Abstract
We evaluated cefepime exposures in patients infected with Pseudomonas aeruginosa to identify the pharmacodynamic relationship predictive of microbiological response. Patients with non-urinary tract P. aeruginosa infections and treated with cefepime were included. Free cefepime exposures were estimated by using a validated population pharmacokinetic model. P. aeruginosa MICs were determined by Etest and pharmacodynamic indices (the percentage of the dosing interval that the free drug concentration remains above the MIC of the infecting organism [fT > MIC], the ratio of the minimum concentration of free drug to the MIC [fCmin/MIC], and the ratio of the area under the concentration-time curve for free drug to the MIC [fAUC/MIC]) were calculated for each patient. Classification and regression tree analysis was used to partition the pharmacodynamic parameters for prediction of the microbiological response. Monte Carlo simulation was utilized to determine the optimal dosing regimens needed to achieve the pharmacodynamic target. Fifty-six patients with pneumonia (66.1%), skin and skin structure infections (SSSIs) (25%), and bacteremia (8.9%) were included. Twenty-four (42.9%) patients failed cefepime therapy. The MICs ranged from 0.75 to 96 μg/ml, resulting in median fT > MIC, fCmin/MIC, and fAUC/MIC exposures of 100% (range, 0.8 to 100%), 4.3 (range, 0.1 to 27.3), and 206.2 (range, 4.2 to 1,028.7), respectively. Microbiological failure was associated with an fT > MIC of ≤60% (77.8% failed cefepime therapy when fT > MIC was ≤60%, whereas 36.2% failed cefepime therapy when fT > MIC was >60%; P = 0.013). A similar fT > MIC target of ≤63.9% (P = 0.009) was identified when skin and skin structure infections were excluded. While controlling for the SSSI source (odds ratio [OR], 0.18 [95% confidence interval, 0.03 to 1.19]; P = 0.07) and combination therapy (OR, 2.15 [95% confidence interval, 0.59 to 7.88]; P = 0.25), patients with fT > MIC values of ≤60% were 8.1 times (95% confidence interval, 1.2 to 55.6 times) more likely to experience a poor microbiological response. Cefepime doses of at least 2 g every 8 h are required to achieve this target against CLSI-defined susceptible P. aeruginosa organisms in patients with normal renal function. In patients with non-urinary tract infections caused by P. aeruginosa, achievement of cefepime exposures of >60% fT > MIC will minimize the possibility of a poor microbiological response.
Cefepime is a commonly used broad-spectrum cephalosporin with potent activity against a wide variety of Gram-negative bacteria, including Pseudomonas aeruginosa (11). Despite its extensive use, its presence in multiple clinical guidelines, and numerous indications for its use (1, 16, 20, 26), a recent meta-analysis found treatment with cefepime to be associated with an increase in the patient mortality rate compared to that obtained by the use of other antimicrobial agents. Importantly, in that study the pharmacokinetics and pharmacodynamics (i.e., cefepime exposure and MIC of the infecting organisms) of cefepime were not included, leaving out a critical part of understanding antibiotic treatment outcomes (25).
The pharmacodynamic relationship historically thought to be predictive of cefepime efficacy, as with all beta-lactams, is the percentage of the dosing interval that the free drug concentration remains above the MIC of the infecting organism (fT > MIC) (24). Numerous in vivo animal studies with various cephalosporins have suggested that an fT > MIC target of 50 to 70% is required to achieve maximal reductions in the numbers of CFU of Gram-negative bacteria (5). However, the data available from evaluations of the clinical pharmacodynamics of cephalosporin have been less decisive and are discordant with the findings of in vivo animal studies. Two recently published reports of studies examining the pharmacodynamics of cefepime in patients with infections caused by various Gram-negative bacteria found the ratio of the minimum cefepime concentration to the MIC (Cmin/MIC) to be the parameter best associated with a microbiological response, while another study defined the ratio of the area under the concentration-time curve (AUC) to the MIC (AUC/MIC) to be the most predictive (13, 17, 23). Moreover, when the T > MIC for total drug was evaluated, those investigators found that targets of 90 to 100% were required for predictable microbiological success (13, 23). Given these discrepancies, coupled with the fact that cefepime is primarily used for the treatment of P. aeruginosa infections, we sought to focus on a pharmacodynamic analysis of patients with severe infections caused by this organism.
MATERIALS AND METHODS
Study design.
A retrospective cohort study was performed with all patients treated with cefepime for non-urinary tract P. aeruginosa infections from February to November 2006 at Hartford Hospital, Hartford, CT. This study was reviewed and approved by the Hartford Hospital Institutional Review Board. Informed consent was not required, as all data were in existence at the time of collection.
Inclusion criteria.
Adult patients (≥18 years old) with a culture positive for P. aeruginosa from a sample from a non-urinary tract source were considered for inclusion in the study. The patients were required to have an active P. aeruginosa infection, as defined by the Centers for Disease Control and Prevention/National Healthcare Safety Network criteria for infection (10), or a simplified Clinical Pulmonary Infection Score of ≥5 for respiratory infections (15). The patients were also required to have undergone treatment with cefepime for ≥3 days if a sample for follow-up culture was available or ≥7 days if a sample for follow-up culture was not available.
Exclusion criteria.
The patients were excluded if any of the following criteria were met: receipt of another antibiotic with activity against P. aeruginosa other than fluoroquinolones or aminoglycosides within 24 h before the start of cefepime treatment or within 72 h after the start of cefepime treatment, receipt of dialysis during cefepime therapy, or a presumed requirement for cefepime for extended durations for indications such as osteomyelitis and endocarditis.
Data collection.
The information extracted from the medical record for each patient included age, race, gender, height, weight, hospital admission and discharge dates, intensive care unit (ICU) admission and discharge dates, infection-related diagnosis, the APACHE II score on the day that P. aeruginosa was cultured (12), the Charlson comorbidity score (3), the serum creatinine concentration at the start of cefepime treatment, microbiological data, and concurrent antimicrobial use data.
Outcome assessments.
The primary outcome assessed was the microbiological response, which was defined as either success or failure. A successful microbiological response included elimination of the infecting P. aeruginosa strain from the original site of isolation upon repeat culture (eradication) or, if a follow-up culture result was unavailable, a positive patient clinical response (presumed eradication). Microbiological failure was defined as recovery of the original infecting P. aeruginosa strain from the original site of isolation upon follow-up culture (persistence) or the absence of an appropriate follow-up culture result coupled with a lack of patient clinical improvement (presumed persistence). Secondary outcomes included death for any reason during the identified hospital admission (all-cause mortality) and the duration of hospital and ICU length of stay (LOS) from the time of P. aeruginosa culture.
Susceptibility testing.
The cefepime MIC for each P. aeruginosa isolate was determined in triplicate by Etest, according to the manufacturer's recommendations. The modal MIC was utilized for pharmacodynamic analyses.
Pharmacokinetics and pharmacodynamics.
Patient-specific cefepime pharmacokinetic parameters were calculated by using the following equations/constants from an independently validated two-compartment population pharmacokinetic study developed at our hospital for patients with lower respiratory tract infections (18): the elimination rate variable (k10), which is equal to 0.0027 × creatinine clearance (CLCR) + 0.071; the volume of distribution (V1), which is equal to 0.21 liter/kg of actual body weight; the intercompartmental transfer rate constant from the central to the peripheral compartments (k12), which is equal to 0.78; and the intercompartmental transfer rate constant from the peripheral to the central compartments (k21), which is equal to 0.472. CLCR was calculated by using an adjusted Cockcroft-Gault equation that excluded patient weight [CLCR = (140 − age)/serum creatinine concentration; the result is multiplied by 0.85 for females].
After estimation of the values of the pharmacokinetic parameters, steady-state concentration-time profiles were simulated for each patient on the basis of the cefepime dose that he or she received as treatment of the P. aeruginosa infection (WinNonlin software; Pharsight Corp., Mountain View, CA). Simulated concentrations were corrected for 15% protein binding. The following pharmacodynamic indices were calculated for each patient by using the cefepime MIC of the infecting P. aeruginosa isolate: fT > MIC, the ratio of the AUC for free drug to the MIC (fAUC/MIC), the ratio of the maximum concentration of free drug to the MIC (fCmax/MIC), and the ratio of the minimum concentration of free drug to the MIC (fCmin/MIC).
Data analysis.
Classification and regression tree (CART) analysis (Salford Systems, San Diego, CA) was used to partition each of the pharmacodynamic indices on the basis of the microbiological response. The statistical significance of CART-derived breakpoints and other categorical variables was determined by Pearson's chi-square test or Fisher's exact test, while continuous variables were analyzed by Student's t test or the Mann-Whitney U test. Multiple logistic regression analysis was performed to determine the pharmacodynamic parameter independently associated with a microbiological response after confounding variables were controlled for. Variables with P values of <0.2 were included in the regression analysis. Statistical analyses were performed with SigmaStat software (SPSS Inc., San Rafael, CA). For all two-tailed tests, a P value of <0.05 was considered statistically significant.
Monte Carlo simulation.
A 5,000-patient Monte Carlo simulation (Crystal Ball; Decisioneering Inc., Denver, CO) was performed as described previously (6) by using the free drug exposure-response target identified from the CART analyses described above. Cefepime steady-state concentration-time profiles were simulated by using the median values of the pharmacokinetic parameter estimates from the two-compartment population pharmacokinetic model described above. All input variables except for the fraction unbound (80 to 90%) and CLCR were assumed to follow a log-Gaussian distribution. The fraction unbound (80 to 90%) and CLCR were assumed to follow a uniform distribution over the specified ranges. Simulations were run over three CLCR ranges: 50 to 120 ml/min, 30 to 49 ml/min, and 10 to 29 ml/min.
RESULTS
A total of 336 P. aeruginosa isolates were identified from individual patients at Hartford Hospital during the time frame of this study, and 119 of these patients were treated with cefepime. Fifty-six of the cefepime-treated patients had active infections, and data for those patients were included in the final analysis. The baseline characteristics of these patients are presented in Table 1. Microbiological success was achieved in 32 (57.1%) patients, and the mortality rate was 14.3%. After the population was divided on the basis of the microbiological responses, the patient demographics did not differ between groups (Table 2). However, a greater percentage of patients with microbiological failure was noted to have respiratory infections, and statistically significantly more patients with microbiological success had skin and skin structure infections (SSSIs). While the incidence of the receipt of combination therapy with either an aminoglycoside or a fluoroquinolone did not differ statistically significantly between the groups, all five patients who received a fluoroquinolone as combination therapy failed microbiologically (P = 0.01). The mortality rate was higher among patients with microbiological failure (20.8% versus 9.4% for patients without microbiological failure), but the difference was not significant (P = 0.2). Both the hospital LOS and the ICU LOS were significantly greater for patients who failed microbiologically than for those who did not: medians for the hospital LOS, 22 days (range, 6 to 94 days) for patients who failed microbiologically versus 14 days (range, 4 to 49 days) for patients who did not (P = 0.033); medians for ICU LOS, 16.5 days (range, 0 to 94 days) for patients who failed microbiologically versus 1 day (range, 0 to 44 days) for patients who did not (P = 0.001).
TABLE 1.
Characteristics of the 56 patients with P. aeruginosa infections
| Characteristic | Valuea |
|---|---|
| No. (%) male patients | 34 (60.7) |
| Mean (SD) age (yr) | 60.9 (19.8) |
| Mean (SD) wt (kg) | 81.2 (21.4) |
| Race | |
| Caucasian | 44 (78.6) |
| Hispanic | 7 (12.5) |
| Other | 5 (8.9) |
| Mean (SD)APACHE II score | 13.8 (7.0) |
| Mean (SD) CLCR (ml/min) | 99.4 (53.9) |
| No. (%) of patients with infection at the following site: | |
| Respiratory | 37 (66.1) |
| Skin and skin structure | 14 (25.0) |
| Bacteremia | 5 (8.9) |
| Mean (SD) Charlson comorbidity score | 3.8 (3.6) |
| No. (%) of patients with the following comorbidities: | |
| Diabetes | 18 (32.1) |
| Cardiovascular disease | 22 (39.3) |
| Chronic renal failure | 11 (19.6) |
| Immunosuppression | 4 (7.1) |
| No. (%) of patients receiving the following cefepime regimen: | |
| 1 g every 24 h | 2 (3.6) |
| 1 g every 12 h | 28 (50) |
| 1 g every 8 h | 12 (21.4) |
| 2 g every 12 h | 2 (3.6) |
| 2 g every 8 h | 4 (7.1) |
| 2 g every 8 h (3-h infusion) | 8 (14.3) |
Data are for 56 patients.
TABLE 2.
Univariate analysis for association with microbiological response
| Variable | Microbiological failure (n = 24) | Microbiological success (n = 32) | P value |
|---|---|---|---|
| Mean (SD) age (yr) | 59.4 (22.4) | 62.0 (17.9) | 0.624 |
| Mean (SD) APACHE II score | 14.6 (6.2) | 13.2 (7.6) | 0.443 |
| Mean (SD) CLCR (ml/min) | 110 (57) | 91.3 (50.5) | 0.199 |
| No. (%) of patients with the following comorbidities: | |||
| Diabetes | 9 (37.5) | 11 (34.4) | 0.968 |
| Cardiovascular disease | 12 (50) | 10 (31.3) | 0.252 |
| Chronic renal failure | 4 (16.7) | 7 (21.9) | 0.741 |
| Immunosuppression | 0 (0) | 4 (12.5) | 0.127 |
| No. (%) of patients with infection at the following site: | |||
| Respiratory | 20 (83.3) | 17 (53.1) | 0.038 |
| Bacteremia | 2 (8.3) | 3 (9.4) | 1.000 |
| Skin and skin structure | 2 (8.3) | 12 (37.5) | 0.029 |
| No. (%) of patients receiving combination therapy | 13 (54.2) | 8 (25.0) | 0.051 |
The cefepime MICs for the P. aeruginosa isolates ranged from 0.75 to 96 μg/ml. The MIC50 was 3 μg/ml, and the MIC90 was 16 μg/ml. The median MIC did not differ between patients with microbiological failure or success: 3 μg/ml (range, 1.5 to 96 μg/ml) for failure and 3 μg/ml (range, 0.75 to 32 μg/ml) for success (P = 0.35). The estimated values of the pharmacodynamic indices at steady state varied greatly among the individuals in our patient population, and a summary of these values is presented in Table 3. When the pharmacodynamic indices were evaluated as continuous variables, no differences in any of these indices were noted between patients when the patients were stratified by their microbiological responses (P ≥ 0.42).
TABLE 3.
Values of pharmacodynamic parameters for 56 patients treated with cefepime for P. aeruginosa infections
| Data type | % fT > MIC | fAUC/MIC | fCmax/MIC | fCmin/MIC |
|---|---|---|---|---|
| Median | 100 | 167 | 20 | 3.1 |
| Range | 0.8-100 | 4.2-1028.7 | 1.1-93.9 | 0-27.3 |
| Mean | 85.2 | 206.2 | 21.7 | 4.3 |
| SD | 29.7 | 194.3 | 18.6 | 4.8 |
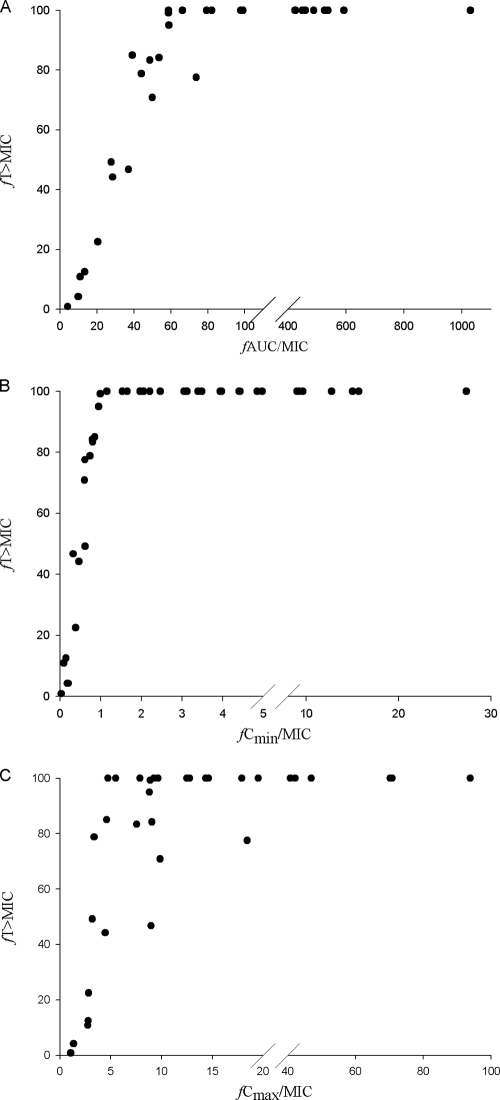
When CART analysis was used to partition fT > MIC at a value predictive of microbiological failure, a fT > MIC breakpoint of 60% was identified. The microbiological failure rate was 77.8% when fT > MIC was ≤60% and only 36.2% when fT > MIC was greater than 60% (P = 0.03). While all pharmacodynamic parameters were tested, the only other statistically significant CART-derived breakpoint was an fAUC/MIC of 41.6 (P = 0.013). As shown in Fig. 1, fT > MIC and fAUC/MIC were highly colinear, as were fT > MIC and fCmin/MIC and, to a lesser degree, fT > MIC and fCmax/MIC. Of note, after exclusion of the data for the 14 patients with SSSIs from this analysis, CART identified a similar fT > MIC breakpoint of 63.9%, in which microbiological success was 100% when fT > MIC was above that value and 0% when it was below that value (P = 0.009).
FIG. 1.
Colinearity between CART-derived breakpoints for fT > MIC and fAUC/MIC (A), fCmin/MIC (B), and fCmax/MIC (C) for patients infected with P. aeruginosa.
The results of the multivariable logistic regression analysis on the basis of the data for all patients are shown in Table 4. The variables included in that analysis were CLCR, immunosuppression, respiratory infection site, SSSI site, combination therapy, and an fT > MIC of ≤60%. After combination therapy and SSSIs were controlled for, patients were found to be eight times more likely to fail microbiologically when the fT > MIC was less than 60%. When we compared patients who achieved 60% fT > MIC to those who did not, the mortality rate (14.9% and 11.1%, respectively; P = 1.0), the median ICU LOS (6 days [range, 0 to 94 days] and 8 days [range, 0 to 28 days], respectively; P = 1) and the median hospital LOS (17 days [range, 4 to 94 days] and 22 days [range, 8 to 41 days], respectively; P = 0.585) were not different between the groups.
TABLE 4.
Results of the multivariate logistic regression analysis for predicting microbiological failure
| Variable | OR (95% CI)a | P value |
|---|---|---|
| ≤60% fT > MIC | 8.10 (1.18-55.57) | 0.033 |
| Combination therapy | 2.15 (0.59-7.88) | 0.247 |
| Skin and skin structure infection | 0.18 (0.03-1.19) | 0.074 |
OR, odds ratio; CI, confidence interval.
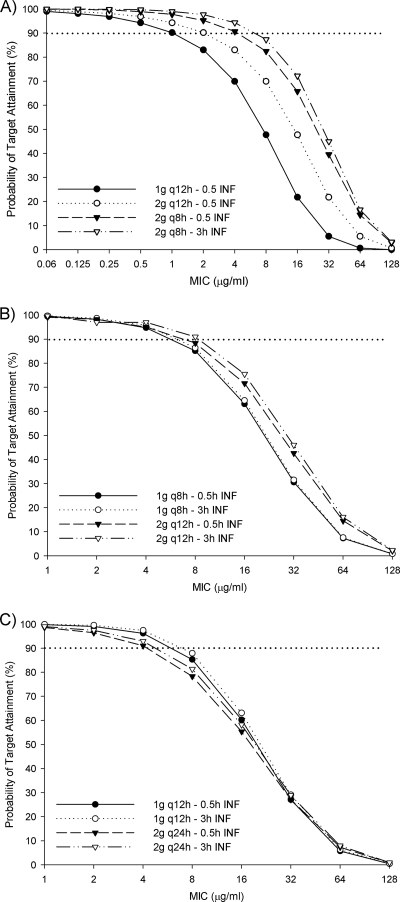
The results of the probability of target attainment from the Monte Carlo simulation are provided in Fig. 2. At the current Clinical and Laboratory Standards Institute (CLSI)-defined breakpoint of 8 μg/ml, only a dose of 2 g every 8 h as either a 30-min or a 3-h infusion has a ≥82% likelihood of achieving at least 60% fT > MIC in patients with normal renal function (Fig. 2A). At this MIC, the labeled dosages for immunocompetent patients with severe P. aeruginosa infections, 30-min infusions of 1 and 2 g every 12 h have only a 47.7% or 65.8% chance, respectively, of achieving adequate pharmacodynamic targets. For patients with CLCRs of 30 to 49 ml/min, only the regimen of 2 g every 12 h as a 3-h infusion has a >90% chance of achieving ≥60% fT > MIC at an MIC of 8 μg/ml, with the same dose given as a 30-min infusion being not far behind with an 88.4% probability (Fig. 2B). At the same MIC, 1 g every 12 h either as a 30-min or a 3-h infusion in patients with CLCRs of 10 to 29 ml/min achieved similar probabilities of pharmacodynamic target attainments rates (85.3 and 87.9%, respectively) (Fig. 2C).
FIG. 2.
Probability that various cefepime doses will achieve 60% fT > MIC in infected patients with creatinine clearances of 50 to 120 ml/min (A), 30 to 49 ml/min (B), and 10 to 29 ml/min (C). q12h, every 12 h; q8h, every 8 h; INF, infusion.
DISCUSSION
While data describing the exposure-response relationship of a given antimicrobial in animal models are often available, clinical data are often lacking. More troubling, however, is when both animal and clinical data are available but do not agree. The latter of these two scenarios has been much the case for the cephalosporins to date (5, 13, 17, 23, 24). Given the potential impact that knowledge of these data can make, it is imperative that the proper relationship be identified and employed to provide patients with optimal drug therapy. In line with previous work in studies with animals, we found the cefepime fT > MIC to be the most predictive of a microbiological response in patients infected with P. aeruginosa.
Studies with neutropenic animal represent the purest form of in vivo assessment of the bug-drug relationship. Dating back upwards of 20 years ago, these types of studies have identified fT > MIC as the cephalosporin pharmacodynamic parameter associated with bacterial killing and have described a target of 50 to 70% to be required for maximal activity against Gram-negative organisms (5). In contrast, when previous clinical assessments of cefepime have evaluated total drug T > MIC, values of 90 to 100% were required for positive outcomes (13, 17, 23). In two of those studies, not only were the concentrations required to be above the MIC for the entire interval but also they needed to be four to seven times the MIC over that time period (i.e., Cmin/MIC) (13, 23). When a standard cefepime regimen of 2 g every 12 h given to a patient with a creatinine clearance of 85 ml/min is considered, these Cmin/MIC targets would be reliably achieved only against organisms with MICs of 0.25 to 0.75 μg/ml (18). The discrepancies noted in the pharmacodynamic targets identified could have been attributed to a number of factors not occurring in the present analysis, e.g., the inclusion of various Gram-negative organisms, the MICs of most isolates residing in the two extremes of the distribution, pharmacokinetic compartment model selection, a narrow range of cefepime exposures, and the inclusion of isolates from the urinary tract. It should be noted that during the analysis of our data, we, too, found an association between fAUC/MIC and the microbiological response. However, fT > MIC and fAUC/MIC were highly colinear, with only one patient having an fAUC/MIC of less than 41.6 not having an fT > MIC of <60%. Given our previous knowledge of the pharmacodynamics of the beta-lactam class of antibiotics, we incorporated only the fT > MIC target into the multivariate analyses.
The observation that the MIC is not the sole driving force in predicting outcomes is one of utmost importance when it comes to clinical practice. The availability of an exposure-response target gives the prescriber a systematic approach to optimizing the only modifiable factor in the host, bug, and drug relationship, i.e., drug exposure. This was evidenced in the current analysis, by evaluating the seven patients with non-SSSIs and isolates with cefepime MICs above the CLSI-defined breakpoint of 8 μg/ml (4). In this subset of patients, only the two patients, both with creatinine clearances greater than 70 ml/min, that received cefepime doses (2 g every 8 h as a 3-h infusion) adequate enough to reach a fT > MIC above 60% (78 and 100%, respectively) achieved microbiological success. Moreover, a recent study evaluating the 28-day mortality rate for cefepime-treated patients with bacteremia caused by Gram-negative bacteria found an increased rate of mortality among those patients infected with isolates with MICs of ≥8 μg/ml (2). While patient-specific pharmacokinetic exposures were not evaluated in that study, protocol doses for patients with normal renal function were 1 to 2 g every 12 h, doses not highly predictive of achieving adequate fT > MIC targets at these MICs (Fig. 2A). On the basis of the results of our Monte Carlo simulation, doses of 2 g every 8 h as either a standard or a prolonged infusion would be required to achieve an appreciable probability of attaining pharmacodynamic targets against organisms with MICs of 8 μg/ml.
During univariate analysis, we found microbiological success to be more likely in patients with SSSIs. These results are not entirely unexpected. Recent clinical guidelines for the treatment of these infections suggest that for certain patients, incision and drainage without antibiotic therapy are suitable management strategies (21). Moreover, a recent review of the literature surrounding this issue found that the available data suggest similar cure rates for patients with and without antibiotic therapy, assuming that proper incision and drainage are performed (9). For this reason, we reanalyzed our data after excluding the data for patients with SSSIs and identified a similar fT > MIC target of 63.9%. Taken collectively, while patients were more likely to achieve microbiological success if their P. aeruginosa infection was of a skin or skin structure source, a dose-response relationship still existed.
Another interesting observation made during the univariate analysis was that all five patients who received a fluoroquinolone as part of combination therapy failed microbiologically. The association between combination therapy and microbiological failure is unclear. Four of the fluoroquinolone-treated patients received ciprofloxacin, while the fifth one received levofloxacin, and all patients had respiratory tract infections. By Etest, the ciprofloxacin MICs for the P. aeruginosa isolates from the four patients were 0.125, 0.125, 0.5, and 6 μg/ml, and the levofloxacin MIC for the remaining isolate was 0.75 μg/ml. According to CLSI-derived breakpoints (4), all but one of these isolates would be considered susceptible; it is of note, however, that this classification may not be an accurate representation of the pharmacodynamics of these agents, as found by Monte Carlo simulation, in addition to a recent clinical evaluation of bacteremic patients (7, 8). Given the retrospective nature of this study, it is impossible to evaluate the prescribing habits of the treating clinicians on the basis of data gathered from previous encounters. It is entirely possible that combination therapy was reserved for patients viewed as being the “sickest” and could have inherently been more likely to fail therapy.
As secondary outcomes, we evaluated both all-cause mortality and both hospital and ICU LOSs. Likely due to the low mortality rate seen among our patients, we found no association between the microbiological response or pharmacodynamic target attainment and mortality. A larger sample size would be needed to confirm these findings. As might be expected, there were significantly greater ICU and hospital LOSs for patients who achieved microbiological failure. However, these differences did not translate to target attainments; again, this is potentially due to the relatively small number of patients with fT > MIC values of ≤60%.
The limitations of this study are directly related to the fact that the design was retrospective in nature. Despite our diligence in critically reviewing all data available from the medical record, prospective data collection is a preferred method. Also, while it would have been optimal to collect cefepime pharmacokinetic data for every patient, this was not possible, given our study design. Instead, we choose to use a validated population pharmacokinetic model to estimate patient-specific pharmacokinetics. The model chosen, through independent validation with data for six patients, displayed a bias, precision, and coefficient of determination of 1.64 μg/ml, 17.1 μg/ml, and 62%, respectively (18). Furthermore, the population model noted was developed by our group by using cefepime concentrations for patients admitted to Hartford Hospital who were similar to the majority of those patients included in the current analysis. This, coupled with the similarities between the model chosen and other population pharmacokinetic models developed elsewhere with data for critically ill patients (14, 19, 22), we believed that this model was an acceptable approach for predicting cefepime exposures. Nevertheless, it is possible that the actual exposure of a given patient could have differed from that predicted by using the population model. Lastly, although a large number of patients were identified, a relatively small sample ultimately met the criteria for inclusion, and all these patients came from a single institution. Despite these potential shortcomings, the pharmacodynamic targets identified in the present study are remarkably similar to those reported in vivo.
As the armamentarium of agents with activity against P. aeruginosa continues to dwindle, knowledge of local or individual MIC data coupled with a pharmacodynamic target can provide clinicians the ability to reinvigorate cefepime, a drug with ample safety data and proven clinical efficacy. Consistent with previous in vivo animal data, we identified the cefepime fT > MIC to be the pharmacodynamic parameter that was the most associated with a microbiological response in patients with non-urinary tract Pseudomonas infections. Moreover, a fT > MIC of >60% minimized the chance of a poor microbiological response in these patients. Given these targets, cefepime doses of 2 g every 8 h in patients with normal renal function are required to achieve adequate exposures and ultimately increase the probability of microbiological success against CLSI-defined susceptible P. aeruginosa isolates.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.American Thoracic Society and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388-416. [DOI] [PubMed] [Google Scholar]
- 2.Bhat, S. V., A. Y. Peleg, T. P. Lodise, Jr., K. A. Shutt, B. Capitano, B. A. Potoski, and D. L. Paterson. 2007. Failure of current cefepime breakpoints to predict clinical outcomes of bacteremia caused by gram-negative organisms. Antimicrob. Agents Chemother. 51:4390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlson, M. E., P. Pompei, K. L. Ales, and C. R. MacKenzie. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40:373-383. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI publication M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Craig, W. A. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1-10. [DOI] [PubMed] [Google Scholar]
- 6.Crandon, J. L., J. L. Kuti, R. N. Jones, and D. P. Nicolau. 2009. Comparison of 2002-2006 OPTAMA programs for US hospitals: focus on gram-negative resistance. Ann. Pharmacother. 43:220-227. [DOI] [PubMed] [Google Scholar]
- 7.Defife, R., M. H. Scheetz, J. M. Feinglass, M. J. Postelnick, and K. K. Scarsi. 2009. Effect of differences in MIC values on clinical outcomes in patients with bloodstream infections caused by gram-negative organisms treated with levofloxacin. Antimicrob. Agents Chemother. 53:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRyke, C. A., J. L. Kuti, and D. P. Nicolau. 2007. Reevaluation of current susceptibility breakpoints for Gram-negative rods based on pharmacodynamic assessment. Diagn. Microbiol. Infect. Dis. 58:337-344. [DOI] [PubMed] [Google Scholar]
- 9.Hankin, A., and W. W. Everett. 2007. Are antibiotics necessary after incision and drainage of a cutaneous abscess? Ann. Emerg. Med. 50:49-51. [DOI] [PubMed] [Google Scholar]
- 10.Horan, T. C., M. Andrus, and M. A. Dudeck. 2008. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 36:309-332. [DOI] [PubMed] [Google Scholar]
- 11.Jones, R. N., J. T. Kirby, and P. R. Rhomberg. 2008. Comparative activity of meropenem in US medical centers (2007): initiating the 2nd decade of MYSTIC program surveillance. Diagn. Microbiol. Infect. Dis. 61:203-213. [DOI] [PubMed] [Google Scholar]
- 12.Knaus, W. A., E. A. Draper, D. P. Wagner, and J. E. Zimmerman. 1985. APACHE II: a severity of disease classification system. Crit. Care Med. 13:818-829. [PubMed] [Google Scholar]
- 13.Lee, S. Y., J. L. Kuti, and D. P. Nicolau. 2007. Cefepime pharmacodynamics in patients with extended spectrum beta-lactamase (ESBL) and non-ESBL infections. J. Infect. 54:463-468. [DOI] [PubMed] [Google Scholar]
- 14.Lipman, J., S. C. Wallis, and C. Rickard. 1999. Low plasma cefepime levels in critically ill septic patients: pharmacokinetic modeling indicates improved troughs with revised dosing. Antimicrob. Agents Chemother. 43:2559-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna, C. M., D. Blanzaco, M. S. Niederman, W. Matarucco, N. C. Baredes, P. Desmery, F. Palizas, G. Menga, F. Rios, and C. Apezteguia. 2003. Resolution of ventilator-associated pneumonia: prospective evaluation of the clinical pulmonary infection score as an early clinical predictor of outcome. Crit. Care Med. 31:676-682. [DOI] [PubMed] [Google Scholar]
- 16.Mandell, L. A., R. G. Wunderink, A. Anzueto, J. G. Bartlett, G. D. Campbell, N. C. Dean, S. F. Dowell, T. M. File, Jr., D. M. Musher, M. S. Niederman, A. Torres, and C. G. Whitney. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 44(Suppl. 2):S27-S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinnon, P. S., J. A. Paladino, and J. J. Schentag. 2008. Evaluation of area under the inhibitory curve (AUIC) and time above the minimum inhibitory concentration (T>MIC) as predictors of outcome for cefepime and ceftazidime in serious bacterial infections. Int. J. Antimicrob. Agents 31:345-351. [DOI] [PubMed] [Google Scholar]
- 18.Nicasio, A. M., R. E. Ariano, S. A. Zelenitsky, A. Kim, J. L. Crandon, J. L. Kuti, and D. P. Nicolau. 2009. Population pharmacokinetics of high-dose, prolonged-infusion cefepime in adult critically ill patients with ventilator-associated pneumonia. Antimicrob. Agents Chemother. 53:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roos, J. F., J. Bulitta, J. Lipman, and C. M. Kirkpatrick. 2006. Pharmacokinetic-pharmacodynamic rationale for cefepime dosing regimens in intensive care units. J. Antimicrob. Chemother. 58:987-993. [DOI] [PubMed] [Google Scholar]
- 20.Solomkin, J. S., J. E. Mazuski, E. J. Baron, R. G. Sawyer, A. B. Nathens, J. T. DiPiro, T. Buchman, E. P. Dellinger, J. Jernigan, S. Gorbach, A. W. Chow, and J. Bartlett. 2003. Guidelines for the selection of anti-infective agents for complicated intra-abdominal infections. Clin. Infect. Dis. 37:997-1005. [DOI] [PubMed] [Google Scholar]
- 21.Stevens, D. L., A. L. Bisno, H. F. Chambers, E. D. Everett, P. Dellinger, E. J. Goldstein, S. L. Gorbach, J. V. Hirschmann, E. L. Kaplan, J. G. Montoya, and J. C. Wade. 2005. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin. Infect. Dis. 41:1373-1406. [DOI] [PubMed] [Google Scholar]
- 22.Tam, V. H., P. S. McKinnon, R. L. Akins, G. L. Drusano, and M. J. Rybak. 2003. Pharmacokinetics and pharmacodynamics of cefepime in patients with various degrees of renal function. Antimicrob. Agents Chemother. 47:1853-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam, V. H., P. S. McKinnon, R. L. Akins, M. J. Rybak, and G. L. Drusano. 2002. Pharmacodynamics of cefepime in patients with Gram-negative infections. J. Antimicrob. Chemother. 50:425-428. [DOI] [PubMed] [Google Scholar]
- 24.Turnidge, J. D. 1998. The pharmacodynamics of beta-lactams. Clin. Infect. Dis. 27:10-22. [DOI] [PubMed] [Google Scholar]
- 25.Yahav, D., M. Paul, A. Fraser, N. Sarid, and L. Leibovici. 2007. Efficacy and safety of cefepime: a systematic review and meta-analysis. Lancet Infect. Dis. 7:338-348. [DOI] [PubMed] [Google Scholar]
- 26.Zanetti, G., F. Bally, G. Greub, J. Garbino, T. Kinge, D. Lew, J. A. Romand, J. Bille, D. Aymon, L. Stratchounski, L. Krawczyk, E. Rubinstein, M. D. Schaller, R. Chiolero, M. P. Glauser, and A. Cometta. 2003. Cefepime versus imipenem-cilastatin for treatment of nosocomial pneumonia in intensive care unit patients: a multicenter, evaluator-blind, prospective, randomized study. Antimicrob. Agents Chemother. 47:3442-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]