters. Black bars, wild-type background; gray bars, misS background. (D) Differential activation of hemO::lacZ (O1101 and O101) and hmbR::lacZ (R1203 and R103) fusions by iron limitation. Induction ratios were determined by dividing the promoter activity of the iron-depleted culture by the promoter activity of the iron-replete culture. Black bars, wild-type background; gray bars, misS background. (Inset) Alignment of the hemO Fur box and the best hmbR Fur box with the Fur box consensus sequence. Mismatched nucleotides are underlined and in bold type.
In order to further understand the regulatory mechanisms of the MisR/S system for controlling hmbR expression, we also compared the extents of hemO-hmbR transcription that were affected by the misS mutation. The reporter fusions were introduced into the misS mutant background. As shown in Fig. 6B, ∼5-fold reductions were seen for all reporter fusions in the ΔmisS background compared to the levels observed for the wild-type background. These reductions correlated with the qRT-PCR data and clearly demonstrated that the MisR/S system functions as a positive regulator of both hemO and hmbR expression. Introduction of the O2002 reporter fusion yielded identical activities in the wild-type strain and the misS mutant because its promoter fragment did not contain the MisR binding site (Fig. 6A) and thus did not respond to the misS mutation.
To map the sequence motif conferring the reduction in transcriptional read-through, the sequence cloned in the R1203 reporter strain was analyzed, but no potential stem-loop-forming secondary structure that could function as a transcriptional terminator was detected (data not shown). A ∼30-bp sequence near the 3′ end of the hemO coding sequence contains a clustered direct repeat, a dyad repeat, and a short inverted repeat, and thus a reporter strain, O305, was constructed to assess the influence of these motifs. Since the MisR and Fur binding motifs located in the hemO-hmbR intergenic region were also present in the R1203 reporter strain, another strain, O501, containing the MisR binding site but not the Fur binding site, was generated to examine the effect of these binding sequences (Fig. 6A). All of these fusions included the strong PhemO1 promoter. As shown in Fig. 6C, both strain O305 and strain O501 displayed activities intermediate between those of strains O1101 and R1203. The activity of the O305 strain was reduced modestly (∼68%) compared to that of strain O1101 and was ∼11-fold higher than that of the R1203 strain, suggesting that the various repeats near the 3′ end of the hemO coding sequence were not the major factors interfering with transcriptional read-through. The activity of the O501 strain was ∼25% of that of strain O1101 and was ∼4-fold higher than that of strain R1203, implying that the MisR binding motif (as reflected in the strain O501 data) and the Fur box motif (as reflected in the strain R1203 data) in the hemO-hmbR intergenic region may have similar negative effects on the overall hmbR expression. However, the fact that a parallel gradual reduction in transcriptional activity from strain O1101s to strain 305s to strain 501s to strain R1203s was detected in the misS background (Fig. 6C) implied that the much reduced transcriptional activity in strain O501 compared to that in strain O1101 was not likely due to the MisR/S system. It is commonly believed that in a histidine kinase mutant a certain amount of nonphosphorylated response regulator is expressed. Thus, the remaining nonphosphorylated MisR in the misS mutant could elicit the negative effect. If this was the case, a difference in hmbR transcription was expected for the misR mutant compared to the misS mutant and the MisR/S system would function as a negative regulator of hmbR. However, the qRT-PCR data showed that the misS and misR mutations resulted in similar decreases in hmbR expression, suggesting that the nonphosphorylated MisR was not likely to contribute to the repression of transcriptional coupling of hemO and hmbR.
Fur elicited stronger iron-mediated repression of hmbR expression than of hemO expression.
Because the absence of the hmbR Fur box in reporter strain O501 resulted in a 4-fold increase in hmbR expression (when O501 was compared to R1203) and another putative Fur box was present upstream of hemO, we also examined the effect of iron availability on the promoter activities of various reporter strains using a 45-min treatment with dipyridyl to generate iron-depleted conditions. As determined by qRT-PCR, the same treatment resulted in 4-fold and 2-fold induction of hmbR and hemO expression, respectively (Fig. 1). Surprisingly, although we indeed detected ∼3-fold induction of the lacZ reporter activities of strains R1203 and R103 upon iron starvation (Fig. 6D), very limited induction was seen in reporter strains probed for hemO expression (O1101 and O101), which was consistent with the qRT-PCR pattern, in which a lower level of iron induction was observed for hemO. The lack of strong iron-dependent induction of hemO suggests that iron-loaded Fur has limited binding affinity for the putative Fur box in the hemO promoter region or, alternatively, that insertion of a Correia element downstream interfered with the regulatory function of Fur. The reduced Fur repression accounts for the significantly higher level of hemO expression in iron-replete growth conditions used in standard reporter assays. In comparison, stronger repression of hmbR transcription that correlated with a probable higher affinity of Fur for the hmbR Fur boxes could result in a higher induction ratio for hmbR upon iron starvation. The diminished transcriptional read-through from hemO could potentially be caused by the binding of Fur to the hmbR Fur boxes. However, it is also possible that the mRNA stability of various reporter transcripts (i.e., posttranscriptional regulation) accounts for the difference in the apparent promoter activities. Alternatively, cryptic premature transcriptional termination may be responsible for the decrease in the amount of hmbR mRNA transcribed from hemO.
To examine whether the level of Fur repression was affected by the MisR/S system due to the close proximity of the binding motifs in the hemO and hmbR promoter regions, reporter strains with the misS background were treated similarly with or without dipyridyl, and the induction ratios (Fe negative/Fe positive) were determined. Reporter fusions for hemO and hmbR at either the native locus or the irrelevant site were examined. If Fur elicited stronger repression in the absence of MisR/S regulation, a higher induction ratio was expected for the misS mutant. As shown in Fig. 6D, there were not significant differences between the induction ratios for the wild-type strains and the misS mutants. Thus, it appeared that the MisR/S system-dependent activation and the iron-mediated Fur repression act independently.
DISCUSSION
Iron acquisition is essential for successful colonization and infection by many bacterial pathogens, and N. meningitidis, an obligate human pathogen, is capable of utilizing a diverse array of host iron sources, including hemoglobin. Two hemoglobin acquisition receptors, HmbR and HpuAB, have been characterized in N. meningitidis. The lack of hmbR in commensal organisms (38) and the characteristics of a pathogenicity island of the hmbR locus (17, 23) suggest that hmbR was acquired more recently by meningococci. It is believed that the functional redundancy and the ability to regulate expression of important virulence factors allow bacterial pathogens to minimize their exposure to immune recognition and responses. Hemoglobin receptors play an important role in the virulence of both Gram-negative and Gram-positive bacterial pathogens. For example, the HgbA hemoglobin receptor of Haemophilus ducreyi is required for virulence in both human and animal models of chancroid (2, 46), and Staphylococcus aureus mutants lacking the IsdB hemoglobin receptor display attenuated virulence in a murine model of abscess formation (49). The important contribution of hmbR and hemoglobin utilization to meningococcal virulence has been experimentally demonstrated by the findings that acquisition of iron via HmbR enhanced the ability of meningococci to replicate in the bloodstream of infant rats and the hmbR mutant was attenuated in an infant rat model of meningococcal infection (47). A recent investigation of the distribution of the hmbR gene among more than 700 disease and carriage meningococcal isolates from three separate isolate collections revealed a statistically significant association between disease and the presence of hmbR (17). In addition, all isolates belonging to six hyperinvasive lineages are hmbR positive (17). Thus, this study provides epidemiological evidence for the importance of hmbR in meningococcal virulence.
The regulation of Hb receptors in meningococci has been observed at two levels; one mechanism is phase variation (38) (a translational on-off switch), and the other is transcriptional regulation by Fur in response to iron availability (48). hmbR is downstream of hemO, and potential Fur box sequences can be identified in both hemO and hmbR promoter regions. Several experimental findings have documented the independent transcription of hmbR and the direct interaction of the hmbR promoter region with Fur. First, a Fur titration assay with E. coli showed that when multiple copies of the hemO-hmbR intergenic region were present, they were able to titrate Fur away from a Fur-regulated reporter gene (47). Second, purified gonococcal Fur proteins bind to the upstream region of hmbR (44), and DNase I protection assays have mapped the Fur binding site of the hmbR promoter region, which overlaps the predicted promoter element (10). Using lacZ reporter assays, we showed that the hemO-hmbR intergenic region has weak promoter activity and that both Fur and the MisR/S system exhibit direct regulation of the promoter. However, hmbR was repressed more strongly than hemO based on the different iron induction ratios (Fig. 6D). Sequence analysis using the Fur binding motif (15) with a maximum of three mismatches identified a single putative Fur box sequence upstream of hemO (Fig. 2B), while four potential Fur boxes clustered upstream of hmbR (Fig. 2C). Alignment of the single hemO Fur box and the best hmbR Fur box with the consensus sequence (Fig. 6D, inset) indicated that the hemO Fur box has an additional mismatch at the center of the motif. It is not clear whether this mismatch contributes to the weak repression of hemO or the clustering of several Fur boxes in the hmbR promoter region enhances the binding affinity of Fur for this region.
As meningococci reside in iron-restricted host environments, repression by Fur would likely be alleviated in many situations, and it is likely that there are other regulatory mechanisms that respond to different host microenvironments to control expression of important virulence factors. In the present study, we demonstrated that the transcription of hmbR and hemO is also regulated by the MisR/S two-component system. The results of the disk diffusion growth assays with Hb as the sole iron source support the hypothesis that the MisR/S system has an important role in Hb utilization. The strains were grown under iron-limiting conditions in the presence of the iron chelator Desferal, and relief of Fur repression is expected under these growth conditions. We detected phenotypic alteration of Hb utilization resulting from mutations in the MisR/S two-component system in meningococcal strain NMB that encodes the HpuAB receptor, as well as in the hmbRoff (NMB and IR3261) and hmbRon (IR3287) backgrounds, indicating that the transcriptional control of hmbR by the MisR/S system is an important regulatory mechanism in meningococcal Hb utilization.
Transcriptional coupling of hemO and hmbR was detected. A full-length Correia element is present upstream of hemO in all sequenced meningococcal genomes, as well as in the genomes of additional 15 strains analyzed by De Gregorio et al. (8); however, a CE is not present in the gonococcal FA1090 genome and in Neisseria lactamica strains (8). Correia elements account for ∼2% of the N. meningitidis genome and are frequently inserted close to open reading frames (ORF). They fall into two major size classes, and a full-length element (154 to 158 bp) contains a 50-bp internal segment. The presence of CE may impact the expression of nearby downstream ORF in several ways. First, insertion of CE introduced a TA duplication and resulted in potential divergent outward promoters at the ends of elements (5). This has been demonstrated for urvB (4) and lst (36) in Neisseria gonorrhoeae, and we also identified a weak hemO promoter potentially associated with the duplicated TA dinucleotides. Second, in the unique 50-bp internal segment of the full-length element, a functional IHF binding site has been shown to interact with purified IHF proteins (5, 40). The IHF binding sequence in the hemO-associated CE is identical to that analyzed by Buisine et al. (5), which has been reported to have an apparent dissociation constant of ∼5 nM (5). IHF is a DNA binding protein that induces DNA bending and is associated with regulation of a broad range of cellular functions, including DNA replication, recombination, and transcription (12). Deletion of the IHF binding site in the CE located upstream of the mtrCDE operon encoding an efflux pump in meningococci (40) enhanced expression of mtrC (40). As the hemO-associated CE is nearly identical to the CE located upstream of mtrC, similar IHF-mediated regulatory controls are likely for hemO expression. Finally, CE-containing transcripts were processed by RNase III, resulting in posttranscriptional regulation of the associated gene. Primer extension identified a strong promoter upstream of the CE, confirming that the CE is cotranscribed with hemO, and similarly processed transcripts were also detected, implying that there is possible posttranscriptional regulation of hemO-hmbR transcripts by RNase III.
We identified two MisR binding motifs, one located upstream of the PhemO1 promoter and the other located upstream of the proximal PhmbR promoter in the hemO-hmbR intergenic region. qRT-PCR experiments with hmbR-specific and hemO-specific primers, as well as lacZ reporter assays for either a permissive chromosomal site or the native locus, clearly showed that inactivation of the MisR/S system resulted in significantly decreased transcription of both hemO and hmbR. The MisR/S two-component system functions as an independent activator because without the positive regulatory input of the MisR/S system, the level of hemO and hmbR transcription did not reach the maximal level when Fur repression was alleviated by iron starvation. When meningococci encounter other iron sources and are able to obtain sufficient iron from the environment to elicit Fur repression of iron assimilation proteins, including the HmbR receptor, the MisR/S two-component system is positioned to upregulate hmbR and hemO expression when the activating signal is sensed, thus ensuring appropriate hemoglobin utilization capacity in certain host environments. Iron acquisition systems with secondary regulatory pathways that respond to particular iron-independent environmental signals and operate in concert with Fur have been described for several bacteria. The PchR regulator, which responds to extracellular pyochelin, regulates siderophore expression in Pseudomonas aeruginosa (18). In addition, the extracytoplasmic sigma factor has been demonstrated to control uptake systems for various iron sources, such as siderophores and hemin, in E. coli (3), Pseudomonas putida (24, 54), and Bordetella pertussis (53), in response to an extracytoplasmic inducing signal.
Interestingly, we observed a significant reduction in transcriptional read-through that minimized hemO-initiated hmbR expression. This was most likely mediated by an unidentified mechanism(s) operating near or in the hemO-hmbR intergenic sequence, since reporters carrying the intergenic sequence showed significantly decreased activities. The activities of various transcriptional lacZ reporter constructs in the wild-type and misS backgrounds suggested that binding of the MisR protein is not involved, while the involvement of Fur has not been tested. Decreased stability of hemO-hmbR mRNA upon RNase III processing might also contribute to or be responsible for the reduction. Another alternative explanation is premature Rho-dependent intracistronic transcription termination that was first described for another meningococcal phase-variable gene, siaD (synD) (26). It was reported that only ∼5% of siaD mRNA was detected in unencapsulated variants (siaD phase off) compared to the encapsulated parental strain (16) because transcription of the out-of-frame siaD gene is prematurely terminated at a cryptic Rho-dependent site, a sequence segment with a higher cytosine content than guanosine content termed the transcription termination element, which is the target of Rho and is usually located upstream of multiple actual termination sites (1, 39). Further, this transcription elongation termination mechanism influences the phase variation frequency (26). As hmbR is phase off in strain NMB, it is probable that a similar Rho-dependent intracistronic termination mechanism operates on hemO-hmbR transcription and affects the overall hmbR expression.
Coordinated regulation of the heme catabolism protein and the heme uptake receptor is beneficial. Since excess heme is potentially toxic, downregulation of heme uptake when the heme catabolism capability is disrupted or reduced may be one of the mechanisms by which meningococci defend against heme toxicity. The transcriptional coupling of hemO and hmbR provides a means for such coordination. hemO is, however, transcribed at a much higher level than hmbR as a result of the reduction in transcriptional read-through. This correlates with the greater need for heme metabolism than for heme uptake. In addition, the immunogenic nature of the HmbR outer membrane receptor requires that it is absent (phase off) or expressed at a low level when it is not needed, while the cells require a certain amount of the heme-degrading enzyme, HemO. Another type of coordinated regulation is iron-mediated Fur-dependent repression, as indicated by the fact that transcription of both genes is induced under iron-limiting conditions and, again, weaker repression by Fur resulted in greater transcription of hemO in the iron-replete environment. This is consistent with the presence of other heme uptake mechanisms in meningococci. Finally, a third type of coordinated control of the two genes is mediated by the MisR/S two-component system since MisR interacts directly with both hemO and hmbR promoters and expression of both genes decreased when the MisR/S system was inactivated. Despite the transcriptional linkage with hemO, the significant reduction in transcriptional read-through into hmbR might necessitate independent activation of the considerably weaker hmbR promoter by the MisR/S system, and the possible different affinities of MisR for hemO and hmbR promoters can differentially activate their expression. One can envision that higher-affinity binding of MisR to hemO than to hmbR ensures the presence of sufficient heme-degrading HemO prior to induction of the HmbR heme-hemoglobin uptake system. Finally, additional regulatory mechanisms due to insertion of the Correia element upstream of hemO, including the potential interaction with IHF and posttranscriptional processing by RNase III, as well as the potential Rho-dependent premature transcriptional termination mechanism influencing the phase variation frequency, further demonstrated that there is a complex regulatory network controlling the expression of hemO and hmbR, which allows regulation by various environmental signal inputs.
As the MisR/S system is not induced by hemoglobin, the rationale for specific positive regulation by the MisR/S two-component system awaits identification of the signal(s) activating this system. The intracellular phase of infection is not likely when enhanced hmbR expression is needed, because a meningococcal hmbR mutant has been shown to replicate more rapidly in epithelial cells than its wild-type parental strain (25). In addition, hemoglobin utilization in gonococci was shown to be dispensable for colonization of the female mouse genital tract (21). Hemoglobin is not readily accessible to meningococci due to its compartmentalization in erythrocytes, and the low level of Hb found in normal human serum as a result of spontaneous hemolysis is rapidly captured by haptoglobin (43). However, during the invasive phase of infection, local hemolysis may make free Hb available for assimilation. Thus, when a particular iron-limiting host environment is reached during infection, derepression of hmbR by Fur after the lack of iron is sensed might not be sufficient, and meningococci need to further increase the expression of hmbR through activation of the MisR/S two-component system, possibly to allow prompt adaptation to the new environment.
Supplementary Material
Acknowledgments
This work was supported by grant R01 AI061031 to Y.-L.T. from the National Institutes of Health.
We thank Shaojia Bao for technical assistance.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 14 December 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alifano, P., F. Rivellini, D. Limauro, C. B. Bruni, and M. S. Carlomagno. 1991. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell 64:553-563. [DOI] [PubMed] [Google Scholar]
- 2.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 3.Angerer, A., S. Enz, M. Ochs, and V. Braun. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol. Microbiol. 18:163-174. [DOI] [PubMed] [Google Scholar]
- 4.Black, C. G., J. A. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buisine, N., C. M. Tang, and R. Chalmers. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52-58. [DOI] [PubMed] [Google Scholar]
- 6.Correia, F. F., S. Inouye, and M. Inouye. 1986. A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J. Bacteriol. 167:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, N. L., and H. A. Nash. 1984. E. coli integration host factor binds to specific sites in DNA. Cell 39:707-716. [DOI] [PubMed] [Google Scholar]
- 8.De Gregorio, E., C. Abrescia, M. S. Carlomagno, and P. P. Di Nocera. 2003. Asymmetrical distribution of Neisseria miniature insertion sequence DNA repeats among pathogenic and nonpathogenic Neisseria strains. Infect. Immun. 71:4217-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Gregorio, E., C. Abrescia, M. S. Carlomagno, and P. P. Di Nocera. 2003. Ribonuclease III-mediated processing of specific Neisseria meningitidis mRNAs. Biochem. J. 374:799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delany, I., R. Grifantini, E. Bartolini, R. Rappuoli, and V. Scarlato. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goosen, N., and P. van de Putte. 1995. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 16:1-7. [DOI] [PubMed] [Google Scholar]
- 13.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 14.Grifantini, R., E. Frigimelica, I. Delany, E. Bartolini, S. Giovinazzi, S. Balloni, S. Agarwal, G. Galli, C. Genco, and G. Grandi. 2004. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 54:962-979. [DOI] [PubMed] [Google Scholar]
- 15.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, O. B., N. J. Evans, J. M. Blair, H. S. Grimes, C. R. Tinsley, X. Nassif, P. Kriz, R. Ure, S. J. Gray, J. P. Derrick, M. C. Maiden, and I. M. Feavers. 2009. Epidemiological evidence for the role of the hemoglobin receptor, HmbR, in meningococcal virulence. J. Infect. Dis. 200:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology Press, Washington, DC.
- 20.Janik, A., E. Juni, and G. A. Heym. 1976. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 4:71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerse, A. E., E. T. Crow, A. N. Bordner, I. Rahman, C. N. Cornelissen, T. R. Moench, and K. Mehrazar. 2002. Growth of Neisseria gonorrhoeae in the female mouse genital tract does not require the gonococcal transferrin or hemoglobin receptors and may be enhanced by commensal lactobacilli. Infect. Immun. 70:2549-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, C. R., J. Newcombe, S. Thorne, H. A. Borde, L. J. Eales-Reynolds, A. R. Gorringe, S. G. Funnell, and J. J. McFadden. 2001. Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39:1345-1355. [DOI] [PubMed] [Google Scholar]
- 23.Kahler, C. M., E. Blum, Y. K. Miller, D. Ryan, T. Popovic, and D. S. Stephens. 2001. exl, an exchangeable genetic island in Neisseria meningitidis. Infect. Immun. 69:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson, J. A., D. L. Higashi, I. Stojiljkovic, and M. So. 2002. Replication of Neisseria meningitidis within epithelial cells requires TonB-dependent acquisition of host cell iron. Infect. Immun. 70:1461-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavitola, A., C. Bucci, P. Salvatore, G. Maresca, C. B. Bruni, and P. Alifano. 1999. Intracistronic transcription termination in polysialyltransferase gene (siaD) affects phase variation in Neisseria meningitidis. Mol. Microbiol. 33:119-127. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct hemogobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977-989. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S. V., N. J. Saunders, A. Jeffries, and R. F. Rest. 2002. Genome analysis and strain comparison of correia repeats and correia repeat-enclosed elements in pathogenic Neisseria. J. Bacteriol. 184:6163-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano, and P. P. Di Nocera. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211-222. [DOI] [PubMed] [Google Scholar]
- 32.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Newcombe, J., L. J. Eales-Reynolds, L. Wootton, A. R. Gorringe, S. G. Funnell, S. C. Taylor, and J. J. McFadden. 2004. Infection with an avirulent phoP mutant of Neisseria meningitidis confers broad cross-reactive immunity. Infect. Immun. 72:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto, B. R., A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18:217-233. [DOI] [PubMed] [Google Scholar]
- 36.Packiam, M., D. M. Shell, S. V. Liu, Y. B. Liu, D. J. McGee, R. Srivastava, S. Seal, and R. F. Rest. 2006. Differential expression and transcriptional analysis of the alpha-2,3-sialyltransferase gene in pathogenic Neisseria spp. Infect. Immun. 74:2637-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins-Balding, D., M. Ratliff-Griffin, and I. Stojiljkovic. 2004. Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 68:154-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson, A. R., and I. Stojiljkovic. 1999. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 181:2067-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivellini, F., P. Alifano, C. Piscitelli, V. Blasi, C. B. Bruni, and M. S. Carlomagno. 1991. A cytosine- over guanosine-rich sequence in RNA activates rho-dependent transcription termination. Mol. Microbiol. 5:3049-3054. [DOI] [PubMed] [Google Scholar]
- 40.Rouquette-Loughlin, C. E., J. T. Balthazar, S. A. Hill, and W. M. Shafer. 2004. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol. Microbiol. 54:731-741. [DOI] [PubMed] [Google Scholar]
- 41.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 42.Sannigrahi, S., X. Zhang, and Y. L. Tzeng. 2009. Regulation of the type I protein secretion system by the MisR/MisS two-component system in Neisseria meningitidis. Microbiology 155:1588-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 44.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens, D. S., J. S. Swartley, S. Kathariou, and S. A. Morse. 1991. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stojiljkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 48.Stojiljkovic, I., J. Larson, V. Hwa, S. Anic, and M. So. 1996. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J. Bacteriol. 178:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres, V. J., G. Pishchany, M. Humayun, O. Schneewind, and E. P. Skaar. 2006. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188:8421-8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzeng, Y. L., A. Datta, K. D. Ambrose, J. K. Davies, R. W. Carlson, D. S. Stephens, and C. M. Kahler. 2004. The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J. Biol. Chem. 279:35053-35062. [DOI] [PubMed] [Google Scholar]
- 51.Tzeng, Y. L., C. M. Kahler, X. Zhang, and D. S. Stephens. 2008. MisR/MisS two-component regulon in Neisseria meningitidis. Infect. Immun. 76:704-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzeng, Y. L., X. Zhou, S. Bao, S. Zhao, C. Noble, and D. S. Stephens. 2006. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J. Bacteriol. 188:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venturi, V., C. Ottevanger, M. Bracke, and P. Weisbeek. 1995. Iron regulation of siderophore biosynthesis and transport in Pseudomonas putida WCS358: involvement of a transcriptional activator and of the Fur protein. Mol. Microbiol. 15:1081-1093. [DOI] [PubMed] [Google Scholar]
- 55.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, D., and M. A. Apicella. 1996. Plasmids with erythromycin resistance and catechol 2,3-dioxygenase- or beta-galactosidase-encoding gene cassettes for use in Neisseria spp. Gene 171:133-134. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, W., D. J. Hunt, A. R. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.