Abstract
In the present study, we successfully cloned a 21-kb DNA fragment containing a d-cycloserine (DCS) biosynthetic gene cluster from a DCS-producing Streptomyces lavendulae strain, ATCC 11924. The putative gene cluster consists of 10 open reading frames (ORFs), designated dcsA to dcsJ. This cluster includes two ORFs encoding d-alanyl-d-alanine ligase (dcsI) and a putative membrane protein (dcsJ) as the self-resistance determinants of the producer organism, indicated by our previous work. When the 10 ORFs were introduced into DCS-nonproducing Streptomyces lividans 66 as a heterologous host cell, the transformant acquired DCS productivity. This reveals that the introduced genes are responsible for the biosynthesis of DCS. As anticipated, the disruption of dcsG, seen in the DCS biosynthetic gene cluster, made it possible for the strain ATCC 11924 to lose its DCS production. We here propose the DCS biosynthetic pathway. First, l-serine is O acetylated by a dcsE-encoded enzyme homologous to homoserine O-acetyltransferase. Second, O-acetyl-l-serine accepts hydroxyurea via an O-acetylserine sulfhydrylase homolog (dcsD product) and forms O-ureido-l-serine. The hydroxyurea must be supplied by the catalysis of a dcsB-encoded arginase homolog using the l-arginine derivative, NG-hydroxy-l-arginine. The resulting O-ureido-l-serine is then racemized to O-ureido-d-serine by a homolog of diaminopimelate epimerase. Finally, O-ureido-d-serine is cyclized to form DCS with the release of ammonia and carbon dioxide. The cyclization must be done by the dcsG or dcsH product, which belongs to the ATP-grasp fold family of protein.
The soil-dwelling genus Streptomyces undergoes a complex morphological differentiation and produces an enormous variety of bioactive secondary metabolites. Because they include clinically useful antibiotics and immunosuppressants, the genus Streptomyces occupies an important position as an industrial microorganism.
d-Cycloserine (DCS), a cyclic structural analogue of d-alanine, is produced by “Streptomyces garyphalus” and Streptomyces lavendulae. This antibiotic is used as an antitubercular agent (21). Since DCS is similar to d-alanine, it prevents the action of both alanine racemase and d-alanyl-d-alanine ligase, which are necessary for the biosynthesis of a bacterial cell wall. Thus, DCS functions as an inhibitor of bacterial cell wall biosynthesis (16, 19). Although the structure of DCS is very simple, the biosynthetic genes for DCS have never been cloned until now.
In general, antibiotic biosynthetic genes form a cluster and are adjacent to their self-resistance genes. We have previously cloned a gene (orfB) that confers resistance to DCS on Streptomyces lividans and Escherichia coli from DCS-producing S. garyphalus (CSH) 5-12 (17). The sequence analysis suggests that the gene may encode a membrane protein that is necessary for the excretion of the DCS outside the cell. We have found that the same gene is also present in S. lavendulae ATCC 25233 (20). We have also previously demonstrated that d-alanyl-d-alanine ligase, which is a target enzyme of DCS, functions as a self-resistance determinant in S. lavendulae ATCC 25233 (20). Interestingly, the gene encoding d-alanyl-d-alanine ligase, designated ddlS, was located just upstream of the putative membrane protein gene, orfB (20). Although the self-resistance genes in the DCS producers have been thoroughly analyzed, no attempt to clone the DCS biosynthetic genes has been carried out yet.
To clone the biosynthetic genes for DCS from another DCS-producing S. lavendulae strain, ATCC 11924, we first investigated whether this strain harbors orfB and ddlS. Since both genes were found to be conserved, the flanking region was cloned from the chromosomal DNA. Here, we hypothesize about the biosynthetic pathway for DCS on the basis of our present data and previous studies by another research group (23, 24). The present study is the first report identifying the biosynthetic gene cluster for DCS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Streptomyces strains were grown at 28°C on an FB agar medium (9). To select objective clones, appropriate antibiotics were added to the medium (14). S. lividans 66 was grown at 28°C in a yeast extract-malt extract liquid medium containing 0.5% glycine for the preparation of protoplasts (14). To produce DCS, Streptomyces strains were cultured at 28°C in liquid medium B (15). Plasmids pSET152 and pKC1132 (2) were used for gene integration and gene disruption, respectively. A Streptomyces expression vector, pIJ8600 (3), which contains a tipA promoter (18), was used for the complementation experiment. E. coli DH5α and plasmids pUC19, pTA2 (Toyobo), and pBluescript II SK(+) (Stratagene) were used for DNA cloning and sequencing. E. coli XL1-Blue MRA(P2) and the lambda FIX II vector (Stratagene) were used for genomic library construction. E. coli ET12567/pUZ8002 was used as a transient host for E. coli-Streptomyces conjugation (5). E. coli BL21(DE3) and a plasmid, pET-28a(+) (Novagen), were used for protein expression. All E. coli strains were cultivated at 37°C or 28°C in Luria broth or on Luria agar supplemented with the appropriate antibiotics when necessary (14, 22).
DNA manipulations.
The genomic DNA of Streptomyces was isolated following a standard protocol (14). The phage DNA was isolated according to a method described elsewhere (22). The plasmid DNA in E. coli was isolated using the Wizard Plus Minipreps DNA purification system (Promega).
Hybridizations.
Southern and plaque hybridizations were performed using a Hybond-N+ membrane (GE Healthcare). Probe labeling, hybridization, and detection were performed with the Alkphos direct labeling and detection System (GE Healthcare) according to the protocol supplied by the manufacturer.
Construction of genomic library.
Genomic DNA of S. lavendulae ATCC 11924 was partially digested with Sau3AI following purification by phenol-chloroform extraction and 2-propanol precipitation. After DNA fragments with a size of 10 to 20 kb were gel purified, a partial fill-in reaction was carried out. Subsequently, the DNA fragments were purified by phenol-chloroform extraction and ethanol precipitation, followed by ligation to the lambda FIX II vector (Stratagene). In vitro packaging was performed using the Gigapack III Gold packaging extract (Stratagene) to generate the genomic library of S. lavendulae ATCC 11924.
Cloning of DNA fragments containing DCS biosynthetic genes.
Using the PCR-amplified ddlS gene of S. lavendulae ATCC 25233 as a probe DNA (20), a genomic library of S. lavendulae ATCC 11924 was screened by the plaque hybridization technique. After the phage DNA was isolated from a positive plaque, an approximately 12-kb DNA fragment (called DCS1) derived from S. lavendulae ATCC 11924 was removed by digestion with XbaI and subcloned into the same site of pUC19 to generate pUC19/DCS1. Since the DCS biosynthetic gene cluster seemed to be incomplete according to the sequence analysis, a DNA fragment adjacent to DCS1 was cloned from the ATCC 11924 genomic library using chromosomal walking. The cloned fragment (called DCS2), with a size of 10 kb, was subcloned into pUC19 to yield pUC19/DCS2. The entire DCS biosynthetic gene cluster was obtained by the sequence analysis of DCS1 and DCS2.
DNA sequencing and analysis.
DNA sequencing was performed using ABI Prism 310 and Avant 3100 sequencers (Applied Biosystems) using the Big Dye terminator cycle sequencing ready reaction kit, ver.1.1. Genetic analysis was performed using GENETYX ver.7 for Windows (Software Development). The open reading frames (ORFs) were predicted using FramePlot ver.2.3.2 (11). Homology searches were conducted with the BLAST algorithm (1).
Heterologous expression of DCS biosynthetic gene cluster in S. lividans.
The putative DCS biosynthetic gene cluster, which consists of 10 genes (dcsA to dcsJ), including two self-resistance genes, was amplified by PCR using PrimeSTAR GXL DNA polymerase (Takara). The primers used were a 5′-phosphorylated sense primer (5′-TCTAGAATGGCGATCATCTGGGCCTGC-3′; XbaI site underlined) and a 5′-phosphorylated antisense primer (5′-GATATCAAGACGGCGGTGCGTGAGGTG-3′; EcoRV site underlined). PCR was carried out under the following conditions: denaturing for 5 min at 98°C, 30 cycles of 10 s at 98°C and 10 min at 70°C, and finally an extension period at 70°C for 15 min. The amplified fragment was cloned into SmaI-digested pUC19 to generate pUC19/dcs. After the fragment was confirmed by DNA sequencing, the DCS biosynthetic gene cluster was cut off from pUC19/dcs by digestion with XbaI and EcoRV and inserted into the same sites of pSET152 to yield pSET152/dcs. The chimeric plasmid was introduced into S. lividans 66 by protoplast transformation (14). For DCS production, S. lividans cells transformed with pSET152 or pSET152/dcs were cultured in medium B at 28°C for 48 h.
Construction of dcsG mutant via chromosomal gene disruption.
A 5.6-kb DNA fragment was cut off from pUC19/DCS1 by digestion with PvuII and ligated to pUC19, which was previously digested with PstI/XbaI and blunt ended by using a DNA blunting kit (Takara), to yield pUC19/DCS11. The internal region of dcsG (636 bp) was removed from pUC19/DCS11 by digestion with SalI, followed by self-ligation to give pUC19/ΔdcsG. A 5.0-kb fragment containing the disrupted dcsG gene was removed from pUC19/ΔdcsG by digestion with EcoRI/HindIII and inserted into the same sites of pKC1132 to generate pKC/ΔdcsG. pKC/ΔdcsG was introduced into methylation-deficient E. coli ET12567/pUZ8002, and the resulting transformant was used for conjugal transfer to S. lavendulae ATCC 11924. Conjugal transfer from E. coli to Streptomyces was carried out according to a method described elsewhere (14). An apramycin-resistant conjugant, obtained by single-crossover homologous recombination, was further cultured on medium without apramycin for several rounds. An apramycin-sensitive dcsG mutant, which contains the in-frame-deleted dcsG generated by double-crossover homologous recombination, was obtained and designated the ΔdcsG mutant.
Complementation of disrupted mutant with dcsG.
The structural gene of dcsG was amplified by PCR using a sense primer, 5′-CATATGGGCATCCTCGCCTTGGTCACCG-3′ (NdeI site underlined), and an antisense primer, 5′-TCTAGACTAGGGCTTGAGCCGTTCGGCCAG-3′ (XbaI site underlined). The amplified gene was cloned into pTA2 to give pTA2/dcsG. After confirmation of its DNA sequence, dscG was removed from pTA2/dcsG by digestion with NdeI/XbaI and inserted into the same sites of pIJ8600 (3), a Streptomyces expression vector, to yield pIJ8600/dcsG. In this construction, dcsG is under the control of the tipA promoter, and its expression is induced by the addition of thiostrepton (18). pIJ8600/dcsG was introduced into the ΔdcsG strain using conjugal transfer.
Assay of DCS production.
DCS production was analyzed by high-performance liquid chromatography (HPLC) (15). After the culture supernatant fluid was filtrated with a membrane filter (0.2-μm pore size; Advantec), aliquots (10 μl) were applied in HPLC (Senshu Pak; Senshu Scientific Co., Ltd.) with 10 mM ammonium acetate (pH 5.0 adjusted with acetate) for elution and detected at 210 nm by comparison with authentic DCS (Wako). Alternatively, the amount of DCS contained in the culture filtrate was colorimetrically estimated with a sodium nitroprusside reagent according to a previously described method (12).
MALDI-TOF mass analysis.
The eluate from HPLC was analyzed using matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry. MALDI-TOF mass analysis was performed using a Voyager RP-3 mass spectrometer (PerSeptive Biosystems) operated in the reflector mode. Mass spectra were obtained under the following conditions: accelerating voltage (ACV), 25 kV; grid voltage, 57.5% of ACV; guide wire voltage, 0.05% of ACV; pulse delay time, 100 ns. CHCA was used for the matrix.
Expression in E. coli and purification of DcsB and DcsE proteins.
The gene dcsB was amplified by PCR using a sense primer, 5′-CATATGATTGATCTGATCGTCTCCCAG-3′ (NdeI site underlined), and an antisense primer, 5′-CTCGAGTCAGGGCCGTGCGGCGGC-3′ (XhoI site underlined). The amplified gene was cloned into pTA2 to generate pTA2/dcsB. After the nucleotide sequence of dcsB was analyzed for confirmation, the dcsB fragment was cut off from pTA2/dcsB by digestion with NdeI/XhoI and inserted into the same sites of pET-28a(+) to yield pET-28a/dcsB. E. coli BL21(DE3) harboring pET-28a/dcsB was grown in Luria broth at 28°C. The expression of dcsB was induced by the addition of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation and disrupted by sonication. N-terminal histidine-tagged DcsB was purified by nickel affinity chromatography using His-Bind resin (Novagen) according to the supplier's instruction manual.
The gene dcsE was amplified by PCR using a sense primer, 5′-GGAATTCCATATGAGGGAATTCATACCCCCG-3′ (NdeI site underlined), and an antisense primer, 5′-CCGCTCGAGTCAGGCGACGATCGCCAGGAACTT-3′ (XhoI site underlined). The amplified gene was digested with NdeI/XhoI, gel purified, and inserted into pET-28a(+) to give pET-28a/dcsE. E. coli BL21(DE3) harboring pET-28a/dcsE was grown in Luria broth at 28°C. Expression of dcsE was induced by the addition of 1 mM IPTG. N-terminal histidine-tagged DcsE was purified by His-Bind resin as described above.
The dcsB and dcsE gene products were purified to near-homogeneity, which was judged by SDS-PAGE analysis. Each protein concentration was determined by measuring the absorbance at 280 nm using the molar extinction coefficients 34,500 M−1 cm−1 and 38,700 M−1 cm−1, respectively.
Analysis of enzymatic activity of DcsE.
The reaction mixture (200 μl), consisting of 4 mM Tris-HCl (pH 7.6), 40 mM NaCl, 1 mM dithiothreitol (DTT), 0.2 mM EDTA, 1 mM l-serine, 1 mM acetyl-coenzyme A (CoA), and 15 μM DcsE, was incubated at 37°C for 2 h. As a control, the same reaction mixture without DcsE was also used. The formation of enzymatic product was analyzed by thin-layer chromatography (TLC) using silica gel 60 F254 (Merck). The solvent for the chromatogram was 2-butanone-pyridine-water (20:5:8). l-Serine and O-acetyl-l-serine were visualized by the acid ninhydrin regent as described previously (26).
Analysis of enzymatic activity of DcsB.
The reaction mixture (90 μl), consisting of 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 1 mM NG-hydroxy-l-arginine or 1 mM l-arginine, was preincubated at 37°C for 10 min. The reaction was initiated by the addition of 10 μl of DcsB at given concentrations. After incubation at 37°C for 10 min, the reaction was terminated by the addition of 100 μl of glacial acetic acid. The amount of ornithine liberated by the catalysis of DcsB was determined by the previously described method (4).
Effect of nitric oxide synthase (NOS) inhibitor on DCS productivity.
S. lavendulae ATCC 11924 was grown in medium B at 28°C for 24 h in the presence or absence of 1 mM nitro-l-arginine methyl ester, known as a NOS inhibitor (13). The inhibitor was added to the medium at the time of inoculation. The amount of DCS produced was determined colorimetrically as described above. The data were confirmed by carrying out three independent experiments.
Nucleotide sequence accession number.
The DNA sequence data for the entire DCS biosynthetic gene cluster of S. lavendulae ATCC 11924 were deposited in the DDBJ database under accession number AB516431.
RESULTS AND DISCUSSION
Cloning of a DCS biosynthetic gene cluster from S. lavendulae ATCC 11924.
We previously found that a DCS resistance gene (orfB), which encodes a putative membrane protein responsible for the excretion of DCS, is harbored in DCS-producing S. garyphalus (CSH) 5-12 (17) and S. lavendulae ATCC 25233 (20). Furthermore, we have shown that a gene (ddlS) encoding d-alanyl-d-alanine ligase, located just upstream of orfB, also plays a role as a self-resistance determinant for DCS in the strain ATCC 25233 (20). A gene corresponding to ddlS is also just upstream of orfB in S. garyphalus but is not complete (17). These results suggest that the chromosomal locus responsible for DCS resistance is conserved between two DCS producers, suggesting the possibility that the biosynthetic genes for DCS are present in close proximity to these genes. To evaluate this possibility, testing by the Southern hybridization technique was undertaken using orfB and ddlS as the probes to determine whether another DCS-producing S. lavendulae strain, ATCC 11924, also harbors these two genes and to confirm that these genes are conserved (data not shown).
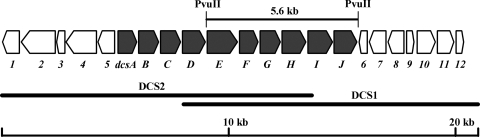
We cloned DNA regions flanking the two DCS resistance genes from the genomic library of S. lavendulae ATCC 11924. By cloning of two successive DNA fragments (DCS1 and DCS2) (Fig. 1), the nucleotide sequences, of approximately 21 kb, were determined. FramePlot analysis (11) suggests the presence of 22 ORFs in the cloned DNA fragment (Fig. 1). Table 1 summarizes the characteristics of ORFs seen in the cloned region. Considering the putative functions of these ORFs and their transcriptional direction, 10 genes, designated dcsA to dcsJ, may be responsible for the DCS biosynthesis (Fig. 1). This hypothesis was confirmed by heterologous expression in S. lividans 66 as a DCS-nonproducing host cell. Computer analysis suggested that the expression of the gene cluster from dcsA to dcsJ may be controlled by only one putative promoter just upstream of dcsA. But at this time, we cannot neglect the possibility that another promoter might be in the intergenic regions. Judging from the high sequence similarity (Table 1), dcsI and dcsJ, contained in the cluster, correspond to ddlS and orfB, respectively. These genes must function as the self-resistance determinants for DCS-producing microorganisms.
FIG. 1.
Gene organization in a 21-kb DNA fragment from S. lavendulae ATCC 11924. The entire nucleotide sequences were determined on the basis of information from two successive clones, DCS1 and DCS2. ORFs were predicted by FramePlot analysis. The 10 genes (dcsA to dcsJ) involved in a putative DCS biosynthetic cluster are indicated by shaded arrows. A bar drawn above ORFs indicates a 5.6-kb DNA fragment used for the gene disruption analysis of dcsG.
TABLE 1.
Summary of ORFs in the cloned DNA
| Protein | No. of aaa | Proposed function | Sequence homolog (protein name, origin) | Identity (%) | Protein accession no. |
|---|---|---|---|---|---|
| ORF1 | 247 | Glycine-sarcosine methyltransferase | SIAM614_20226, Stappia aggregata IAM12614 | 37 | EAV43187 |
| ORF2 | 515 | Peptide synthase | Ppro3117, Pelobacter propionicus DSM2397 | 31 | ABL00711 |
| ORF3 | 156 | Cyanate hydratase | MAB0054c, Mycobacterium abscessus | 77 | CAM60154 |
| ORF4 | 273 | Unknown | SSCG01501, Streptomyces clavuligerus ATCC 27064 | 39 | EDY48220 |
| ORF5 | 459 | Proline permease | SAV5161, Streptomyces avermitilis MA-4680 | 61 | BAC72873 |
| DcsA | 265 | Unknown | Csal1088, Chromohalobacter salexigens DSM3043 | 50 | ABE58444 |
| DcsB | 273 | Arginase | RocF, Erwinia tasmaniensis | 43 | CAO97798 |
| DcsC | 287 | Diaminopimelate epimerase | DapF, Xanthomonas campestris pv. campestris | 60 | AAM42769 |
| DcsD | 324 | O-Acetylserine sulfhydrylase | Csal1086, Chromohalobacter salexigens DSM3043 | 73 | ABE58442 |
| DcsE | 374 | Homoserine O-acetyltransferase | Smal2708, Stenotrophomonas maltophila R551-3 | 63 | ACF52408 |
| DcsF | 267 | Short-chain dehydrogenase | XCV1542, Xanthomonas campestris pv. vesicatoria | 59 | CAJ23174 |
| DcsG | 299 | Unknown | Csal1083, Chromohalobacter salexigens DSM3043 | 39 | ABE58440 |
| DcsH | 337 | Unknown | Haur2023, Herpetosiphon aurantiacus ATCC 23779 | 30 | ABX04666 |
| DcsI | 345 | d-alanyl-d-alanine ligase | DdlS, Streptomyces lavendulae ATCC 25233 | 93 | BAD60786 |
| DcsJ | 299 | Membrane protein | OrfB, Streptomyces garyphalus (CSH) 5-12 | 97 | BAD05865 |
| ORF6 | 147 | Acetyltransferase | OrfIII, Streptomyces lavendulae ATCC 25233 | 89 | BAD60788 |
| ORF7 | 261 | Aminoglycoside phosphotransferase | Sare1186, Salinispora arenicola CNS-205 | 40 | ABV97094 |
| ORF8 | 217 | Methyltransferase | OrfD, Streptomyces garyphalus (CSH) 5-12 | 93 | BAD05867 |
| ORF9 | 85 | Unknown | OrfE, Streptomyces garyphalus (CSH) 5-12 | 94 | BAD05868 |
| ORF10 | 260 | Lipoprotein | OrfF, Streptomyces garyphalus (CSH) 5-12 | 88 | BAD05869 |
| ORF11 | 242 | Transcriptional regulator | SSAG06618, Streptomyces sp. Mg1 | 49 | EDX26827 |
| ORF12 | 197 | Unknown | SSEG02386, “Streptomyces sviceus” ATCC 29083 | 47 | EDY55086 |
aa, amino acids.
Heterologous expression of DCS biosynthetic gene cluster.
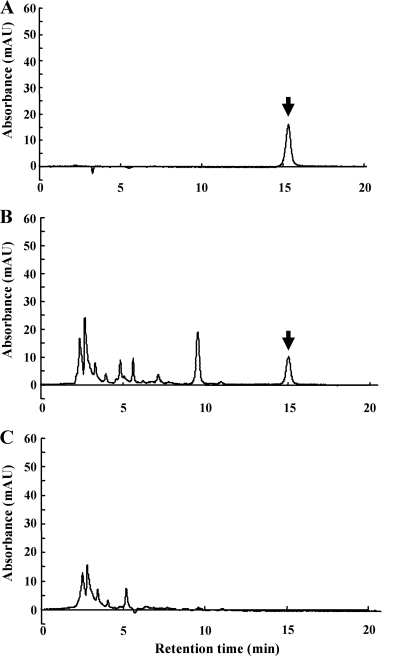
To clarify whether the putative gene cluster consisting of the 10 genes is responsible for DCS biosynthesis, the DNA fragment, including the region from dcsA to dcsJ, was PCR amplified, subcloned into an integrating vector, pSET152 (2), and introduced into S. lividans 66 by the protoplast transformation method. To facilitate its expression in the regenerated cells, a putative promoter located upstream of dcsA was included in the process of PCR amplification. S. lividans 66 cells transformed with pSET152/dcs, which contains the cluster integrated in the chromosome, clearly produced DCS, while the same host transformed with pSET152 did not (Fig. 2). The molecular mass of DCS was also confirmed by MALDI-TOF mass analysis (data not shown), indicating that the cluster is actually responsible for DCS biosynthesis.
FIG. 2.
Heterologous expression of a putative DCS biosynthetic gene cluster. S. lividans 66 transformed with pSET152/dcs or pSET152 was cultivated at 28°C in liquid medium B for 48 h. The amount of DCS produced in the culture supernatant of the transformant was analyzed by HPLC. (A) Authentic DCS. (B) S. lividans 66 transformed with pSET152/dcs. (C) S. lividans 66 transformed with pSET152. An arrow indicates a peak corresponding to DCS.
Loss of DCS production by dcsG disruption and rescue by its complementation.
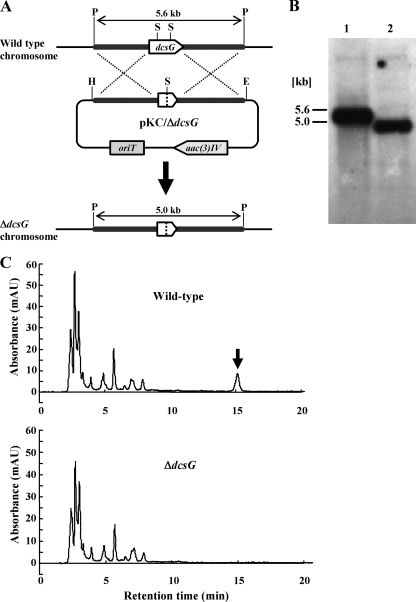
To clarify further that the gene cluster is responsible for the biosynthesis of DCS, the dcsG gene located chromosomally in S. lavendulae ATCC 11924 was disrupted by the homologous recombination method (Fig. 3A). The function of the dcsG product could not be annotated by the homology search, but it seemed to catalyze the final step of DCS biosynthesis, because DcsG has the ATP-grasp fold motif. Thus, we selected dcsG as a candidate for gene disruption analysis. Since dcsG seems to be transcribed polycistronically with other genes, the in-frame deletion method (14) was adopted for the creation of the dcsG mutant to avoid the polar effect. Through two steps of homologous recombination, a desired in-frame deletion mutant, designated the ΔdcsG strain, was obtained. In this experiment, the authenticity of the ΔdcsG mutant was confirmed by Southern blot analysis. As estimated in Fig. 3A, by Southern analysis, 5.6-kb and 5.0-kb fragments were detected in the wild-type and the ΔdcsG mutant, respectively. (Fig. 3B).
FIG. 3.
Strategy for creation of a dcsG-disrupted mutant by an in-frame deletion method and analysis of DCS productivity. (A) Schematic representation of the construction of a dcsG mutant via chromosomal gene disruption. The restriction sites are as follows: E, EcoRI, H, HindIII; P, PvuII; S, SalI. (B) Southern blot analysis of wild-type and ΔdcsG genomes. The probe used was a PCR-amplified dcsG gene. Lane 1, PvuII-digested wild-type genome; lane 2, PvuII-digested ΔdcsG genome. (C) DCS production in the ΔdcsG mutant. The wild type and ΔdcsG mutant of S. lavendulae ATCC 11924 were cultured at 28°C in liquid medium B for 48 h. The detection of DCS produced in the culture supernatant was confirmed by HPLC analysis. Upper chart, wild type; lower chart, ΔdcsG mutant. An arrow shows a peak for DCS.
As expected, an in-frame deletion mutant, the ΔdcsG strain, did not produce DCS, whereas the wild-type strain clearly produced it (Fig. 3C). To determine whether this disappearance of DCS productivity is due to the loss of dcsG function, the gene was complemented in trans using an expression vector, pIJ8600. The dcsG gene, located under the control of the tipA promoter in pIJ8600, was introduced into the ΔdcsG mutant cell. The complemented cell restored the production of DCS in the presence but not in the absence of thiostrepton (20 μg/ml) as an inducer (data not shown). These results show that the loss of DCS production in the ΔdcsG mutant is due to the disruption of dcsG and suggest that dcsG is necessary for DCS biosynthesis.
Putative pathway for DCS biosynthesis.
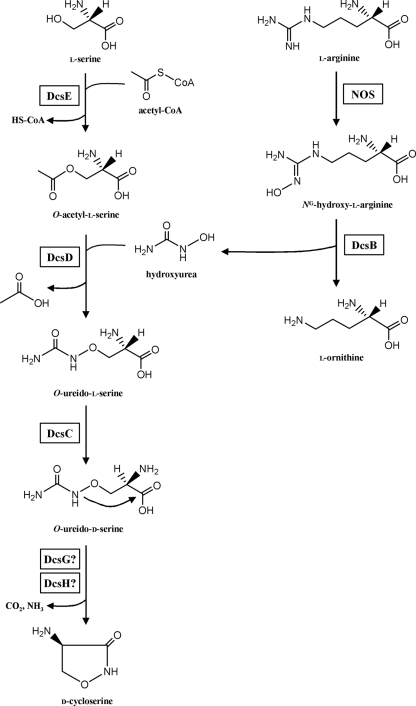
Based on the sequence information of ORFs found in the DCS biosynthetic gene cluster (encoding DcsA to DcsJ; Table 1) and previous reports (23, 24) by another group, we propose a putative biosynthetic pathway for DCS in S. lavendulae ATCC 11924, as shown in Fig. 4.
FIG. 4.
Predicted biosynthetic pathway of DCS in S. lavendulae ATCC 11924.
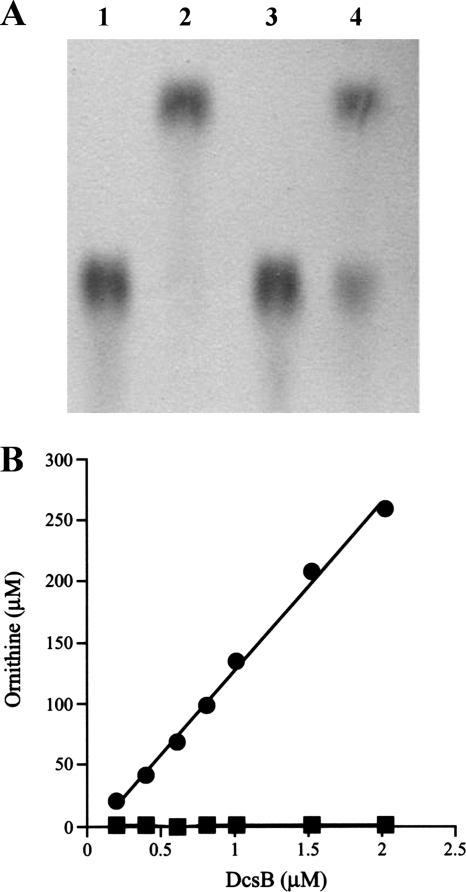
l-Serine and hydroxyurea have been proposed previously to be components of DCS (23, 25). In order for l-serine to react with hydroxyurea, it must first be activated. Indeed, in S. garyphalus, O-acetyl-l-serine has been suggested to be the activated form of l-serine in DCS biosynthesis (24). The O acetylation of l-serine may be catalyzed by DcsE (Fig. 4), which is a homolog of homoserine O-acetyltransferase (Table 1), as shown in a report that a homolog of homoserine O-acetyltransferase in Aspergillus nidulans can contribute to the formation of O-acetyl-l-serine (7). In bacteria, O-acetyl-l-serine is used in cysteine biosynthesis and its formation is generally catalyzed by serine O-acetyltransferase. To investigate the function of DcsE, the protein was expressed in E. coli and purified. As shown in Fig. 5A, the chromatographic profile in TLC indicates that O-acetyl-l-serine is formed when l-serine is incubated with DcsE in the presence of acetyl-CoA. This result demonstrates that DcsE functions as a serine O-acetyltransferase, suggesting that DcsE contributes to the formation of O-acetyl-l-serine in DCS biosynthesis.
FIG. 5.
Analysis of the recombinant DcsE and DcsB proteins. (A) Serine O-acetyltransferase activity of DcsE. After the reaction mixture containing l-serine (1 mM) was incubated with or without DcsE (15 μM) in the presence of acetyl-CoA (1 mM), an aliquot of the mixture was analyzed by TLC. Lane 1, l-serine; 2, O-acetyl-l-serine; 3, the reaction mixture without DcsE; 4, the reaction mixture with DcsE. (B) Hydrolysis of NG-hydroxy-l-arginine by DcsB. NG-hydroxy-l-arginine (1 mM) or l-arginine (1 mM) was incubated with the given concentration of DcsB for 10 min at 37°C. After the incubation, the amount of ornithine liberated by the catalysis of DcsB was determined. Closed circles or squares indicate the data obtained with NG-hydroxy-l-arginine or l-arginine, respectively.
O-Acetyl-l-serine is suggested to be converted to O-ureido-l-serine when incubated with hydroxyurea (24). Although we have not yet obtained the experimental evidence, this reaction may be catalyzed by a homolog of O-acetylserine sulfhydrylase encoded by dcsD (Table 1 and Fig. 4), judging from a report that O-ureido-l-serine is generated by the catalysis of O-acetylserine sulfhydrylase when O-acetyl-l-serine and hydroxyurea were used as the substrates (10). Generally, the O-acetylserine sulfhydrylase catalyzes the formation of l-cysteine using O-acetyl-l-serine and H2S as the substrates. To investigate the function of DcsD in vitro, the expression of recombinant DcsD in E. coli is now under way.
Hydroxyurea, a supplier of the nitrogen in the isoxazolidone structure of DCS, was suggested to be derived from l-arginine (23). It is thought that the DcsB protein, which is similar to arginase (Table 1), may be involved in the formation of hydroxyurea from l-arginine (Fig. 4) because arginase hydrolyzes l-arginine to yield urea and l-ornithine. There are two possibilities for the generation of hydroxyurea: (i) DcsB forms urea and l-ornithine by hydrolyzing l-arginine, and the resulting urea is converted to hydroxyurea; (ii) after l-arginine is first hydroxylated to NG-hydroxy-l-arginine, the hydroxylated substance is hydrolyzed by DcsB to form hydroxyurea and l-ornithine. The former case may be less likely, because urea did not stimulate the production of DCS in the washed cells of S. garyphalus but l-arginine did (23). Therefore, the latter case is likely to be involved in hydroxyurea formation. To support this hypothesis, we tested whether the recombinant DcsB protein, which was produced in E. coli and purified, hydrolyzes NG-hydroxy-l-arginine. Figure 5B shows that DcsB actually hydrolyzes NG-hydroxy-l-arginine. However, l-arginine was not a substrate of the enzyme. This result strongly suggests that hydroxyurea is formed from NG-hydroxy-l-arginine by the enzymatic reaction of DcsB (Fig. 4). This finding is surprising, because NG-hydroxy-l-arginine is known as an inhibitor of arginase (6). In the latter case, we hypothesize that NG-hydroxy-l-arginine is formed by the action of NOS from l-arginine (Fig. 4), because NOS is known to catalyze the generation of NO from l-arginine through forming NG-hydroxy-l-arginine as an intermediate. Indeed, we observed that 1 mM nitro-l-arginine methyl ester, a NOS inhibitor, prevents the production of DCS by S. lavendulae ATCC 11924 (75% ± 2.9% of the control). Since the candidate gene encoding NOS is not seen in the DCS biosynthetic gene cluster (Table 1), the corresponding gene may be located on another region of the chromosome. It has been reported that some Streptomyces strains possess the NOS gene (8, 13).
O-Ureido-l-serine, presumably generated by the catalysis of DcsD, should be racemized to its d form, O-ureido-d-serine (Fig. 4). A homolog of diaminopimelate epimerase encoded by dcsC (Table 1) could participate in this reaction. Although DcsC exhibits a similarity to diaminopimelate epimerase, which catalyzes the interconversion of l,l-diaminopimelate and its meso isomer, it could function as racemase when O-ureido-l-serine is used as a substrate. This hypothesis should be confirmed by in vitro analysis using the recombinant DcsC protein.
Finally, O-ureido-d-serine is cyclized to form DCS with the release of ammonia and carbon dioxide (Fig. 4). Indeed, when O-[14C]ureido-d-serine was incubated with the cell extract of S. garyphalus, the formation of DCS was observed with the release of 14CO2 (24). The cyclization of O-ureido-d-serine could be catalyzed by DcsG or DcsH (Fig. 4), because both proteins have an ATP-grasp fold motif and belong to the ATP-grasp fold family of proteins, which activate the substrate by phosphorylating its carboxyl group using ATP and form a C-N bond. This hypothesis may be supported by an observation by another research group that ATP is required to form DCS from O-ureido-l-serine (24). At this moment, DcsG may be a more potent candidate than DcsH since it is necessary for DCS biosynthesis as revealed by gene disruption analysis. The question of when the hydrolysis of the carbamoyl derivative occurs remains to be answered. It may occur at the time of cyclization catalyzed by DcsG or DcsH or after it. In vitro analysis using recombinant DcsG and DcsH with the substrate O-ureido-d-serine will clarify which protein catalyzes the cyclization of O-ureido-d-serine and when the hydrolysis of carbamoyl derivative occurs. We are currently attempting to express DcsG and DcsH in E. coli.
Based on our proposal (Fig. 4), the dcsA and dcsF products might not be involved in DCS biosynthesis. To investigate this possibility, the creation of in-frame deletion mutants for both genes is now ongoing. Furthermore, the creation of the dcsB, dcsC, dcsD, dcsE, and dcsH in-frame deletion mutants is also in progress. These mutants will contribute to a near-perfect understanding of the DCS biosynthetic pathway.
Acknowledgments
We thank Haruyasu Kinashi, Hiroshima University, for providing the vector pIJ8600. DNA sequence determination was carried out with the kind cooperation of the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University.
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 3.Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp level. Microbiology 149:2173-2182. [DOI] [PubMed] [Google Scholar]
- 4.Chinard, F. P. 1952. Photometric estimation of proline and ornithine. J. Biol. Chem. 199:91-95. [PubMed] [Google Scholar]
- 5.Choi, S. U., C. K. Lee, Y. I. Hwang, H. Kinoshita, and T. Nihira. 2004. Intergeneric conjugal transfer of plasmid DNA from Escherichia coli to Kitasatospora setae, a bafilomycin B1 producer. Arch. Microbiol. 181:294-298. [DOI] [PubMed] [Google Scholar]
- 6.Colleluori, D. M., and D. E. Ash. 2001. Classical and slow-binding inhibitors of human type II arginase. Biochemistry 40:9356-9362. [DOI] [PubMed] [Google Scholar]
- 7.Grynberg, M., J. Topczewski, A. Godzik, and A. Paszewski. 2000. The Aspergillus nidulans cysA gene encodes a novel type of serine O-acetyltransferase which is homologous to homoserine O-acetyltransferase. Microbiology 146:2695-2703. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda, K., T. Masujima, K. Suzuki, and M. Sugiyama. 1996. Cloning and sequence analysis of the highly expressed melanin-synthesizing gene operon from Streptomyces castaneoglobisporus. Appl. Microbiol. Biotechnol. 45:80-85. [DOI] [PubMed] [Google Scholar]
- 10.Ikegami, F., M. Mizuno, M. Kihara, and I. Murakoshi. 1990. Enzymatic synthesis of the thyrotoxic amino acid mimosine by cysteine synthase. Phytochemistry 29:3461-3465. [Google Scholar]
- 11.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 12.Jones, L. R. 1956. Colorimetric determination of cycloserine, a new antibiotic. Anal. Chem. 28:39-41. [Google Scholar]
- 13.Kers, J. A., M. J. Wach, S. B. Krasnoff, J. Widom, K. D. Cameron, R. A. Bukhalid, D. M. Gibson, B. R. Crane, and R. Loria. 2004. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429:79-82. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces Genetics. The John Innes Foundation, Norwich, United Kingdom.
- 15.Kitani, S., Y. Yamada, and T. Nihira. 2001. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J. Bacteriol. 183:4357-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of D-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuo, H., T. Kumagai, K. Mori, and M. Sugiyama. 2003. Molecular cloning of a D-cycloserine resistance gene from D-cycloserine-producing Streptomyces garyphalus. J. Antibiot. 56:762-767. [DOI] [PubMed] [Google Scholar]
- 18.Murakami, T., T. G. Holt, and C. J. Thompson. 1989. Thiostrepton-induced gene expression in Streptomyces lividans. J. Bacteriol. 171:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuhaus, F. C., and J. L. Lynch. 1964. The enzymatic synthesis of D-alanyl-D-alanine. III. On the inhibition of D-alanyl-D-alanine synthetase by the antibiotic D-cycloserine. Biochemistry 3:471-480. [DOI] [PubMed] [Google Scholar]
- 20.Noda, M., Y. Kawahara, A. Ichikawa, Y. Matoba, H. Matsuo, D.-G. Lee, T. Kumagai, and M. Sugiyama. 2004. Self-protection mechanism in D-cycloserine-producing Streptomyces lavendulae: gene cloning, characterization, and kinetics of its alanine racemase and D-alanyl-D-alanine ligase, which are target enzymes of D-cycloserine. J. Biol. Chem. 279:46143-46152. [DOI] [PubMed] [Google Scholar]
- 21.Pinsker, K. L., and S. K. Koerner. 1976. Chemotherapy of tuberculosis. Am. J. Hosp. Pharm. 33:275-283. [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Svensson, M.-L., K. Valerie, and S. Gatenbeck. 1981. Hydroxyurea, a natural metabolite and an intermediate in D-cycloserine biosynthesis in Streptomyces garyphalus. Arch. Microbiol. 129:210-212. [Google Scholar]
- 24.Svensson, M.-L., and S. Gatenbeck. 1982. The pathway of D-cycloserine biosynthesis in Streptomyces garyphalus. Arch. Microbiol. 131:129-131. [Google Scholar]
- 25.Tanaka, N., and K. Sashikata. 1963. Biogenesis of D-4-amino-3-isoxazolidone and O-cabamyl-D-serine. J. Gen. Appl. Microbiol. 9:409-414. [Google Scholar]
- 26.Work, E. 1957. Reaction of ninhydrin in acid solution with straight-chain amino acids containing two amino groups and its application to the estimation of alpha epsilon-diaminopimelic acid. Biochem. J. 67:416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]