Abstract
Plasmodium vivax Duffy binding protein (DBP) is a merozoite microneme ligand vital for blood-stage infection, which makes it an important candidate vaccine for antibody-mediated immunity against vivax malaria. A differential screen with a linear peptide array compared the reactivities of noninhibitory and inhibitory high-titer human immune sera to identify target epitopes associated with protective immunity. Naturally acquired anti-DBP-specific serologic responses observed in the residents of a region of Papua New Guinea where P. vivax is highly endemic exhibited significant changes in DBP-specific titers over time. The anti-DBP functional inhibition for each serum ranged from complete inhibition to no inhibition even for high-titer responders to the DBP, indicating that epitope specificity is important. Inhibitory immune human antibodies identified specific B-cell linear epitopes on the DBP (SalI) ligand domain that showed significant correlations with inhibitory responses. Affinity-purified naturally acquired antibodies on these epitopes inhibited the DBP erythrocyte binding function greatly, confirming the protective value of specific epitopes. These results represent an important advance in our understanding of part of blood-stage immunity to P. vivax and some of the specific targets for vaccine-elicited antibody protection.
Plasmodium vivax is the major cause of malaria in most regions where this disease is endemic outside Africa, and it causes substantial morbidity worldwide (17). Plasmodium microneme proteins, such as Duffy binding protein (DBP), have important roles in the merozoite invasion of reticulocytes during asexual blood-stage infection (1, 5). DBP is a member of the Duffy binding-like erythrocyte binding protein (DBL-EBP) family expressed in the micronemes and on the surface of P. vivax merozoites and is associated with the decisive junction formation step during the invasion process (1). It is this critical interaction of DBP with its cognate receptor that makes DBP an important antimalaria vaccine candidate. The critical erythrocyte binding motif of DBP is in a 330-amino-acid cysteine-rich domain referred to as DBP region II (DBPII) or the DBL domain, which is the minimal domain responsible for binding to Duffy-positive human erythrocytes (2, 6). The central portion of the DBP domain is hypervariable compared to other DBP regions, and polymorphisms occur frequently at certain residues in a pattern consistent with selection pressure on DBP, suggesting that allelic variation functions as a mechanism for immune evasion (9, 15, 24).
Naturally acquired antibodies to DBP are prevalent in residents of areas where malaria is highly endemic, but individuals show significant quantitative and qualitative differences in their anti-DBP serological responses (10, 12, 27, 28). Generally, serological responses to DBP and the inhibition of DBP-erythrocyte binding activity increase with a person's age, suggesting that there is a boosting effect due to repeated exposure through recurrent infection (13, 16, 18). The initial antibody response to a single P. vivax infection is a response to conformational epitopes and is not broadly protective, while an immunity that transcends strain specificity develops only after repeated exposure (10, 28). Repeated exposure of residents of the areas of Papua New Guinea (PNG) where P. vivax is endemic was observed to correlate with development of antibodies that are reactive to linear epitopes in the critical binding region of DBP. In this study, we compared the reactivity of inhibitory human immune sera to the reactivity of noninhibitory immune sera to identify linear epitopes in DBPII that may serve as a target for vaccine-induced protective humoral immunity.
MATERIALS AND METHODS
Sample collection.
Blood samples were collected from March to July 2001 from 38 volunteers selected from a previously surveyed population in Liksul, a village northwest of Madang, Papua New Guinea (27). The individuals selected ranged from 9 to 73 years old and represented high-responder, low-responder, and nonresponder groups as classified in a previous study (18). Blood was collected by venipuncture in Vacutainer tubes without anticoagulant. Approximately 8 ml was taken from each individual, kept at the ambient temperature (30 to 35°C) for 30 min, and then incubated at 4°C overnight. Serum was removed, decomplemented at 56°C for 30 min, and stored at −80°C. Cryopreserved samples were shipped to the United States for analysis. All human blood samples used in this study were collected after consent was obtained from study participants under protocols approved by the Ethical Review Board of the Cleveland Veteran's Administration Medical Center, the Papua New Guinea Medical Research Advisory Committee, and the University of Notre Dame Institutional Review Board.
Measurement of serological responses to DBP.
Anti-DBP responses were quantified by an enzyme-linked immunosorbent assay (ELISA) using recombinant DBP regions II to IV (rDBPII-IV) as described previously (12, 18, 27). Briefly, rDBPII-IV was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli, affinity purified with glutathione, and cleaved from GST with thrombin using standard methods (12, 19). Purified rDBPII-IV was added to 96-well plates at a concentration of 2 μg/ml, incubated for 30 min at room temperature, and washed three times with wash buffer (0.2% Tween 20 in phosphate-buffered saline [PBS]). Wells were incubated with 200 μl blocking buffer (1% bovine serum albumin in PBS) for 30 min, washed three times with wash buffer, allowed to dry, and stored overnight at 4°C. Serum diluted 1:400 in blocking buffer was added to prewetted wells and incubated for 90 min at 37°C. Plates were rinsed three times in wash buffer, incubated with a 1:400 dilution of goat anti-human IgG-alkaline phosphatase, and rinsed three times, and substrate was added. Absorbance at 405 nm was recorded 45 min after addition of a developer reagent.
A baseline value was established using control sera from nonexposed North Americans, and this control value was subtracted from the test optical density (OD) values. Based on ELISA data preparations were classified into the following three groups: high responders (OD, 1.5 to 3.85); low responders (OD, 0.5 to 1.5); and nonresponders (OD, <0.5).
Antibody purification.
Antibodies to B-cell epitopes were affinity purified from human sera containing high titers of DBPII inhibitory antibodies from individuals from PNG exposed to P. vivax or from rabbit sera raised against peptides corresponding to selected B-cell epitopes. Pooled human sera from a separate study in PNG were used for affinity purification because of the limited quantity of experimental samples. Using the manufacturer's recommended protocol, diluted serum was passed over an affinity column prepared by coupling 3 mg of each peptide to a Sulfur Link coupling resin (Thermo Scientific, Rockford, IL). After the column was washed three times with PBS (pH 7.4), the bound antibody was eluted with 0.1 M glycine-HCl (pH 3.0) and immediately neutralized with 1 M Tris-HCl (pH 8.5). Antibodies were dialyzed against PBS before they were stored at −20°C until they were needed.
COS7 cell expression of DBP and inhibition assays.
COS7 cells were transfected with plasmid pEGFP-DBPII-SalI, which allows expression of DBPII as a fusion to the N terminus of enhanced green fluorescent protein, which was used as a transfection marker as previously described (18). The inhibition assay was performed 42 h after the initial transfection. Serum at a 1:10 dilution or different concentrations of the affinity-purified antibodies were incubated for 60 min with transfected COS7 cells, and this was followed by incubation with Duffy-positive human erythrocytes. Unbound erythrocytes were removed by three washes with PBS. Binding was quantified by counting rosettes observed in 30 fields of view at a magnification of ×200. In this assay rosettes were defined as COS7 cells covered by bound erythrocytes on 50% or more of their surface area. An inhibition percentage was calculated for each serum sample by comparison to binding in the presence of nonexposed North American (NA) control serum. Each assay included a duplicate test of each sample, and the results were expressed as the averages for three independent assays.
Synthesized peptide array.
A peptide array consisting of 178 overlapping 12-mer peptides displaced by 2 amino acids, spanning the entire DBPII-SalI molecule, was generated on “gear” attached pins in a 96-well format (Chiron Mimotopes). The peptide purity was >90% as determined by high-performance liquid chromatography. Additional peptides that corresponded to the different variants for peptides 179 to 205 were also synthesized. Sequence variations were based on the most common alleles identified in the study population (11).
Assessment of antibody responses to DBPII peptides.
Serological responses to each peptide were measured using a modified ELISA method. Coating buffer (0.1 M PBS, pH 7.2) was dispensed into each well of a microtiter plate, and the peptide gears were placed in the well and incubated for 1 h at room temperature. The gears were washed in 0.01 M PBS (pH 7.2) for 30 min, incubated with a 1:400 dilution of human sera overnight at 4°C, and washed in wash buffer for 10 min. To detect primary antibody reactivity to each peptide, secondary antibodies and goat anti-human IgG-peroxidase (KPL Inc., MD) were incubated at room temperature for 1 h with agitation and washed, 2,2′-azino-di-(3-ethylbenzthiazolinsulfonate) (ABTS) substrate was added to each well and incubated for 45 min, and the optical density at 405 nm (OD405) was determined.
Data analysis.
Classification of high responders, low responders, and nonresponders to DBP was based on averaged OD values for three wells per individual; a baseline value was created using data for nonexposed North Americans (NA) and was subtracted from test OD values to standardize the ELISA results (12). A cluster analysis was performed with the ELISA values using SYSTAT (version 6.0); individual values clustered in the three distinct groups. High responders were defined as individuals whose serum OD values were equal to or more than the mean plus 2 standard deviations of the values for the North American controls. The OD values for nonresponder sera were equal to or less than the mean plus 1 standard deviation of the value for the control serum. For the peptide scan, B-cell epitopes were identified by comparison of the specific antibody reactivities (average OD values) of inhibitory and noninhibitory sera for each specific peptide. A baseline OD value was established using serum from nonexposed North Americans. Peptide array data were analyzed using SPSS (version 11.5). Nonparametric analysis (two independent samples; Mann-Whitney test) was used for comparison of the peptide reactivities of high and noninhibitory sera. A P value of <0.05 was considered statistically significant.
RESULTS
Serological response to DBP.
Naturally acquired anti-DBP responses of 38 residents living in an area of Papua New Guinea where malaria is highly endemic were evaluated by ELISA at the end of the 2001 high-transmission season for malaria. None of these individuals recalled experiencing a clinical episode of malaria during the previous 6 months, and none was positive for Plasmodium parasitemia as determined by a blood smear at the time of collection. The antigen-specific titers were surprisingly variable and could be assigned to three distinct antibody response groups: nonresponders (N), low responders (L), and high responders (H) (Table 1). A comparison of the responder classifications to the results for samples collected during the previous year revealed that for 50% of the volunteers the responder classification changed between the 2000 and 2001 transmission seasons (Table 1). Twelve of the original high responders were low responders in 2001, and two were nonresponders. The classifications of the seven low-responder individuals were unchanged, but four of the nonresponders became members of the low-responder group and one nonresponder became a high responder. Changes in the response category were not observed to correlate with either the age or the gender of the individual.
TABLE 1.
Naturally acquired anti-DBP serum antibody levels for residents in an area of Papua New Guinea where P. vivax malaria is highly endemic during two transmission seasons
| Subject | Age in 2001 (yr) | Gendera | Type of responder in 2000b | Type of responder in 2001b |
|---|---|---|---|---|
| 1 | 44 | M | H | H |
| 2 | 69 | M | H | H |
| 3 | 14 | F | H | H |
| 4 | 59 | M | H | H |
| 5 | 35 | M | H | H |
| 6 | 9 | F | N | H |
| 7 | 19 | M | H | H |
| 8 | 33 | F | H | H |
| 9 | 29 | M | H | H |
| 10 | 13 | M | H | H |
| 11 | 23 | M | L | L |
| 12 | 17 | F | L | L |
| 13 | 63 | M | H | L |
| 14 | 23 | M | H | L |
| 15 | 23 | M | H | L |
| 16 | 13 | F | H | L |
| 17 | 29 | M | H | L |
| 18 | 17 | F | L | L |
| 19 | 69 | M | H | L |
| 20 | 27 | M | L | L |
| 21 | 31 | M | N | L |
| 22 | 46 | M | H | L |
| 23 | 16 | F | H | L |
| 24 | 13 | M | H | L |
| 25 | 33 | M | N | L |
| 26 | 73 | M | L | L |
| 27 | 16 | F | H | L |
| 28 | 33 | F | L | L |
| 29 | 42 | M | N | L |
| 30 | 13 | M | H | L |
| 31 | 12 | M | L | L |
| 32 | 9 | F | N | L |
| 33 | 26 | F | H | L |
| 34 | 28 | F | H | N |
| 35 | 8 | M | N | N |
| 36 | 17 | F | H | N |
| 37 | 22 | F | N | N |
| 38 | 31 | M | N | N |
M, male; F, female.
The high-responder (H), low-responder (L), and nonresponder (N) groups are described in the text.
Measurement of functional inhibition of DBP-erythrocyte binding.
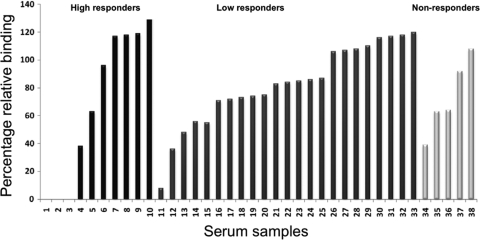
To evaluate the correlation between the anti-DBP titer and functional inhibition of the DBP-erythrocyte interaction, serum samples were tested individually to examine in vitro inhibition of DBPII binding to Duffy-positive erythrocytes, using a test similar to tests used in a previous study (18). Unexpectedly, the functional inhibition by the individual samples did not correlate with their anti-DBP antibody titer levels (Fig. 1). Only 3 of the 10 high-responder samples and 1 of the 23 low-responder samples (in a total of 38 samples) inhibited binding completely or almost completely. Most high-titer serum samples had a moderate inhibitory effect or no inhibitory effect on DBP binding to erythrocytes. None of the highly inhibitory sera changed responder classification between the 2000 and 2001 sample collections. Only sera with high titers of anti-DBPII antibodies completely inhibited DBPII binding, but not all high-titer sera were inhibitory. This indicates that titer alone is not an indicator of functional inhibition, a characteristic consistent with the predominant strain-specific responses observed in PNG residents. The low levels of inhibition in the functional assays observed for the nonresponder sera may reflect differences in presentation of epitopes (such as conformational epitopes not present on the recombinant DBPII used in the ELISA) on functional DBPII expressed on COS7 cells.
FIG. 1.
Inhibition of DBPII binding to Duffy-positive erythrocytes by human sera from Papua New Guinea. Individual sera were tested at a dilution of 1:10 for inhibition of binding in an in vitro assay (COS7 cell assay). Sera were classified based on their responses to recombinant DBP as determined by ELISA, as follows: high responders (black bars), low responders (dark gray bars), and nonresponders (light gray bars). Sera showed marked differences in inhibitory efficacy, and the differences in inhibition did not correlate with antibody titers. Binding inhibitory effects were classified as follows: major effect, ≥80% inhibition; moderate effect, 40 to 80% inhibition; and minor or no effect, 0 to 40% inhibition. The sample numbers used are not the same as those in Table 1.
Mapping of the dominant epitopes of the DBP ligand domain.
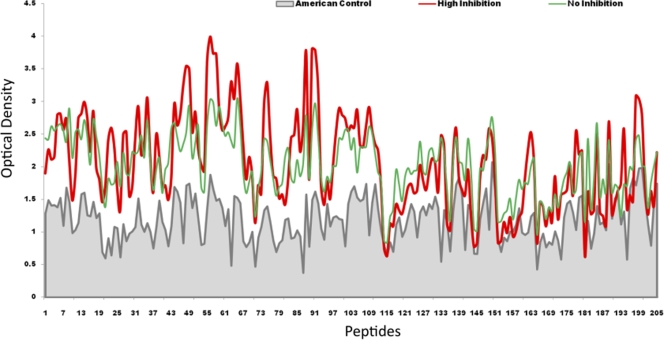
To identify potential DBPII-neutralizing B-cell linear epitopes, we compared the reactivities of inhibitory high-titer human sera (high-inhibition [HI] group that inhibited DBPII-erythrocyte binding) and noninhibitory high-titer sera (no-inhibition [NI] group that did not inhibit DBPII-erythrocyte binding) to 178 peptides spanning the 330-residue ligand domain of DBP. The HI group included three samples (samples 1, 2, and 3) that completely inhibited DBPII-erythrocyte binding and one sample (sample 11) that almost completely inhibited DBPII-erythrocyte binding, and the NI group included 22 of the remaining high- and low-titer positive samples (Fig. 1). The average responses of nonexposed NA residents (n = 6) were used as the control for background reactivity. A positive reaction with a peptide was defined as an OD reactivity that was more than the mean plus 2 standard deviations of the value for NA controls (Fig. 2, gray line). A peptide was considered part of a potential epitope if there was significantly greater antibody reactivity of HI group antibodies than of NI group antibodies. Potential inhibitory B-cell epitopes were identified as shared sequences present in two or more than two contiguous overlapping peptides with significantly more reactivity with HI group sera but not with NI samples. An isolated positive peptide was not considered a B-cell epitope.
FIG. 2.
Reactivity of human sera to overlapping DBPII peptide array. The average OD values for the peptides with highly inhibitory sera (n = 4) and noninhibitory sera (n = 22) are indicated by red and green lines, respectively, while the gray line indicates the average OD values for the nonexposed North American sera (n = 6) that served as a background control. An epitope is considered a B-cell epitope when the difference in OD value between the highly inhibitory sera (HI) and the noninhibitory sera (NI) is greater than the mean plus 2 standard deviations of the values for the North American control sera.
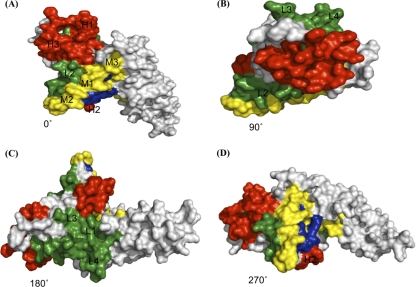
Comparative analysis of inhibitory and noninhibitory sera revealed strong differential binding to 10 putative continuous B-cell epitopes centered at peptides 22, 31, 46, 56, 63, 76, 83, 90, 98, and 109 (Table 2). Statistical analysis indicated that some of these presumptive linear targets of protective immune responses had significantly stronger correlations with the inhibitory responses. Based on the significance of the difference (P value) from the average NI group reactivity, the inhibitory epitopes were placed into three groups, the high (H), medium (M), and low (L) epitopes (Table 2). When mapped on the three-dimensional (3D) model of DBPII, most of these linear epitopes were localized in subdomain II, while two of them were present in subdomain I of DBPII and no epitopes preferentially correlating with inhibitory sera were identified in subdomain III (Fig. 3; see Fig. S1 and S2 in the supplemental material). Of greatest interest as targets are the H epitopes, especially the conserved H2 epitope that overlaps an area implicated as part of the DBPII receptor recognition site associated with binding a putative sulfated tyrosine of the Duffy antigen/receptor for chemokines (DARC) (8, 20).
TABLE 2.
Potential inhibitory B-cell epitopes in DBPII ligand domain
| Peptide | Sequence | Difference in OD value from NI (mean ± SD) | Pa |
|---|---|---|---|
| H1 | FHRDITFRKLYLKRKL | 0.55 ± 0.28 | 0.007 |
| H2 | EGDLLLKLNNYRYN | 0.95 ± 0.15 | 0.008 |
| H3 | DEKAQQRRKQWWNESK | 0.94 ± 0.21 | 0.021 |
| M1 | FCKDIRWSLGDF | 0.66 ± 0.21 | 0.031 |
| M2 | LKGNFIWICKLNVAVN | 0.45 ± 0.07 | 0.032 |
| M3 | PQIYRWIREWGRDYVS | 0.56 ± 0.45 | 0.050 |
| L1 | CIPDRRYQLCMK | 0.46 ± 0.31 | 0.333 |
| L2 | AQIWTAMMYSVK | 0.38 ± 0.26 | 0.333 |
| L3 | MEGIGYSKVVENNL | 0.72 ± 0.24 | 0.343 |
| L4 | DWDCNTKKDVCIPD | 0.58 ± 0.21 | 0.394 |
P values were obtained by comparison of average values for the HI and NI peptides using a nonparametric Mann-Whitney test. Differences were considered statistically significant if the P value was <0.05. H1 to H3 are high-inhibition B-cell epitopes, M1 to M3 are moderate-inhibition B-cell epitopes, and L1 to L4 are low-inhibition B-cell epitopes.
FIG. 3.
3D model of DBPII. The molecule is depicted as a space-filling model showing localization of B-cell epitopes. The structure is rotated in the panels by 90° around a horizontal axis. (A) Front (0°). (B) Top (90°). (C) Back (180°). (D) 270°. High-inhibition (H1 to H3), medium-inhibition (M1 to M3), and low-inhibition (L1 to L4) B-cell epitopes are indicated by red, yellow, and green, respectively, against a gray background, while residues of the predicted DARC binding site are indicated by dark blue.
Allelic variation is concentrated in the inhibitory B-cell epitopes.
Previous studies showed that DBP polymorphisms are concentrated in DBPII subdomain II, suggesting that there are positive changes driven by immune selection similar to those in other functional microbial ligands. Consistent with this, we found numerous polymorphisms in the strongest B-cell epitopes identified in this study. The variant epitopes include four of the peptides that are most highly reactive with the inhibitory immune sera, H1, H3, M2, and M3 (Table 2). These B-cell epitopes contain polymorphic residues in alleles identified in the PNG population (26), and the variant residues tend to have a central location in the B-cell epitopes, as they are clustered in the 3D DBPII structure.
Antibodies to B-cell epitopes significantly reduce DBPII binding to erythrocytes.
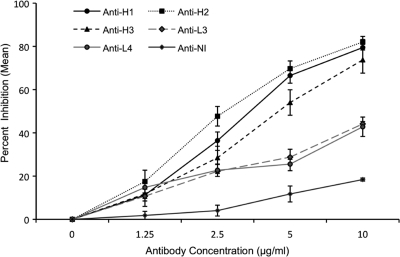
To determine whether antibodies against the identified B-cell epitopes inhibit DBPII binding to Duffy-positive erythrocytes, affinity-purified antibodies against the H epitopes (H1, H2, and H3) and two L epitopes (L3 and L4), as well as an NI peptide (CDGKINYTDKKVCKVP), from an epitope recognized by the noninhibitory sera (between peptides 121 and 124 [Fig. 2]), were affinity purified from a pool of high-titer anti-DBPII inhibitory sera from individuals exposed to P. vivax or rabbit sera raised against epitopes H1 and H3. The purified antibodies were tested to examine their abilities to inhibit DBPII-SalI binding to human Duffy-positive erythrocytes in a COS7 cell assay. The results obtained with the antibodies showed that there was a dose-dependent inhibitory response of DBPII binding to the erythrocytes (Fig. 4). At the highest concentration of antibody tested (10 μg/ml), we observed 73 to 82% inhibition of binding with the affinity-purified anti-H epitope antibodies (50% inhibitory concentration, 2.5 to 5 μg/ml), compared to 42 to 43% inhibition with the purified anti-L epitope antibodies (P < 0.0001, Student's t test) and less than 18% inhibition with the purified anti-NI peptide epitope antibody (P < 0.0001, Student's t test). Antibodies from similarly purified rabbit sera had a smaller inhibitory effect than the naturally acquired human antibodies (see Fig. S3a in the supplemental material), and when purified total IgG from the same pool of inhibitory human sera was tested for inhibition, the 50% inhibitory concentration was at least 2- to 4-fold greater than that observed for the affinity-purified antibodies (data not shown). This indicates that multiple epitopes in the DBPII ligand domain contribute to the overall induction of protective immunity to P. vivax infection. Control experiments with total IgG from sera of nonexposed North Americans and total IgG from preimmune rabbit sera showed binding inhibition of ≤4% (data not shown).
FIG. 4.
Inhibition of DBPII binding to human erythrocytes. Human antibodies specific to the H1, H2, H3, L3, L4, and NI peptides were tested to determine their abilities to inhibit DBP binding to Duffy-positive erythrocytes. Different concentrations of the antibodies were preincubated with transfected COS7 cells prior to addition of erythrocytes, and the numbers of rosettes in 30 microscopic fields at a magnification of ×200 were determined. The symbols indicate the mean percentages of binding for three independent experiments compared to the results of a control experiment with no antibody, and the error bars indicate the standard deviations. For an antibody concentration of 10 μg/ml the P value was <0.0001 for comparisons of H and L peptides, H and NI peptides, and L and NI peptides. P values were adjusted for multiple comparisons.
Affinity-purified antibody activity is inhibited by homologous peptide.
To confirm that the purified antibodies against the synthetic linear peptides mediated the observed inhibitory responses shown in Fig. 4, 10 μg/ml of the affinity-purified human antibodies to the H1, H3, L3, L4, and NI epitopes were first incubated with the corresponding peptides at different concentrations for 1 h, and the binding experiment was repeated. Preincubation of the purified antibodies with the matching peptides at a concentration of 20 μg/ml blocked the functional inhibitory effects of the antibodies substantially (see Fig. S3b in the supplemental material), confirming that these antibodies are highly enriched against the specific peptides.
DISCUSSION
P. vivax depends on the interaction of the DBP with its cognate receptor for efficient invasion of human reticulocytes. The vital nature of this interaction makes DBP an ideal target for vaccine development. Potential impediments to the use of DBP as an effective vaccine have been attributed to the low frequency of development of naturally acquired DBP binding inhibitory antibodies, the lack of knowledge of DBP contact residues necessary for receptor recognition, and the polymorphic nature of the DBP ligand. Understanding the specificity of the protective anti-DBP responses is critical for effective vaccine development. Therefore, two purposes of this study were to obtain a clearer understanding of anti-DBP serological responses from natural exposure and to initiate identification of epitopes targeted by functionally inhibitory anti-DBP antibodies. Here we present evidence identifying potential inhibitory B-cell linear epitopes in the critical receptor-binding region of the DBP ligand.
When we screened residents of an area of PNG where malaria is highly endemic, we discovered that naturally acquired antibodies that effectively inhibit DBPII-erythrocyte binding are infrequent. There was great variability among these residents, and for a substantial proportion (∼50%) there was a significant change in the titer from one transmission season to the next (Table 1). The observation that there are relatively rapid changes in serological responses is similar to the previous observation that in PNG temporal variation was the primary source of variation in the IgG response to Plasmodium antigens (22). The small number of high-titer, highly inhibitory sera remained relatively stable, which is consistent with the results of another study performed in this area where malaria is endemic (16), but low-responder individuals whose responses were mostly ineffective for functional inhibition of DBP binding to red blood cells also exhibited a poor-quality antibody response in all sampling periods.
In the high-titer group only 3 of 10 individuals exhibited effective inhibition of DBP function, while only 1 of the 23 low responders exhibited a high level of inhibition. Such marked individual differences in the naturally acquired anti-DBP responses and the functional inhibition of DBP-erythrocyte binding indicated that epitope specificity and antibody avidity are both critically important for effective inhibition. In addition, our results indicate that DBP inhibitory antibodies are relatively infrequent and most antibody responses are relatively unstable. These discoveries for anti-DBP serological responses contrast with acquisition of immunity to the homologous VAR2CSA DBL domain, where there is a better correlation between development of protective immunity and repeated exposure (21). However, a longitudinal study to closely monitor the development of antibodies and B-cell memory for DBP is necessary to adequately determine the stability of naturally acquired anti-DBP antibodies.
A comparison of the epitope specificity of the highly inhibitory anti-DBP sera with that of the noninhibitory immune sera identified potential targets associated with protective anti-DBP immunity. Nearly all of the putative inhibitory B-cell linear peptides identified in this study are localized in the central region of the DBP ligand domain. This finding is similar to the finding for naturally acquired antibodies to VAR2CSA-DBL that are directed toward surface-exposed epitopes in the S1/S2 domains, a region homologous to the central region of DBPII, and the F1/F2 domains of EBA-175 (3). This central region is known to be important for receptor recognition, but precisely how the Duffy receptor (DARC), interacts with this region is not yet understood. Mutagenesis studies have identified numerous residues in this region as residues that are important or essential for receptor recognition, including many residues absent from the 3D model based on the crystal structure of Plasmodium knowlesi DBPα (14, 20, 24, 25). Much attention has been focused on a receptor-binding pocket that is proposed to recognize a sulfated tyrosine thought to be part of the N-terminal domain of the DARC receptor similar to CXC-chemokine receptors (8, 20). This DARC binding site is on the surface opposite the residues of the homologous DBL domains of Plasmodium falciparum EBA-175 identified as important for interactions with its receptor, glycophorin A. Even though these homologous domains are structurally conserved, this finding highlights a significant difference in how these ligands bind their cognate receptors; for EBA-175 the mechanism is “handshake” dimerization of the tandem F1/F2 DBL to form central channels that bind glycophorin A (23), compared to a proposed monomeric interaction for DBP (20).
The most significant neutralizing epitopes identified in our study occur in two areas of the DBL domain that are potentially important for receptor recognition (H1 and H3) and binding the DARC sulfated tyrosine (H2). Epitopes H1 and H3 are in regions containing variant residues with radical substitutions, while the position of H2 varies minimally. Epitopes that are considered moderately inhibitory (M1, M2, and M3) occupy a region parallel to the H2 epitope flanking the other side of the putative tyrosine recognition motif. The location of multiple putative epitopes (H2 and M1 to M3) that are targets of protective immunity does not support the “just-in-time” release model of immune evasion (7, 20), since identification of multiple targets for neutralizing antibodies in the area flanking the DARC sulfated tyrosine-binding site shows that this site is accessible to inhibitory antibodies. The relative lack of variability in this area may be a consequence of other functional requirements necessary for receptor recognition and a somewhat weaker effect of immune selection on these epitopes.
Antibody reactivity to the H1, H2, and H3 epitopes, which contain clusters of polymorphic residues, was significantly correlated with inhibition of DBPII-erythrocyte binding. For this reason, these epitopes do not appear to be simply associated with a nonprotective immune evasion mechanism misdirecting antibodies away from crucial functional sites elsewhere in the ligand domain. Instead, the pattern of polymorphisms of DBPII is similar to the pattern of variability observed for the P. falciparum vaccine candidate AMA1, where polymorphisms are concentrated adjacent to the putative receptor-binding site (4). It appears likely that the receptor-binding site of DBPII is larger than that identified so far for the DARC sulfated tyrosine.
Our results identified critical DBP epitopes that are the targets of inhibitory antibodies naturally acquired in P. vivax infection and represent an important advance in understanding part of blood-stage immunity to P. vivax. These linear epitopes, which are recognized as targets of a functionally inhibitory antibody, may represent potential correlates of immunity for a DBP vaccine, although further characterization is necessary to confirm their relative importance compared to the importance of other epitopes (e.g., conformational epitopes). The identification of specific epitope targets of inhibitory immunity against DBP opens the way for optimization of DBP immunogenicity for protection against diverse P. vivax strains.
Supplementary Material
Acknowledgments
We thank Ren Chen, Biostatistics Core Research, College of Medicine, University of South Florida, and Samantha Jones for help with statistical analyses. We also thank Amy M. McHenry for critical discussions. Finally, we are grateful to participants in Papua New Guinea.
This study was supported by National Institutes of Health grant R01AI064478 (to J.H.A.).
We have no commercial or other association that poses a conflict of interest.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 14 December 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Adams, J. H., D. E. Hudson, M. Torii, G. E. Ward, T. E. Wellems, M. Aikawa, and L. H. Miller. 1990. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63:141-153. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. H., B. K. Sim, S. A. Dolan, X. Fang, D. C. Kaslow, and L. H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 89:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, P., M. A. Nielsen, M. Resende, T. S. Rask, M. Dahlback, T. Theander, O. Lund, and A. Salanti. 2008. Structural insight into epitopes in the pregnancy-associated malaria protein VAR2CSA. PLoS Pathog. 4:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, T., M. Becker, A. Gupta, P. Strike, V. J. Murphy, R. F. Anders, and A. H. Batchelor. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. U. S. A. 102:12736-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnwell, J. W., and M. R. Galinski. 1995. Plasmodium vivax: a glimpse into the unique and shared biology of the merozoite. Ann. Trop. Med. Parasitol. 89:113-120. [DOI] [PubMed] [Google Scholar]
- 6.Chitnis, C. E., and L. H. Miller. 1994. Identification of the erythrocyte-binding domain of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis, C. E., and A. Sharma. 2008. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 24:29-34. [DOI] [PubMed] [Google Scholar]
- 8.Choe, H., M. J. Moore, C. M. Owens, P. L. Wright, N. Vasilieva, W. Li, A. P. Singh, R. Shakri, C. E. Chitnis, and M. Farzan. 2005. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC). Mol. Microbiol. 55:1413-1422. [DOI] [PubMed] [Google Scholar]
- 9.Cole-Tobian, J., and C. L. King. 2003. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol. Biochem. Parasitol. 127:121-132. [DOI] [PubMed] [Google Scholar]
- 10.Cole-Tobian, J. L., A. Cortes, M. Baisor, W. Kastens, J. Xainli, M. Bockarie, J. H. Adams, and C. L. King. 2002. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J. Infect. Dis. 186:531-539. [DOI] [PubMed] [Google Scholar]
- 11.Cole-Tobian, J. L., P. A. Zimmerman, and C. L. King. 2007. High-throughput identification of the predominant malaria parasite clone in complex blood stage infections using a multi-SNP molecular haplotyping assay. Am. J. Trop. Med. Hyg. 76:12-19. [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, T., P. Michon, J. W. Barnwell, A. R. Noe, F. Al-Yaman, D. C. Kaslow, and J. H. Adams. 1997. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect. Immun. 65:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimberg, B. T., R. Udomsangpetch, J. Xainli, A. McHenry, T. Panichakul, J. Sattabongkot, L. Cui, M. Bockarie, C. Chitnis, J. Adams, P. A. Zimmerman, and C. L. King. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hans, D., P. Pattnaik, A. Bhattacharyya, A. R. Shakri, S. S. Yazdani, M. Sharma, H. Choe, M. Farzan, and C. E. Chitnis. 2005. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol. Microbiol. 55:1423-1434. [DOI] [PubMed] [Google Scholar]
- 15.Kho, W. G., J. Y. Chung, E. J. Sim, D. W. Kim, and W. C. Chung. 2001. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J. Parasitol. 39:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, C. L., P. Michon, A. R. Shakri, A. Marcotty, D. Stanisic, P. A. Zimmerman, J. L. Cole-Tobian, I. Mueller, and C. E. Chitnis. 2008. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc. Natl. Acad. Sci. U. S. A. 105:8363-8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendis, K., B. J. Sina, P. Marchesini, and R. Carter. 2001. The neglected burden of Plasmodium vivax malaria. Am. J. Trop. Med. Hyg. 64:97-106. [DOI] [PubMed] [Google Scholar]
- 18.Michon, P., T. Fraser, and J. H. Adams. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michon, P. A., M. Arevalo-Herrera, T. Fraser, S. Herrera, and J. H. Adams. 1998. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am. J. Trop. Med. Hyg. 59:597-599. [DOI] [PubMed] [Google Scholar]
- 20.Singh, S. K., R. Hora, H. Belrhali, C. E. Chitnis, and A. Sharma. 2006. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439:741-744. [DOI] [PubMed] [Google Scholar]
- 21.Staalsoe, T., R. Megnekou, N. Fievet, C. H. Ricke, H. D. Zornig, R. Leke, D. W. Taylor, P. Deloron, and L. Hviid. 2001. Acquisition and decay of antibodies to pregnancy-associated variant antigens on the surface of Plasmodium falciparum-infected erythrocytes that protect against placental parasitemia. J. Infect. Dis. 184:618-626. [DOI] [PubMed] [Google Scholar]
- 22.Stirnadel, H. A., F. Al-Yaman, B. Genton, M. P. Alpers, and T. A. Smith. 2000. Assessment of different sources of variation in the antibody responses to specific malaria antigens in children in Papua New Guinea. Int. J. Epidemiol. 29:579-586. [PubMed] [Google Scholar]
- 23.Tolia, N. H., E. J. Enemark, B. K. Sim, and L. Joshua-Tor. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122:183-193. [DOI] [PubMed] [Google Scholar]
- 24.Tsuboi, T., S. H. Kappe, F. al-Yaman, M. D. Prickett, M. Alpers, and J. H. Adams. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect. Immun. 62:5581-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanBuskirk, K. M., E. Sevova, and J. H. Adams. 2004. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. U. S. A. 101:15754-15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xainli, J., J. H. Adams, and C. L. King. 2000. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111:253-260. [DOI] [PubMed] [Google Scholar]
- 27.Xainli, J., M. Baisor, W. Kastens, M. Bockarie, J. H. Adams, and C. L. King. 2002. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J. Immunol. 169:3200-3207. [DOI] [PubMed] [Google Scholar]
- 28.Xainli, J., J. L. Cole-Tobian, M. Baisor, W. Kastens, M. Bockarie, S. S. Yazdani, C. E. Chitnis, J. H. Adams, and C. L. King. 2003. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect. Immun. 71:2508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.