Abstract
We studied the biochemical mechanisms associated with inhibition and resistance to a 4,5-dihydroxypyrimidine carboxylate that inhibits the hepatitis C virus (HCV) RNA-dependent RNA polymerase NS5B. On the basis of the structure of the pharmacophore, it has been suggested that these compounds may act as pyrophosphate (PPi) mimics. We monitored nucleotide incorporation events during the elongation phase and showed that the polymerase activity of wild-type NS5B was inhibited by the dihydroxypyrimidine at a 50% inhibitory concentration (IC50) of 0.73 μM. Enzymes with the G152E or P156L mutation, either of which confers resistance to this compound, showed four- to fivefold increases in IC50s. The inhibitor was competitive with respect to nucleotide incorporation. It was likewise effective at preventing the PPi-mediated excision of an incorporated chain terminator in a competitive fashion. In the absence of the dihydroxypyrimidine, the reaction was not significantly affected by the G152E or P156L mutation. These data suggest that the resistance associated with these two mutations is unlikely due to an altered interaction with the pyrophosphate-mimicking domain of the compound but, rather, is due to altered interactions with its specificity domain at a region distant from the active site. Together, our findings provide strong experimental evidence that supports the notion that the members of this class of compounds can act as PPi mimics that have the potential to mechanistically complement established nucleoside and nonnucleoside analogue inhibitors.
Hepatitis C virus (HCV) is a major public health problem, with an estimated 170 million people worldwide being infected with the virus (9). Chronic infection with HCV can lead to the development of severe liver disease, including cirrhosis and hepatocellular carcinoma (HCC) (17). The current standard of care for those who are in need of antiviral therapy consists of a combination of pegylated alpha interferon and the nucleoside analogue ribavirin (21). However, the clinical use of both components is associated with toxic side effects, and by far, not everyone benefits from treatment (4, 11).
Nonstructural proteins NS2 through NS5B represent important targets for current drug discovery and development efforts aimed at improving anti-HCV therapy. Various classes of inhibitors of the HCV RNA-dependent RNA polymerase NS5B have been developed. These compounds can be further grouped into nonnucleoside analogue inhibitors (NNIs) and nucleoside analogue inhibitors (NIs). The two classes of inhibitors target different stages of RNA synthesis. The HCV NS5B protein is capable of initiating RNA synthesis de novo, i.e., in the absence of a primer (12, 14, 23, 25, 33). At this early stage, productive initiation complexes are fragile and RNA synthesis is distributive. After four to six nucleotide incorporation events, at which point the enzyme switches to the elongation stage, conformational changes render the polymerization process highly processive (18). NNIs were shown to interfere with steps prior to or during the initiation of RNA synthesis (7, 10, 19, 30, 31). Although the binding sites and the detailed mechanisms of action can differ among the various families of these compounds, they do not appear to affect RNA elongation. In contrast, the triphosphate form of nucleoside analogue inhibitors binds to the active site and competes with the natural counterpart for incorporation into the growing RNA chain, preferentially during elongation. Several NIs have advanced into clinical trials, including prodrugs of 2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and 2′-C-methylcytidine (NM283) (16). While the former is currently being evaluated in phase II clinical trials, the development of the latter has been placed on hold.
Derivatives of 4,5-dihydroxypyrimidine carboxylic acid and α,γ-diketoacid are commonly referred to as pyrophosphate (PPi) mimics. These compounds may likewise act at the active site of the HCV RNA-dependent RNA polymerase NS5B (15, 29). The structures suggest that the members of the two families of compounds are able to coordinate the two divalent metal ions in a fashion similar to that which one would predict for the PPi product that is released from the complex following nucleotide incorporation (5). However, it remains to be studied how these compounds interfere with nucleotide binding and nucleotide incorporation and whether or how the reverse reaction, i.e., pyrophosphorolysis, might be affected. Our recent studies have shown that the NS5B protein of HCV is capable of excising chain-terminating nucleotides in the presence of physiologically relevant concentrations of PPi (6). The excision reaction can compromise the efficacy of nucleotide analogues, and PPi mimics may have the potential to counteract these effects.
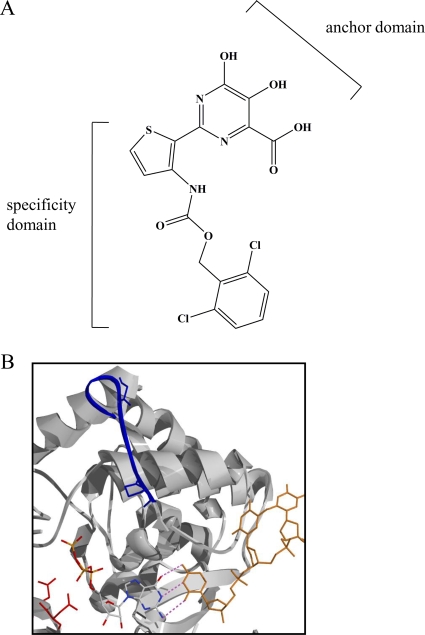
To address these questions, we have studied the biochemical mechanisms associated with both the inhibition of RNA synthesis and resistance to a prototype pyrimidine, herein referred to as compound A (Fig. 1A). The inhibitor contains two distinct domains: the anchor domain, which is implicated in interactions with the two catalytic metal ions at the active site, and the specificity domain, which provides additional contacts at distant regions of the enzyme (28). Here we demonstrate that compound A is able to inhibit both nucleotide incorporation and the PPi-mediated excision of nucleotide analogues. Increasing concentrations of natural nucleotides and PPi, respectively, counteract these inhibitory effects, which points to a competitive mechanism of action. Moreover, the P156L and G152E mutations in HCV NS5B (Fig. 1B), which were previously shown to decrease susceptibility to dihydroxypyrimidines in cell-based assays, also compromise the inhibitory effects of compound A in cell-free assays (32). Both the unique resistance profile and the ability of the compound to mechanistically complement other classes of NS5B inhibitors warrant further investigation in the development of PPi mimics as antiviral agents.
FIG. 1.
(A) Structure of compound A. (B) Model of the HCV NS5B active site with bound template (orange) and NTP. Active-site residues D318 and D319 are shown in red, while the G152 and P156 residues are highlighted in blue.
MATERIALS AND METHODS
Nucleic acid, nucleotides, and inhibitors.
The RNA sequence used in this study as a template for NS5B-mediated RNA synthesis was as follows: 5′AACAGUUUCCUUUUCUCUCC-3′ (Trilink). The sequence contained a single site for the incorporation of a cytidine analogue at position +16 (opposite G, underlined). The RNA was purified on a 12% polyacrylamide-7 M urea gel containing 50 mM Tris-borate (pH 8) and 1 mM EDTA. It was then eluted overnight from gel slices in a buffer containing 500 mM ammonium acetate and 0.1% SDS. A 5′-GG-3′ dinucleotide (Trilink) was used as a primer. Labeling of the 5′ end of the primer with [γ-32P]ATP was carried out with T4 polynucleotide kinase (Invitrogen). Nucleoside triphosphates (NTPs) were treated with inorganic pyrophosphatase to prevent PPi contamination. The nucleotide analogue 2′-C-methyl-CTP and the dihydroxypyrimidine carboxylate (compound A) were provided by Merck.
Expression and purification of HCV NS5B.
The HCV NS5B sequence was inserted into the expression vector pET-21 (Novagen), in which the C-terminal 21 amino acids were deleted and a His tag was added to the C terminus. The plasmid encoding the C-terminal truncated enzyme (Δ21) was transformed into Escherichia coli BL21(DE3) cells. The induction of protein expression was undertaken with the addition of 0.25 mM isopropyl-β-d-thiogalactopyranoside at 25°C. Protein was isolated by using an Ni-chelating Sepharose column. The generation of mutant enzymes with the P156L and G152E mutations was accomplished through site-directed mutagenesis with a Stratagene Quick-Change kit, according to the manufacturer's directions. The presence of the mutations was confirmed by sequencing at the Genome Quebec Innovation Center.
RNA synthesis.
The reaction mixtures contained 1 μM RNA template, 2 μM HCV NS5B, 0.2 μM radiolabeled GG primer, and 10 μM NTPs. For reactions requiring chain termination at position +16, 10 μM 2′-C-methyl-CTP was included. Reactions were carried out in a buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 0.2 mM MnCl2, and 6 mM MgCl2. The incubation time for the synthesis experiments was 40 min. The reactions were stopped by the addition of 100 mM EDTA in formamide. The samples were heat denatured at 95°C for 5 min and resolved on 12% polyacrylamide-7 M urea gels. The concentration of dihydroxypyrimidine carboxylate required to inhibit 50% of full-length product formation (IC50) was calculated by using Prism software (GraphPad, Inc). All assays were repeated a minimum of three times.
Pyrophosphorolysis and rescue of RNA synthesis.
After completion of the chain-termination reaction, pyrophosphorolytic excision of 2′-C-methyl-CTP was carried out with the addition of 1.25 mM PPi. This removes the chain terminator, but in the absence of the competing natural nucleotide CTP and additional nucleotide pools, no further RNA synthesis occurs, although the chain terminator may be reincorporated at the same position. The rescue reaction included 300 μM PPi to excise the chain terminator as well as 10 μM UTP and 100 μM the competing natural nucleotide CTP to displace the chain terminator and rescue RNA synthesis, resulting in a full-length product.
Competition with inhibitor.
To determine whether compound A can act competitively with either nucleotide substrates or PPi, the IC50 of the PPi analogue was determined in the presence of different concentrations of nucleotides or PPi. Of note, the enzyme does not dissociate from the stalled elongation complex; thus, the rates are independent of the enzyme concentration, and consequently, reactions cannot be carried out under conditions that allow multiple turnovers (steady-state conditions) (6). To look at competition with nucleotides, synthesis to position +16 of the template was promoted with the addition of 3 μM each GTP and ATP and 16 nM CTP. Further synthesis requires the addition of UTP, which yields a full-length product at position +20. The IC50 of compound A during the elongation reaction was determined in the presence of 0.1, 1, and 5 μM UTP. Inhibitor concentrations ranged up to 1,120 nM. To study competition with PPi, a similar assay was carried out. Briefly, different concentrations of PPi (50 μM, 100 μM, 500 μM) were used for a rescue reaction, and the IC50 of the inhibitor was determined at each concentration. The inhibitor concentrations in the rescue assay ranged from 0 to 2.25 μM.
RESULTS
Inhibition of RNA synthesis.
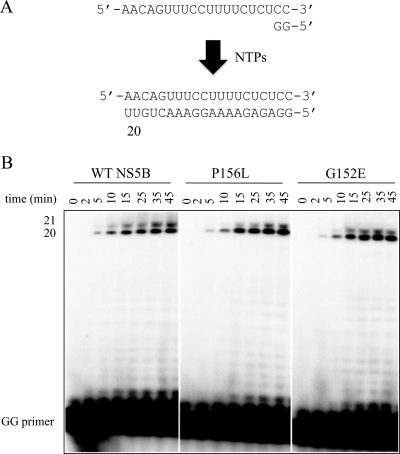
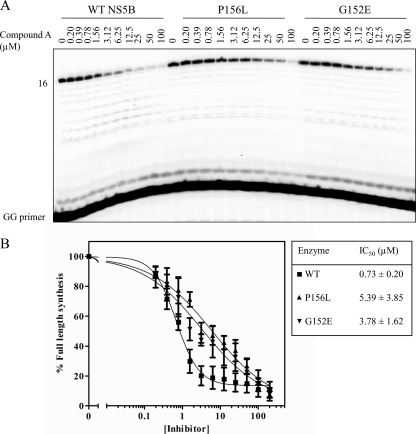
We monitored RNA synthesis in the absence and presence of compound A with purified HCV NS5B using a 20-mer heteropolymeric model template and a dinucleotide primer (5′-GG-3′) (Fig. 2A). The template was designed to allow incorporation of the nonobligate chain terminator 2′-C-methyl-CMP (2′-C-Me-CMP) at a single position, which, in turn, facilitates the study of PPi-mediated excision and its inhibition. Initially, we studied RNA synthesis by wild-type NS5B and enzymes containing the G152E and P156L mutations in the absence of any inhibitor. Extension of the GG primer leads to a full-length product as well as a product further extended by a single position. This extended product may arise through the terminal transferase activity of HCV NS5B, as has been reported previously (2, 24). The level of full-length product formed over a 45-min period was similar for all enzymes, indicating that the mutations do not affect activity (Fig. 2B). We then examined the effects of increasing concentrations of compound A on RNA synthesis in the context of both wild-type HCV NS5B and the mutant enzymes to test whether the resistant phenotype can be translated in biochemical terms (Fig. 3A). With wild-type HCV NS5B, we measured IC50s of approximately 700 nM on the basis of decreases in the formation of the full-length product compared to the level of formation for the control in the absence of inhibitor. The lack of additional pausing sites in the presence of inhibitor suggests that RNA synthesis is affected throughout the process of RNA synthesis, during initiation and elongation. The G152E mutation increased the IC50 approximately fourfold, while the P156L mutation increased the values fivefold (Fig. 3B).
FIG. 2.
Synthesis of RNA by wild-type (WT) HCV NS5B and mutants with the G152E or P156L mutation. (A) Schematic of RNA synthesis reaction. (B) RNA synthesis by wild-type NS5B and the enzymes with the G152E or P156L mutation was monitored over a period of 45 min.
FIG. 3.
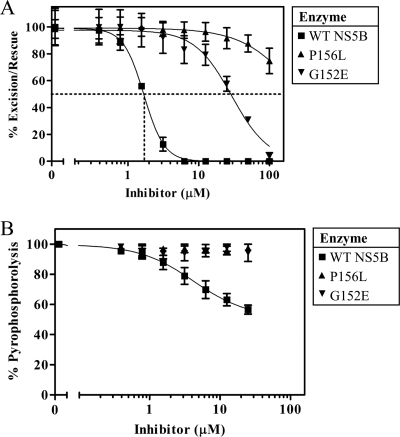
Inhibition of RNA synthesis by compound A. (A) RNA synthesis of a chain-terminated product (position +16) by wild-type NS5B or the mutant enzymes with the G152E or P156L mutation was examined in the presence of increasing concentrations of compound A up to 200 μM. (B) Graphic representation of the data shown in panel A. IC50s for inhibition of the wild-type and mutant enzymes are based on full-length RNA synthesis. Data points show the means and standard deviations determined from three independent experiments.
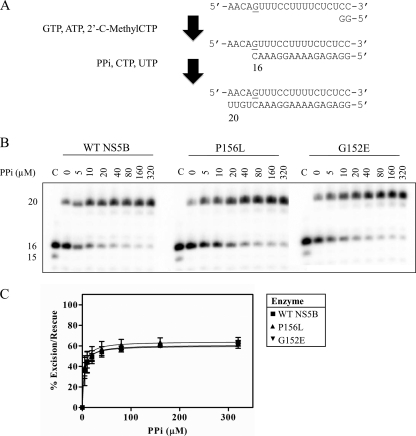
To provide further insight into the mechanism of resistance, we studied the potential effects of the two mutations on the PPi-mediated excision of incorporated 2′-C-Me-CMP. RNA synthesis with limited nucleotide pools and the nucleotide analogue caused chain termination at position +16. The reaction was allowed to proceed to near completion, and the subsequent addition of PPi and a complete set of natural nucleotides could then initiate excision and the rescue of RNA synthesis (Fig. 4A). Increasing concentrations of PPi resulted in increases in excision and the rescue of RNA synthesis with each of the three enzymes (Fig. 4B). The amount of excision was similar for wild-type NS5B and the two mutant enzymes (Fig. 4C). These findings suggest that neither G152E nor P156L affects the binding of PPi. By extension, the two mutations do not appear to interfere with the anchor domain of compound A; rather, they may affect contacts with the specificity domain.
FIG. 4.
PPi-dependent excision of a chain terminator and rescue of RNA synthesis. (A) The schematic depicts a chain-terminated product after addition of only GTP, ATP, and 2′-C-methyl-CTP. Following this reaction, the excision of the chain terminator and the rescue of RNA synthesis are initiated with the addition of PPi, UTP, and CTP. (B) Excision and rescue occur in a dose-dependent manner with the addition of up to 320 μM PPi. The reaction is shown for wild-type NS5B and the mutant enzymes. Lanes C, the excision reaction alone, which produces a product at position +15. (C) The amount of excised and rescued product was quantified as the amount of product at position +20 as a percentage of the total amount of product in the lane. Data points are the means ± standard deviations for three independent experiments.
Inhibition of PPi-mediated excision of nucleotide analogues.
To study whether compound A is able to inhibit the combined excision and rescue reaction, we added increasing concentrations of the inhibitor to the reaction mixture containing PPi and nucleotides. Wild-type HCV NS5B was inhibited during excision and rescue, with the IC50s being approximately 1.7 μM, which is similar to the inhibition values measured during the RNA synthesis reaction (Fig. 5A). The two mutant enzymes again showed increases in IC50s of at least an order of magnitude. To test whether the inhibitor can directly affect pyrophosphorolytic cleavage, we assayed the PPi-mediated excision of the chain terminator in the absence of the nucleotide substrates that are required for the rescue reaction. Of note, excision in the absence of competing nucleotides is generally inefficient compared with the efficiency of excision achieved in the combined excision-rescue reaction. The facile reincorporation of the chain terminator diminishes the net formation of the shorter reaction product. As a consequence, the two mutant enzymes appear to be completely resistant to the inhibitor under these conditions. However, compound A is able to inhibit the reaction with wild-type NS5B, which demonstrates that the inhibitor can interfere directly with pyrophosphorolysis (Fig. 5B).
FIG. 5.
Inhibition of excision and rescue by compound A. (A) Increasing concentrations of compound A up to 100 μM were added during a rescue reaction. The compound inhibited rescue by the wild-type NS5B enzyme with an IC50 of 1.7 ± 0.3 μM, while the enzymes with the G152E and P156L mutations required doses of compound A that were at least an order of magnitude higher. (B) Compound A was also added during an excision reaction to determine its effects directly on pyrophosphorolysis. Data points are the means ± standard deviations for three independent experiments.
Mechanism of inhibition.
The observation that compound A inhibits both RNA synthesis and pyrophosphorolysis provides support for the notion that the inhibitor can coordinate the divalent metal ions at the active site in a fashion similar to that for the PPi product or the phosphates of an incoming nucleotide. Hence, the binding of compound A and the nucleotide substrate is likely to be competitive in nature. To test this hypothesis, we measured the effect of the pyrimidine at different concentrations of the nucleotide substrate. RNA synthesis was halted at position 16, by omitting the required UTP from the reaction mixture. In contrast to previously described experiments, chain-terminating nucleotides were not included in the reaction mixture. The continuation of RNA synthesis in the presence of three different concentrations of UTP and increasing concentrations of compound A was then monitored (Fig. 6A). We measured increasing IC50s at higher concentrations of the nucleotide substrate, which suggests a competitive mechanism of inhibition of RNA synthesis (Table 1). We obtained essentially the same result when we looked at the inhibition of the combined excision-rescue reaction with different concentrations of PPi, which also confirms a competitive mechanism of inhibition of the reverse reaction (Fig. 6B).
FIG. 6.
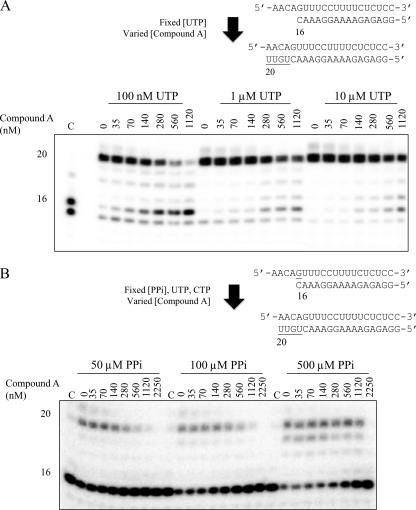
Competition of compound A with nucleotides or PPi. (A) The concentration of the PPi analogue required to inhibit 50% of an elongation reaction was determined in the presence of different concentrations of UTP (100 nM, 1 μM, 10 μM). Lane C, a chain-terminated product showing the results for position +16. (B) Similarly, IC50s were calculated during a rescue reaction with various concentrations of PPi (50 μM, 100 μM, 500 μM). Lane C, CTP and UTP without PPi.
TABLE 1.
IC50s of compound A required to inhibit nucleotide incorporation or PPi-mediated excision/rescue
| Nucleotide or PPi (concn [μM]) | Compound A IC50 (nM)a |
|---|---|
| UTP (0.1) | 257.7 ± 174.6 |
| UTP (1.0) | >500 |
| UTP (5.0) | >1,000 |
| PPi (50) | 173.6 ± 16.6 |
| PPi (100) | 302.0 ± 41.4 |
| PPi (500) | 881.9 ± 93.5 |
Data are means ± standard deviations determined from three independent experiments.
DISCUSSION
In the study described here, we elucidated the possible mechanisms of action and resistance associated with the inhibition of HCV NS5B by a prototype dihydroxypyrimidine carboxylate, referred to as compound A. Our findings provide strong evidence that members of this class of compounds act as PPi mimics. We show that compound A interferes directly with the PPi-mediated excision of incorporated chain terminators. The excision of an incorporated nucleotide analogue is inhibited, with the IC50s being in the low-micromolar range. Higher concentrations of PPi lead to higher IC50s, which is indicative of a steric conflict. The geometry of the dihydroxypyrimidine carboxylate anchor region may interact with the two divalent metal ions at the active site in a fashion similar to that described in crystallographic models for PPi (8, 27). Together, the data point to overlapping binding sites; however, the properties of PPi and compound A are entirely different. PPi, as the product of a nucleotidyl transfer reaction, needs to be released from the complex to facilitate the binding to and the incorporation of the next nucleotide. Studies with other RNA and DNA polymerases, including the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1), have shown that the affinity to PPi is generally low, with the Kd (dissociation constant) values being in the millimolar range (1, 13, 26). The excision of the incorporated nucleotide analogues with HCV NS5B requires concentrations of PPi in the high-micromolar range, which is consistent with the notion that PPi is preferentially released from the complex rather than being used as a substrate. In contrast, the relatively low IC50s of compound A point to the improved binding characteristics of compound A compared with those of PPi. The specificity domain of the inhibitor likely increases the affinity between the enzyme and the inhibitor (29). At the same time, our data suggest that the P156L and G152E mutations, which are associated with resistance to pyrimidines, can diminish the binding of the inhibitor through an altered interaction with the specificity domain. Neither P156L nor G152E affects pyrophosphorolysis; it is therefore unlikely that these mutations alter the interaction with the anchor region of the inhibitor.
There are also several potential differences when the properties of the dihydroxypyrimidine are compared with those of the PPi analogue phosphonoformic acid (PFA or foscarnet), which shows activity against various members of the Herpesviridae family and HIV (22). PFA is a simple small molecule that does not contain a specificity domain, yet this compound efficiently binds to viral DNA polymerases. We have recently shown that PFA stabilizes HIV-1 RT-nucleic acid complexes in the pretranslocational state (3, 20). In this conformation, the nucleotide binding site is occupied by the 3′ terminus of the primer, which provides a mechanism for the inhibition of DNA synthesis. The increase in the stability of the complex is likely to be a result of a conformational change that traps the inhibitor at the site that normally releases the PPi product. As a consequence, PFA is able to inhibit RT translocation, which, in turn, prevents the binding of the next nucleotide. At this point it is unknown whether dihydroxypyrimidines may bind to pre- and/or posttranslocational conformations of NS5B-RNA complexes; techniques that allow a distinction between the two complexes have yet to be developed. However, the observation that increasing concentrations of the nucleotide substrate cause increases in IC50s likewise point to a competitive mechanism of inhibition of RNA synthesis. Hence, the binding of compound A to the pretranslocated complex and the inhibition of HCV NS5B translocation or the binding of compound A to the posttranslocated complex, which prevents nucleotide binding directly, are two possible scenarios that help to explain the inhibitory effects of dihydroxypyrimidines.
Like the structurally related α,γ-diketoacids, dihydroxypyrimidines can interfere with nucleotide incorporation and excision during elongation. These findings are potentially relevant in the development of new strategies to block HCV replication. HCV NS5B is capable of excising 2′-methylated nucleotide analogues, which is detrimental to their inhibitory effects (6). Although the significance of these findings has yet to be demonstrated in biologically relevant settings, our data from cell-free assays demonstrate that the incorporation of chain-terminating nucleotides is reversible. Unlike current NNIs, which appear to act exclusively at the level of initiation, potent PPi mimics may counteract the excision reaction and may in turn increase the potency of nucleoside analogue-containing drug regimens. The differences in the mechanisms of action of PPi mimics compared to those of established NIs and NNIs, along with the different resistance profile, warrant further investigation into the development of dihydroxycarboxylate pyrimidines as inhibitors of the HCV NS5B polymerase.
Acknowledgments
This study was supported by the Cancer Research Society (CRS). M.G. is the recipient of a national career award from the Canadian Institutes for Health Research (CIHR). M.H.P. is the recipient of a student stipend from the National Canadian Research Training Program in Hepatitis C (NCRTP).
Footnotes
Published ahead of print on 22 December 2009.
REFERENCES
- 1.Anand, V. S., and S. S. Patel. 2006. Transient state kinetics of transcription elongation by T7 RNA polymerase. J. Biol. Chem. 281:35677-35685. [DOI] [PubMed] [Google Scholar]
- 2.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 3.Beilhartz, G. L., M. Wendeler, N. Baichoo, J. Rausch, S. Le Grice, and M. Gotte. 2009. HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J. Mol. Biol. 388:462-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornberg, M., H. Wedemeyer, and M. P. Manns. 2002. Treatment of chronic hepatitis C with PEGylated interferon and ribavirin. Curr. Gastroenterol. Rep. 4:23-30. [DOI] [PubMed] [Google Scholar]
- 5.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Deval, J., M. H. Powdrill, C. M. D'Abramo, L. Cellai, and M. Gotte. 2007. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 51:2920-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 8.Doublie, S., M. R. Sawaya, and T. Ellenberger. 1999. An open and closed case for all polymerases. Structure 7:R31-R35. [DOI] [PubMed] [Google Scholar]
- 9.Global Burden of Hepatitis C Working Group. 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20-29. [DOI] [PubMed] [Google Scholar]
- 10.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 11.Heathcote, J., and J. Main. 2005. Treatment of hepatitis C. J. Viral Hepat. 12:223-235. [DOI] [PubMed] [Google Scholar]
- 12.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh, J. C., S. Zinnen, and P. Modrich. 1993. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 268:24607-24613. [PubMed] [Google Scholar]
- 14.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch, U., B. Attenni, S. Malancona, S. Colarusso, I. Conte, M. Di Filippo, S. Harper, B. Pacini, C. Giomini, S. Thomas, I. Incitti, L. Tomei, R. De Francesco, S. Altamura, V. G. Matassa, and F. Narjes. 2006. 2-(2-Thienyl)-5,6-dihydroxy-4-carboxypyrimidines as inhibitors of the hepatitis C virus NS5B polymerase: discovery, SAR, modeling, and mutagenesis. J. Med. Chem. 49:1693-1705. [DOI] [PubMed] [Google Scholar]
- 16.Kronenberger, B., C. Welsch, N. Forestier, and S. Zeuzem. 2008. Novel hepatitis C drugs in current trials. Clin. Liver Dis. 12:529-555, viii. [DOI] [PubMed] [Google Scholar]
- 17.Levrero, M. 2006. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 25:3834-3847. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y., W. W. Jiang, J. Pratt, T. Rockway, K. Harris, S. Vasavanonda, R. Tripathi, R. Pithawalla, and W. M. Kati. 2006. Mechanistic study of HCV polymerase inhibitors at individual steps of the polymerization reaction. Biochemistry 45:11312-11323. [DOI] [PubMed] [Google Scholar]
- 19.Ma, H., V. Leveque, A. De Witte, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 20.Marchand, B., E. P. Tchesnokov, and M. Gotte. 2007. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J. Biol. Chem. 282:3337-3346. [DOI] [PubMed] [Google Scholar]
- 21.National Institutes of Health. 2002. NIH consensus statement on management of hepatitis C, 2002. NIH Consens. State Sci. Statements 19:1-46. [PubMed] [Google Scholar]
- 22.Oberg, B. 1982. Antiviral effects of phosphonoformate (PFA, foscarnet sodium). Pharmacol. Ther. 19:387-415. [DOI] [PubMed] [Google Scholar]
- 23.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 26.Reardon, J. E. 1993. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J. Biol. Chem. 268:8743-8751. [PubMed] [Google Scholar]
- 27.Steitz, T. A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395-17398. [DOI] [PubMed] [Google Scholar]
- 28.Summa, V., A. Petrocchi, V. G. Matassa, M. Taliani, R. Laufer, R. De Francesco, S. Altamura, and P. Pace. 2004. HCV NS5b RNA-dependent RNA polymerase inhibitors: from alpha, gamma-diketoacids to 4,5-dihydroxypyrimidine- or 3-methyl-5-hydroxypyrimidinonecarboxylic acids. Design and synthesis. J. Med. Chem. 47:5336-5339. [DOI] [PubMed] [Google Scholar]
- 29.Summa, V., A. Petrocchi, P. Pace, V. G. Matassa, R. De Francesco, S. Altamura, L. Tomei, U. Koch, and P. Neuner. 2004. Discovery of alpha, gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 47:14-17. [DOI] [PubMed] [Google Scholar]
- 30.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomei, L., S. Altamura, G. Paonessa, R. De Francesco, and G. Migliaccio. 2005. HCV antiviral resistance: the impact of in vitro studies on the development of antiviral agents targeting the viral NS5B polymerase. Antivir. Chem. Chemother. 16:225-245. [DOI] [PubMed] [Google Scholar]
- 33.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]