Abstract
R207910 (also known as TMC207) is an investigational drug currently in clinical studies for the treatment of multidrug-resistant (MDR) tuberculosis. It has a high degree of antimycobacterial activity and is equally effective against drug-susceptible and MDR Mycobacterium tuberculosis isolates. In the present study, we characterized the development of resistance to R207910 in vitro. Ninety-seven independent R207910-resistant mutants were selected from seven different clinical isolates of M. tuberculosis (three drug-susceptible and four MDR isolates) at 10×, 30×, and 100× the MIC. At a concentration of 0.3 mg/liter (10× the MIC), the mutation rates ranged from 4.7 × 10−7 to 8.9 × 10−9 mutations per cell per division, and at 1.0 mg/liter (30× the MIC) the mutation rate ranged from 3.9 × 10−8 to 2.4 × 10−9. No resistant mutants were obtained at 3 mg/liter (100× the MIC). The level of resistance ranged from 0.12 to 3.84 mg/liter for the mutants identified; these concentrations represent 4- to 128-fold increases in the MICs. For 53 of the resistant mutants, the atpE gene, which encodes a transmembrane and oligomeric C subunit of the ATP synthase and which was previously shown to be involved in resistance, was sequenced. For 15/53 mutants, five different point mutations resulting in five different amino acid substitutions were identified in the atpE gene. For 38/53 mutants, no atpE mutations were found and sequencing of the complete F0 ATP synthase operon (atpB, atpE, and atpF genes) and the F1 ATP synthase operon (atpH, atpA, atpG, atpD, and atpC genes) from three mutants revealed no mutations, indicating other, alternative resistance mechanisms. Competition assays showed no measurable reduction in the fitness of the mutants compared to that of the isogenic wild types.
Drug-susceptible (DS) tuberculosis (TB) is curable but requires a long treatment period and multidrug regimen (36). In light of the increasing prevalence of antibiotic resistance among Mycobacterium tuberculosis strains and the challenge with the effective control of the TB epidemic, there is an urgent need for new antituberculosis drugs. Furthermore, the drastic rise in the incidence of multidrug-resistant (MDR) strains (strains resistant to at least rifampin and isoniazid) and, now, extensively drug-resistant (XDR) strains (MDR strains as well as strains resistant to any fluoroquinolone and at least one of three injectable second-line drugs) further underlines this need (35). R207910 is a compound that belongs to the diarylquinolines. It has a high degree of specificity for mycobacteria and specifically targets the C subunit of ATP synthase in replicating as well as dormant mycobacteria (4, 17, 20). We have previously shown that R207910 is equally effective against drug-susceptible and MDR M. tuberculosis isolates (median MIC, 0.03 mg/liter) (4, 17). R207910 has a strong bactericidal effect in a mouse model, being more effective than rifampin when R207910 is given alone. Furthermore, the use of R207910 in combination with first-line or second-line drugs in the murine model accelerates the bactericidal effect (4, 21), supporting the potential for the use of R207910 to treat both drug-susceptible and MDR TB. With few to no adverse effects being seen in healthy individuals (4, 32), the compound has undergone a phase IIa clinical trial for the treatment of treatment-naïve patients with drug-susceptible TB (29), and initial results from phase II trials of the use of R207910 for the treatment of MDR TB have confirmed its significant bactericidal effect in patients (11).
M. tuberculosis develops antibiotic resistance exclusively through the acquisition of spontaneous chromosomal mutations (7). For most bacterial species, resistance-conferring mutations often confer a biological cost that presents a selective growth disadvantage relative to the growth capability of drug-susceptible isogenic strains in the absence of the drug (1-3). In M. tuberculosis, such fitness costs have previously been demonstrated for in vitro-selected rifampin-resistant mutants (5, 25), streptomycin-resistant mutants (7, 8, 15, 30), and isoniazid-resistant mutants (27). On the basis of the isolation in vitro of a limited number of mutants, mutations conferring resistance to R207910 were previously measured to occur at a frequency of 2 × 10−8, and resistance-conferring mutations were identified within the atpE gene (4, 26). R207190 binds to the C subunit of ATP synthase and potently inhibits its ATP-synthesizing activity in a highly stereospecific manner, with the less potent enantiomer having about a 10-fold lower inhibitory activity against ATP synthase (20). This suggests that R207190 has a high level of target selectivity. Furthermore, computational models suggest that R207910 blocks the transfer of hydrogen ions between the A and C subunits of ATP synthase (10). This blockage shuts the motor or rotational activity of ATP synthase, thereby inhibiting the activity of the enzyme and, concurrently, ATP production (10). The resistance-conferring mutations identified within the atpE gene suggest that resistance is conferred by preventing the compound from binding to the C subunit, thus maintaining H+ transfer and ATP production (10, 20).
The aim of the study described here was to further characterize the development of resistance to R207910 in vitro in M. tuberculosis with regard to mutation rates, mechanisms of resistance, and the potential impacts of these resistance mechanisms on bacterial fitness. By selecting several independent and spontaneous R207910-resistant mutants in vitro and characterizing them with regard to their resistance mechanisms and growth rates, we show that R207910 resistance development occurs at a rate similar to the rates for the two most effective anti-TB drugs, isoniazid and rifampin, and that resistance confers no apparent biological cost in vitro.
MATERIALS AND METHODS
Strains.
Six clinical M. tuberculosis isolates and the H37Rv reference strain (hereafter denoted parent strains) were selected from our previous study of R207910 and belonged to the strain collection of the Swedish Institute for Infectious Disease Control (17). Three strains were fully drug susceptible and four were MDR, as determined with a Bactec 460 radiometric system (31). All seven isolates were susceptible to R207910 (MIC = 0.03 mg/liter), as previously determined by MIC determinations (17). Prior to all experiments, the strains were cultured on Lowenstein-Jensen slants for 3 to 4 weeks at 37°C.
Mutation rate measurements.
A Luria-Delbrück fluctuation assay was used for the in vitro selection of independent, spontaneous R207910-resistant mutants and determination of the mutation rates (22). Suspensions of each parent strain were prepared in Middlebrook 7H9 broth (M7H9) containing oleic acid-albumin-dextrose-catalase (OADC) and 0.05% Tween 80, and about 103 CFU was distributed into each of 12 independent culture flasks. The use of this low number of cells ensured that no preexisting R207910-resistant mutants were present in the inoculum. The cultures were incubated for 3 to 4 weeks until the optical density (OD; 600 nm) reached 0.8 (approximately 108 CFU/ml). From each independent culture, 108 bacteria were plated onto selective Middlebrook 7H10 (M7H10) agar (with OADC and glycerol) containing 0.3 mg/liter, 0.9 mg/liter, or 3 mg/liter of R207910; and mutant colonies were counted following 4 weeks of incubation. To determine the total number of cells plated, 3 of each of the 12 cultures were serially diluted and plated in duplicate on nonselective M7H10 (with OADC and glycerol).
For each parent strain, the mutation rate from the 12 independent cultures was calculated by the Lea-Coulson method (mutation rate = m/Nt, where m is the number of mutations per culture and Nt is the final number of cells in the culture). On the basis of the frequency of mutants in each parent group, m was calculated by the use of either (i) the Poisson distribution, when more than one culture had zero mutant colonies [m = ln(P0/Ptot), where P0 is the number independent cultures with zero mutant colonies, and Ptot is the total number of independent cultures]; (ii) the Lea-Coulson method of the median [r/m − ln(m) = 1.24, where r is the median number of mutants per culture]; or (iii) the Drake formula of the median [r/m − ln(m) = 0] (28). From each independent culture spread on a selective plate, a single mutant colony was picked and subcultured in M7H9 broth with OADC for further characterization.
MIC determinations.
The level of resistance of each mutant clone was determined by running MIC determinations on solid medium with a replicator system, as described previously (17). Briefly, 20 μl of bacteria was resuspended in 3 ml phosphate-buffered saline (PBS) and homogenized by sonication, and 100 μl was distributed into a 96-well plate. By using a 96-stick replicator, the bacteria were inoculated onto M7H10 agar (with OADC and glycerol) containing dilutions of R207910 (range, 0.015 to 3.84 mg/liter), as well as two nonselective control plates. A rifampin plate was included as a control for growth and nongrowth. The R207910-susceptible parent strains and a bacterium-negative control (PBS) were also plated. MICs were determined after 4 weeks of incubation and were defined as the lowest concentration at which no growth was visible.
DNA sequencing.
Bacteria were killed by heating at 85°C for 30 min, and the DNA was extracted with chloroform-water, as described earlier (19). The quantity and quality of the DNA were examined spectrophotometrically by determining the absorbance at 260/280 nm, and working solutions of 5 ng/μl were prepared.
To amplify and sequence the 246-bp atpE gene, forward primer atpEforward (5′-TGT ACT TCA GCC AAG CGA TGG-3′) and reverse primer atpEreverse (5′-CCG TTG GGA ATG AGG AAG TTG-3′) were used. The PCR was run with an initial denaturation of 10 min, followed by denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s for 30 cycles. The reaction was terminated by use of a 20-min final elongation at 72°C. The PCR products were purified with a GFX PCR DNA and gel band purification kit (Amersham Bioscience). Sequencing was performed with the primers described above and an ABI Prism BigDye Terminator cycle sequencing ready reaction kit in an ABI Prism 3700 DNA analyzer (Applied Biosystems). Blast2 sequence software was used to align the wild-type H37Rv atpE gene (GenBank accession number Rv1305) with the sequences obtained. The base pairs and amino acids were enumerated by using M. tuberculosis.
Preparation of growth curves.
One week prior to use, start cultures of each mutant strain, as well as their corresponding R207910-susceptible parent strain, were prepared in M7H9 (with OADC and 0.05% Tween 80). On day 0, the start cultures were homogenized by sonication for 5 min, and the ODs (600 nm) were adjusted to 0.1 (approximately106 to 107 CFU/ml). Low-density cultures were then prepared by adding 60 μl of the culture (OD, 0.1) to 20 ml fresh M7H9 broth (with OADC and 0.05% Tween 80). The cultures were incubated with regular shaking at 37°C in 5% CO2. On days 0, 2, 4, 6, 8, and 10, serial dilutions of each culture were prepared and plated in duplicate on nonselective M7H10 agar (with OADC and glycerol). Following 3 to 4 weeks of incubation, the numbers of CFU were counted and growth curves were plotted.
Competition assays.
One week prior to use, start cultures of each mutant strain were prepared as described above for preparation of the growth curves. For each competition assay, equal volumes (30 μl) of each mutant start culture and its corresponding parent culture were inoculated in 20 ml M7H9 broth (with OADC and 0.05% Tween 80). The cultures were incubated with regular shaking at 37°C. On days 0, 2, 4, 6, 8, and 10, serial dilutions of each competition culture were prepared and plated in duplicate on nonselective M7H10 agar (with OADC), as well as on selective agar (with 0.3 mg/liter R207910). For three of the mutant classes, selective plates with a lower concentration (0.08 mg/liter) were used, because although these mutants were resistant to R207910, they showed inhibited growth when they were plated with the higher concentrations used in the competition assays. Following 3 to 4 weeks of incubation, the numbers of CFU on both the selective and the nonselective plates were counted, the mutant and parent growth dynamics were plotted, and the relative fitness of the mutants was calculated as the ln numbers of CFU for the mutant/ln numbers of CFU for the parent).
RESULTS
Rates of mutation to R207910 resistance vary by strain and drug concentration.
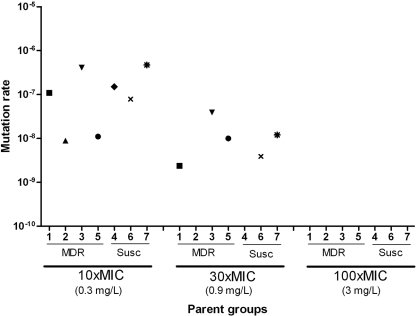
Mutants resistant to R207910 were selected from 12 independent cultures from each of three drug-susceptible strains (including H37Rv) and four MDR clinical strains at three different selective concentrations: 0.3 mg/liter (10× MIC for R207910-susceptible M. tuberculosis), 0.9 mg/liter (30× MIC), and 3 mg/liter (100× MIC). The mutation rates were calculated; and at the lowest selection concentration (10× MIC), the mutation rate ranged from 4.7 × 10−7 and 8.9 × 10−9 mutations per cell division (mean 1.8 × 10−7), depending on the clinical isolate used. At the selection concentration of 30× MIC, the mutation rate ranged from 3.9 × 10−8 and 2.4 × 10−9 mutations per cell division (mean, 1.3 × 10−8) (Fig. 1). At the selection concentration of 30× MIC, one strain (parent strain 2) showed no mutants and one strain (parent strain 4) showed very few mutants, preventing the accurate calculation of their mutation rates. The mean mutation rates were similar for the DS and MDR parent strains at 10× MIC (means, 2.0 × 10−7 and 1.3 × 10−7, respectively), although the MDR parent groups showed a wider range of rates (Fig. 1). At the selection concentration of 30× MIC, the mean mutation rates differed twofold between the DS and MDR groups (means, 7.9 × 10−9 and 1.7 × 10−8, respectively). At the highest selective concentration (100× MIC), no mutant colonies were obtained for any of the seven parent groups (i.e., from 84 independent cultures with 108 cells plated from each culture, not a single mutant was obtained).
FIG. 1.
Mutation rates for seven different M. tuberculosis strains at three different drug concentrations: 10×, 30×, and 100× the MIC of the susceptible parent strains. Independent mutants were selected from six clinical isolates (isolates 1 to 6) and reference strain H37Rv (isolate 7) at three selection concentrations. Parent strains 4, 6, and 7 were drug susceptible (Susc) and strains 1 to 3 and 5 were MDR, as determined with a Bactec radiometric system (29).
Level of resistance in mutants.
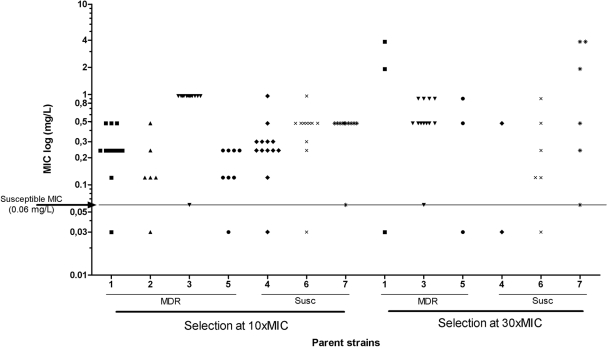
The resistance levels of 69 independent mutants selected at a concentration of 10× MIC and 27 mutants selected at a concentration of 30× MIC were determined by determination of the MICs on solid medium (Fig. 2). Two mutants exhibited a susceptible phenotype (MIC level, 0.06 mg/liter) and were excluded from the study. For the remainder of the strains, the levels of resistance ranged from 0.12 to 3.84 mg/liter (median, 0.48 mg/liter), which represent 4- to 128-fold increases in the MICs. No systematic differences in MICs were seen between mutants selected at the two selection concentrations, apart from the fact that all five mutants exhibiting an MIC of >1 mg/liter were selected at 30× MIC. No difference in the MIC level was seen between mutants from the drug-susceptible parent strains and the MDR parent strains. All parent isolates showed susceptible MIC levels (0.03 to 0.06 mg/liter).
FIG. 2.
MICs of R207910-resistant mutants. The MICs of 69 independent mutants selected with R207910 at 10× MIC and 27 selected with R207910 at 30× MIC were determined by the use of serial dilutions (0.015 to 3.84 mg/liter) on solid Middlebrook 7H9 agar. The MIC was defined as the first concentration with no visible growth. The MIC50 for all mutants was 0.48 mg/liter, and the highest MIC was 3.84 mg/liter. The arrow and line mark the MIC level of the R207910-susceptible parent isolates (0.03 to 0.06 mg/liter). MDR, mutants selected from MDR parent strains; Susc, mutants selected from drug-susceptible parent strains.
Identification of resistance mutations.
Having previously identified resistance mutations in the atpE gene, which encodes the membrane-bound C subunit of the bacterial ATP synthase, this gene was sequenced from 53 mutants (33 isolated at 10× MIC and 20 isolated at 30× MIC). Five different classes of mutations were observed (Table 1). Fifteen mutants had one of five single point mutations within the atpE gene: Asp28 → Val, Ala63 → Pro, Ile66 → Met, Asp28 → Pro, or Glu61 → Asp. The first three point mutations have previously been reported in R207910-resistant mutants (4, 26), whereas the last two have not previously been observed. No mutation within the sequenced atpE gene (Table 1) was identified in the remaining 38 resistant clones. The F0 operon (which includes the atpB, atpE, and atpF genes, whose gene products form the entire membrane-bound F0 unit of the ATP synthase) of 11 of the clones was subsequently sequenced, as was the entire ATP synthase encoded by the F1 operon (including the atpH, atpA, atpG, atpD, and atpC genes, whose gene products form the catalytic, cytoplasmic F1 unit) and the F0 operons of three mutants, but no mutations could be identified in any of these mutants.
TABLE 1.
Resistance mutations in the atpE gene and MIC levelsa
| Parent strain group and strain | Selection concn (multiple of MIC) | No. of mutants sequenced (total no. obtained) | atpE, F0 operon, or ATP synthase operon mutation | No. of independent mutants | MIC range (mg/liter) |
|---|---|---|---|---|---|
| MDR parent strains | |||||
| 1 | 10 | 6 (12) | Glu61 → Asp (GAG-GAC) | 1 | 0.48 |
| WT atpE | 5 | 0.12-0.48 | |||
| 30 | 2 (2) | Ala63 → Pro (GCA-CCA) | 2 | 1.92-3.84 | |
| 2 | 10 | 3 (5) | Asp28 → Val (GAC-GTC) | 1 | 0.48 |
| Glu61 → Asp (GAG-GAC) | 1 | 0.24 | |||
| WT atpE | 1 | 0.12 | |||
| 30 | 0 (0) | ||||
| 3 | 10 | 6 (12) | WT atpE | 2 | 0.9 |
| WT atpE and F0 operon | 1 | 0.9 | |||
| WT atpE and ATP synthase | 3 | 0.9 | |||
| 30 | 5 (11) | WT atpE | 3 | 0.48-0.9 | |
| WT atpE and F0 operon | 2 | 0.9 | |||
| 5 | 10 | 3 (7) | WT atpE | 2 | 0.12-0.24 |
| WT atpE and F0 operon | 1 | 0.24 | |||
| 30 | 2 (2) | Ala63 → Pro (GCA-CCA) | 1 | 0.9 | |
| Ile66 → Met (ATC-ATG) | 1 | 0.48 | |||
| Drug-susceptible parent strains | |||||
| 4 | 10 | 7 (12) | Asp28 → Pro (GAC-GGC) | 1 | 0.3 |
| Glu61 → Asp (GAG-GAC) | 1 | 0.96 | |||
| WT atpE | 3 | 0.12-0.24 | |||
| WT atpE and F0 operon | 2 | 0.3-0.48 | |||
| 30 | 1 (1) | Glu61 → Asp (GAG-GAC) | 1 | 0.48 | |
| 6 | 10 | 5 (10) | WT atpE | 5 | 0.24-0.96 |
| 30 | 5 (5) | Glu61 → Asp (GAG-GAC) | 1 | 0.48 | |
| Ala63 → Pro (GCA-CCA) | 1 | 0.9 | |||
| WT atpE | 3 | 0.12-24 | |||
| 7 | 10 | 3 (10) | WT atpE | 1 | 0.48 |
| WT atpE and F0 operon | 2 | 0.48 | |||
| 30 | 5 (5) | Ala63 → Pro (GCA-CCA) | 3 | 3.84 | |
| WT atpE | 2 | 0.24-0.48 |
The atpE genes of 53 mutants (33 selected at 10× MIC [0.3 mg/liter] and 20 selected at 30× MIC [0.9 mg/liter]) were sequenced, and six different classes of mutations were identified. The first five classes represented single point mutations within the atpE gene; and the last group, which consisted of 38 mutant clones, represented mutants in which no mutation within the sequenced gene was identified. Eleven of the mutants exhibited wild-type (WT) F0 operons, and three of these had wild-type ATP synthase (F0 and F1 operons).
Fitness effects of resistance.
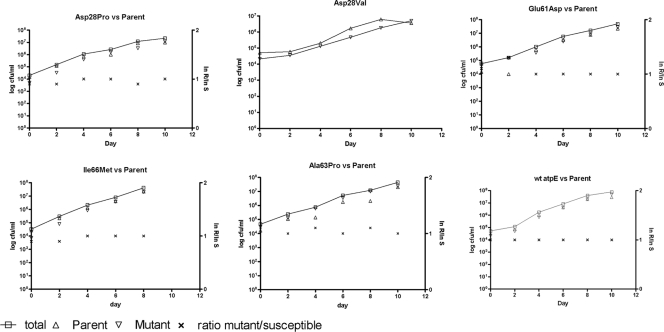
The relative fitness of six mutants (five with point mutations identified in the atpE gene and one with no mutations identified in either the F0 or the F1 operon) was determined by the preparation of growth curves for single cultures in vitro and competition assays. For these assays, equal amounts of each mutant clone and its corresponding parent strain were inoculated into broth. Their growth dynamics relative to each other were then examined by plating culture aliquots on selective and nonselective media and determining the changes in the ratio of the mutant strains/susceptible parent strains as a function of time (Fig. 3). In vitro, none of the resistant mutants showed a decrease in fitness relative to that of the respective parent strain. In fact, for each competition assay, similar growth dynamics were seen between the mutant and the parent strain, resulting in a relative fitness of 1 for each class of mutant. The Asp28→Val mutant class showed a reduced growth rate on the selective agar in the competition assay, and the number of resistant colonies could thus not be reliably counted. Growth curves for the mutant alone, however, did not show a difference in growth rate compared to that for the parent strain (data not shown).
FIG. 3.
Relative fitness of six different mutant classes. Competition assays between each mutant and its corresponding isogenic, R207910-susceptible parent strain were conducted in Middlebrook 7H9 broth; and the colonies were counted by plating on selective and nonselective Middlebrook 7H10 agar on days 0, 2, 4, 6, 8, and 10. The left y axis depicts the growth of the mutant and parent strains as the total numbers of CFU/ml. The right y axis depicts the ratio of ln numbers of CFU of resistant mutants (R)/ln numbers of CFU of susceptible parents (S). Due to difficulties with assessment of the Asp28Val mutation in the competition assays, single-culture growth curves were determined for the mutant and its isogenic parent strain.
DISCUSSION
Our results show that the apparent rate of mutation to high-level resistance against the novel anti-TB drug R207910 decreases with an increase in the antibiotic concentration. Thus, at 0.3 mg/liter (10× MIC of R207910-susceptible M. tuberculosis), the mutation rate ranged from 4.7 × 10−7 to 8.9 × 10−9 mutations per cell per division, at 0.9 mg/liter (30× MIC) the mutation rate ranged from 3.9 × 10−8 to 2.4 × 10−9 mutants per cell per division, and at the highest concentration tested, 3 mg/liter (100× MIC), no resistant mutants were found. The mean mutation rate at 10× MIC was approximately 10-fold higher that that at 30× MIC (Fig. 1). Furthermore, the mutation rate measured at 10× MIC was comparable to the mutation rate for resistance to isoniazid and rifampin (10−8), the two most important anti-TB drugs, in M. tuberculosis (9, 18, 34). The levels of resistance of the mutants that were selected ranged from 0.12 to 3.84 mg/liter for the 53 mutants analyzed (these represent 4- to 128-fold increases in the MICs), with no systematic differences between the two selection regimens being detected (Fig. 2), except that all the mutants with the highest level of resistance (3.84 mg/liter) were selected at 30× MIC.
The fact that no single resistant mutant was observed at the highest selective concentration suggests that R207910 has a mutant preventive concentration (MPC) below 3 mg/liter for M. tuberculosis. The MPC has been defined as the concentration threshold at which the least susceptible bacteria (in general, this means first-step resistant bacteria) present in a population of 1010 susceptible cells are inhibited (12, 13). Since approximately 1010 cells were plated in our mutant selection assays (i.e., 84 independent cultures, with approximately 108 CFU being plated from each culture) without yielding a single mutant colony at 3 mg/liter but yielding several mutant colonies at 0.9 mg/liter, this suggests that 0.9 mg/liter < MPC ≤ 3 mg/liter. The mutant selection window is the drug concentration range above the MIC level of the drug-susceptible population and below the MPC. Antimicrobial regimens have often been chosen on the basis of the MIC level, thus typically remaining within the selection window for resistance (14). However, if therapeutic doses above the MPC level for the drug are attainable without being toxic to the patient, the rate of clinical resistance development could be reduced (14, 37). In a phase IIa trial with R207910, when 400 mg/day of drug was administered for 7 days, R207910 was seen to reach an average steady-state plasma concentration of 2.7 ± 0.8 mg/liter (maximum serum concentration, 5.5 ± 2.9 mg/liter) (29). If it is assumed that the bacteria within infected patients are exposed to antibiotic levels corresponding to those determined by the plasma concentration, this concentration is within the postulated MPC range (0.9 mg/liter < MPC ≤ 3 mg/liter), and it is conceivable that a therapeutic dose above the mutant selection window might be attainable for R207910.
With regard to the mechanism of resistance in the isolated mutants, five of the mutations observed in 15/53 of the selected mutants were within the previously identified atpE gene. Complementation assays of the Asp28→ Val or Glu32 → Val mutation in M. smegmatis have previously confirmed the role of this gene in conferring R207910 resistance (4, 20), making it likely that the atpE mutations identified in this study cause the resistance observed in the mutants. Furthermore, binding studies with wild-type and mutant subunit C (AtpEA63P) proteins suggested that the point mutation in the atpE gene significantly influences its binding to R207190 (20). Structural simulations have suggested that R20910 binds in the vicinity of amino acid 61 (Glu) in the C subunit of the ATP synthase, interacting with the amino acid to block the passage of H+ and thus blocking ATP synthesis (10). The point mutations causing the substitutions Ala63 → Pro and Asp28→ Val are thought to affect the spatial area around amino acid 61, thereby interfering with the access of R207910 to its binding site (10). The substitution of Glu for the equally negatively charged but smaller Asp seen in our mutants at position 61 (Glu61 → Asp) might also confer a similar effect. Thus, the shorter carbon chain of Asp might prevent R207910 from interfering with the passage of H+, thereby maintaining the function of ATP synthase and conferring resistance. As Asp is itself negatively charged, the C subunit most likely maintains its function, enabling continued H+ transfer. This could explain why the relative fitness of the mutant is not measurably impaired (see below). For 38/53 mutants, we could not identify any mutations within the atpE gene. Furthermore, for 3 of these 38 mutants (parent strain 3, selected on 10× MIC), the complete F0 and F1 operons were sequenced buy no mutations were identified. These results suggest that at least one additional, ATP synthase-independent mechanism of resistance exists.
For the six classes of in vitro-selected R207910-resistant mutants observed in this study, resistance to R207910 in M. tuberculosis did not affect the relative fitness in vitro. Thus, mutants from each mutation class grew equally well as the corresponding isogenic parent strain, as shown by both the growth curves for a single culture (data not shown) and competition assays. It is unclear how well one can extrapolate in vitro fitness to the expected fitness of the mutants in an animal model or humans (1-3), but previous studies of streptomycin and isoniazid resistance in M. tuberculosis suggest that the growth rates measured in vitro are well correlated with the frequencies at which these resistant mutants are recovered from patients in clinical settings. For example, streptomycin-resistant mutants with high fitness costs (slow growth) in vitro are rare clinically, whereas mutants with cost-free mutations are commonly isolated from patients (7, 8, 30). Similarly, the low-cost/cost-free Ser315Thr isoniazid resistance-conferring mutation in the katG gene is the mutation type most commonly detected in isolates recovered from patients (27, 33). However, it is still possible that these mutations could confer a fitness cost in vivo without showing a measurable cost in vitro, as has been seen for other types of resistance (6, 23), and this question remains to be further explored by, for example, experiments with animals to determine in vivo growth rates and survival (24, 27) or epidemiological studies to determine the basic reproductive numbers of susceptible and R207910-resistant mutants in human populations (16, 33).
In conclusion, our results suggest that resistance to the novel compound R207910 develops at a rate similar to that observed for rifampin without causing any obvious associated fitness costs during growth in vitro. Importantly, a concentration of 3 mg/liter of drug will prevent the selection of one-step resistant mutants in vitro, and previous pharmacokinetic studies of the compound show that therapeutic doses above our postulated MPC are attainable, without being toxic to the host. Thus, it is conceivable that dosing regimens that effectively reduce the risk of resistance development can be developed for R207910 (in combination with other drugs). The data from this study provide important knowledge for the design of the optimal R207910 treatment regimens for the prevention of the emergence of resistance in clinical settings.
Acknowledgments
This work was partly supported by grants to D.I.A. from the Swedish Research Council and the 7th Framework Program of the European Union.
We thank Luc Vranckx and Brenda Molenberghs for technical support.
Footnotes
Published ahead of print on 28 December 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Andersson, D. I. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9:461-465. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452-456. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 4.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 5.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottger, E. C., and B. Springer. 2008. Tuberculosis: drug resistance, fitness, and strategies for global control. Eur. J. Pediatr. 167:141-148. [DOI] [PubMed] [Google Scholar]
- 8.Bottger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343-1344. [DOI] [PubMed] [Google Scholar]
- 9.David, H. L. 1970. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 20:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jonge, M. R., L. H. Koymans, J. E. Guillemont, A. Koul, and K. Andries. 2007. A computational model of the inhibition of Mycobacterium tuberculosis ATPase by a new drug candidate R207910. Proteins 67:971-980. [DOI] [PubMed] [Google Scholar]
- 11.Diacon, A. H., A. Pym, M. Grobusch, R. Patientia, R. Rustomjee, L. Page-Shipp, C. Pistorius, R. Krause, M. Bogoshi, G. Churchyard, A. Venter, J. Allen, J. C. Palomino, T. De Marez, R. P. van Heeswijk, N. Lounis, P. Meyvisch, J. Verbeeck, W. Parys, K. de Beule, K. Andries, and D. F. Mc Neeley. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N. Engl. J. Med. 360:2397-2405. [DOI] [PubMed] [Google Scholar]
- 12.Dong, Y., X. Zhao, J. Domagala, and K. Drlica. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1756-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, Y., X. Zhao, B. N. Kreiswirth, and K. Drlica. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2581-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein, B. J., J. G. Gums, and K. Drlica. 2004. The changing face of antibiotic prescribing: the mutant selection window. Ann. Pharmacother. 38:1675-1682. [DOI] [PubMed] [Google Scholar]
- 15.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Bottger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9:1239-1246. [DOI] [PubMed] [Google Scholar]
- 16.Gagneux, S., C. D. Long, P. M. Small, T. Van, G. K. Schoolnik, and B. J. Bohannan. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944-1946. [DOI] [PubMed] [Google Scholar]
- 17.Huitric, E., P. Verhasselt, K. Andries, and S. E. Hoffner. 2007. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 51:4202-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, R., E. M. Streicher, G. E. Louw, R. M. Warren, P. D. van Helden, and T. C. Victor. 2006. Drug resistance in Mycobacterium tuberculosis. Curr. Issues Mol. Biol. 8:97-111. [PubMed] [Google Scholar]
- 19.Jureen, P., J. Werngren, and S. E. Hoffner. 2004. Evaluation of the line probe assay (LiPA) for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb.) 84:311-316. [DOI] [PubMed] [Google Scholar]
- 20.Koul, A., N. Dendouga, K. Vergauwen, B. Molenberghs, L. Vranckx, R. Willebrords, Z. Ristic, H. Lill, I. Dorange, J. Guillemont, D. Bald, and K. Andries. 2007. Diarylquinolines target subunit C of mycobacterial ATP synthase. Nat. Chem. Biol. 3:323-324. [DOI] [PubMed] [Google Scholar]
- 21.Lounis, N., N. Veziris, A. Chauffour, C. Truffot-Pernot, K. Andries, and V. Jarlier. 2006. Combinations of R207910 with drugs used to treat multidrug-resistant tuberculosis have the potential to shorten treatment duration. Antimicrob. Agents Chemother. 50:3543-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luria, S., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macvanin, M., J. Bjorkman, S. Eriksson, M. Rhen, D. I. Andersson, and D. Hughes. 2003. Fusidic acid-resistant mutants of Salmonella enterica serovar Typhimurium with low fitness in vivo are defective in RpoS induction. Antimicrob. Agents Chemother. 47:3743-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manca, C., L. Tsenova, C. E. Barry III, A. Bergtold, S. Freeman, P. A. Haslett, J. M. Musser, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740-6746. [PubMed] [Google Scholar]
- 25.Mariam, D. H., Y. Mengistu, S. E. Hoffner, and D. I. Andersson. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrella, S., E. Cambau, A. Chauffour, K. Andries, V. Jarlier, and W. Sougakoff. 2006. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob. Agents Chemother. 50:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rustomjee, R., A. H. Diacon, J. Allen, A. Venter, C. Reddy, R. F. Patientia, T. C. Mthiyane, T. De Marez, R. van Heeswijk, R. Kerstens, A. Koul, K. De Beule, P. R. Donald, and D. F. McNeeley. 2008. Early bactericidal activity and pharmacokinetics of the diarylquinoline TMC207 in treatment of pulmonary tuberculosis. Antimicrob. Agents Chemother. 52:2831-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander, P., B. Springer, T. Prammananan, A. Sturmfels, M. Kappler, M. Pletschette, and E. C. Bottger. 2002. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 46:1204-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqi, S. H., J. P. Libonati, and G. Middlebrook. 1981. Evaluation of rapid radiometric method for drug susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 13:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibotec Pharmaceuticals Limited, I. 2009. TMC207-TiDP13-C208: anti-bacterial activity, safety, and tolerability of TMC207 in patients with multi-drug resistant Mycobacterium tuberculosis. http://clinicaltrials.gov/ct2/show/NCT00449644.
- 33.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 34.Werngren, J., and S. E. Hoffner. 2003. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 41:1520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WHO. 2008. Anti-tuberculosis drug resistance in the world. Report no. 4. WHO Press, Geneva, Switzerland.
- 36.WHO. 2003. Treatment of tuberculosis: guidelines for national programmes, 3rd ed. WHO, Geneva, Switzerland.
- 37.Zhao, X., and K. Drlica. 2008. A unified anti-mutant dosing strategy. J. Antimicrob. Chemother. 62:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]