Abstract
Macrolide resistance in Streptococcus pneumoniae has emerged as an important clinical problem worldwide over the past decade. The aim of this study was to analyze the phenotypes (serotype and antibiotic susceptibility), genotypes (multilocus sequence type [MLST] and antibiotic resistance gene/transposon profiles) among the 31% (102/328) of invasive isolates from children in New South Wales, Australia, in 2005 that were resistant to erythromycin. Three serotypes—19F (47 isolates [46%]), 14 (27 isolates [26%]), and 6B (12 isolates [12%])—accounted for 86 (84%) of these 102 isolates. Seventy four (73%) isolates had the macrolide-lincosamide-streptogramin B (MLSB) resistance phenotype and carried Tn916 transposons (most commonly Tn6002); of these, 73 (99%) contained the erythromycin ribosomal methylase gene [erm(B)], 34 (47%) also carried the macrolide efflux gene [mef(E)], and 41 (55%) belonged to serotype 19F. Of 28 (27%) isolates with the M phenotype, 22 (79%) carried mef(A), including 16 (57%) belonging to serotype 14, and only six (19%) carried Tn916 transposons. Most (84%) isolates which contained mef also contained one of the msr(A) homologues, mel or msr(D); 38 of 40 (95%) isolates with mef(E) (on mega) carried mel, and of 28 (39%) isolates with mef(A), 10 (39%) carried mel and another 11(39%) carried msr(D), on Tn1207.1. Two predominant macrolide-resistant S. pneumoniae clonal clusters (CCs) were identified in this population. CC-271 contained 44% of isolates, most of which belonged to serotype 19F, had the MLSB phenotype, were multidrug resistant, and carried transposons of the Tn916 family; CC-15 contained 23% of isolates, most of which were serotype 14, had the M phenotype, and carried mef(A) on Tn1207.1. Erythromycin resistance among S. pneumoniae isolates in New South Wales is mainly due to the dissemination of multidrug-resistant S. pneumoniae strains or horizontal spread of the Tn916 family of transposons.
Streptococcus pneumoniae is an important cause of respiratory tract infections, bacteremia, and meningitis, for which antibiotic treatment is often difficult because of resistance to penicillin and other antibiotics, especially macrolides. During the last decade, macrolide resistance among S. pneumoniae isolates has increased, with considerable geographical variation among the genotypes and phenotypes involved (3, 14, 17, 34, 35).
Macrolide resistance in S. pneumoniae is mediated by two main mechanisms. Target modification due to a ribosomal methylase, encoded by erm(B) confers high-level resistance to macrolides, lincosamides, and streptogramin B (MLSB phenotype). In S. pneumoniae and related Streptococcus spp., the frequent association of erythromycin and tetracycline resistance is often related to insertion of erm(B) into a conjugative transposon of the Tn916 family that harbors tet(M) and carries integrase (int) and excisase (xis) genes. Members of this family, which carry erm(B), include Tn6002, Tn1545 (which also carries the kanamycin resistance gene aphA3), and Tn3872 (which also carries transposase genes tnpA and tnpR) (2, 8).
The second macrolide resistance mechanism is an efflux pump system encoded by mef which confers resistance to 14- and 15-member macrolides only (M phenotype) (22). The two main subclasses of mef in S. pneumoniae, mef(E) and mef(A), are carried on different, but related elements: mef(A) on the defective transposon Tn1207.1 (32, 33) or the closely related Tn1207.3 and mef(E) on an element named “macrolide efflux genetic assembly” (mega) (11, 19). Both of these elements carry an open reading frame downstream of mef, designated msr(D) (9, 10) or mel (1, 11, 19), which are homologues of msr(A), which codes for an ATP-dependent efflux pump in Staphylococcus spp. (31). msr(D) and mel are cotranscribed with mef and contribute to an erythromycin-inducible dual efflux system in S. pneumoniae (1, 9, 19). Different investigators have reported and named these msr(D) homologues separately, and it is not clear whether or not they are identical.
The prevalence of isolates carrying both mef and erm(B) has reportedly increased as a result of the worldwide spread of a limited number of multidrug-resistant clonal complexes (CCs), of which the most prevalent is Taiwan19F-14 (CC-271) (15). Recently, two new composite elements of the Tn916 family, containing tet(M) plus mega (Tn2009) and tet(M), erm(B), and mega (Tn2010), have been described (10, 12). The distribution of these transposons and the genes they carry also vary in different geographic regions (4, 9) and provide a clue to the origins of antibiotic-resistant strains of S. pneumoniae.
This study is the first to analyze the distribution of antibiotic susceptibility patterns and phenotypic and genotypic characteristics of erythromycin-resistant invasive S. pneumoniae isolates in Australia, including the transposons on which resistance genes are carried. The isolates studied had been referred to the Pneumococcal Reference Laboratory at the Centre for Infectious Diseases and Microbiology, which receives all sterile-site isolates from patients with invasive pneumococcal disease, in New South Wales, for serotyping (30). The 7-valent pneumococcal conjugate vaccine (PCV7) first became available in Australia in 2001, with limited uptake. It was introduced into the routine infant immunization schedule in January 2005 as a 3-dose regimen given at 2, 4, and 6 months.
MATERIALS AND METHODS
Isolates, identification, and antibiotic susceptibility testing.
As part of a detailed study of antibiotic resistance in invasive S. pneumoniae, before the widespread use of conjugate vaccine, all 328 invasive (mainly blood culture) isolates, from children less than 5 years old referred in 2005, were tested for susceptibility to 15 antibiotics.
S. pneumoniae isolates were identified by bile solubility and optochin susceptibility and were serotyped by the Quellung reaction, using antisera provided by the Statens Serum Institute (Copenhagen, Denmark). Antimicrobial susceptibility testing was performed by the broth microdilution method using Sensititre Microtiter Trays (Trek Diagnostics Systems, West Sussex, England) and cation-adjusted Mueller-Hinton broth supplemented with 3 to 5% lysed horse blood (28). The results were interpreted according the Clinical and Laboratory Standards Institute (CLSI) criteria, using penicillin breakpoints in place before they were changed in 2008 (susceptible, ≤0.06 mg/liter; intermediate, 0.12 to 1 mg/liter; and resistant, ≥2 mg/liter) (6). S. pneumoniae ATCC 49619 was used for quality control. Isolates for which the erythromycin MIC was ≥2 mg/liter were selected for further study.
Phenotypic characterization of macrolide resistance, for erythromycin-resistant/clindamycin-susceptible (MIC, <2 mg/liter) isolates was performed by the double-disk diffusion method. The clindamycin disk was placed approximately 22 mm from the edge of the erythromycin disk (7); after incubation, organisms that showed flattening of the clindamycin zone adjacent to the erythromycin disk (“D zone”) were interpreted as having inducible resistance (iMLSB phenotype), whereas those with a conserved inhibition zone around the clindamycin disk were considered to have the M phenotype. Isolates resistant to both erythromycin and clindamycin by broth dilution had the constitutive (cMLSB) phenotype.
Detection of resistance and transposon genes.
We used a multiplex PCR-based reverse line blot (mPCR/RLB) assay (21) to identify the presence of antibiotic resistance and transposon genes [tet(M), tet(O), int, xis, tnpR, tnpA, erm(B), mef(A), mef(E), mef(I), mel, msr(D), cat, and aphA3], as shown in Table 1. DNA was prepared as described previously (21). Briefly, five individual S. pneumoniae colonies were sampled using a disposable loop and resuspended in 0.2 ml digestion buffer (10 mM Tris-HCl, pH 8.0, 0.45% Triton X-100, and 0.45% Tween 20) in 2-ml Eppendorf tubes. The tubes containing S. pneumoniae suspension were heated at 100°C (dry block heater) for 10 min and then cooled on ice and centrifuged for 2 min at 13,000 × g to pellet the cell debris. A 2-μl aliquot of each supernatant containing extracted DNA was used as a template for mPCR.
TABLE 1.
Oligonucleotide primers used in this study
| Primera | GenBank accession no. | Tm (°C)b | Sequence (5′→3′)c | Source or reference |
|---|---|---|---|---|
| tetm-Sb | X90939 | 60.69 | 2668TCCGGTAAATCATTAGAAGCATT2690 | This study |
| tetm-Ab | X90939 | 61.62 | 3101TGTGGCAATAGYTTTGTATCTCC3079 | This study |
| teto-Sb | Y07780 | 61.34 | 121TCACATGAAAATAATTAACTTAGGCA146 | This study |
| teto-Ab | Y07780 | 60.02 | 683CTAATAGTTCATCGTTTCCCATAAT659 | This study |
| int-Sb | U09422 | 60.52 | 16854CATGATGGTATTGATGTTGTAGG16876 | 37 |
| int-Ab | U09422 | 60.41 | 17386TGATGGTCTATATTGACAAGACG17364 | 37 |
| tnpR-01-Upb | AM490850 | 64.94 | 6642CCAAGGAGCTAAAGAGGTCCC6662 | 2 |
| tnpR-01-Dnb | AM490850 | 62.46 | 6937TACTCACTCGAGCATAGCCAA6917 | This study |
| tnpR-02-Upb | AM490850 | 61.61 | 7936TTTCCATCTATAGCTACACTTGAAGA7962 | This study |
| tnpR-02-Dnb | AM490850 | 70.18 | 8189GTCCCGAGTCCCATGGAAGC8170 | 2 |
| tnpA-01-Upb | AM490850 | 61.67 | 8190AATTAATGTCTCCCATATTAATCGG8214 | This study |
| tnpA-02-Dnb | AM490850 | 62.37 | 8476CATCAATTAAAGAAGCATAATGTTCC8451 | This study |
| tnpA-02-Upb | AM490850 | 63.06 | 10031CAGATAGTGAGCTACGGCGA10050 | This study |
| tnpA-02-Dnb | AM490850 | 63.87 | 10318CAAGAAAAGTGATATGCTCCCAA10296 | This study |
| xis-forb | X61025 | 60.29 | 230ATGAAGCAGACTGACATTCCTA251 | This study |
| xis-revb | X61025 | 61.03 | 433CTAGATTGCGTCCAATGTATCTATAA408 | This study |
| ermb-Sb | M11180 | 59.84 | 828GGTAAAGGGCATTTAACGAC847 | 37 |
| ermb-Ab | M11180 | 59.44 | 1321CGATATTCTCGATTGACCC1303 | 37 |
| mefE/A-Sb | AF227521 | 63.41 | 3314GGCAGGGCAAGCAGTATC3331 | 37 |
| mefE/A-Ab | AF227521 | 59.76 | 3674CTGTTCTTCTGGTACTAAAAGTGG3651 | 37 |
| msrD-Sb | AF227520 | 62.98 | 5463CCATAATCCATACCCTATAGTCGG5486 | This study |
| msrD-Ab | AF227520 | 60.65 | 5645GAAATAGAAATTCCTTCTTCATGG5622 | This study |
| mel/mefI-Sb | AF376746 | 61.77 | 3306GAACAATTTATTGCGGAACG3325 | This study |
| mel/mefI-Ab | AF376746 | 61.90 | 3661TGAAAAGATGCATTTTCAAACA3640 | This study |
| aphA3-Sb | AF060241 | 63.77 | 854TGCCTGTTCCAAAGGTCC871 | 38 |
| aphA3-Ab | AF060241 | 59.41 | 1407TTTTATTTTCTCCCAATCAGG1387 | 38 |
| cat-Sb | V01277 | 61.16 | 1318ATTTGAACCAACAAACGACTTT1339 | This study |
| cat-Ab | V01277 | 60.08 | 1657GGTGTTTTGGGAAACAATTT1638 | This study |
Biotin-labeled primer.
Tm, melting temperatures provided by manufacturer.
Numbers relate to positions in GenBank sequences.
Some primers and probes from a previous study (37) were used, and new primers and probes were designed or modified based on published sequences in GenBank (2) so that they could be amplified, without interference, in a single mPCR reaction (21) (Tables 1 and 2). The mPCR system contained the following: 2 μl template DNA, 0.1 μl each forward (50 pmol/μl) and reverse (50 pmol/μl) primers, 2 μl deoxynucleoside triphosphates (dNTPs; 2.5 mM each dNTP), 2.5 μl 10× PCR buffer, 0.2 μl Qiagen HotStar Taq polymerase (5 U/μl), and water to 25 μl. PCR was performed according to the Qiagen Hotstar Taq polymerase kit instructions: 95°C for 15 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 72°C for 10 min; and 22°C hold.
TABLE 2.
Oligonucleotide probes used in this study
| Probea | GenBank accession no. | Tm (°C)b | Sequence (5′→3′)c | Source or reference |
|---|---|---|---|---|
| tetm-AP | X90939 | 61.70 | 2711CTTTCCTCTTGTTCGAGTTCC2691 | This study |
| tetm-SP | X90939 | 61.46 | 3049AATGAGTTTTTGAAGTTAAATAGTGTTCTT3078 | This study |
| teto-AP | Y07780 | 63.87 | 164CGTCAACGTGAGCCAGAA147 | This study |
| teto-SP | Y07780 | 62.20 | 641GAACAGTGGGATGCGGTA658 | This study |
| int-AP | U09422 | 60.64 | 16905CGTAAAGCTGGCAGAGTGT16887 | 37 |
| int-SP | U09422 | 59.02 | 17321AGAGTTTGGTGGTTTGACAC17340 | 37 |
| xis-AP | X61025 | 63.49 | 274GGTTAGGGTATAACGTTCCCAAA252 | This study |
| xis-SP | X61025 | 63.14 | 368GGCAATCGTATTCAGATTAAACG390 | This study |
| tnpR-AP | AM490850 | 67.79 | 6682CAAATTCCCCGTAGGCGCTA6663 | This study |
| tnpR-SP | AM490850 | 61.75 | 8149TGTAAAAAAGTGGAAGTGATTGC8171 | This study |
| tnpA-AP | AM490850 | 62.05 | 8239GTCTAGCTAGCTGAAGAAACCTGTT8215 | This study |
| tnpA024-SP | AM490850 | 61.54 | 10261GTAACTAAGTATATGAAGCATGTATCTCCT10290 | This study |
| ermb-AP | M11180 | 58.50 | 876TTACCTGTTTACTTATTTTAGCCAG852 | 37 |
| ermb-SP | M11180 | 61.98 | 1236CTTACCCGCCATACCACA1253 | 37 |
| mef(A/E)-AP | AF227521 | 60.04 | 3353CAAGATGGCACTAGTGATTAATG3331 | 37 |
| mef(A)-SP | AF227521 | 60.04 | 3632GGCTCTCAATGCGGTTAC3649 | 37 |
| msrD-AP | AF227520 | 61.99 | 5524ACAGTGCCTTATCCCCAAATA5504 | This study |
| msrD-SP | AF227520 | 61.61 | 5580TAATGGAACCGGAAAAACAA5599 | This study |
| mel-AP | AF376746 | 61.05 | 3493AGGGTTTTAGCAGCATTATACATC3470 | This study |
| mel-SP | AF376746 | 60.58 | 3601TCGGTGCAGAAATTAATAAAGTATT3625 | This study |
| mefI-AP | AJ971089 | 62.80 | 2397GGGATTTAACGGCATTATGC2378 | This study |
| mefI-SP | AJ971089 | 60.64 | 2506TCGGTACGGAAATTACTAAAATATTT2531 | This study |
| aphA3-SP | AF060241 | 59.63 | 891ATCATGCCGTTCAAAGTG874 | 38 |
| aphA3-AP | AF060241 | 59.72 | 1341GGAAGAACAGTATGTCGAGC1360 | 38 |
| cat-AP | V01277 | 60.72 | 1372AAAACACTAATATCAATTTCTGTGGTT1346 | This study |
| cat-232-SP | V01277 | 65.55 | 1616GGTTATTGGGATAAGTTAGAGCCACTTTAT1637 | This study |
Amine-labeled probe.
Tm, melting temperatures provided by manufacturer.
Numbers relate to positions in GenBank sequences.
Reverse line blot (RLB) hybridization.
The RLB hybridization assay was based on a method described previously (21), except that the hybridization temperature was 60°C and the time of exposure to X-ray film (Hyperfilm; Amersham) was 15 min. RLB results were regarded as positive when both probes for a particular sequence gave positive signals. To optimize hybridization conditions, the probes were tested at several 2-fold dilutions, starting at a concentration of 1.2 pM and with final labeling concentrations of between 5.0 and 10 pM (21). Any discrepancies between phenotypic susceptibility to erythromycin or tetracycline and RLB results were confirmed by Etest (AB Biodisk, Solna, Sweden) and single-gene-specific PCR [for erm(B), mef(A), and mef(E) or tet(M) and tet(O), respectively] (37).
MLST analysis.
Multilocus sequence typing (MLST) was performed as described previously (37). The seven genes targeted were aroE, gdh, gki, recP, spi, xpt, and ddl. Clusters of related sequence types (STs) were grouped into clonal complexes (CCs) using the eBURST program (http://www.mlst.net).
Statistical analysis.
All statistical analyses were performed using SPSS version 11.0 (SPSS, Chicago, IL). The χ2 test was used to compare dichotomous variables.
RESULTS
Susceptibility phenotypes of erythromycin-resistant S. pneumoniae.
One hundred two of the 328 (31%) S. pneumoniae isolates tested were resistant to erythromycin (MIC, ≥2 mg/liter). A summary of susceptibilities of these isolates to the 15 antibiotics tested is shown in Table 3. All isolates were susceptible to the three fluoroquinolones levofloxacin, gatifloxacin, and moxifloxacin and to vancomycin and linezolid, but the majority were resistant (MIC, ≥2 mg/liter) or of intermediate susceptibility (MIC, 0.12 to 1 mg/liter) to penicillin (80 isolates [78%]), tetracycline (75 isolates [74%]), and trimethoprim-sulfamethoxazole (79 isolates [77%]). Eighty-four isolates (82%) were multidrug resistant (i.e., resistant to three or more classes of antibiotic), which is significantly higher than the proportion (50/226 [22%]) which was resistant to penicillin among erythromycin-susceptible isolates in this set (P < 0.0001) (data not shown).
TABLE 3.
Antimicrobial susceptibilities of 102 erythromycin-resistant S. pneumoniae strains
| Antibiotic | MIC (μg/ml)a |
No. (%) of strains |
||||
|---|---|---|---|---|---|---|
| 50% | 90% | Range | Susceptible | Intermediate | Resistant | |
| Penicillinb | 2 | 4 | <0.03->8 | 25 (24.5) | 7 (6.9) | 70 (68.6) |
| Amoxicillin-clavulanic acid | <2/1 | >8/4 | <2/1-8/4 | 68 (66.7) | 5 (4.9) | 29 (28.4) |
| Cefotaxime | 1 | 2 | <0.06->2 | 64 (62.7) | 32 (31.4) | 6 (5.9) |
| Tetracycline | ≥8 | ≥8 | <0.5->8 | 25 (24.5) | 1 (1.0) | 76 (74.5) |
| Chloramphenicol | 4 | 8 | <2->16 | 68 (66.7) | 0 (0) | 34 (33.3) |
| Levofloxacin | 1 | 1 | <0.5-1 | 102 (100.0) | 0 (0) | 0 (0) |
| Gatifloxacin | <0.5 | <0.5 | <0.5 | 102 (100.0) | 0 (0) | 0 (0) |
| Moxifloxacin | <0.25 | <0.25 | <0.25-1 | 102 (100.0) | 0 (0) | 0 (0) |
| Erythromycin | >2 | >2 | >2 | 0 (0) | 0 (0) | 102 (100.0) |
| Azithromycin | >2 | >2 | >2 | 0 (0) | 0 (0) | 102 (100.0) |
| Linezolid | 1 | 1 | 0.5-2 | 102 (100.0) | 0 (0) | 0 (0) |
| Clindamycin | >2 | >2 | <0.06->2 | 31 (30.4) | 2 (2.0) | 69 (67.6) |
| Vancomycin | <0.5 | <0.5 | <0.5-1 | 102 (100.0) | 0 (0) | 0 (0) |
| SXTc | >4/76 | >4/76 | <0.5/9.5-4/76 | 23 (22.6) | 18 (17.6) | 61 (59.8) |
| Meropenem | 0.5 | 1 | <0.25-1 | 34 (33.3) | 33 (32.4) | 35 (34.3) |
50% and 90%, MIC50 and MIC90, respectively.
Susceptible, intermediate, and resistant MIC breakpoints for penicillin were ≤0.06, 0.12-1, and ≥2 mg/liter, respectively.
SXT, trimethoprim-sulfamethoxazole.
Seventy-four (73%) isolates expressed the MLSB phenotype, including 69 that were constitutively resistant to clindamycin and five that demonstrated inducible resistance in the D-test (Table 4). Of these 67 (91%) and 68 (92%) were resistant to penicillin and tetracycline, respectively. There were nine different resistance profiles among these 74 isolates, but two (Penr Eryr Clir Chlr SXTr Tetr [n = 23] and Penr Eryr Clir Chls SXTr Tetr [n = 31]) were represented by 54 (73%) isolates; of these, 36 (67%) belonged to serotype 19F.
TABLE 4.
Distribution of serotypes and sequence types among different antibiotic resistance phenotypic categories of 102 erythromycin-resistant S. pneumoniae isolates
| Phenotype (n) | Penicillin resistance/susceptibility (n)a | Serotype (n) | ST (n) |
|---|---|---|---|
| Constitutive MLSB (69) | R (65) | 19F (39) | 236 (1), 242 (1), 271 (1), 320 (24), 352 (10), 4234 (1), 4235 (1) |
| 14 (11) | 15 (6), 143 (3), 230 (1), 1492 (1) | ||
| 6B (6) | 315 (4), 322 (1), 4233 (1) | ||
| 19A (2) | 320 (2) | ||
| 23F (3) | 242 (1), 342 (1), 880 (1) | ||
| 6A (2) | 81 (1), 320 (1) | ||
| NT (1) | 90 (1) | ||
| 9V (1) | 156 (1) | ||
| S (4) | 6B (3) | 90 (1), 315 (1), 4236 (1) | |
| NT (1) | 344 (1) | ||
| Inducible MLSB (5) | R (2) | 19F (2) | 271 (1), 320 (1) |
| S (3) | 6B (3) | 1645 (3) | |
| M (28) | R (10) | 19F (5) | 9 (1), 81 (1), 242 (1), 651 (1), 2477 (1) |
| 23F (3) | 81 (2), 242 (1) | ||
| 14 (1) | 242 (1) | ||
| 19A (1) | 156 (1) | ||
| S (18) | 14 (15) | 9 (15) | |
| 15B (1) | 411 (1) | ||
| 19F (1) | 33 (1) | ||
| 4 (1) | 205 (1) |
S, susceptible; R, resistant. MLSB resistance is either constitutive (always produced) or inducible following exposure to a macrolide (as shown by D-test [see text]).
Twenty-eight (27%) isolates expressed the M phenotype. Of these, 10 (37%) were resistant to penicillin and nine (32%) to tetracycline. There were seven different antibiotic resistance profiles: one, Pens Eryr Clis Chls SXTs Tets, accounted for 15 (54%) isolates, 14 of which were serotype 14.
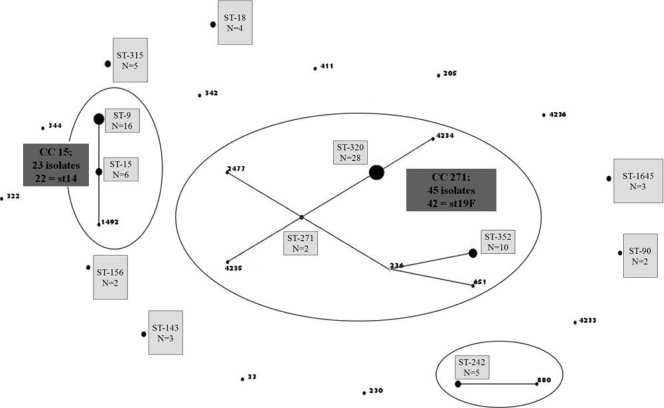
There were 9 serotypes and 28 sequence types (STs) among these 102 erythromycin-resistant isolates. Three serotypes, 19F (n = 47), 14 (n = 27), and 6B (n = 12), accounted for 84% of isolates. Among isolates with the MLSB phenotype, serotypes 19F (41/74 [55%]) and 6B (12/74 [16%]) were predominant, whereas serotype 14 was the most common among those with the M phenotype (16/28; 57%). Three STs (ST-320 [n = 28], ST-352 [n = 10], and ST-9 [n = 16]) accounted for 53% of the erythromycin-resistant isolates (Fig. 1). Among 41 penicillin-resistant, serotype 19F isolates with the MLSB phenotype, ST-320 (25 isolates [61%]) and ST-352 (10 isolates [24%]) were predominant. ST-9 was predominant (15/16 isolates [94%]) among serotype 14 isolates with the M phenotype. The distribution of serotypes and sequence types between phenotypic categories is shown in Table 4.
FIG. 1.
Population structure of 102 erythromycin-resistant S. pneumoniae isolates. Relationships between sequence types (ST) of 102 erythromycin-resistant S. pneumoniae isolates were demonstrated using eBURST v3. Three groups were defined using stringent criteria (6/7 shared alleles). Two were identified as clonal complexes (CC): CC-271 and CC-15. The predicted clonal ancestor of CC-271 is shown in blue (ST-271), and a subgroup founder is shown in yellow (ST-236). Most of the isolates in each of the CCs belonged to single serotypes (st): CC-271, serotype 19F, and CC-15, serotype 14.
Antibiotic resistance and transposon-related genes of erythromycin-resistant S. pneumoniae.
Among 78 (76%) of 102 erythromycin-resistant isolates that were also tetracycline resistant, four strains contained neither tet(M) nor tet(O): all were of the MLSB phenotype—two were serotype 14 (ST-15 and ST-143) and two serotype 6B (ST-322 and ST-1645). One phenotypically susceptible isolate (MLSB, serotype 19F, ST-271) carried tet(M). None of the 102 isolates contained mef(I), cat, or tet(O); four isolates contained apha3 (serotype 14, ST-143 [3 isolates]; and nontypeable, ST-344 [1 isolate]).
erm(B) was identified in 73 of 74 (99%) isolates with the MLSB phenotype, and of these, 40 (55%) also carried mef—34 mef(E) and 6 mef(A). Neither erm(B) nor mef was detected in the other isolate with the MLSB phenotype (serotype 19F, ST-352). All isolates with the M phenotype carried mef—22 mef(A) and 6 mef(E). Three also carried erm(B). Sixteen of 22 (79%) M phenotype isolates with mef(A) belonged to serotype 14. Overall, 36 of 102 (35%) isolates carried both erm(B) and mef(E). Of these isolates, 29 (81%) belonged to serotype 19F and 33 (92%) carried Tn6002.
All 74 isolates with the MLSB phenotype and nine of 28 isolates with the M phenotype carried transposons belonging to the Tn916 family: 49 carried Tn6002 [characterized by int, xis, tet(M), and erm(B)], and four carried a variant without tet(M). Fifty-one of these 53 isolates had the MLSB phenotype: 31 carried mef(E), and 30 belonged to serotype 19F. Three isolates carried Tn1545 [int, xis, tet(M), and erm(B), plus aphA3], 15 carried Tn3872 [int, xis, tnpA, tnpR, tet(M), and erm(B)], and another 5 carried a variant without tet(M)—all but one of these 20 isolates had the MLSB phenotype.
Of 68 isolates with either mef(A) or mef(E), 59 (87%) carried one of the msr(A) homologues: 38/40 (95%) with mef(E) and 10/28 (36%) with mef(A) carried mel, and 11/28 (39%) with mef(A) carried msr(D). The distributions of antibiotic resistance genes and transposon markers among different serotypes and sequence types are shown in Table 5.
TABLE 5.
Antibiotic resistance and transposon genes identified among 102 erythromycin-resistant S. pneumoniae isolates
| Isolate type | Antibiotic resistance and transposon gene profilea |
Presumed transposonb | Serotype (n) and ST/CC (n)c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tet(M)/int/xis | tnpA/tnpR | erm(B) | mef(E) | mef(A) | msr(D) | mel | aphA3 | |||
| MLSB | −/+/+ | +/+ | + | − | − | − | − | − | Serotype 14 (5), ST-15 (5) | |
| +/+/+ | −/− | + | − | − | − | − | − | Tn6002 | Serotype 6B (8), ST-1645 (2), ST-4236 (1), and ST-315 (5); serotype 23F (1), ST-342 (1); serotype NT (1), ST-90 (1); serotype 14 (2), ST-230 (1) and ST-1492 (1) | |
| +/+/+ | −/− | + | − | − | − | − | + | Tn1545 | Serotype 14 (2), ST-143 (2); serotype NT (1), ST-344 (1) | |
| +/+/+ | +/+ | + | - | - | - | - | - | Tn3872 | Serotype 19F (7), ST-352 (7); serotype 23F (1), ST-242 (1) | |
| +/+/+ | +/+ | + | − | − | + | − | − | Tn3872 | Serotype 6B (1), ST-4233 (1) | |
| −/+/+ | −/− | + | − | − | − | − | − | Serotype 6B (2), ST-1645 (1) and ST-322 (1); serotype 14 (1), ST-15 (1) | ||
| −/+/+ | −/− | + | − | − | − | − | + | Serotype 14 (1), ST-143 (1) | ||
| +/+/+ | −/− | − | − | − | − | − | − | Tn916 | Serotype 19F (1), ST-352 (1) | |
| +/+/+ | −/− | + | − | + | − | + | − | Tn6002 + Tn1207.1 | Serotype 19A (1), ST-320 (1); serotype 19F (3), ST-320 (2) and ST-4234 (1) | |
| +/+/+ | +/+ | + | − | + | − | − | − | Tn3872 + Tn1207.1 | Serotype 19F (2), ST-352 (2) | |
| +/+/+ | −/− | + | + | − | − | + | − | Tn6002 + mega | Serotype 6A (2), ST-320 (1) and ST-81 (1); serotype 19A (1), ST-320 (1); serotype 19F (27), CC-271 (27) | |
| +/+/+ | −/− | + | + | − | − | − | − | Tn6002 + mega | Serotype 6B (1), ST-90 (1) | |
| +/+/+ | +/+ | + | + | − | − | + | − | Tn3872 + mega | Serotype 9V (1), ST-156 (1); serotype 19F (1), ST-242 (1) | |
| +/+/+ | +/+ | + | + | − | − | − | − | Tn3872 + mega | Serotype 23F (1), ST-880 (1) | |
| M | +/+/+ | −/− | + | + | − | − | + | − | Tn6002 + mega | Serotype 19F (1), ST-2477 (1); serotype 23F (1), ST-242 (1) |
| +/+/+ | −/− | − | + | − | − | + | − | Tn916 + mega or Tn2009 | Serotype 19F (1), ST-651 (1); serotype 23F (1), ST-81 (1) | |
| −/−/− | −/− | − | + | − | − | + | − | mega | Serotype 19F (2), ST-33 (1) and ST-9 (1) | |
| −/−/− | −/− | − | − | + | + | − | − | Tn1207.1 | Serotype 14 (11), ST-9 (11) | |
| −/−/− | −/− | − | − | + | − | − | − | Tn1207.1 | Serotype 14 (4), ST-9 (4) | |
| −/−/− | −/− | − | − | + | − | + | − | Tn1207.1 | Serotype 19A (1), ST-156 (1); serotype 15B (1), ST-411 (1) | |
| +/+/+ | +/+ | + | − | + | − | + | − | Tn3872 + Tn1207. 1 | Serotype 4 (1), ST-205 (1) | |
| +/+/+ | −/− | − | − | + | − | + | − | Tn916 + Tn1207.1 | Serotype 19F (2), ST-242(1) and ST-81 (1); serotype 23F (1), ST-81 (1) | |
| +/+/+ | −/− | − | − | + | − | − | − | Tn916 + Tn1207.1 | Serotype 14 (1), ST-242 (1) | |
+, positive; −, negative.
Transposons were identified by the following markers: Tn916, tet(M), int, and xis; Tn6002, tet(M), int, xis, and erm(B); Tn1545, tet(M), int, xis, erm(B), and aphA3; Tn3872, tet(M), int, xis, tnpA, tnpR, and erm(B); Tn1207.1, mef(A); mega, mef(E); and Tn2009, tet(M), int, xis, and mef(E).
NT, nontypeable; ST, sequence type; CC, clonal cluster.
MLST of erythromycin-resistant S. pneumoniae.
The distribution of STs among isolates with different antibiotic resistance phenotypes and genotypes is shown in Tables 4 and 5. Among 74 isolates with the MLSB phenotype, which carried erm(B) and mef(E) on Tn6002 or Tn3872, 37 (50%) belonged to ST-320 and ST-352, and of these, 34 belonged to serotype 19F (Table 5). Among the remaining 13 serotype 19F isolates, there were 11 STs, including two new ones, ST-4234 and ST-4235, which were single-nucleotide variants of ST-320 and ST-236, respectively.
Twelve serotype 6B isolates were distributed among six STs: five isolates for ST-315; three isolates for ST-1645; one isolate each for ST-322 and ST-90; and one isolate each for the two new STs, ST-4233 and ST-4236.
Among 28 M phenotype isolates, there were nine sequence types; one (ST-9), which carries mef(A)—and usually msr(D)—on Tn1207.1 and belongs to serotype 14, accounted for 15 (57%) of these isolates. Among 27 serotype 14 isolates, 15 belonged to ST-9, six to ST-15, and three to ST-143, and the remaining three isolates each belonged to a different ST.
The 28 STs generated in this data set were separated by eBURST into two CCs, one doublet, and 15 singletons (Fig. 1). The largest clonal cluster was CC-271; 45 of 102 (44%) isolates belonged to this cluster, and of these, 42 belonged to serotype 19F, corresponding with the widely distributed antibiotic-resistant strain, Taiwan19F-14 (CC-271). This CC was represented by eight STs (ST-271, ST-236, ST-320, ST-2477, ST-352, ST-651, ST-4234, and ST-4235, of which ST-271 was defined by eBURST as the founder of the CC). The next largest CC was CC-15, comprising 23 isolates, of which 22 belonged to serotype 14. This CC comprised three STs (ST-15, ST-9, and ST-1492), of which ST-15 was identified as the founder. There was one doublet containing five isolates belonging ST-242 and ST-880. All of the remaining isolates belonged to individual STs that were not closely related to others identified in this study.
Antibiotic resistance genes and transposons carried by erythromycin-susceptible S. pneumoniae.
Of the original 328 S. pneumoniae isolates tested, 226 were phenotypically sensitive to erythromycin. Of these, four were tetracycline resistant and all carried tet(M), int, and xis. Their serotypes were (one each of) 6B, 19A, 23F, and 14; one isolate (serotype 14) was also clindamycin resistant and carried erm(B). Another four erythromycin- and clindamycin-sensitive isolates carried mef(E) or mef(A), and one carried msr(D); of these isolates, two belonged to serotype 14 and one each belonged to serotypes 6B and 6A.
DISCUSSION
Erythromycin-resistant S. pneumoniae strains are already distributed worldwide: their numbers appear to be increasing rapidly, and resistance is expanding to include multiple antimicrobial agents (20). There is considerable geographic variation in prevalence, from 30 to 55% in France, Spain, South Africa, the United States, and Asia to as low as 4 to 7% in parts of northern and western Europe (e.g., the Czech Republic, the Netherlands, and Sweden) (35). Differences in erythromycin resistance are believed to reflect variations in macrolide consumption (European Surveillance of Antibiotic Consumption; http://www.ua.ac.be/esac) and the spread of multidrug-resistant clones.
This is the first study of genotypes among invasive pneumococcal isolates from young children in Australia. In 2005, before the widespread use of pneumococcal conjugate vaccine, 31% of invasive isolates (mainly from blood cultures), were erythromycin resistant; 82% of erythromycin-resistant isolates were multidrug resistant, compared with only 22% of those that were erythromycin susceptible.
The MLSB phenotype is predominant in most European countries, whereas the M phenotype predominates in North America, England, and Germany (13, 18, 24, 29). Although the MLSB phenotype was the more common in New South Wales, a substantial minority of isolates had the M phenotype (27%). Serotypes 19F and 14 accounted for more than half of the MLSB and M phenotype isolates, respectively.
Worldwide, the proportion of S. pneumoniae isolates harboring both erm(B) and mef(E) genes is increasing (15, 25) and is associated with the clonal dissemination of the Taiwan19F-14 (ST-237) clone and, less frequently, the Taiwan23F-15 and Spain23F-1 clones (16). None of the isolates in our study belonged to ST-237, but two single-locus variants, ST-320 and ST-352, together represented 51% of isolates, and of these, most belonged to serotype 19F and carried erm(B) and mef(E) on Tn6002 or Tn3872. eBURST analysis identified them as belonging to CC-271, which is closely related to ST-237. Two isolates carrying both erm(B) and mef(E) showed unusual serotype-sequence type combinations—6A and ST-81 and 6A and ST320. The serotype 6A-ST-81 combination has been described previously in Korea (http://www.mlst.net), but we have been unable to find any previous examples of the 6A-ST-320 combination. We assume this represents a “serotype switch” from 19F to 6A.
Another of the international clones identified by the Pneumococcal Molecular Epidemiology Network (26), which carries mef(A) and belongs to serotype 14 and ST-9 (13, 15), is a major contributor to the worldwide dissemination of M phenotype erythromycin resistance (http://www.mlst.net). Isolates with the same allelic profile and resistance pattern as England14-9 were the most frequently identified among isolates with the M phenotype in this study. eBURST analysis showed that they were in CC-15. ST-9 is a single-locus variant of ST-15.
Most of the isolates that contained either mef(E) or mef(A) contained one of the msr(A) homologues msr(D) or mel. These elements appear to be closely related variants with about 98% homology, but based on sequence heterogeneity between available sequences in GenBank, we were able to design primers and probes specific for each. There was a difference in their distribution between isolates carrying mef(E) (carried on mega), most of which were associated with mel, and those carrying mef(A) (carried on Tn1207.1), which were associated with msr(D) (all serotype 14-ST-9 isolates) and mel (belonging to various serotypes and STs); mel and msr(D) were not identified together. Presumably this indicates that mel, at least, can be carried on either mega, with mef(E), or Tn1207.1 with mef(A). However, the differences in distribution between genotypes and transposons suggest that they are distinct. Previous studies have shown that they are cotranscribed with mef, including the newly described variant mef(I) (27), which together act as an erythromycin-inducible dual-efflux system (19). However, it appears that the msr(D)/mel-encoded efflux system can also function independently (1, 9).
Most S. pneumoniae isolates with erm(B)-mediated erythromycin resistance are also tetracycline resistant, because they result from the insertion of erm(B) into conjugative transposons of the Tn916 family, which typically carry tet(M) (5, 8, 23). For example, the composite transposon Tn3872 is formed by the insertion of the erm(B) and mef(E) on transposable element Tn917 (which also carries tnpA and tnpR) (10, 23). Among the 102 isolates studied, 66% harbored both erm(B) and tet(M). Most of these carried either Tn6002 or Tn3872. In common with others (23), we found a low prevalence of Tn1545, a Tn916 transposon that, in addition to erm(B), also carries aphA3 (36).
The finding of one clindamycin-resistant and erythromycin-susceptible isolate is unusual. The isolate carried erm(B), and we assume that erythromycin resistance was not expressed, but there are other uncommon mechanisms of clindamycin resistance in Streptococcus agalactiae isolates (37), which, presumably, could also occur in S. pneumoniae.
In conclusion, the high prevalence of erythromycin-resistant S. pneumoniae strains among isolates from children in New South Wales, before the widespread use of pneumococcal conjugate vaccine, was mainly due to the dissemination of multiresistant S. pneumoniae strains and to the horizontal spread of the Tn916 family of transposons. This study is the first of its kind in Australia and will provide a valuable baseline from which to monitor changes in the prevalence of vaccine serotypes and antibiotic resistance following introduction of the vaccine.
Acknowledgments
This study was funded, in part, by NHMRC grant 358351 (2005).
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Ambrose, K. D., R. Nisbet, and D. S. Stephens. 2005. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob. Agents Chemother. 49:4203-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenciani, A., A. Bacciaglia, M. Vecchi, L. A. Vitali, P. E. Varaldo, and E. Giovanetti. 2007. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. D., D. J. Farrell, and I. Morrissey. 2004. Prevalence and molecular analysis of macrolide and fluoroquinolone resistance among isolates of Streptococcus pneumoniae collected during the 2000-2001 PROTEKT US Study. J. Clin. Microbiol. 42:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calatayud, L., C. Ardanuy, E. Cercenado, A. Fenoll, E. Bouza, R. Pallares, R. Martin, and J. Linares. 2007. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob. Agents Chemother. 51:3240-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.CLSI. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth international supplement M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Courvalin, P., and C. Carlier. 1987. Tn1545: a conjugative shuttle transposon. Mol. Gen. Genet. 206:259-264. [DOI] [PubMed] [Google Scholar]
- 9.Daly, M. M., S. Doktor, R. Flamm, and D. Shortridge. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J. Clin. Microbiol. 42:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Grosso, M., R. Camilli, F. Iannelli, G. Pozzi, and A. Pantosti. 2006. The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50:3361-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Grosso, M., J. G. Northwood, D. J. Farrell, and A. Pantosti. 2007. The macrolide resistance genes erm(B) and mef(E) are carried by Tn2010 in dual-gene Streptococcus pneumoniae isolates belonging to clonal complex CC271. Antimicrob. Agents Chemother. 51:4184-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias, R., and M. Canica. 2004. Emergence of invasive erythromycin-resistant Streptococcus pneumoniae strains in Portugal: contribution and phylogenetic relatedness of serotype 14. J. Antimicrob. Chemother. 54:1035-1039. [DOI] [PubMed] [Google Scholar]
- 14.Doern, G. V., S. S. Richter, A. Miller, N. Miller, C. Rice, K. Heilmann, and S. Beekmann. 2005. Antimicrobial resistance among Streptococcus pneumoniae in the United States: have we begun to turn the corner on resistance to certain antimicrobial classes? Clin. Infect. Dis. 41:139-148. [DOI] [PubMed] [Google Scholar]
- 15.Farrell, D. J., S. G. Jenkins, S. D. Brown, M. Patel, B. S. Lavin, and K. P. Klugman. 2005. Emergence and spread of Streptococcus pneumoniae with erm(B) and mef(A) resistance. Emerg. Infect. Dis. 11:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell, D. J., I. Morrissey, S. Bakker, L. Morris, S. Buckridge, and D. Felmingham. 2004. Molecular epidemiology of multiresistant Streptococcus pneumoniae with both erm(B)- and mef(A)-mediated macrolide resistance. J. Clin. Microbiol. 42:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felmingham, D., R. Canton, and S. G. Jenkins. 2007. Regional trends in beta-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J. Infect. 55:111-118. [DOI] [PubMed] [Google Scholar]
- 18.Fotopoulou, N., P. T. Tassios, D. V. Beste, S. Ioannidou, A. Efstratiou, E. R. Lawrence, J. Papaparaskevas, R. C. George, and N. J. Legakis. 2003. A common clone of erythromycin-resistant Streptococcus pneumoniae in Greece and the UK. Clin. Microbiol. Infect. 9:924-929. [DOI] [PubMed] [Google Scholar]
- 19.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 20.Klugman, K. P., and J. R. Lonks. 2005. Hidden epidemic of macrolide-resistant pneumococci. Emerg. Infect. Dis. 11:802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668-2680. [DOI] [PubMed] [Google Scholar]
- 22.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougal, L. K., F. C. Tenover, L. N. Lee, J. K. Rasheed, J. E. Patterson, J. H. Jorgensen, and D. J. LeBlanc. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEllistrem, M. C., J. M. Adams, K. Shutt, L. T. Sanza, R. R. Facklam, C. G. Whitney, J. H. Jorgensen, and L. H. Harrison. 2005. Erythromycin-nonsusceptible Streptococcus pneumoniae in children, 1999-2001. Emerg. Infect. Dis. 11:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGee, L., K. P. Klugman, A. Wasas, T. Capper, and A. Brink. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mingoia, M., M. Vecchi, I. Cochetti, E. Tili, L. A. Vitali, A. Manzin, P. E. Varaldo, and M. P. Montanari. 2007. Composite structure of Streptococcus pneumoniae containing the erythromycin efflux resistance gene mefI and the chloramphenicol resistance gene catQ. Antimicrob. Agents Chemother. 51:3983-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Trallero, E., J. M. Marimon, M. Ercibengoa, M. J. Gimenez, P. Coronel, and L. Aguilar. 2007. Antimicrobial susceptibilities of amoxycillin-non-susceptible and susceptible isolates among penicillin-non-susceptible Streptococcus pneumoniae. Clin. Microbiol. Infect. 13:937-940. [DOI] [PubMed] [Google Scholar]
- 29.Rikitomi, N., P. S. Sow, K. Watanabe, D. S. Nunez, G. Martinez, and T. Nagatake. 1996. Rapid increase of pneumococcal resistance to beta-lactam and other antibiotics in isolates from the respiratory tract (Nagasaki, Japan: 1975-1994). Microbiol. Immunol. 40:899-905. [DOI] [PubMed] [Google Scholar]
- 30.Roche, P. W., V. L. Krause, M. Bartlett, D. Coleman, H. Cook, C. Davis, J. E. Fielding, R. Holland, C. Giele, R. Gilmour, R. Kampen, M. Brown, L. Gilbert, G. Hogg, D. Murphy, and a Pneumococcal Working Party of the Communicable Diseases Network. 2006. Invasive pneumococcal disease in Australia, 2004. Commun. Dis. Intell. 30:80-92. [PubMed] [Google Scholar]
- 31.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in Staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 32.Santagati, M., F. Iannelli, C. Cascone, F. Campanile, M. R. Oggioni, S. Stefani, and G. Pozzi. 2003. The novel conjugative transposon tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9:243-247. [DOI] [PubMed] [Google Scholar]
- 33.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schito, G. C. 2004. Resistance trends in Streptococcus pneumoniae (PROTEKT years 1-3 [1999-2002]). J. Chemother. 16(Suppl. 6):19-33. [DOI] [PubMed] [Google Scholar]
- 35.Schito, G. C., and D. Felmingham. 2005. Susceptibility of Streptococcus pneumoniae to penicillin, azithromycin and telithromycin (PROTEKT 1999-2003). Int. J. Antimicrob. Agents 26:479-485. [DOI] [PubMed] [Google Scholar]
- 36.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 37.Zeng, X., F. Kong, H. Wang, A. Darbar, and G. L. Gilbert. 2006. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob. Agents Chemother. 50:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]