Abstract
Iron (Fe) in soluble elemental form is found in the tissues and fluids of animals at concentrations insufficient for sustaining growth of bacteria. Consequently, to promote colonization and persistence, pathogenic bacteria evolved a myriad of scavenging mechanisms to acquire Fe from the host. Bordetella bronchiseptica, the etiologic agent of upper respiratory infections in a wide range of mammalian hosts, expresses a number of proteins for acquisition of Fe. Using proteomic and genomic approaches, three Fe-regulated genes were identified in the bordetellae: bfrH, a gene encoding a putative siderophore receptor; ecfI, a gene encoding a putative extracellular function (ECF) sigma factor; and ecfR, a gene encoding a putative EcfI modulator. All three genes are highly conserved in B. pertussis, B. parapertussis, and B. avium. Genetic analysis revealed that transcription of bfrH was coregulated by ecfI, ecfR, and fur1, one of two fur homologues carried by B. bronchiseptica. Overexpression of ecfI decoupled bfrH from Fe-dependent regulation. In contrast, expression of bfrH was significantly reduced in an ecfI deletion mutant. Deletion of ecfR, however, was correlated with a significant increase in expression of bfrH, due in part to a cis-acting nucleotide sequence within ecfR which likely reduces the frequency of readthrough transcription of bfrH from the Fe-dependent ecfIR promoter. Using a murine competition infection model, bfrH was shown to be required for optimal virulence of B. bronchiseptica. These experiments revealed ecfIR-bfrH as a locus encoding a new member of the growing family of Fe and ECF sigma factor-modulated regulons in the bordetellae.
Due to its involvement in various metabolic processes, iron (Fe) is an essential growth factor for both nonpathogenic and pathogenic bacteria. Enzymes involved in processes of glycolysis, ATP synthesis, and DNA replication in bacteria require Fe as a cofactor or as a requisite prosthetic group (60). Sequestration of the metal by high-affinity Fe-binding proteins, such as transferrin, lactoferrin, ferritin, and hemoproteins, decreases the concentration of elemental Fe in mammalian tissues and fluids to <10−18 M, a level which is insufficient to support the growth of invading microorganisms (19, 60). Thus, to thrive and elicit an infection in this hostile environment, pathogenic microorganisms evolved a wide range of regulated mechanisms for scavenging Fe from the host. In Neisseria spp. and Haemophilus influenzae, for example, expression of specific surface proteins involved in acquiring Fe from heme, xenosiderophores, or ferriproteins is upregulated when the bacteria encounter Fe-limiting environments (19, 53, 60, 65). Bacteria in various genera, including Escherichia, Klebsiella, and Shigella, obtain Fe from the local environment by expression and secretion of Fe-scavenging siderophores (19, 53, 60, 65). Binding of the siderophores to specific Fe-regulated outer membrane (OM) receptors initiates uptake of the molecules into the cytoplasm for utilization of the bound Fe.
Commonly, expression of Fe-responsive genes in bacteria is tightly regulated. Global Fe-responsive regulatory proteins, such as Fur (Fe uptake regulator), expressed by numerous types of bacteria, and DtxR (diphtheria toxin regulator) of Corynebacterium diphtheriae, have essential regulatory roles in maintaining Fe homeostasis in the cell (38, 40, 58, 65). Both Fur and DtxR mediate expression of their regulons by repressing expression from promoters (7, 58). The Fe-complexed form of Fur, a regulatory protein found almost ubiquitously in Gram-negative and Gram-positive bacteria, exhibits binding affinity for a 19-bp consensus nucleotide sequence (Fur box) located near or within the promoters of many Fe-responsive genes (40, 58). Binding of Fur to a Fur box represses expression of the fur-regulated genes, ostensibly by excluding RNA polymerase from the promoter sequences (40). During periods of Fe stress, loss of Fe from Fur (apo-Fur) reduces Fur's binding affinity for the Fur box. Dissociation of apo-Fur from the Fur box derepresses the promoter and permits transcription of the Fur-regulated gene (40).
A more complicated signal transduction cascade for controlling expression of Fe-dependent genes, however, has been described for Escherichia coli (3), Pseudomonas spp. (37, 39, 54), Bordetella spp. (36), and several other bacteria (10, 56). In these systems, a special class of sigma factor, the extracellular function (ECF) sigma factor, is activated by extracellular signals and interacts with specific promoters to drive expression of one or more genes involved in Fe acquisition (11, 29, 36, 37, 53). Like other sigma factors, ECF sigma factors reversibly interact with core RNA polymerase and specific nucleotide sequences to recruit the holoenzyme to the cognate ECF sigma factor-regulated promoters. Unlike members of the sigma 70 family, which routinely control expression of housekeeping genes, ECF sigma factors commonly regulate the expression of genes involved in adaptation of the bacterium to particular environments (29, 47).
ECF sigma factors implicated in the regulation of Fe acquisition genes have been denoted Fe starvation ECF sigma factors (71). The molecular cascade by which an Fe starvation ECF sigma factor regulates gene expression has been studied most extensively with the fecIR-fecABCD system of E. coli (9, 11). Under Fe-stressed growth conditions, FecA, the OM receptor for iron citrate, is basally expressed by readthrough transcription originating upstream from PfecIR, a promoter which controls expression of σFecI and FecR (9). Upon binding to extracellular ferric citrate, FecA undergoes a conformational change which transmits a signal across the periplasm to FecR, which in turn transmits an activation signal across the plasma membrane and into the cytoplasm to σFecI. Once activated, σFecI associates with the core RNA polymerase, which induces transcription from PfecABCD to drive prolific expression of the polycistronic fecABCD operon (9, 11). ECF sigma factors are usually coexpressed with a second regulator which, in turn, modulates the activity of the cognate ECF sigma factor. FecR of E. coli appears to positively modulate σFecI when the bacterium encounters the specific extracellular inducing ligand ferric citrate (9, 11). In Pseudomonas putida, however, PupR inhibits the activity of σPupI in the absence of the extracellular signal and functions as a sequestering type of anti-sigma factor (37). In Bordetella bronchiseptica, σHurI regulates expression of the bhuRSTUV operon, which encodes receptor and transport proteins required for acquisition of exogenous heme (67). HurR of B. bronchiseptica positively modulates σHurI in the presence of heme (67), and hurP, a gene encoding a prospective plasma membrane-associated protease, is also required to mediate the heme-dependent expression of BhuR, the outer membrane heme receptor (34).
Four members of the genus Bordetella have been described as causative agents of several upper respiratory diseases in a variety of animals. B. pertussis and B. parapertussis are obligate human pathogens which elicit whooping cough and a whooping cough-related disease, respectively (45). B. avium, a major environmental and agricultural pathogen, infects a number of species of wild and domestic birds (36). B. bronchiseptica exhibits the broadest host range, having the capacity to infect mice, dogs, pigs, birds, porcupines, rabbits, horses, monkeys, and humans (45, 72). It is believed that B. bronchiseptica is the evolutionary progenitor of B. pertussis and B. parapertussis (51). To date, three Fe-scavenging systems have been characterized for the pathogenic bordetellae (2, 6, 14-16, 67): (i) the BhuR system for acquisition of heme (12, 16, 67); (ii) the FauA system for uptake of alcaligin, the endogenous siderophore of the bordetellae (14-16); and (iii) the BfeA system for retrieval of the xenosiderophore enterobactin (2, 6, 12, 16). Each of these systems has an important role in pathogenesis. Mixed-infection experiments using a murine infection model demonstrated that these three acquisition systems are essential for optimal virulence of B. pertussis (12, 16-18).
To identify additional proteins likely to be involved in Fe uptake and pathogenesis, a proteomic approach was employed to analyze OM proteins isolated from several different species of Bordetella. Herein we characterize the Fe-dependent regulatory mechanisms by which BfrH, a prospective OM protein expressed by Fe-stressed B. bronchiseptica, is expressed. Genetic experiments revealed that expression of bfrH depends upon (i) the Fe environment, (ii) one of two fur genes carried by B. bronchiseptica, and (iii) genes encoding a cognate ECF sigma factor and sigma factor regulator. In combination with results obtained from an experimental infection model employing a bfrH-deficient mutant, these data provide strong evidence that the ecfIR-bfrH locus encodes a highly conserved, ECF-regulated Fe retrieval system in B. bronchiseptica which is important for optimal virulence of the bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids utilized in this study are described in Table 1. Unless otherwise stated, strains of B. bronchiseptica RB50 were maintained on brain heart infusion (BHI) agar plates or in BHI broth (Difco Laboratories, Detroit, MI). E. coli strains were maintained on Luria-Bertani (LB) agar or in LB broth. Bacteria utilized for the murine infection assay were maintained on Bordet Gengou (BG) plates supplemented with 15% sheep blood (Crane Laboratories Inc., Syracuse, NY) and cultured in modified Stainer Scholte (SS) broth (30, 46), which contained the following (per liter): 10.72 g glutamic acid (monosodium salt), 1.525 g Tris, 2.5 g NaCl, 3.67 ml 1 M KH2PO4, 2.68 ml 1 M KCl, 0.490 ml 1 M MgCl2, 0.136 ml 1 M CaCl2, 10 ml proline supplement, 10 ml SS supplement, and 15 ml 10% Casamino Acids. To establish Fe-replete conditions, BHI and SS cultures were supplemented with 36 μM FeSO4. Fe-stressed growth conditions were instituted by supplementing BHI cultures with 25 to 50 μM ethylenediamine di-o-hydroxyphenylacetic acid (EDDHA; Sigma Biochemicals, St. Louis, MO). Fe-stressed growth of bacteria in SS cultures was achieved by not supplementing the medium with Fe. Antibiotics were obtained from Sigma Biochemicals and Amresco (Solon, OH). Unless otherwise noted, ampicillin was utilized at 200 μg/ml, streptomycin at 200 μg/ml, tetracycline at 10 μg/ml, gentamicin at 40 μg/ml, and kanamycin at 50 μg/ml.
TABLE 1.
Plasmids and strains used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| B. bronchiseptica strains | ||
| RB50 | Wild type | 22 |
| RB50ΔbfrH | Chromosomal deletion of bfrH in RB50 | This study |
| RB50ΔbfrH::Kan | Kanr cassette insertion into bfrH of RB50 | This study |
| RB50ΔecfR | Chromosomal deletion of ecfR in RB50 | This study |
| RB50ecfRK-I | Knock-in of ecfR at the ecfR locus | This study |
| RB50ΔecfI | Chromosomal deletion of ecfI in RB50 | This study |
| RB50Δfur1 | Chromosomal deletion of fur1 in RB50 | This study |
| E. coli strains | ||
| DH5αF′kan | χ80dlacZM15 Δ(lacZYA-argF)U169 deoR recA1 phoA hsdR17(rK− mK+) supE44λ thi-1 gyrA96 relA1 [F′ proAB lacqZΔM15 Tn5(Kanr)] | Invitrogen |
| DH5αF′tet | DH5α transconjugant with [F′ proAB lacqZΔM15 Tn10(Tetr)] | Invitrogen |
| HB101(pRK2013) | F− Δ(gpt-proA)62leuB6 glnV44 ara-14 galK2 lacY1Δ(mcr-mrr) rpsL20 xyl-5 mtl-1 recA13; Stpr | 23 |
| MC4100λpir | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR (λpir); pRK2013 | 64 |
| SM10λpir | Conjugation helper strain; RP4 plasmid integrated into the chromosome | 49 |
| BL21(DE3)(pLysS) | F−ompT hsdSB (rB− mB−) dcm gal λ(DE3); pLysS; Cmr | Promega |
| Plasmids | ||
| pTOPO | Cloning vector; Ampr | Stratagene |
| pBluescriptKS(+) | Cloning vector; Ampr | Stratagene |
| pET21a | Protein expression vector; Ampr | Novagen |
| pGEM-T | Cloning vector; Ampr | Promega |
| pCVD442tet | Allelic exchange vector; λpirsacB Ampr Tetr | 49 |
| pRK415 | Mobilizable broad-host-range expression vector; Tetr | 32 |
| pRK415Δ | Gene encoding the LacZ alpha peptide in pRK415 frameshifted at the XhoI site | 36 |
| pRK2013 | Conjugative helper plasmid; Kanr | 23 |
| pKSAC | Kanr cassette | Pfizer |
| pDJM41Δ | Promoterless lacZ from pRS415 in pUFR047 | 36 |
| pTOPOKan | Kanr cassette from pKSAC ligated into pTOPO | This study |
| pJB3 | 166-bp fragment containing 3′ region of ecfR (30 bp), ecfR-bfrH intergenic region (93 bp), and 5′ region of bfrH (43 bp) in pTOPO | This study |
| pJB3.1 | 166 bp from pJB3 in pDJM41Δ | This study |
| pJB7 | RB50 bfrH-His with pET21a-derived RBS in pTOPO | This study |
| pJB7.1 | RB50 bfrH-His with pET21a-derived RBS in pET21a | This study |
| pJB8 | Fragment containing 612 bp upstream of bfrH, bfrH, and 587 bp downstream of bfrH in pTOPO | This study |
| pJB8.1 | Self-ligated inverse PCR fragment generated from pJB8 containing 612 bp upstream of bfrH and 587 bp downstream of bfrH | This study |
| pJB8.2 | Fragment from pJB8.1 containing 612 bp upstream of bfrH and 587 bp downstream of bfrH in pCVD442tet | This study |
| pJB8.3 | Kanr cassette from pTOPOKan ligated to inverse PCR fragment generated from pJB8, containing 612 bp upstream of bfrH and 587 bp downstream of bfrH | This study |
| pJB8.4 | Fragment from pJB8.3 containing 612 bp upstream of bfrH, Kanr cassette, and 587 bp downstream of bfrH in pCVD442tet | This study |
| pJB10 | Fragment in pGEM-T containing 643 bp upstream of ecfR, the ecfR ORF, and 603 bp downstream of ecfR | This study |
| pJB10.1 | Self-ligated inverse PCR fragment generated from pJB10, containing 643 bp upstream of ecfR and 603 bp downstream of ecfR | This study |
| pJB10.3 | Generated from pJB10.1, containing 643 bp upstream of ecfR and 603 bp downstream of ecfR in pTOPO | This study |
| pJB10.4 | Fragment from pJB10.3 containing 643 bp upstream of ecfR and 603 bp downstream of ecfR in pCVD442tet | This study |
| pJB10XbaI | Fragment containing 643 bp upstream of ecfR, ecfR, and 603 bp downstream of ecfR in pTOPO | This study |
| pJB10.5 | Fragment from pJB10XbaI in pCVD442tet | This study |
| pJB11 | Fragment containing 600 bp upstream of fur1, fur1, and 600 bp downstream of fur1 in pTOPO | This study |
| pJB11.1 | Self-ligated inverse PCR fragment generated from pJB11, containing 600 bp upstream of fur1 and 600 bp downstream of fur1 | This study |
| pJB11.2 | Fragment from pJB11.1 containing 600 bp upstream of fur1 and 600 bp downstream of fur1 in pCVD442tet | This study |
| pJB16 | Fragment containing 604 bp upstream of ecfI, ecfI, and 600 bp downstream of ecfI in pGEM-T | This study |
| pJB16.1 | Self-ligated inverse PCR fragment generated from pJB16, containing 604 bp upstream of ecfI and 100 bp upstream of the TGA stop codon of ecfI | This study |
| pJB16.2 | Fragment from pJB16.1 containing 604 bp upstream of ecfI and 100 bp upstream of the TGA stop codon of ecfI in pCVD442tet | This study |
| pGEM-fur1-1 | fur1 in pGEM-T | This study |
| pRK-fur1-1 | RB50 fur1 in pRK415 | This study |
| pGEMTecfI | RB50 ecfI in pGEM-T | This study |
| pecfI | RB50 ecfI in pRK415 | This study |
Escherichia coli strains DH5αF′kan and DH5αF′tet were used as host strains for recombinant engineering. E. coli MC4100λpir and HB101(pRK2013) were employed as helper strains for triparental matings, while SM10λpir was used as a donor strain for biparental matings.
Synthetic oligonucleotides and PCR amplification of DNA from B. bronchiseptica.
All synthetic oligonucleotides (Integrated DNA Technologies, Coralville, IA) used for PCR, reverse transcription-PCR (RT-PCR), or quantitative RT-PCR (qRT-PCR) or for engineering recombinant strains and plasmids are listed in Table 2. PCR was used to generate DNA from B. bronchiseptica for use in the construction of plasmids and for engineering of mutants. Unless otherwise indicated, DNA was amplified using either EasyA polymerase or Pfu Turbo polymerase (Stratagene, La Jolla, CA). PCR mixtures contained 1 unit polymerase, 1× EasyA or Pfu Turbo buffer (Stratagene), 10% dimethyl sulfoxide (DMSO), a 62.5 μM or 125 μM concentration of each deoxynucleoside triphosphate, respectively, and a 0.4 μM concentration of each oligonucleotide primer. PCR products were resolved by agarose gel electrophoresis, purified using a GFX purification kit (GE Healthcare, Piscataway, NJ), and cloned into pGEM-T (Promega, Madison, WI), pTOPO (Invitrogen, Carlsbad, CA), or pBluescript KS(+) (Stratagene), as indicated. All PCR-amplified DNAs were confirmed by nucleotide sequencing (Roswell Park Cancer Institute, Buffalo, NY).
TABLE 2.
Synthetic oligonucleotides used in this study
| Primer | Amplified region | Sequence (5′-3′a) |
|---|---|---|
| Primers for allelic exchange | ||
| Δfur1-5 | 600 bp upstream of fur1, fur1, and 600 bp downstream of fur1 | CTAGTCTAGACCTTGTCGATGCCTTCCTG |
| Δfur1-3 | CGAGCTCGCAGACCCTGTCCATGCAG | |
| fur1invPCR1 | 600 bp upstream of fur1 and 600 bp downstream of fur1 | GGTGTAATCGCTCTCCATC (5′ phosphorylated) |
| fur1invPCR2 | TCCTCCGGCTCTACCAACC (5′ phosphorylated) | |
| ΔbfrH-5 | 612 bp upstream of bfrH, bfrH, and 587 bp downstream of bfrH | CTAGTCTAGAGCCATAGCGCTGTTGCGTG |
| ΔbfrH-3 | CGAGCTCCGCATCAGCAGCTTGGACC | |
| bfrHinvPCR1 | 612 bp upstream of bfrH and 587 bp downstream of bfrH | GGGAACTCTCCTATGCGTTAG (5′ phosphorylated) |
| bfrHinvPCR2 | ATCCGGGATTTGGTCGGAAATT (5′ phosphorylated) | |
| pTOPOKan5 | 5′ end of Kanr cassette in pKSAC | CCGGAATTCGATATCAAAGCCACGTTGTGTCTCA |
| pTOPOKan3 | 3′ end of Kanr cassette in pKSAC | CGCGGATCCGATATCTTAGAAAAACTCATCGAGC |
| ΔecfI-5 | 604 bp upstream of ecfI, ecfI, and 600 bp downstream of ecfI | CTAGTCTAGACGAGCCCAGGGGAGGCTC |
| ΔecfI-3 | CTAGTCTAGACTGCCCGCATACACGCCG | |
| ecfIinvPCR1 | 604 bp upstream of ecfI and 600 bp downstream of ecfI | AACGGCAATACCATCGGGGC (5′ phosphorylated) |
| ecfIinvPCR2 | CTATGCCGAGATCGCGCAA (5′ phosphorylated) | |
| ΔecfR-5 | 643 bp upstream of ecfR, ecfR, and 603 bp downstream of ecfR | GGAAGATCTGCCTTGGGCGGTTTGCGG |
| ΔecfR-3 | AAAAGTACTGCGAGTCACCACCGAAATC | |
| ecfRinvPCR1 | 643 bp upstream of ecfR and 603 bp downstream of ecfR | CTACTCATGGAATGCCATGTA (5′ phosphorylated) |
| ecfRinvPCR2 | GCGCGGTTACAAAACAATCGC (5′ phosphorylated) | |
| pJB10.1ecfRUpXbaI | 643 bp upstream of ecfR and 603 bp downstream of ecfR | CTAGTCTAGAGCCTTGGGCGGTTTGCGC |
| pJB10.1ecfRdXbaI-2 | CTAGTCTAGAGCGAGTCACCACCGAAATC | |
| Primers for recombinant plasmids | ||
| fur1HIII | 547-bp ORF of fur1 | CGCAAGCTTGATAGTTCCGTGGGGCAG |
| fur1ERI3 | GGGGAATTCGGTAGAGCCGGAGGATCA | |
| bfrHwt5′pET21 | 2,505-bp ORF of bfrH | CTAGTCTAGAAAGGAGATATACATATGTTTTCTCGCAGTCAGAA |
| bfrHWT3′HispET21′ | CGAGCTCTCACACCACCACCACCACCACCACTTATCTCAGTCTTTCCCCTG | |
| BpbfrI5′ | 534-bp ORF of ecfI | GGTCTAGAGAAGGAGATATACATATGCCTGCCAGCCTGATCACTTCG |
| BpbfrI3′ | GGGAATTCCTACTCATGGAATGCCATGTA | |
| Primers for end-point RT-PCR | ||
| RTbhuR5 | 513-bp internal region of bhuR | TGTTCGACAACCGCTACCAGAACT |
| RTbhuR3 | GCACGTTGATGGCTTCCCAGTATT | |
| RTbfrH5 | 598-bp internal region of bfrH | TACGGCGCCAGCAACAACAATTAC |
| RTbfrH3 | ATTTCACGCCGACCTCATACTGCT | |
| RPAecfI5 | 218-bp internal region of ecfI | CCCAAGCTTACGCAGGATGTGTTCGTGC |
| RPAecfI3 | GGATCCCAGCAGCTCGGCCTGCTGT | |
| RTecfR5 | 488-bp internal region of ecfR | TGGCGAAATGCTGGTGGATGT |
| RTecfR3 | GCGTCACGCTGAGCCAGTAA | |
| RPAecfIR5 | 288-bp overlapping region of ecfIR | CCCAAGCTTACGCAGGATGTGTTCGTGC |
| RPAecfIR3 | CGCGGATCCCGCCAGCCAGGCGCGAC | |
| bfrHPromRPA5 | 237-bp intergenic region of ecfR-bfrH | CCCAAGCTTCCGCCGTGCGCCAGAGC |
| bfrHPromRPA3 | CGCGGATCCATGGCGACAGGCGCCAGG | |
| RTrecA5 | 402-bp internal region of recA | GCACCAACTGCATGGTCATCTTCA |
| RTrecA3 | 5CGATGGCCATTTCCTTGTGCTCTT | |
| Primers for qRT-PCR | ||
| qRTbfrH5 | 11-bp internal region of bfrH | ATCAACTGGAGCATCGCTTCAACG |
| qRTbfrH3 | TAGCCGTAATTGTTGTTGCTGGCG | |
| qRTPbfrH1 | 98-bp region of ecfR-bfrH | CCGTCCTTGCAGATTCGGCTTT |
| qRTPbfrH2 | AACACGGGAACTCTCCTATGCGTT | |
| qRTrecA5 | 118-bp internal region of recA | GCAACGCGCTCAAGTTCTATTCCT |
| qRTrecA3 | ACCTTGTTCTTGACCACCTTGACG |
Various restriction sites incorporated into the oligonucleotides are underlined.
Construction of pRK-fur1-1.
The 547-bp fur1 open reading frame (ORF) (BB3942) (Sanger Institute Wellcome Trust Genome Campus, Cambridge, United Kingdom) (51) was amplified from RB50 chromosomal DNA by PCR using synthetic oligonucleotides (fur1HIII and fur1ERI3), EasyA polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 1 min 30 s. The purified PCR product was cloned into pGEM-T (Promega) to produce pGEM-fur1-1. Subsequently, fur1 was directionally cloned into pRK415 (32) at the HindIII/EcoRI sites to orient the ORF with Plac of the vector to produce pRK-fur1-1.
Construction of pJB7.1.
The 2,505-bp bfrH open reading frame (Sanger annotation, BB3658) (51) was amplified from RB50 chromosomal DNA by PCR using synthetic oligonucleotides (bfrHwt5′pET21 and bfrHWT3′HispET21′), EasyA polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 3 min. The purified PCR product was ligated into pTOPO (Invitrogen) to produce pJB7. The DNA fragment carrying bfrH was then directionally cloned into pET21a (Promega) at the XbaI and SacI sites to produce pJB7.1.
Construction of pecfI.
The 534-bp ecfI open reading frame (Sanger annotation, BB3942) (51) was amplified from RB50 chromosomal DNA by PCR, using synthetic oligonucleotides (BpbfrI5′ and BpbfrI3′), EasyA polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 53°C for 45 s, and 72°C for 1 min. The purified PCR product was ligated into pGEM-T (Promega) to produce pGEMTecfI. The DNA fragment carrying ecfI was then directionally cloned into pRK415 (32) at the BamHI and EcoRI sites to produce pecfI.
Construction of pJB3.1.
A region encompassing the last 30 bp of ecfR, the 93-bp ecfR-bfrH intergenic region, and the first 43 bp of bfrH was amplified from RB50 chromosomal DNA by PCR, using synthetic oligonucleotides (BpbfrHprom5 and BpbfrHprom3), EasyA polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 1 min. The purified PCR product was ligated into pTOPO (Invitrogen) to produce pJB3. The DNA fragment carrying the putative bfrH promoter was liberated from pJB3 by digestion with EcoRI and BamHI and directionally cloned into pDJM41Δ (36) to produce pJB3.1.
Engineering of RB50Δfur1.
The fur1 gene (51) was deleted from the chromosome of RB50 by use of allelic exchange. Nucleotide sequences encoding the fur1 ORF and the respective 5′- and 3′-flanking regions were amplified from RB50 chromosomal DNA by a PCR utilizing oligonucleotide primers (Δfur1-5 and Δfur1-3) whose nucleotide sequences were homologous to regions located 601 bp upstream of the fur1 ATG start codon and 600 bp downstream of the fur1 TGA stop codon, respectively. PCR was performed using EasyA polymerase (Stratagene) under the following conditions: 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 4 min 30 s. The purified PCR product was ligated into pTOPO (Invitrogen) to produce pJB11. To remove the nucleotide sequences carrying the fur1 ORF from the amplified fragment, inverse PCR was performed, with pJB11 as the template and with 5′-phosphorylated oligonucleotide primers (fur1invPCR1 and fur1invPCR2) whose sequences were homologous to nucleotide sequences located immediately upstream of the fur1 ATG start codon and immediately downstream of the fur1 TGA stop codon, respectively. PCR was performed using Pfu Turbo polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 4 min 30 s. Purified PCR products were self-ligated to produce pJB11.1. The 1.2-kb DNA fragment containing the fur1 flanking sequences was removed from pJB11.1 and ligated into the allelic exchange vector pCVD442tet (49) at the XbaI and SacI sites to produce pJB11.2, which was introduced by conjugation into the chromosome of RB50 at the fur1 locus.
Engineering of RB50ΔbfrH and RB50ΔbfrH::Kan.
Allelic exchange was employed to remove the bfrH gene (Sanger annotation, BB3658) (51) from the RB50 chromosome. DNA including the 5′- and 3′-flanking sequences and bfrH was amplified from RB50 chromosomal DNA by a PCR utilizing oligonucleotide primers (ΔbfrH-5 and ΔbfrH-3) with nucleotide sequences which were homologous to regions located 612 bp upstream of the bfrH GTG start codon and 590 bp downstream of the bfrH TGA stop codon, respectively. PCR was performed using Pfu Turbo polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 52°C for 45 s, and 72°C for 5 min 30 s. Purified PCR products were digested with XbaI and SacI, and the digested fragment was directionally cloned into pBluescriptKS(+) (Stratagene) to produce pJB8. To remove nucleotide sequences carrying the bfrH ORF from the amplified fragment, inverse PCR was performed, utilizing 5′-phosphorylated oligonucleotide primers (bfrHinvPCR1 and bfrHinvPCR2) with homology to nucleotide sequences located immediately upstream of the GTG start codon and directly downstream of the TAA stop codon of bfrH, respectively. The amplicon was generated using pJB8.0 as a DNA template, and PCR was performed using Pfu Turbo polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 4 min 30 s. Purified PCR products were self-ligated to produce pJB8.1. The 1.2-kb fragment of pJB8.1 containing the flanking sequences of bfrH was removed from pJB8.1 and ligated into pCVD442tet (49) at the XbaI and SacI sites to produce pJB8.2, which was introduced into RB50 by conjugation.
A second bfrH deletion mutant of RB50 (RB50ΔbfrH::Kan) was engineered by replacing the chromosomal copy of the bfrH gene with a nonpolar kanamycin resistance cassette. A 926-bp cassette encoding resistance to kanamycin from Tn903 was obtained from pKSAC (Pfizer, New York, NY) by PCR (50). Synthetic oligonucleotides pTOPOKan5 and pTOPOKan3 were used as PCR primers. The amplicon was generated using EasyA polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 2 min. Purified PCR products were ligated into pTOPO (Invitrogen) to produce pTOPOKan. To engineer a bfrH::Kan mutant of RB50, nucleotide sequences flanking bfrH were amplified via inverse PCR by utilizing the oligonucleotide primers bfrHinvPCR1 and bfrHinvPCR2. The amplicon was generated using pJB8 as a template, Pfu Turbo polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 4 min 30 s. pJB8.3 was produced by ligating the purified PCR product to the 927-bp DNA fragment encoding the Kanr determinant, which was obtained from pTOPOKan via digestion with EcoRV. The 2.1-kb DNA fragment of pJB8.3 containing the bfrH flanking sequences and the Kanr gene was ligated into pCVD442tet (49) at the XbaI and SacI sites to produce pJB8.4, which was introduced into RB50 by conjugation.
Engineering of RB50ΔecfI.
Allelic exchange was employed to delete ecfI (51) from the RB50 chromosome. DNA including the 5′- and 3′-flanking sequences and ecfI was amplified from RB50 chromosomal DNA by utilizing oligonucleotides (ΔecfI-5 and ΔecfI-3) which mapped 604 bp upstream of the ATG start codon and 600 bp downstream of the TGA stop codon of ecfI, respectively. DNA was amplified using Pfu Turbo polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 2 min 30 s. pJB16 was produced by ligating the purified PCR product into pBluescript KS(+) (Invitrogen) at the XbaI site. To remove ecfI from the amplified fragment, inverse PCR was performed, utilizing 5′-phosphorylated oligonucleotides (ecfIinvPCR1 and ecfIinvPCR2) which mapped immediately upstream of the ATG start codon and 100 bp upstream of the TGA stop codon of ecfI, respectively. pJB16 was used as a template for a PCR with Pfu Turbo polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 5 min 30 s. pJB16.1 was produced by self-ligating the purified PCR product. The 1.2-kb DNA fragment containing the ecfI flanking sequences in pJB16.1 was ligated into pCVD442tet (49) at the XbaI site to produce pJB16.2, which was introduced into RB50 by conjugation.
Engineering of RB50ΔecfR.
Methods similar to those used to delete ecfI were employed to engineer an ecfR mutant of RB50. DNA including the 5′- and 3′-flanking sequences and ecfR was amplified from RB50 chromosomal DNA by utilizing oligonucleotide primers (ΔecfR-5 and ΔecfR-3) homologous to nucleotide sequences located 643 bp upstream of the ecfR ATG start codon and 603 bp downstream of the TGA stop codon of ecfR, respectively. The amplicon was generated by a PCR with EasyA polymerase (Stratagene) and the following parameters: 30 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 2 min 30 s. pJB10 was produced by ligating the purified amplicon into pGEM-T (Promega). To remove ecfR from the amplified fragment, two 5′-phosphorylated oligonucleotides (ecfRinvPCR1 and ecfRinvPCR2), which mapped 5 bp downstream of the ATG start codon of ecfR and directly downstream of the gene's TGA stop codon, respectively, were employed in an inverse PCR with the template, pJB10, Pfu Turbo polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 4 min 30 s. pBJ10.1 was engineered by self-ligating the purified PCR product. The 1.2-kb fragment containing the ecfR flanking sequences was reamplified utilizing the oligonucleotides pJB10.1ecfRUpXbaI and pJB10.1ecfRdXbaI-2, EasyA polymerase (Stratagene), and the following parameters: 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 2 min. pJB10.3 was produced by ligating the purified amplicon into pTOPO (Invitrogen). The 1.2-kb DNA fragment containing the ecfR flanking sequences was removed from pJB10.3 and ligated into pCVD442tet (49) at the XbaI site to produce pJB10.4, which was introduced into RB50 by conjugation.
Engineering of RB50ecfRK-I.
To generate an ecfR knock-in strain, a wild-type (wt) copy of ecfR was reintroduced into the genome at the gene's original locus. DNA including the 5′- and 3′-flanking sequences and ecfR was amplified from RB50 chromosomal DNA by utilizing oligonucleotide primers pJB10.1ecfRUpXbaI and pJB10.1ecfRdXbaI-2. The amplicon was generated using Taq polymerase (1 unit), 1× EasyA polymerase buffer (Stratagene), 10% DMSO, a 62.5 μM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each oligonucleotide primer, and the following parameters: 30 cycles of 95°C for 45 s, 64°C for 45 s, and 72°C for 3 min 30 s. pJB10XbaI was produced by ligating the purified amplicon into pTOPO (Invitrogen). The plasmid pJB10.5 was produced by ligating the 2.2-kb DNA fragment containing ecfR and the flanking sequences within pJB10XbaI into pCVD442tet (49) at the XbaI site for conjugation into RB50ecfR.
Conjugation and sucrose selection of engineered mutants.
pCVD442tet-derived plasmids were introduced into the B. bronchiseptica chromosome by use of conjugation, employing E. coli SM10λpir (49). Transconjugants were selected on BHI agar supplemented with streptomycin and tetracycline. To select for plasmid disintegration, transconjugants were cultured in BHI broth in the presence of streptomycin, and dilutions were plated onto BHI agar supplemented with streptomycin and 20% sucrose. Sucrose-resistant colonies were screened for plasmid disintegration by replica patching on BHI agar plates containing either streptomycin or streptomycin and tetracycline. Sucrose-sensitive colonies were further analyzed by colony lift hybridization, colony PCR, and Southern hybridization to confirm integration and subsequent disintegration of the plasmid.
Isolation of total RNA.
Total RNA was harvested from bacteria by a modification of the acid phenol method (36, 73). Bacterial strains cultured in Fe-replete or Fe-limiting BHI broth were grown to an optical density at 600 nm (OD600) of 0.6. Twenty-five ml of culture was mixed with 5 ml of ice-cold RNA Protect (Qiagen, Valencia, CA). The cells were pelleted at 3,220 × g and 4°C for 30 min. The supernatant was discarded, and the cell pellets were quickly frozen in liquid nitrogen. Cells were resuspended in 1.5 ml ice-cold buffer A (0.02 M sodium acetate [pH 5.3], 1 mM EDTA [pH 8.0]) and 200 μl of RNA Protect (Qiagen). The cell suspension was transferred to a solution containing 160 μl 10% SDS, 2 ml buffer A, and 3.5 ml acid phenol (pH 6.6) which was preheated at 65°C. The mixture was incubated at 65°C for 5 min and vortexed for 30 s, followed by incubation for 2 min at 65°C. The mixture was vortexed for 1 min, followed by incubation for 5 min at 65°C. Cells were centrifuged at 5,927 × g and 4°C for 5 min. The aqueous phase was extracted once with 3 ml of acid phenol-chloroform-isoamyl alcohol (25:24:1) and once with 3 ml of chloroform. For each extraction, phases were separated by centrifugation at 5,927 × g and 4°C for 5 min. RNA was precipitated by adding a 1/10 volume of 3 M sodium acetate and 2 volumes of 100% ethanol and was kept overnight at −80°C. Precipitated RNA was pelleted by centrifugation at 12,096 × g and 4°C for 45 min. RNA pellets were washed with 1 ml of 70% ethanol, air dried, and resuspended in 160 μl diethyl pyrocarbonate (DEPC)-treated water. Total RNA preparations were treated with DNase I to remove DNA contamination. After treatment, DNase I was removed from the RNA by extraction using acid phenol-chloroform-isoamyl alcohol and chloroform. RNA was reprecipitated as described above. The precipitated RNA was pelleted by centrifugation at 13,900 × g and 4°C for 30 min and then washed with 1 ml of 70% ethanol. After removal of all residual ethanol, RNA was resuspended in 100 μl of DEPC-treated water and stored at −80°C.
End-point RT-PCR.
RT-PCR was performed using a One Step RT-PCR kit (Qiagen) according to the vendor's suggested reaction conditions. Initially, 10 ng total RNA was reverse transcribed into cDNA (one 30-min cycle at 50°C, 15 min at 95°C). As a control to check for possible genomic DNA contamination, identical reaction mixtures were prepared and kept on ice for 30 min, followed by 95°C for 15 min to inactivate reverse transcriptase. Secondly, cDNA was amplified by PCR (30 cycles of 45 s at 95°C, 45 s at 50°C, and 1 min at 72°C). Amplified DNAs were resolved by electrophoresis using 2% agarose gels.
qRT-PCR.
Transcription of bfrH and the ecfR-bfrH intergenic region was measured by use of a modified qRT-PCR protocol (59). Briefly, cDNA was reverse transcribed from 500 ng of total RNA by use of an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Synthesized cDNA was utilized as the PCR template. IQ SYBR green supermix (Bio-Rad) and an iCycler thermal cycler (Bio-Rad) were used for the reactions. PCR mixtures initially heated to 95°C for 3 min were subjected to 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. RB50 chromosomal DNA was used as a PCR template to generate a standard curve for each gene. Starting quantities (SQs) of mRNAs for bfrH, the ecfR-bfrH intergenic region, and recA were calculated from the corresponding standard curves. Transcription of recA was employed as an internal control, and the quantities of bfrH and the ecfR-bfrH intergenic region were normalized to the quantity of recA detected for each experimental condition.
β-Galactosidase assay.
Expression of the lacZ reporter gene in pJB3.1 was determined by performing a β-galactosidase assay as previously described (36). Briefly, overnight Fe-replete or Fe-stressed cultures were pelleted, resuspended in Z buffer (60 mM Na2HPO4-7H2O, 40 mM NaH2PO4-H2O, 10 mM KCl, 1 mM MgSO4-H2O, 38 mM β-mercaptoethanol), and adjusted to an OD600 of 0.30 to 0.70. After dilution, 400 μl of the suspension was permeabilized with 45 μl of 0.1% SDS and 90 μl of chloroform, vortexed for 10 s, and incubated at 30°C for 30 min. Enzymatic reactions were initiated by adding 160 μl of 4-mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) and incubating the mixtures at room temperature. The reactions were allowed to continue until a yellow pigment was observed, at which point 400 μl of 1 M Na2CO3 was added to terminate the reaction. To pellet the cellular debris and chloroform, the reactions were centrifuged for 2 min at 16,000 × g at room temperature, the OD420 and OD550 were obtained, and the β-galactosidase activity was calculated by use of the following formula: 1,000[OD420 − 1.75(OD550)]/(t)(0.4)(OD600).
Isolation of OMs.
OMs were isolated using previously described methods (36, 49). Strains were cultured in Fe-replete and Fe-stressed BHI broth to a final OD600 of 0.6. Cells pelleted by centrifugation at 3,000 × g for 20 min at 4°C were washed three times with (and resuspended in) 20 ml of 1 mM ice-cold 10 mM HEPES (pH 7.4) supplemented with the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (0.1 mM). Cells were frozen and kept at −80°C overnight. After thawing on ice, cells were disrupted by sonication (four 2-min pulses at 50% duty cycle), using a sonifier (Branson Ultrasonics Corp., Dansbury, CT) fitted with a microtip. Unbroken cells were removed from the lysate by centrifugation at 3,000 × g for 20 min at 4°C. The lysate was centrifuged at 100,000 × g at 4°C, and the pellet containing the total membrane fraction was resuspended in 10 ml of 1% Sarkosyl in 10 mM HEPES (pH 7.4) supplemented with PMSF (0.1 mM) and mixed at room temperature for 1 h by gentle agitation. After centrifugation at 100,000 × g and 4°C for 1 h, the pellet, containing the OM fraction, was retreated with the solution of 1% Sarkosyl in 10 mM HEPES (pH 7.4) supplemented with PMSF (0.1 mM). The remaining OMs were pelleted by centrifugation at 100,000 × g and 4°C for 1 h, resuspended in 200 μl of deionized water, and stored at −80°C. Total proteins in the OMs were measured using a Micro BCA protein assay kit (Thermo Scientific, Rockford, IL), with bovine serum albumin as the standard.
Production of rabbit anti-BfrH-peptide antiserum and purification of polyclonal antibody.
Production of rabbit anti-BfrH-peptide antiserum and purification of IgG antibody from the antiserum were performed as previously described (48). Briefly, a synthetic peptide (N-LGPNHYDGDADFEKS-C) with an amino acid sequence identical to an internal amino acid sequence of BfrH predicted by Psipred (31), Yaspin (42), and PhD (57) to be antigenic was chosen as the immunogen. Antipeptide antiserum was produced commercially by Sigma Genosys (St. Louis, MO). The peptide-specific polyclonal antibodies in the antiserum were purified by affinity chromatography, using a Sulfolink peptide affinity column (Pierce, Rockford, IL) to which the immunizing peptide had been coupled. After adding 1 ml of antiserum, the column was washed with 12 ml of phosphate-buffered saline (PBS) (pH 7.4), and the bound peptide-specific antibodies were eluted in 1-ml fractions, using 3.0 M glycine (pH 2.0). Eluates were immediately neutralized by the addition of 1 ml of 1 M Tris-HCl (pH 8.0) and were analyzed spectrophotometrically (OD488) for protein. Peak fractions were combined and dialyzed exhaustively against PBS (pH 7.4).
Immunodetection of BfrH.
Aliquots of OMs containing 200 μg of total protein were resolved using SDS-PAGE with 7.5% (wt/vol) polyacrylamide gels. Proteins were electrotransferred to Optitran BA-S 83 nitrocellulose membranes (Whatman-Schleicher and Schuell, Dassel, Germany), and the membranes were probed overnight at 4°C with purified anti-BfrH-peptide polyclonal antibodies (1:50). After being washed, the membranes were probed with a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:8,000) (Southern Biotech, Birmingham, AL). Immunoreactive proteins were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
In vitro growth analysis.
Initial broth cultures of bacteria were prepared by subculturing single colonies taken from BG agar plates into SS broth. After overnight incubation at 37°C, cultures were standardized to an OD600 of 1.0 and 250 μl of the culture was utilized to inoculate 25 ml of Fe-stressed or Fe-replete SS broth. All cultures were prepared in triplicate and incubated at 37°C in an Innova 4330 incubator shaker (New Brunswick Scientific, Edison, NJ). The OD600 values for samples taken at various time points were measured using a Beckman Coulter DU640 spectrophotometer (Fullerton, CA).
Murine infection assay.
Female BALB/c mice (4 to 6 weeks) were obtained from Harlan Sprague, Inc. (Indianapolis, IN). RB50 and RB50ΔbfrH::Kan were maintained on BG agar plates and subsequently cultured in SS broth for infection studies. A total of nine female mice lightly sedated with 5% isoflurane were inoculated intranasally with a 1:1 mixture of 2.5 × 105 CFU of RB50 and 2.5 × 105 CFU of RB50ΔbfrH::Kan in a 20-μl PBS resuspension or with 20 μl of PBS. The bvg+ status of both strains was confirmed by analyzing actively growing cultures for the ability to bind Congo red (25). Colonization was assessed at 5 and 10 days postinfection. At each time point, mice were euthanized and the nasal cavity, tracheal, and lung tissues were separately harvested and homogenized in 1 ml of PBS. Serial dilutions of the homogenates were plated onto BHI agar plates supplemented with streptomycin to select for total bacteria and onto BHI agar plates supplemented with streptomycin and kanamycin to select for mutant bacteria. Bacterial colony counts were enumerated after 2 days of incubation at 37°C. The competitive index (CI) was calculated as the mutant/wild-type CFU ratio for each tissue at the indicated time point divided by the mutant/wild-type ratio of the starting inoculums.
Statistical methods.
Student's t test (paired, two-tailed) was used to determine whether the mean CI (n = 9) for each tissue at each time point examined in the murine infection assay differed significantly from a hypothetical value of 1.00 (the mutant/wild-type ratio predicted given no difference in the ability of either strain to successfully colonize the host). P values of ≤0.05 were considered significant. All other experimental results were compared statistically using unpaired t tests (InStat, version 3.00; GraphPad Software Inc., San Diego, CA).
RESULTS
Identification of bfrH.
To identify Fe-regulated membrane proteins in the pathogenic bordetellae, proteomic investigations using two-dimensional difference in-gel electrophoresis (2D-DIGE) (Bio-Rad) were initiated using B. pertussis and B. avium, the two most distantly related pathogenic members of the genus (61). Total OMs from these bacteria, each cultured under Fe-replete or Fe-limiting conditions, were separately labeled with different fluorescent dyes, combined, and simultaneously resolved by 2D gel electrophoresis (data not shown). Two polypeptides with similar electrophoretic mobilities were identified which were highly expressed when the bacteria were cultured under Fe-limiting conditions but were absent or barely detectable when the bacteria were cultured under Fe-replete conditions. Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry identified the polypeptides as the product of bfrH, a gene predicted in the annotated genomes of B. avium, B. pertussis, B. bronchiseptica, and B. parapertussis (51) to encode a putative Fe-regulated OM protein (N. D. King-Lyons and T. D. Connell, unpublished data). While all four pathogenic Bordetella species carried the gene, subsequent investigations to reveal the mechanisms by which bfrH was regulated in response to Fe and the role(s) of BfrH in pathogenesis were performed with B. bronchiseptica, the member of the genus which has the broadest host range and for which facile genetics and a mouse challenge model are available.
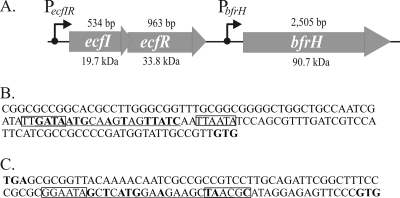
The ORF of bfrH is composed of 2,505 bp with a coding capacity for a polypeptide of ∼90 kDa (∼87 kDa in the absence of the predicted signal peptide) (Fig. 1A). BfrH exhibits significant homology to FhuA, a ferrichrome receptor of E. coli (38% identity, 56% similarity), and to FpvA, a pyoverdine receptor of Pseudomonas aeruginosa (37% identity, 55% similarity) (9, 21, 52, 62). Pfam protein software predicted that BfrH contains an N-terminal TonB-binding domain similar to the TonB-binding domains located in PupB, a pseudobactin receptor of P. putida (37), and in FecA, a ferric citrate receptor in E. coli (24). Structural analysis indicated that BfrH likely possesses a Plug domain, which in FecA and FhuA is believed to interact specifically with the receptors' cognate ligands (24). These in silico data provided strong support for a model in which BfrH of B. bronchiseptica was likely a new Fe-regulated siderophore receptor.
FIG. 1.
Genetic organization of the ecfIR-bfrH locus. (A) Schematic arrangement of the ecfIR-bfrH locus in B. bronchiseptica. ecfI and ecfR encode a putative ECF sigma factor and a putative sigma factor regulator, respectively. bfrH exhibits homology to genes for OM proteins involved in siderophore uptake. The length of each ORF is denoted above the gene; the molecular mass of each of the predicted polypeptides is denoted below the gene. Positions of the two putative promoters, PecfIR and PbfrH, and the direction of transcription are denoted by arrows. (B) Sequences at PecfIR. This promoter contains regions homologous to σ70-type promoters. Putative −10 and −35 regions of PecfIR are boxed. Sequences homologous to the consensus Fur box of E. coli are underlined; nucleotides in bold are perfectly conserved in RB50 and in the consensus Fur box of E. coli. The translational GTG start codon of ecfI is denoted in bold. (C) Sequences at PbfrH. This region contains homology to other ECF sigma factor-regulated promoters. Putative −10 and −35 regions of PbfrH are boxed. Sequences homologous to the consensus Fur box of E. coli are underlined; nucleotides in bold are perfectly conserved in RB50 and in the consensus Fur box of E. coli. The translational stop codon of ecfR and the translational start codon of bfrH are denoted in bold.
ecfI and ecfR.
Further analysis of the genome of B. bronchiseptica in the vicinity of bfrH revealed the presence of two smaller ORFs (ecfI and ecfR) which were located 93 bp immediately upstream of bfrH (Fig. 1A).
The 534-bp ORF located most distal to bfrH, designated ecfI, encoded a polypeptide of 19.7 kDa. Database comparisons revealed that EcfI exhibited strong homology to several Fe starvation-induced ECF sigma factors, including HurI of B. bronchiseptica (44% identical), PupI of P. putida (39% identical), and FecI of E. coli (39% identical). Several recognizable domains are routinely found in ECF sigma factors. Region 2 interacts with RNA core polymerase; region 4 binds the ECF sigma factor to the −35 hexameric region of ECF sigma factor-responsive promoters (3, 43); and region 1, a domain found within the σ70 family of sigma factors which regulates binding of the sigma factor to promoters only when the sigma factor is associated with RNA polymerase, is shortened or missing in ECF sigma factors (43). Amino acid analysis indicated that regions 2 and 4 were significantly conserved in EcfI. Furthermore, a shortened region 1 was recognizable in EcfI. From these observations, it was determined that ecfI likely encoded a new ECF sigma factor of B. bronchiseptica.
Of the two ORFs, ecfR, which was comprised of 963 bp encoding a polypeptide of 33.8 kDa, was located proximal to bfrH (Fig. 1A). The 5′ end of ecfR (TAG) overlapped the 3′ end of ecfI (ATG) by 8 bp (5′-ATGAGTAG-3′), and the 3′ end of ecfR was positioned 93 bp upstream of the 5′ end of bfrH. EcfR exhibited 29% identity to FecR of E. coli (69) and was 34% identical to HurR of B. bronchiseptica (66) and 34% identical to PupR of P. aeruginosa (37). Amino acid sequences consistent with a transmembrane region and a signal peptide indicated that EcfR was likely integrated into the plasma membrane. Collectively, these data were consistent with EcfR having the major attributes of an ECF sigma factor regulator. Since ecfR is genetically linked to ecfI, it was hypothesized that EcfR was a ligand-dependent regulator of EcfI.
PecfIR and PbfrH.
An evaluation of the region located immediately upstream of ecfIR revealed the occurrence of nucleotides consistent with a promoter sequence (PecfIR) (Fig. 1A). Putative −35 (5′-TTGATA-3′) and −10 (5′-TTAATA-3′) hexameric regions of PecfIR exhibited similarities to promoters usually regulated by σ70 (Fig. 1B) (36). Further inspection indicated a region near PecfIR which was homologous to the consensus Fur box of E. coli (14 of 19 nucleotides [nt]) (Fig. 1B) (58).
Putative promoter sequences (PbfrH) were located within the region flanked by ecfR and bfrH (Fig. 1A). These sequences exhibited characteristics common to promoters controlling the expression of other ECF sigma factor-responsive genes (Fig. 1C) (68). Nucleotide sequences within the prospective PbfrH included a −35 hexameric region (5′-GGAATA-3′) located 40 bp upstream of the prospective GTG start codon and a region located 15 bp downstream from the putative −35 hexamer sequence with features similar to a −10 consensus promoter sequence (5′-TAACGC-3′) (Fig. 1C). A nucleotide sequence with homology to a consensus Fur box (9 of 19 nt) was identified within the putative PbfrH (Fig. 1C).
Overall, the architecture of the ecfIR-bfrH locus was reminiscent of the arrangement of genes encoding other ECF sigma factor-regulated bacterial Fe uptake systems, including fecIR-fecABCDE of E. coli (11), pupIR-pupB of P. putida (37), rhuIR-bhuRSTUV of B. avium (49), and hurIR-bhuRSTUV of B. bronchiseptica and B. pertussis (67). These observations were consistent with a model in which (i) genes within the ecfIR-bfrH locus were coordinately regulated by Fur and (ii) EcfI and EcfR were likely involved in the coregulated expression of bfrH in response to an extracellular ligand.
Transcriptional characterization of the ecfIR-bfrH locus.
To ascertain whether ecfI, ecfR, and bfrH were expressed in B. bronchiseptica, end-point RT-PCR was performed using total RNA obtained from B. bronchiseptica RB50 cells that had been cultured under Fe-replete and Fe-stressed growth conditions.
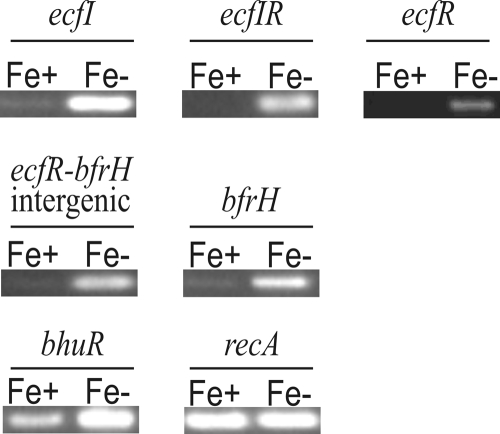
Strong transcriptional signals for ecfI and ecfR were observed only when cells had been cultured under Fe-stressed conditions (Fig. 2). An identical transcriptional profile was detected for the ecfI-ecfR overlapping region (ecfIR), the ecfR-bfrH intergenic region, and bfrH. Expression of bhuR, a previously described Fe-regulated OM protein (66), was employed as a positive control. As expected, bhuR exhibited an enhanced transcriptional signal when cells were cultured under Fe-stressed conditions. Transcription of recA, employed as an internal control, remained unchanged regardless of the presence of Fe. No transcriptional signals were observed when reverse transcriptase was omitted from the reaction mixtures (data not shown).
FIG. 2.
Fe-dependent transcription of the ecfIR-bfrH locus. Total RNAs were isolated from Fe-replete and Fe-stressed RB50 cultures. Oligonucleotide primer sets used in the reaction mixtures targeted a 218-bp internal region of ecfI, a 288-bp overlap region containing the 3′ end of ecfI and the 5′ end of ecfR (ecfIR), a 488-bp internal region ecfR, a 237-bp region overlapping ecfR, bfrH, and a 93-bp region between ecfR and bfrH (ecfR-bfrH intergenic), a 598-bp internal region of bfrH, a 513-bp internal region of bhuR, and a 402-bp internal region of recA. Amplified DNA from each RT-PCR was resolved in a 2% agarose gel and visualized by ethidium bromide staining. Fe+, Fe-replete conditions; Fe−, Fe-stressed conditions.
Collectively, these data indicated that ecfI, ecfR, and bfrH were transcribed when B. bronchiseptica was cultured under Fe-stressed conditions. Moreover, these data illustrated that ecfI and ecfR were likely cotranscribed and that, in Fe-stressed cells, bfrH was polycistronically transcribed with ecfIR, most likely by a process of readthrough transcription from PecfIR. A similar polycistronic mRNA derived from readthrough transcription was described for B. bronchiseptica for the ECF sigma factor-regulated heme acquisition locus (hurIR-bhuR) (68).
Fe-dependent regulation of ecfIR-bfrH by fur1.
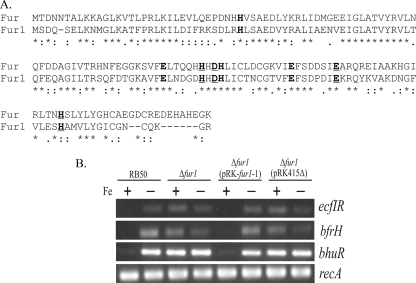
Since the RT-PCR experiments demonstrated that ecfI, ecfR, and bfrH were transcribed in an Fe-dependent manner and that both PecfIR and PbfrH contained a potential Fur box (Fig. 1A), it was surmised that expression of the ecfIR-bfrH locus was likely to be influenced by a Fur homologue. In silico analysis of the RB50 chromosome (51) revealed two genes (BB3942 and BB0215) which were homologous to fur of E. coli. BB3942 (fur1) encoded a polypeptide with 54% amino acid sequence homology to the Fur protein of E. coli, and BB0215 (fur2) encoded a polypeptide with only 42% homology to the Fur protein of E. coli. In most Fur proteins, two metal binding sites, composed of discontinuous amino acid sequences and containing histidines and glutamates, are conserved (Fig. 3A) (26, 44). Amino acid sequence alignment with the Fur protein of E. coli revealed that both sites are conserved in Fur1 (Fig. 3A). Neither of these two sites is conserved in Fur2 (data not shown).
FIG. 3.
Regulation of the ecfIR-bfrH locus by fur1. (A) Alignment of Fur1 sequence with sequence of Fur of E. coli. Amino acids within metal binding site 1 are indicated in bold and underlined, and amino acids within metal binding site 2 are denoted only in bold. (B) Fur1-dependent transcription of the ecfIR-bfrH locus. RT-PCR was performed using total RNAs obtained from Fe-replete and Fe-stressed cells, utilizing oligonucleotide primers which targeted the overlap region encompassing the 3′ end of ecfI and the 5′ end of ecfR (ecfIR), bfrH, bhuR, and recA. Amplified DNA from each RT-PCR was resolved in a 2% agarose gel and visualized by ethidium bromide staining. +, Fe-replete conditions; −, Fe-stressed conditions.
To determine if either fur1 or fur2 influenced expression of ecfIR-bfrH, attempts were made to engineer isogenic mutants for each of the two genes in RB50. Unfortunately, repeated attempts to engineer a mutant of fur2 were unsuccessful, thereby suggesting that this gene is critical for survival of RB50. In contrast, an isogenic fur1 mutant of RB50 was obtained.
The effects of the fur1 mutation on the regulation of ecfIR and bfrH were assessed by end-point RT-PCR, using total RNAs isolated from wt and fur1-deficient cells, with each strain cultured under Fe-replete and Fe-stressed conditions (Fig. 3B). For RB50, transcription of ecfI, ecfR, and bfrH was observed only in Fe-stressed cells. In contrast, all three genes were transcribed in RB50Δfur1 and in RB50Δfur1(pRK415Δ), the vector control strain, regardless of the Fe environment. An identical Fe-dependent transcriptional profile was observed in B. pertussis for bhuR, a gene known to be controlled by fur (only a single fur gene is carried by B. pertussis) (66). To ensure that overall transcription was not altered in RB50 by the fur1 deficiency, the level of transcription of recA, a gene which is not regulated by Fe, was determined. Expression of recA was equivalent in all strains, regardless of the Fe status of the cells. Complementation of RB50Δfur1 with pRK-fur1-1, a plasmid carrying a wt copy of fur1, reestablished Fe-dependent regulation of ecfI, ecfR, and bfrH. No transcriptional signals were observed when reverse transcriptase was omitted from the reaction mixtures (data not shown). These data suggested that expression of ecfI, ecfR, and bfrH was regulated, at least in part, by fur1 and that Fe-dependent repression of ecfIR and bfrH was fur dependent.
Regulation of bfrH by ecfI.
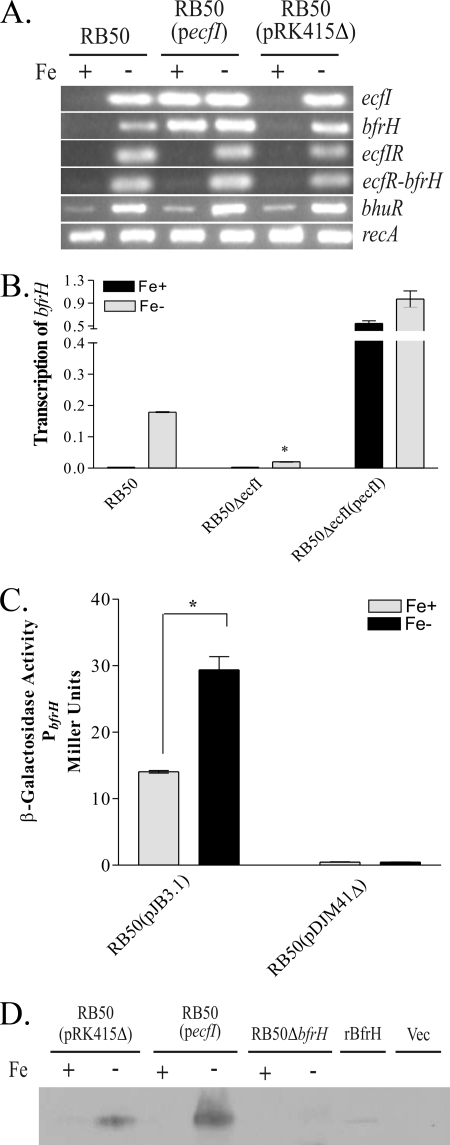
To promote transcription of the cognate gene in ECF-regulated loci, ECF sigma factors must be either liberated from or activated by the associated ECF sigma factor modulators after induction by the extracellular ligand (36, 37, 67). Unfortunately, the inducing ligand for bfrH has not yet been discovered, which limited the types of experiments which could be performed to characterize ecfI. But an alternative approach revealed in other ECF-regulated systems was employed. In several other systems, overexpression of the cognate ECF sigma factor decouples ECF sigma factor-regulated systems from ligand-dependent induction. When the ECF sigma factor is overexpressed, the regulated gene is constitutively expressed (36, 37, 53). To determine if EcfI regulated expression of bfrH, an expression plasmid carrying a wt copy of ecfI (pecfI) was introduced by conjugation into RB50 and the level of transcription of bfrH was evaluated after growth of the bacterium under various Fe conditions. As expected, introduction of pecfI into RB50 promoted high-level transcription of the recombinant ecfI gene, regardless of the Fe environment in which the cells were cultured (Fig. 4A). Constitutive expression of bfrH was also observed in RB50(pecfI) when cells were cultured under conditions in which Fe was either plentiful or limiting. In contrast, transcription of the region of ecfI-ecfR overlap and the intergenic region located between ecfR and bfrH was unaltered by overexpression of ecfI. No transcriptional signals were observed when RNA was not reverse transcribed before PCR amplification (data not shown). These data demonstrated that constitutive expression of bfrH in RB50(pecfI) was not mediated by modulation of PecfIR by EcfI. Rather, the data are consistent with a model in which expression of bfrH is promoted by modulation of PbfrH by EcfI. Since overexpression of ecfI did not alter the transcription profile of the polycistronic ecfIR mRNA, these data also indicated that EcfI is not an autoregulatory ECF sigma factor.
FIG. 4.
Regulation of bfrH by EcfI. (A) Effects of overexpression of EcfI on expression of bfrH. RT-PCR was performed using total RNAs isolated from Fe-replete and Fe-stressed cells. Oligonucleotide primer sets used in the reaction mixtures targeted a 218-bp region of ecfI, a 288-bp overlap region encompassing the 3′ end of ecfI and the 5′ end of ecfR (ecfIR), a 237-bp region overlapping ecfR, bfrH, and a 93-bp region between ecfR and bfrH (ecfR-bfrH intergenic), a 598-bp internal region of bfrH, a 513-bp internal region of bhuR, and a 402-bp internal region of recA. Amplified DNA from each RT-PCR was resolved in a 2% agarose gel and visualized by ethidium bromide staining. +, Fe-replete conditions; −, Fe-stressed conditions. (B) Expression of bfrH in RB50ΔecfI. qRT-PCR was performed on total RNAs isolated from Fe-replete and Fe-stressed cells, using oligonucleotides targeting sequences within bfrH. Data are expressed as means and standard errors and were obtained by calculating the relative SQ of the respective mRNA after normalizing to the amount of recA mRNA expressed by the cell. *, statistically significantly different from Fe-stressed RB50 (P < 0.05). (C) β-Galactosidase activities of RB50(pJB3.1) and RB50(pDJM41Δ) cultured under Fe-repelete (Fe+) and Fe-stressed (Fe−) conditions. *, statistically significantly different from Fe-replete RB50(pJB3.1) (P < 0.05). (D) Western immunoblot of BfrH. OMs were prepared from Fe-replete and Fe-stressed cells. OMs were resolved in a 7.5% SDS-PAGE gel and immunoblotted with anti-BfrH-peptide rabbit polyclonal antibodies. +, Fe-replete conditions; −, Fe-stressed conditions; rBfrH, whole-cell extract of IPTG-induced BL21(DE3)(pLysS)(pJB7.1); Vec, whole-cell extract of the IPTG-induced vector control [BL21(DE3)(pLysS)(pET21a)].
To further establish that EcfI regulated the expression of bfrH, transcriptional patterns were evaluated with RB50ΔecfI, a mutant in which ecfI was deleted from the chromosome (Fig. 4B). qRT-PCR was performed utilizing total RNAs obtained from cells after culture under Fe-replete and Fe-stressed growth conditions. As expected, an increase in transcription of bfrH was observed when RB50 was cultured under Fe-stressed conditions in comparison to transcription of bfrH in RB50 cultured under Fe-replete conditions. Deletion of ecfI produced a remarkable decrease, but not a total abrogation, of transcription of bfrH in cells cultured under Fe-stressed conditions. A significant increase in transcription of bfrH was observed when RB50ΔecfI was complemented with pecfI [RB50ΔecfI(pecfI)], regardless of the Fe environment. Collectively, the results from these experiments strongly indicate that EcfI regulates Fe-dependent transcription of bfrH from PbfrH.
End-point RT-PCR data (Fig. 2 and 3B) suggested that ecfI, ecfR, and bfrH were all transcribed from a putative promoter (PecfI) located upstream of ecfI (Fig. 1A). When ecfI was constitutively expressed in trans, however, transcription of bfrH was decoupled from transcription of either ecfI or ecfR (Fig. 4A). These data suggested that EcfI-dependent transcription of bfrH is likely driven from a promoter located downstream of ecfR. To determine if the ecfR-bfrH intergenic region contained a functional promoter, a PbfrH:lacZ promoter-reporter plasmid (pJB3.1) was introduced into RB50, and the transcriptional activity of PbfrH was determined by measuring the β-galactosidase activity of bacteria cultured under Fe-replete and Fe-stressed conditions (Fig. 4C). pDJM41Δ, a vector control plasmid containing a promoterless lacZ gene, exhibited no detectable β-galactosidase activity when expressed in RB50, regardless of the Fe content of the culture medium. Fe-replete cultures of RB50(pJB3.1), however, displayed a small but significant amount of β-galactosidase activity that increased ∼3-fold when measured from bacteria cultured under conditions of Fe stress. These data indicated that the ecfR-bfrH intergenic region contains a functional promoter (PbfrH) which is responsive to Fe and which, in addition to PecfIR, drives expression of bfrH.
To ascertain whether transcriptional regulation of bfrH by EcfI was reflected in synthesis of BfrH and to confirm that BfrH is an OM protein, whole-cell lysates or OMs from bacteria cultured under Fe-replete and Fe-stressed conditions were immunoblotted using a peptide-specific polyclonal antibody to an internal region of BfrH. The antiserum was validated using BL21DE3(pLysS)(pJB7.1), a strain of E. coli which encodes recombinant BfrH (rBfrH). A faint band corresponding to an 87-kDa polypeptide was detected by the anti-BfrH antiserum in whole-cell extracts obtained from BL21DE3(pLysS)(pJB7.1); this band was absent in whole-cell extracts of BL21DE3(pLysS)(pET21a), the vector control strain (Fig. 4D).
Using the anti-BfrH antiserum, a polypeptide of identical size to that of rBfrH was detected in OMs isolated from Fe-stressed RB50(pRK415Δ) (Fig. 4D). A stronger signal from a band of equivalent size was observed for OM proteins obtained from Fe-stressed RB50(pecfI). This immunoreactive polypeptide was not evident in OMs obtained from Fe-stressed RB50ΔbfrH (Fig. 4D) or in OMs obtained from RB50(pRK415Δ) cultured under Fe-replete conditions. Surprisingly, an immunoreactive polypeptide was not observed in OMs obtained from RB50(pecfI) cultured under Fe-replete conditions; transcripts of bfrH were evident in RB50(pecfI) cultured under the same conditions (Fig. 4A). The absence of BfrH expression in RB50(pecfI) could be due to unknown posttranscriptional processes which inhibit translation of bfrH mRNA. Alternatively, BfrH may be rapidly degraded when expressed under conditions where Fe is plentiful. Nonetheless, these immunoblotting experiments indicated that BfrH is an Fe-regulated, EcfI-dependent OM protein.
Regulation of bfrH by ecfR.
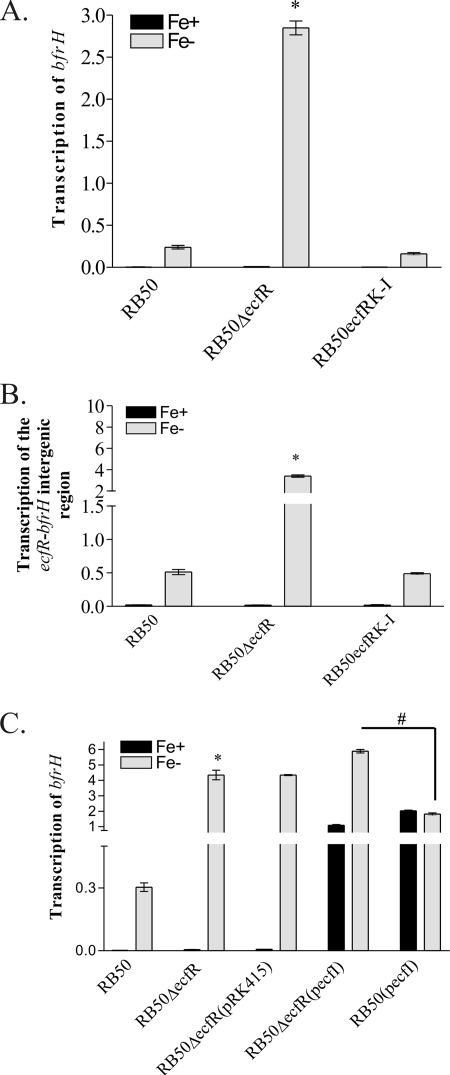
In other ECF-regulated systems, an ECF sigma factor regulator located in the cytoplasmic membrane (e.g., HurR, RhuR, PupR, and FecR) is essential for modulating expression of the regulated protein (BhuR, PupB, and FecA, respectively) (11, 35, 37, 67). To begin to elucidate the function of EcfR, the prospective EcfI regulator, transcription of bfrH was examined in an isogenic ecfR mutant of RB50 (Fig. 5A). As expected, RB50 and RB50ΔecfR did not produce a bfrH transcript when cells were cultured under Fe-replete conditions (Fig. 5A). In contrast, transcription of bfrH was evident in RB50 and RB50ΔecfR when those strains were cultured under conditions of Fe stress. Surprisingly, transcription of bfrH was significantly elevated in RB50ΔecfR in comparison to transcription of the gene in RB50 (Fig. 5A). Attempts were made to genetically complement RB50ΔecfR by introducing pecfR, a plasmid carrying a wt copy of ecfR, into the mutant strain. Unfortunately, the expected wt phenotype of Fe-dependent expression of bfrH was not reestablished in RB50ΔecfR(pecfR) (data not shown). In contrast, the expected wt phenotype was reestablished in RB50ecfRK-I, a knock-in strain in which a wt copy of ecfR was reinserted into the original chromosomal locus. These results were consistent with a model in which EcfR behaves as a typical sequestering anti-sigma factor and its absence enables unsequestered EcfI to drive transcription from PbfrH.
FIG. 5.
Regulation of bfrH by ecfR. qRT-PCR analysis was performed using total RNAs isolated from Fe-replete and Fe-stressed cells and oligonucleotides targeting an internal sequence of bfrH (A and C) or targeting the intergenic region between ecfR and bfrH (B). Data are expressed as means and standard errors and were obtained by calculating the relative SQ of the respective mRNA after normalizing to the amount of recA mRNA expressed by the cell. Fe+, Fe-replete conditions; Fe−, Fe-stressed conditions. *, statistically significantly different from Fe-stressed RB50 (P < 0.05); #, statistical significance between Fe-stressed RB50ΔecfR(pecfI) and Fe-stressed RB50(pecfI) (P < 0.05).
While it was clear that ecfR exerted an influence on expression of bfrH, subsequent experiments suggested an additional form of control exerted by ecfR in modulating transcription of the OM protein. Earlier experiments showed that a polycistronic transcript containing ecfR and bfrH was expressed in Fe-stressed RB50 (Fig. 2). Some transcription of bfrH, therefore, likely occurred by readthrough transcription from the Fe-dependent PecfIR (Fig. 2). Since RB50ΔecfR differed from RB50 not only in the absence of EcfR but also in the absence of the ecfR ORF, it was deemed feasible that the increased transcription of bfrH observed in RB50ΔecfR was due to the absence of a cis-acting sequence within ecfR in the mutant strain. This cis-acting factor(s) was hypothesized to reduce readthrough transcription from PecfIR. To elucidate whether ecfR contained a cis-acting factor, additional qRT-PCR experiments were performed to measure the amount of readthrough transcription in RB50, RB50ΔecfR, and RB50ecfRK-I by evaluating transcription of the ecfR-bfrH intergenic region (Fig. 5B). Transcription of ecfR-bfrH in RB50ΔecfR was dramatically increased in comparison to transcription in RB50 when cells were cultured under Fe-stressed conditions. In RB50ecfRK-I, transcription of the ecfR-bfrH intergenic region was restored to wt levels (Fig. 5B). Since the ecfR-bfrH transcript appears to be derived from PecfIR, these data indicated the presence of sequences within ecfR which suppress the level of readthrough transcription from that promoter.
To further analyze the effects of EcfR on the activity of EcfI with respect to transcription of bfrH, qRT-PCR was conducted on the ecfR mutant in which ecfI was overexpressed. Total RNAs for qRT-PCR were obtained from RB50, RB50ΔecfR, RB50ΔecfR(pRK415Δ), RB50ΔecfR(pecfI), and RB50(pecfI) cultured under Fe-replete and Fe-stressed conditions. Again, in comparison to RB50, a significant increase in transcription of bfrH was observed in RB50ΔecfR and RB50ΔecfR(pRK415Δ) when these strains were cultured under Fe-stressed conditions, but not when the cells were cultured under Fe-replete conditions (Fig. 5C). Consistent with the effects of pecfI on bfrH expression in RB50, a significant increase in bfrH transcription was observed in Fe-replete cultures of RB50ΔecfR(pecfI), while only a modest increase in bfrH transcription was observed in Fe-stressed cultures of RB50ΔecfR(pecfI) compared to that in RB50ΔecfR or RB50ΔecfR(pRK415Δ) (Fig. 5C). Expression of bfrH in RB50ΔecfR(pecfI) was significantly elevated in comparison to expression of bfrH in RB50(pecfI) when these strains were cultured under Fe-stressed conditions (Fig. 5C). These data suggested that the effects of pecfI and the ecfR deletion on Fe-regulated transcription of bfrH were independent.
Collectively, these data indicated that ecfR had a unique role in regulating transcription of bfrH. Although difficult to confirm in the absence of the inducing ligand, it is likely that EcfR directly or indirectly regulates the activity of EcfI. At the genetic level, however, ecfR harbors a cis-acting element which hinders maximal readthrough transcription of bfrH in the absence of the unknown extracellular inducing ligand. Efforts are currently being made to further understand this dual regulatory mechanism.
BfrH is required for optimal virulence.
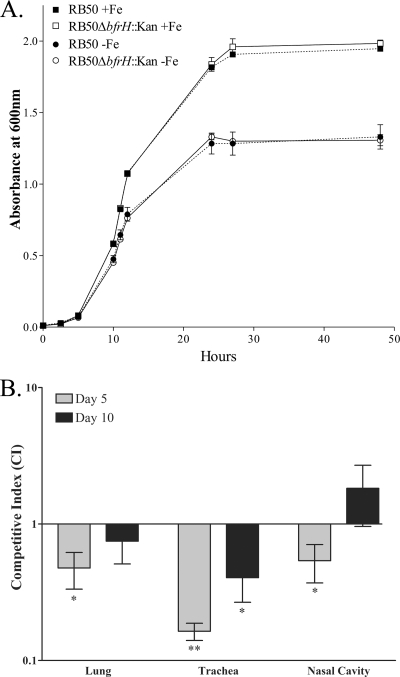
To determine whether BfrH had an effect on virulence of RB50, a well-established mouse competition model (28) was employed, using RB50 and RB50ΔbfrH::Kan, an isogenic mutant of RB50 in which a nonpolar kanamycin cassette was substituted for bfrH. Deletion of bfrH had no evident effect on growth of the bacterium in vitro. RB50 and RB50ΔbfrH::Kan exhibited identical patterns of growth (P = 0.98 in unpaired t test; n = 3) when cultured in either Fe-replete or Fe-stressed SS broth (Fig. 6A). Congo red binding of the actively growing cultures was employed to confirm that cells used for the inoculums were primarily in the bvg+ state (data not shown).
FIG. 6.
BfrH is required for optimal virulence of RB50. (A) Growth of RB50 (solid symbols, dotted lines) and RB50ΔbfrH::Kan (open symbols, solid lines) cultured in SS liquid medium supplemented with 36 μM FeSO4 (+Fe; squares) or left unsupplemented (−Fe; circles). Samples of each culture were taken at time points between 0 and 50 h, and the OD600 was used as a measure of growth. (B) Two groups (n = 9) of female BALB/c mice (4 to 6 weeks old) were inoculated intranasally with 20 μl of PBS or with PBS containing a 1:1 mixture of 2.5 × 105 CFU of RB50 and 2.5 × 105 CFU of RB50ΔbfrH::Kan. The amount of colonization by each strain in the lungs, trachea, and nasal cavity of each mouse was determined on days 5 and 10 postinfection, and the CFU of RB50 and RB50ΔbfrH::Kan were used to determine the mean CI ± SEM. A paired two-tailed t test was used to determine statistical significance. *, P < 0.05; **, P < 0.001.
To determine whether bfrH was required for optimal virulence, BALB/c mice were inoculated intranasally with a mixture of equal numbers of RB50 and RB50ΔbfrH::Kan. As a measure of colonization capacity, bacterial counts of RB50 and RB50ΔbfrH::Kan were obtained from nasal cavity, trachea, and lung tissues at two different time points (Fig. 6B). The CFU of RB50 and RB50ΔbfrH::Kan were used to calculate the mean CI ± standard error of the mean (SEM). On day 5, a significant difference in the levels of colonization was observed in the tracheal tissues, with a CI of 0.164 ± 0.024 (P < 0.0001). CI values of 0.476 ± 0.143 (P = 0.006) and 0.539 ± 0.169 (P = 0.025) were determined for the lung and nasal cavity, respectively. By day 10, there was no significant difference in colonization between RB50 and RB50ΔbfrH::Kan in either the lung or nasal cavity (CI value = 0.749 ± 0.239 [P = 0.324] and CI value = 1.827 ± 0.868 [P = 0.368], respectively). In the tracheal tissues, however, RB50 continued to exhibit a significant competitive advantage over RB50ΔbfrH::Kan (CI = 0.404 ± 0.137 [P = 0.003]), although the differences were less compelling than that on day 5. These experiments indicated that B. bronchiseptica requires bfrH for optimal virulence during early stages of infection. The importance of BfrH in infection, however, appears to diminish in the later stages of infection.
DISCUSSION
A variety of well-conserved Fe uptake mechanisms have been characterized for the bordetellae. Two of those systems have been implicated in the transport of siderophores such as enterobactin (2, 6, 12, 16) and alcaligin (14-16). A third system is required for acquisition of heme (12, 16, 66). Although several other OM proteins (BfrA, BfrB, BfrC, and BfrZ) have been shown to be expressed in response to Fe, the specific role(s) of those proteins in Fe uptake has yet to be elucidated (4, 5, 53). Possessing a variety of Fe retrieval systems may engender optimal flexibility of microorganisms for acquiring a variety of Fe-containing molecules, which in turn provides the microorganism the capacity to successfully colonize different niches in the infected host.
It is a growing trend that ECF sigma factors regulate a variety of critical functions in bacteria. ECF sigma factors have been identified in a number of bacterial species, and many bacteria encode more than one ECF sigma factor, with each regulating expression of a specific protein or family of proteins. For example, 7 ECF sigma factors have been described for Bacillus subtilis, with 10 ECF sigma factors for Mycobacterium tuberculosis, 19 ECF sigma factors for Pseudomonas aeruginosa, and, incredibly, 50 ECF sigma factors for Streptomyces coelicolor (29). Genomic analysis indicates that B. bronchiseptica encodes eight different ECF sigma factors (N. King-Lyons, unpublished data). Two of these ECF sigma factors, HurI and BupI, have been implicated as modulators of Fe acquisition systems. Acquisition of heme by B. bronchiseptica requires HurI (66, 67); expression of BfrZ, a putative xenosiderophore receptor, requires participation of BupI (53). In this study, a third ECF sigma factor-modulated system was described, comprised of a putative ECF sigma factor encoded by ecfI, a sigma factor regulator encoded by ecfR, and a prospective xenosiderophore receptor encoded by bfrH. What features of ecfIR-bfrH initially supported that model? First, the genetic architecture of the ecfIR-bfrH locus is identical to that of other well-described ECF sigma factor-regulated loci in Gram-negative bacteria, including the fecIR-fecABCDE locus of E. coli, the pupIR-pupB locus of P. putida, and the hurIR-bhuRSTUV locus of B. bronchiseptica (Fig. 1A). Second, a hexameric sequence with homology to ECF-regulated −35 promoter regions (5′-GGAATA-3′) was identified at a position located 40 bp upstream of the GTG start codon for bfrH (Fig. 1B) (68). Based upon these factors, it was hypothesized that EcfI and EcfR were components of an ECF sigma factor-regulated system for modulating expression of bfrH.
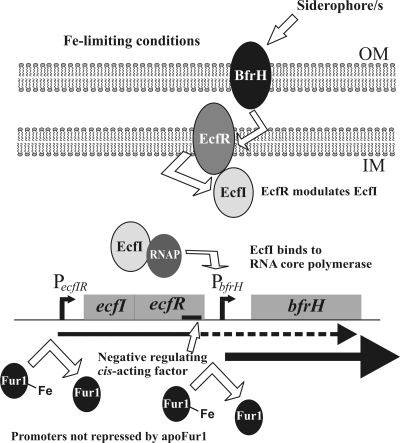
Transcriptional analysis demonstrated that expression of ecfI, ecfR, and bfrH was inversely correlated with the local concentration of Fe (Fig. 2). Since the ORFs of ecfI and ecfR overlap by 8 bp, it was not surprising to find that these two genes were cotranscribed from PecfIR, which was confirmed by detection of a transcript that contained the 3′ terminus of ecfI and the 5′ end of ecfR (Fig. 2). Furthermore, under Fe-limiting growth conditions, a polycistronic transcript containing bfrH likely originated by readthrough transcription from PecfIR; a transcript encompassing the ecfR-bfrH intergenic region was observed when cells were cultured under Fe stress but not when cells were grown under Fe-sufficient conditions. This process of readthrough transcription within an ECF locus is similar to the pattern of transcription described for the rhuIR-bhuR locus in B. avium and the hurIR-bhuR locus of B. bronchiseptica (33, 68). It is proposed that readthrough transcription from the Fe-dependent (fur-regulated) PecfIR promoter enables B. bronchiseptica to preposition EcfI in the cytoplasm, EcfR in the plasma membrane, and small amounts of BfrH in the OM to quickly detect and respond to the inducing extracellular ligand. Once bound to the prepositioned BfrH, the inducing ligand (here predicted to be one or more xenosiderophores) would activate a signal transduction event from BfrH to EcfR and from EcfR to EcfI. EcfI, in combination with RNA polymerase, would promote high-level expression of BfrH to enable maximal acquisition of the Fe-bound xenosiderophore (Fig. 7).
FIG. 7.
Genetic and expression model of the ecfIR-bfrH locus. Under conditions of Fe sufficiency, Fur1 represses transcription of the ecfIR-bfrH locus. Upon encountering Fe-limiting conditions, Fur1 dissociates from PecfIR and possibly also from PbfrH, thus derepressing expression of ecfI and ecfR and enabling readthrough transcription from PecfIR into bfrH. At the genetic level, a cis-acting element (solid black line) in ecfR limits readthrough transcription of bfrH from PecfIR. A transduction signal initiated by BfrH after binding of the extracellular inducing ligand (an unknown siderophore) is transmitted through EcfR and EcfI to highly upregulate expression of bfrH from PbfrH. The extracellular inducing ligand for the ecfIR-bfrH locus is believed to be an unknown xenosiderophore. OM, outer membrane; IM, cytoplasmic membrane; RNAP, RNA core polymerase.
In silico scanning of the nucleotide sequences in the ecfIR-bfrH locus suggested that expression from PecfIR and PbfrH was likely influenced by a Fur-like regulator. Both prospective promoters contain regions which are homologous to the consensus Fur box of E. coli (40, 58). Surprisingly, searches of the B. bronchiseptica genome for a gene with homology to the E. coli fur gene revealed the presence of two separate ORFs (BB3942 and BB0215). The predicted amino acid sequences of BB3942, which we denoted fur1, and BB0215, which we denoted fur2, exhibited 54% and 42% identities, respectively, to the amino acid sequence of Fur of E. coli (55). Moreover, Fur1 was 100% identical in amino acid sequence to the previously published Fur sequence from B. pertussis (13). It is not uncommon, however, for a bacterium to contain multiple genes encoding Fur-like proteins. Three Fur homologues are expressed by B. subtilis, while S. coelicolor carries four fur-like genes (27). The functions of the homologues in these two bacteria are diverse. In B. subtilis, one of the Fur proteins regulates the uptake of Fe; the other homologue, denoted Zur, regulates acquisition of zinc (27). PerR, the third Fur homologue in B. subtilis, represses the peroxide regulon (27). In S. coelicolor, FurA and CatR control expression of peroxidase and catalase A, respectively (1, 27, 63). Nur, which is implicated in nickel homeostasis, and Zur were recently described for S. coelicolor (1, 63). To determine the roles of fur1 and fur2 in expression of ecfIR-bfrH in B. bronchiseptica, attempts were made to engineer deletion mutants of each of the genes. Although it is perilous to interpret negative data, the failure to obtain a mutant of fur2 suggested that fur2 may have one or more essential functions in B. bronchiseptica. Efforts to establish the role of fur1 in regulation of bfrH were more successful. Transcription of ecfIR and bfrH was decoupled from Fe dependency in a mutant deficient in expression of fur1. Likewise, expression of bhuR, a gene which has previously been shown to be Fur regulated in B. avium and B. bronchiseptica, was also rendered constitutive in the fur1 mutant. These experiments clearly implicated Fur1 as the global Fe-regulatory protein in B. bronchiseptica. Experiments are under way to determine the functional role(s) of fur2 in the biology of the pathogen.
ECF sigma factor-regulated systems are defined by their response to cognate extracellular ligands, which, upon binding to the surface receptor, initiate a signal transduction cascade required for inducing additional expression of the receptor. Since BfrH had homology to siderophore receptors, it was hypothesized that the inducing ligand for the ecfIR-bfrH locus would be a xenosiderophore. Although a number of xenosiderophores were screened, a siderophore with the capacity to induce expression of bfrH above the level induced solely by Fe stress has not been identified (data not shown). Therefore, in the absence of the inducing ligand, other strategies were employed to confirm the role of EcfI in regulating expression of bfrH. Overexpression of an ECF sigma factor often abrogates the need for an inducing ligand to upregulate expression of the cognate receptor (36, 37, 53). For example, overexpression of RhuI in B. avium elicited high-level expression of BhuR and decoupled expression of that heme receptor from Fe dependency (36). A similar approach was employed to determine if overexpression of EcfI in B. bronchiseptica would stimulate expression of bfrH. Introduction of a plasmid carrying a wt copy of ecfI into B. bronchiseptica elicited constitutive expression of bfrH regardless of the Fe status of the culture medium (Fig. 4A). In contrast, constitutively expressed transcripts carrying either ecfIR or the ecfR-bfrH intergenic region were not detected in the RNA of either strain (Fig. 4A). Fe starvation ECF sigma factors, including FecI, PupI, and HasI, are also nonautoregulatory (8, 20). The transcriptional profile noted above indicated that EcfI also did not regulate its own promoter (PecfIR). It is likely that EcfI-dependent transcription of bfrH is driven from the PbfrH promoter. A lacZ reporter plasmid was also utilized to confirm the promoter activity of the ecfR-bfrH intergenic region. The results of those experiments suggested that PbfrH is located within this region (Fig. 4C).
To further elucidate the transcriptional effects of EcfI on the expression of bfrH, a mutant deficient in expression of EcfI was engineered. In comparison to transcription of bfrH in RB50 cultured under Fe-stressed conditions, RB50ΔecfI exhibited a significantly lower level of transcription of the gene. These data indicated that EcfI is required for low-level transcription of bfrH and that this level of transcription occurred in the absence of an inducing ligand. It is surmised, therefore, that EcfI has the capacity to interact with PbfrH in a ligand-independent manner. Similar regulatory effects were demonstrated for the ECF sigma factor-regulated rhuIR-bhuRSTUV locus in B. avium (33).
Again, in the absence of the inducing ligand, experiments to precisely determine the role of ecfR in regulating expression of bfrH were not feasible. Yet, even in the absence of an inducing ligand, investigations employing an ecfR-deficient mutant revealed a regulatory mechanism which has yet to be described for other ECF sigma factor-regulated systems. Deletion of ecfR was correlated with a remarkable increase in transcription of bfrH in comparison to transcription in RB50 when cells were cultured in Fe-limiting conditions (Fig. 5A). The amount of polycistronic transcript containing both ecfR and bfrH, however, was significantly increased in the ecfR mutant (Fig. 5B). This result suggested that the increase in bfrH expression was due to increased transcription from the upstream PecfIR promoter, rather than from PbfrH. Furthermore, since EcfI is not autoregulatory, the data indicated that this increase in expression of bfrH was EcfI independent. These experiments, therefore, do not support the model that EcfR is a typical anti-sigma factor that sequesters EcfI in the absence of an inducing ligand. Reinsertion of ecfR into the ecfR locus of RB50ΔecfR reestablished a wt level of expression of bfrH.
Readthrough transcription into bfrH from PecfIR is also modulated by a cis-acting factor which is situated within ecfR. In the absence of the inducing ligand, only small amounts of BfrH are needed in the OM to serve as a trigger for ligand-specific signal transduction. Thus, this cis-acting factor may have evolved to suppress the amount of BfrH produced by readthrough transcription from PecfIR to optimize the levels of the receptor in the OM under noninducing conditions. We are not aware of any other ECF sigma factor-regulated system which employs a similar cis-acting level of control. Experiments are ongoing to identify the cis-acting sequences in ecfR.
A failure to visualize the OM protein by SDS-PAGE and Coomassie blue staining demonstrated the apparent low level of expression of BfrH in the absence of inducing conditions. To assess the translational profile of BfrH, a higher-resolution method was utilized. Western immunoblotting using a polyclonal anti-BfrH-peptide antibody revealed recognized a polypeptide of ∼87 kDa in OMs isolated from RB50 which was absent in OMs isolated from RB50ΔbfrH (Fig. 4D). Surprisingly, BfrH was detected in RB50(pecfI) only when the cells were grown under Fe-stressed conditions. This pattern of expression was in contrast to the transcriptional studies in which high levels of bfrH transcripts were observed in RB50(pecfI) cultured under either Fe-replete or Fe-stressed conditions. It was surmised that under Fe-replete environments, the bfrH transcript may undergo posttranscriptional modifications which hinder translation of BfrH. Alternatively, BfrH may be quickly degraded when cells have sufficient Fe for growth. Additional experiments must be performed to distinguish between those two possibilities.
To establish infection, bacteria must successfully colonize the host. Genes encoding proteins for Fe acquisition, therefore, are likely to be important in establishing a foothold in the Fe-limited tissues and fluids of the prospective host. A typical strategy to test that hypothesis is to delete the gene and ask if the mutant has decreased virulence in an infection model. Unfortunately, the effects of Fe uptake systems on virulence are often subtle. Furthermore, frequent redundancy in the systems often confounds the usefulness of such genetic strategies to probe the importance of the systems in virulence. For example, wt B. avium and an isogenic bhuR-deficient mutant were not distinguishably different in virulence in a direct challenge virulence model (49). In contrast, the wt parent outcompeted the bhuR mutant in a competition virulence model. Likewise, mixed-infection experiments were successful in demonstrating that BfeA, the enterobactin receptor, and FauA, the alcaligin receptor, were required for optimal virulence of B. pertussis (12, 16-18). Using a similar competition challenge model, RB50ΔbfrH::Kan was shown to colonize less well than RB50 in the nasal cavity, trachea, and the lungs (Fig. 6B).
From these data, it was surmised that the xenosiderophores available to B. bronchiseptica via BfrH are likely found in these tissues. Alternatively, BfrH may have a dual function, first as a siderophore receptor and also as an accessory adherence factor for ligands on the surfaces of cells located in the trachea. Such a situation has been described for E. coli. The Iha receptor has been implicated in utilization of catecholate as a nutrient and in adherence of the bacterium to epithelial cells (41). B. pertussis cultured under Fe-stressed conditions exhibited enhanced attachment to epithelial cells (70). The Fe-dependent adhesin which mediates this attachment has not been firmly identified.
Differences in colonization between the wt and the bfrH mutant strain were reduced by day 10 postinfection (Fig. 6B). These data suggested that BfrH was important in virulence only during the early stages of infection. BfrH is one of many putative siderophore receptors encoded in the B. bronchiseptica genome. Therefore, it is likely that the mutant eventually supports colonization by acquiring Fe from other sources, possibly by the activities of other receptors, such as BfeA, FauA, or BhuR (16-18). A model for Bordetella virulence was proposed in which BfeA and FauA, two siderophore receptors of B. pertussis, were deemed to be important only during the early phases of infection (16, 17). In contrast, BhuR, the heme receptor, appeared to be more important in later stages after colonization, when the pathological effects of released toxins increase heme availability from damaged host cells (18). These observations suggest a general trend with regard to the roles of siderophore receptors and virulence in the pathogenic bordetellae.
In summary, ecfIR-bfrH was revealed as a locus encoding three new members of the growing family of Fe-regulated proteins of B. bronchiseptica. This locus is highly conserved in B. pertussis, B. parapertussis, B. avium, and B. bronchiseptica, implying the evolutionary importance of this system in Bordetella virulence. Regulation of expression of bfrH is controlled on several levels (Fig. 7): (i) the ecfIR-bfrH locus is regulated by Fur1, one of two Fur homologues encoded by B. bronchiseptica; (ii) ecfI, a gene encoding a new ECF sigma factor in the genus, modulates expression of BfrH from PbfrH; and (iii) a novel cis-acting element, possibly located in ecfR, suppresses readthrough transcription of bfrH from PecfIR and reduces expression of the receptor in the absence of the inducing ligand.
As additional members of the ECF sigma factor-regulated systems are described for B. bronchiseptica, it will be intriguing to determine if cross talk between the systems is employed to better coordinate the various mechanisms for Fe acquisition.
Acknowledgments
Funds to support this investigation were contributed to T.D.C. by The Department of Microbiology and Immunology and The Office of the Dean, The School of Medicine and Biomedical Sciences at The University at Buffalo, and by The Research Foundation of the State of New York.
We thank Peggy Cotter for providing helpful information on the B. bronchiseptica murine model of infection and for providing B. bronchiseptica ΔcyaA (28), which was employed as a control to validate the infection studies. We also thank Anders Hakansson and Lorrie Mandell for their generous technical assistance in the murine model of infection and Jean-Marie Meyer for the gift of purified siderophores.
Editor: S. M. Payne
Footnotes
Published ahead of print on 14 December 2009.
REFERENCES
- 1.Ahn, B. E., J. Cha, E. J. Lee, A. R. Han, C. J. Thompson, and J. H. Roe. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59:1848-1858. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. T., and S. K. Armstrong. 2006. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J. Bacteriol. 188:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angerer, A., S. Enz, M. Ochs, and V. Braun. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol. Microbiol. 18:163-174. [DOI] [PubMed] [Google Scholar]
- 4.Beall, B. 1998. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res. Microbiol. 149:189-201. [DOI] [PubMed] [Google Scholar]
- 5.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 6.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 7.Bibb, L. A., C. A. Kunkle, and M. P. Schmitt. 2007. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biville, F., H. Cwerman, S. Letoffe, M. S. Rossi, V. Drouet, J. M. Ghigo, and C. Wandersman. 2004. Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 53:1267-1277. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., and F. Endriss. 2007. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20:219-231. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., and S. Mahren. 2005. Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol. Rev. 29:673-684. [DOI] [PubMed] [Google Scholar]
- 11.Braun, V., S. Mahren, and A. Sauter. 2006. Gene regulation by transmembrane signaling. Biometals 19:103-113. [DOI] [PubMed] [Google Scholar]
- 12.Brickman, T. J., M. T. Anderson, and S. K. Armstrong. 2007. Bordetella iron transport and virulence. Biometals 20:303-322. [DOI] [PubMed] [Google Scholar]
- 13.Brickman, T. J., and S. K. Armstrong. 1995. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J. Bacteriol. 177:268-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman, T. J., and S. K. Armstrong. 2007. Impact of alcaligin siderophore utilization on in vivo growth of Bordetella pertussis. Infect. Immun. 75:5305-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickman, T. J., and S. K. Armstrong. 2009. Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals 22:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickman, T. J., T. Hanawa, M. T. Anderson, R. J. Suhadolc, and S. K. Armstrong. 2008. Differential expression of Bordetella pertussis iron transport system genes during infection. Mol. Microbiol. 70:3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2006. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect. Immun. 74:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullen, J. J., H. J. Rogers, P. B. Spalding, and C. G. Ward. 2005. Iron and infection: the heart of the matter. FEMS Immunol. Med. Microbiol. 43:325-330. [DOI] [PubMed] [Google Scholar]
- 20.Cescau, S., H. Cwerman, S. Letoffe, P. Delepelaire, C. Wandersman, and F. Biville. 2007. Heme acquisition by hemophores. Biometals 20:603-613. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty, R., E. Storey, and D. van der Helm. 2007. Molecular mechanism of ferric siderophore passage through the outer membrane receptor proteins of Escherichia coli. Biometals 20:263-274. [DOI] [PubMed] [Google Scholar]
- 22.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman, L. E., B. N. de Rossi, M. T. Messina, and M. A. Franco. 2001. Phenotype evaluation of Bordetella bronchiseptica cultures by urease activity and Congo red affinity. Lett. Appl. Microbiol. 33:285-290. [DOI] [PubMed] [Google Scholar]
- 26.Friedman, Y. E., and M. R. O'Brian. 2004. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J. Biol. Chem. 279:32100-32105. [DOI] [PubMed] [Google Scholar]
- 27.Hahn, J. S., S. Y. Oh, K. F. Chater, Y. H. Cho, and J. H. Roe. 2000. H2O2-sensitive fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254-38260. [DOI] [PubMed] [Google Scholar]
- 28.Harvill, E. T., P. A. Cotter, M. H. Yuk, and J. F. Miller. 1999. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect. Immun. 67:1493-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 30.Imaizumi, A., Y. Suzuki, S. Ono, H. Sato, and Y. Sato. 1983. Effect of heptakis (2,6-O-dimethyl) beta-cyclodextrin on the production of pertussis toxin by Bordetella pertussis. Infect. Immun. 41:1138-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 32.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 33.King, N. D., A. E. Kirby, and T. D. Connell. 2005. Transcriptional control of the rhuIR-bhuRSTUV heme acquisition locus in Bordetella avium. Infect. Immun. 73:1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King-Lyons, N. D., K. F. Smith, and T. D. Connell. 2007. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J. Bacteriol. 189:6266-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirby, A. E., N. D. King, and T. D. Connell. 2004. RhuR, an extracytoplasmic function sigma factor activator, is essential for heme-dependent expression of the outer membrane heme and hemoprotein receptor of Bordetella avium. Infect. Immun. 72:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J. W., and J. D. Helmann. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485-499. [DOI] [PubMed] [Google Scholar]
- 41.Leveille, S., M. Caza, J. R. Johnson, C. Clabots, M. Sabri, and C. M. Dozois. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, K., V. A. Simossis, W. R. Taylor, and J. Heringa. 2005. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21:152-159. [DOI] [PubMed] [Google Scholar]
- 43.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucarelli, D., M. L. Vasil, W. Meyer-Klaucke, and E. Pohl. 2008. The metal-dependent regulators FurA and FurB from Mycobacteriumtuberculosis. Int. J. Mol. Sci. 9:1548-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18:326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mielcarek, N., A. S. Debrie, D. Raze, J. Bertout, C. Rouanet, A. B. Younes, C. Creusy, J. Engle, W. E. Goldman, and C. Locht. 2006. Live attenuated B. pertussis as a single-dose nasal vaccine against whooping cough. PLoS Pathog. 2:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 48.Mocny, J. C., J. S. Olson, and T. D. Connell. 2007. Passively released heme from hemoglobin and myoglobin is a potential source of nutrient iron for Bordetella bronchiseptica. Infect. Immun. 75:4857-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 51.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 52.Poole, K., S. Neshat, K. Krebes, and D. E. Heinrichs. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pradel, E., and C. Locht. 2001. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J. Bacteriol. 183:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redly, G. A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossi, M. S., A. Paquelin, J. M. Ghigo, and C. Wandersman. 2003. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol. Microbiol. 48:1467-1480. [DOI] [PubMed] [Google Scholar]
- 57.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 58.Rudolph, G., H. Hennecke, and H. M. Fischer. 2006. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol. Rev. 30:631-648. [DOI] [PubMed] [Google Scholar]
- 59.Sangwan, I., S. K. Small, and M. R. O'Brian. 2008. The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J. Bacteriol. 190:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 61.Sebaihia, M., A. Preston, D. J. Maskell, H. Kuzmiak, T. D. Connell, N. D. King, P. E. Orndorff, D. M. Miyamoto, N. R. Thomson, D. Harris, A. Goble, A. Lord, L. Murphy, M. A. Quail, S. Rutter, R. Squares, S. Squares, J. Woodward, J. Parkhill, and L. M. Temple. 2006. Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188:6002-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen, J. S., V. Geoffroy, S. Neshat, Z. Jia, A. Meldrum, J. M. Meyer, and K. Poole. 2005. FpvA-mediated ferric pyoverdine uptake in Pseudomonas aeruginosa: identification of aromatic residues in FpvA implicated in ferric pyoverdine binding and transport. J. Bacteriol. 187:8511-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin, J. H., S. Y. Oh, S. J. Kim, and J. H. Roe. 2007. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 189:4070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 65.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 66.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vanderpool, C. K., and S. K. Armstrong. 2004. Integration of environmental signals controls expression of Bordetella heme utilization genes. J. Bacteriol. 186:938-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Hove, B., H. Staudenmaier, and V. Braun. 1990. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J. Bacteriol. 172:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vidakovics, M. L., Y. Lamberti, D. Serra, G. A. Berbers, W. L. van der Pol, and M. E. Rodriguez. 2007. Iron stress increases Bordetella pertussis mucin-binding capacity and attachment to respiratory epithelial cells. FEMS Immunol. Med. Microbiol. 51:414-421. [DOI] [PubMed] [Google Scholar]
- 71.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 72.Woolfrey, B. F., and J. A. Moody. 1991. Human infections associated with Bordetella bronchiseptica. Clin. Microbiol. Rev. 4:243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, J., I. Sangwan, A. Lindemann, F. Hauser, H. Hennecke, H. M. Fischer, and M. R. O'Brian. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]