Abstract
AcrAB-TolC imparts a strong intrinsic resistance phenotype to many clinically significant molecules in Escherichia coli. This complex is composed of a pump, AcrB, and a periplasmic protein, AcrA, that exports substrates through a common outer membrane porin, TolC. A sequence survey of the pump-specific components, acrA and acrB, was conducted on three discrete animal reservoirs: rodents, bovines, and catfish. Although two of the reservoirs (bovine and catfish) were agrarian, and antibiotic use (ceftiofur and oxytetracycline/Romet 30, respectively) was reported for them, the vast majority of structural polymorphisms were silent except for T104A (AcrA) and Q733R (AcrB), found in certain bovine-derived strains. Overall, the genes were well conserved, with high ratios of synonymous to nonsynonymous substitutions (dS/dN ratios), consistent with or, in the case of acrB, better than those of standard multilocus sequence typing (MLST) loci. Furthermore, predicted recombination points from single nucleotide polymorphism (SNP) patterns in acrB support a modular evolution of transporter proteins, consistent with an ancient origin. However, functional studies with clones representing the major silent SNPs and the nonsilent mutation in acrB failed to generate significant differences in resistance to a range of common efflux pump substrates. Interestingly, a comparison between log-phase acrA and acrB expression profiles yielded inconsistent trends, with acrB expression increasing modestly (<1.8-fold) in many strains from the antibiotic-enriched pools. Our results suggest that structural polymorphisms in this major efflux pump system may not contribute significantly to adaptive resistance by altering function or substrate specificity but may have a potential use in improving phylogenetic relationships and/or source tracking.
Bacterial resistance to single drugs or their defined drug classes has traditionally been linked with transmissible and hence acquired gene traits that can be tracked and monitored using a variety of techniques. However, there is an increasing trend to focus on intrinsic resistance measures that are encoded a priori by a respective organism but may arise through mutation or changes in expression profiles of its native genetic complement (25). Such resistances are not typically transmissible but could more accurately be regarded as clonal and can be equally important in a dissemination scheme. Certain structural mutations related, for instance, to cell permeability or transport can generally alter the profiles of resistance to a wide spectrum of drugs (22). A growing collective body of evidence suggests these are major resistance mechanisms that have been inadequately tracked and monitored.
Bacterial multidrug efflux pumps most likely provide the largest degree of active intrinsic resistance in both quantitative and qualitative measure and are found in virtually every genome sequence (6, 19). In Escherichia coli, there are some 37 putative or functionally described efflux pumps representing five different phylogenetic classifications: the ATP-binding cassette (ABC) superfamily, the major facilitator superfamily (MFS), the multidrug and toxic compound extrusion (MATE) family, the small multidrug resistance (SMR) family, and the resistance nodulation division (RND) superfamily (24). Of these representatives, E. coli encodes at least two multidrug efflux pumps that are constitutively expressed: AcrAB-TolC and EmrAB-TolC (19). Nevertheless, the AcrAB-TolC system provides the “overriding” phenotype of intrinsic drug resistance to several structurally unrelated compounds. Other multidrug pump systems may have overlapping substrate repertoires but are generally silent and under tight regulatory control by various local and global regulatory genes, some of which can be induced by drugs (19).
AcrAB-TolC is a tripartite system composed of an integral inner membrane protein and pump proper (AcrB), a periplasmic membrane fusion protein (AcrA), and an outer membrane porin (TolC). Structural studies have debunked the idea of AcrA-mediated fusion of inner and outer membranes (at this point a misnomer) and have rather assigned AcrA a stabilizing role in pump architecture at the junction between ends of AcrB and TolC that protrude well into the periplasmic space (6). The three components function in a coordinated fashion to expel substrates directly across the dual membrane of the Gram-negative envelope and into the extracellular milieu. In this manner, the organism can significantly retard the passive reentry of generally hydrophobic substrates through the cytoplasmic membrane by requiring them to traverse the outer membrane permeability barrier (19, 22).
In this model, substrate specificity is determined by the AcrB pump and has been generally attributed to the two large extracytoplasmic loops that extend into the periplasm (10, 30). Interestingly, under drug-selective laboratory conditions, point mutations that can broaden drug substrate specificity can arise in the loop regions of the pump (20). Regardless of this observation, “natural pools” of isolates studied thus far generally have undefined regulation-based adaptations that alter the expression levels of homologous pump systems (4, 19, 26, 33). In the current study, we addressed the question of whether natural polymorphisms occur in bacterial pumps to promote drug resistance per se as do mutations affecting pump expression, and if so, at what frequency. Our investigation focused on three distinct pools of isolates from rodents, bovines, and catfish. For two of the isolate pools, antibiotic use was reported in the respective farm environment.
(A preliminary account of this study was presented at the 108th and 109th General Meetings of the American Society for Microbiology.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strains were obtained from three different environmental reservoirs. “S-series” strains were isolated nonselectively from nasopharyngeal swabs or intestinal contents of mouse and rat colonies on MacConkey agar medium by our in-house Microbiological Surveillance and Diagnostic Group. “N-series” strains, preenriched from gut contents of farm-raised catfish in Luria broth and subsequently isolated on MacConkey agar medium with tetracycline (30 μg/ml) selection (21), were obtained from Mohamed S. Nawaz (National Center for Toxicological Research [NCTR]). Isolates from both series were identified to species level with an in-house Vitek 2 Compact system, version 3.01 (bioMérieux). “P-series” strains, isolated with ceftiofur selection (8 μg/ml) from fecal samples of healthy dairy heifer calves in central Pennsylvania, were obtained from Bhushan M. Jayarao (Pennsylvania State University). These isolates were previously identified to species level using an API 20E identification kit and were further characterized as clonally distinct based on phenotypic and genotypic characterization and serotype analysis (5). Strain AG100 is an E. coli K-12 derivative described previously (11), and AG102MB is a marR1 acrB::kan derivative of AG100 (10). All routine handling of E. coli was conducted on Luria-Bertani (LB) medium at 37°C with long-term storage in 10 to 15% glycerol stocks at −80°C.
PCR amplification and DNA sequencing of acrAB.
Pump components acrB and acrA were amplified independently under standard conditions from respective genomic DNAs with Klentaq LA proofreading polymerase (Clontech), which is capable of long and accurate processivity. Primer pairs for each gene were designed to obtain full-length amplification with engineered restriction enzyme sites for directional cloning (AcrB[+] and AcrB[−]; AcrA[+] and AcrA[−] [Table 1]). Additional internal walking primers (not shown) were synthesized (MWG Biotech) to generate full-length sequences of both genes. PCR products were sequenced bidirectionally by the University of Arkansas for Medical Sciences (UAMS) core facility using an automated fluorescent method. In a majority of cases, single nucleotide polymorphisms (SNPs) were confirmed by further rounds of sequencing using independent preparations of template DNA.
TABLE 1.
Oligonucleotide primers used in this study
| Name | Use | Sequence (5′ to 3′) |
|---|---|---|
| AcrB[+] | SNP sequencing, cloning | GCC TGA ACA GTC CAA GTC TAG ACT TAA ACA GGA GCCa |
| AcrB[−] | SNP sequencing, cloning | GGT TTT CGT ATG GGA TCC TGA GTT GGT GGb |
| AcrA[+] | SNP sequencing | GCA CGA CGA TAA TCT AGA CGC AGC AAT GG |
| AcrA[−] | SNP sequencing | CCG GCA GTT TGA GGA TCC CCA GCC CCC CTG CC |
| AcrBqRT[+] | Expression | CTG ATC ATC GTG GTC GGC ATG GC |
| AcrBqRT[−] | Expression | CCA GTC CTT CAA GGA AAC GAA CGC |
| AcrAqRT[+] | Expression | CCA ATA TCG CGC AAT TGA CGG TGA ATC G |
| AcrAqRT[−] | Expression | GCG CAG GAA GTC GTT GCT GGA CTG G |
| GyrBqRT[+] | Expression normalization | CAG ACT GCC AGG AAC GCG AT |
| GyrBqRT[−] | Expression normalization | AGC CAA GCG CGG TGA TAA GC |
Underscored and boldface letters represent the XbaI site used for directional cloning into the pSportI plasmid vector.
Underscored and boldface letters represent the BamHI site used for directional cloning into the pSportI plasmid vector.
DNA sequence analyses.
The acrA and acrB sequences from isolates were initially compared to the respective E. coli K-12 loci with the nucleotide BLAST (NCBI website; http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify SNP pools. Phylogenetic and molecular evolutionary relationships, displayed as unrooted neighbor-joining trees, were constructed with MEGA4 (29) using the maximum composite likelihood model of nucleotide substitution. The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by the modified Nei-Gojobori method using MEGA4. Phylogenetic network analysis was conducted with the SplitsTree4 program (13) using the neighbor-net algorithm (2) and untransformed distances (p distances). The φw recombination test (1) as implemented by SplitsTree4 was used to distinguish recurrent mutation from recombination in generating genotypic diversity. Allelic sequences of acrB were also fitted to a nucleotide substitution model, and potential recombinational breakpoints were identified using the genetic algorithm recombination detection (GARD) method (17) as implemented on the Datamonkey website (http://www.datamonkey.org) using the β-Γ model with 3 rate classes.
Cloning and expression of acrB alleles.
The acrB sequences containing SNPs of interest were amplified with high-fidelity polymerase and primers containing BamHI and XbaI sites (AcrB[+] and AcrB[−] [Table 1]). The two restriction enzymes provided directional cloning under the control of the lac-inducible promoter of the high-copy-number plasmid pSportI (Gibco-BRL). DNAs were ligated and electroporated into high efficiency ElectroMAX DH5-αE cells (Invitrogen) and were selected on LB agar medium containing 100 μg of ampicillin (AMP)/ml. The plasmid purified from the respective transformants was digested and screened for inserts of approximately 3.2 kbp. The cloned DNA was expressed with 0.1 mM (final culture concentration) isopropyl-β-d-thiogalactopyranoside (IPTG; Promega).
Drug susceptibilities.
MICs of various compounds for strains harboring acrB allele clones were determined essentially as described previously (9). Briefly, serial 2-fold microdilutions (150 μl) containing a fixed IPTG concentration as appropriate were prepared in 96-well flat-bottom microtiter plates (Falcon; Becton Dickinson). Dilution series were inoculated with a 2% (vol/vol) overnight culture of interest and were incubated statically at 37°C. The MIC was defined as the lowest dilution producing no visible signs of growth after 24 h.
Transcriptional profiling.
Expression profiles of acrA and acrB were obtained by quantitative real-time PCR (qRT-PCR) using the Bio-Rad CFX96 system. Briefly, mRNA was purified from mid-log-phase and stationary-phase growth of the respective isolates in LB medium by using an RNeasy kit (Qiagen). RNA concentrations were quantified spectrophotometrically (NanoDrop) and were standardized to 20 ng/μl. One-step qRT-PCR was conducted on 100 ng template RNA per reaction in a 96-well format using the Express One-Step SYBR GreenER kit (Invitrogen) under the standard cycling and reaction conditions recommended by the manufacturer. Primers for acrB and acrA (AcrBqRT[+] and AcrBqRT[−]; AcrAqRT[+] and AcrAqRT[−] [Table 1]) were designed to target conserved regions lacking SNPs in order to generate short products of 266 and 319 bp, respectively. Standard curves of serial template dilutions were prepared in order to estimate the threshold cycle (CT) range as a function of template concentration. Expression was normalized (by the ΔΔCT method) to that of the control (AG100) by measuring parallel expression of gyrB with primers (GyrBqRT[+] and GyrBqRT[−] [Table 1]) designed to amplify 203 bp of this conserved housekeeping gene (33).
RESULTS
SNP identification in AcrAB-TolC pump components.
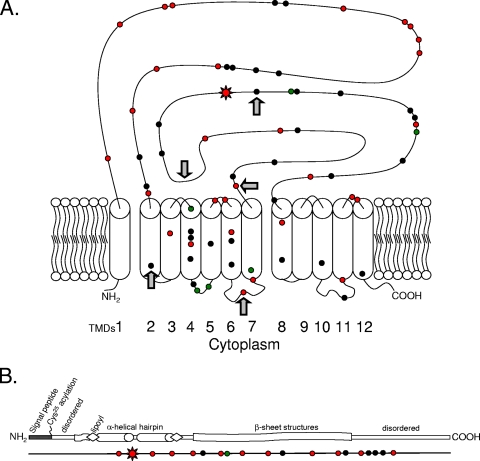
The acrB locus represents a common chromosomal gene archetype of the hydrophobe/amphiphile efflux 1 (HAE1) family of RND transporters (see the Transport Classification Database [http://www.tcdb.org/]) with additional homologs within E. coli and other diverse Gram-negative bacteria. We hypothesized that polymorphisms may segregate with bias to periplasmic loop regions of acrB to generate new or stronger adaptive resistances. However, the study evolved to address the gene in its entirety with a comparative assessment of its system component, acrA, which is not known to participate in substrate specificity. Thus, our initial hypothesis lacked support when we assembled the sequences of acrA and acrB loci from 45 strains of rodent (n = 13), bovine (n = 25), and catfish (n = 7) origin. Polymorphisms did not appear to segregate in the substrate binding regions of acrB but were consistent throughout the gene (Fig. 1A). A total of 69 different polymorphisms in the strain pool (2.2% variation from K-12) were identified, with only 6 strains identical to K-12. Similar results were obtained with acrA, which had 19 polymorphisms (1.6% variation from K-12, with only 8 strains identical [Fig. 1B]), and each of the genes contained only one example of a nonsynonymous mutation (AcrA had T104A in P15 and P18; AcrB had Q733R in P7 and P10). As expected, the vast majority of the polymorphisms were transitions (78% of the polymorphisms in acrA and 84% of those in acrB), of which the majority were G↔ A (66% and 55%, respectively). Interestingly, a majority of the polymorphisms were common only to antibiotic-exposed pools (73% in acrA and 53% in acrB), and none were unique to the (unexposed) rodent pool (Fig. 1; see Tables S1 and S2 in the supplemental material for individual SNPs, their locations, and strain carriage).
FIG. 1.
SNP locations relative to the primary protein sequence and secondary structures using E. coli K-12 as a reference. Symbols: black circles, SNP observed in more than one strain pool; red or green circles, SNP observed in one or more strains but unique to P-series or N-series pools, respectively; red starbursts, SNP resulting in a predicted amino acid change. (A) AcrB with predicted recombination points (arrows). (B) Linear protein sequence schematic of AcrA, with corresponding SNP locations shown below and in scale.
Phylogenetic relationships of pump components.
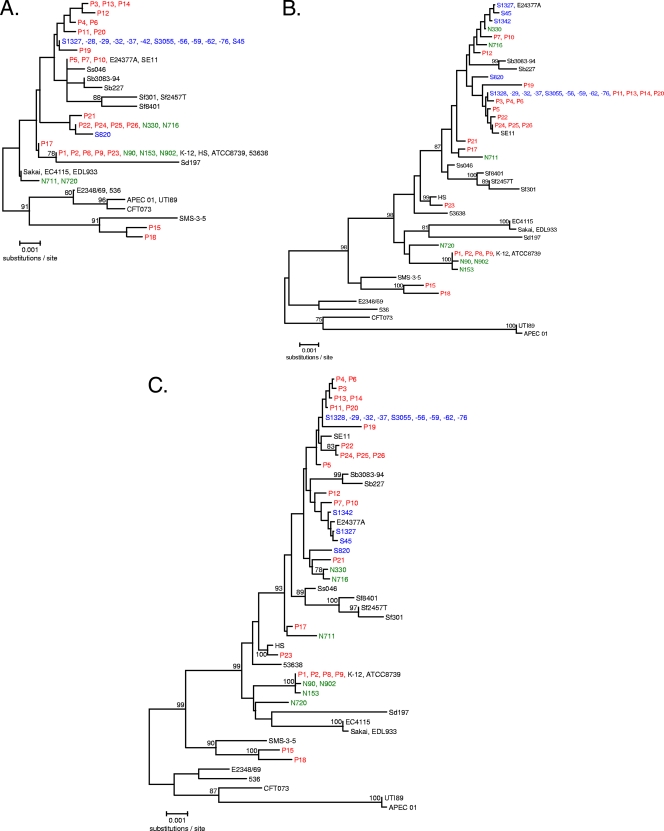
Unrooted neighbor-joining trees of acrA, acrB, and their concatenated sequences revealed some interesting progressive trends in the absence of strong bootstrap values for sequences derived in this study. First, the relationships observed for strains in an acrA-generated tree (Fig. 2A) generally seemed to be reinforced in the acrB-specific tree (Fig. 2B). While this may not be unexpected for sequences derived from a single strain, it suggests a limited role for SNPs in acrB-mediated adaptive drug resistance, which would be displayed as divergent from acrA relationships. Second, using the most informative composite tree (Fig. 2C), some source-specific clustering/relatedness was evident. It should be noted specifically for the “P-series” isolates that some branches include several strains producing identical sequences yet are clearly clonally distinct isolates (5). Third, when analyzed in concert with E. coli and Shigella sp. pathogens with similar levels of sequence variation (<3%), the strains from our study generally grouped only with other commensals, except for E24377A (enterotoxigenic E. coli [ETEC]) and possibly SMS-3-5 (an environmental isolate). However, some established relationships among pathogens were supported with high bootstrap values for acrAB sequences at their respective nodes (or parent nodes). Interestingly, some interrelatedness between pathogens and commensal strains is observed across the tree topologies.
FIG. 2.
Phylogenetic relationships of acrA (A), acrB (B), and concatenated acrAB (C) alleles generated in this study and derived from genome sequences available in public sequence repositories with <3% divergence from E. coli K-12. Bootstrap values of >75 from 500 replicates are shown. N-, P-, and S-series strains are shown in green, red, and blue, respectively, while genomic sequences are in black. Genome sequences are as follows: human commensal E. coli strains ATCC 8739 (also referred to as “C”), HS, K-12, and SE11; environmental E. coli strain SMS-3-5 (from an industrial-metal-contaminated coastal environment); enterohemorrhagic E. coli (EHEC) strains EC4115, EDL933, and Sakai; enteroinvasive E. coli (EIEC) strain 53638; enteropathogenic E. coli (EPEC) strain E2348/69; ETEC strain E24377A; uropathogenic E. coli (UPEC) strains 536, CFT073, and UTI89; avian-pathogenic E. coli (APEC) strain APEC O1; Shigella boydii serotype 4 strain Sb227; S. dysenteriae serotype 1 strain Sd197; S. flexneri serotype 2a strains 2457T and 301; S. flexneri serotype 5b strain Sf8401; and S. sonnei strain Ss046.
Mutation rates.
Synonymous (dS) and nonsynonymous (dN) mutation rates were calculated with the newly derived data sets and were compared between the respective pump constituents from genome repositories and multilocus sequence typing (MLST) alleles for E. coli and Shigella spp. (Table 2). Surprisingly, acrB alleles displayed a higher level of sequence conservation than concatenated MLST loci with comparable lengths of sequence interrogation (3 to 4 kbp) in E. coli and Shigella spp. Hence, the dS/dN ratio for acrB revealed an approximately 49-fold preference for synonymous polymorphisms, compared with approximately 35-fold for E. coli and Shigella spp. MLST. However, this comparison is ultimately biased due to the number of commensal sequences generated in this study, whereas, in a more conservative comparison, the acrB sequences from available sequenced genomes still reveal a slightly higher preference (35.2-fold) for synonymous mutations than their corresponding MLST profiles (Table 2). An inherent sampling bias also exists with one gene locus versus a less restricted approach using multiple geographically distinct segments of the genome (as with MLST). Comparatively, acrA scored lower against both acrB and MLST loci yet included only approximately one-third of the sites in the sequence interrogation. Overall, the generally high dS/dN values of acrA and acrB were indicative of strong genetic conservation under purifying selection.
TABLE 2.
Comparative mutation rates and predicted recombination sites for acrA, acrB, and MLST alleles
| Locus | No. of: |
dSa | dNb | dS/dN | φwc | GARD breakpointsd | |||
|---|---|---|---|---|---|---|---|---|---|
| Loci | Unique alleles | Sites | Variable sites | ||||||
| acrAe | 1 | 26 | 1,194 | 43 | 0.0180 ± 0.0033 | 0.0014 ± 0.0005 | 12.86 | 0.0464 | None |
| acrA (genome)f | 1 | 13 | 1,194 | 36 | 0.0198 ± 0.0039 | 0.0024 ± 0.0008 | 8.25 | 0.1310 | |
| acrB | 1 | 42 | 3,150 | 170 | 0.0295 ± 0.0029 | 0.0006 ± 0.0001 | 49.17 | 1.8E-04 | 1065, (1563),g 2064 |
| acrB (genome) | 1 | 20 | 3,150 | 154 | 0.0388 ± 0.0038 | 0.0011 ± 0.0003 | 35.27 | 8.8E-05 | 1065, 1701, 2238 |
| MLSTh (genome) | 7 | 19 | 3,732 | 243 | 0.0554 ± 0.0041 | 0.0016 ± 0.0003 | 34.63 | N/A | N/A |
Mean synonymous mutation rate per synonymous site ± standard error.
Mean nonsynonymous mutation rate per nonsynonymous site ± standard error.
Calculated using SplitsTree4, with a value of <0.05 suggesting potential recombination. N/A, not applicable.
Nucleotide locations of recombination breakpoints in the acrB coding sequence (see also Fig. 1A, arrows).
Complete allele group consisting of isolates in this study and sequences from genome repositories (see the legend to Fig. 2).
Allele group consisting only of available sequences from genome repositories.
(1563), additional breakpoint identified in intragenic fragment 1065 to 2064 when analyzed independently.
Concatenated E. coli MLST alleles: aspC, clpX, fadD, icdA, lysP, mdh, and uidA (18).
Recombination analysis.
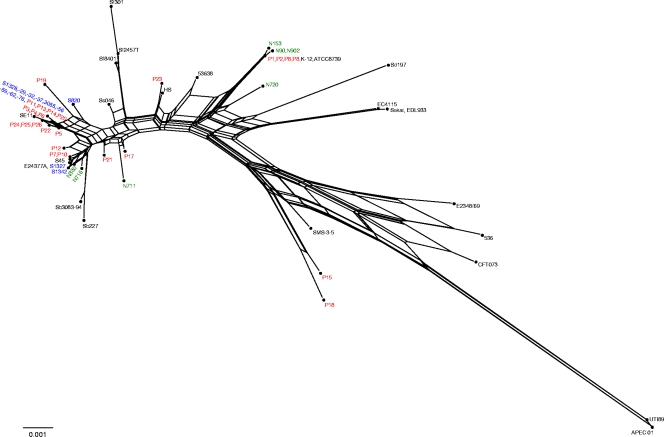
The acrB SNP patterns were further analyzed for hybrid alleles, which are indicative of recombination events (or, alternatively, of recurrent mutations producing parallel evolutionary pathways). This is most evident when one is viewing phylogenetic relationships in a neighbor-net analysis (shown in Fig. 3 for acrB) as opposed to forcing relationships into the traditional bifurcating tree. As with the trees, the low bootstrap values contribute to the number of large parallelograms (conflicting phylogenetic signals) but also underscore the generally well conserved nature of the gene. However, the φw recombination test indicated that some hybrid species also exist (Table 2). The recombination breakpoints within acrB (none are indicated for acrA) were calculated using two different allele groups that divided the gene similarly into four sections at nucleotides 1065, 1563, and 2232 or at nucleotides 1065, 1701, and 2238 (Table 2; Fig. 1A).
FIG. 3.
A neighbor-net analysis of acrB alleles reveals several large parallel paths, indicative of phylogenetic incompatibilities, that are the result of recombination and/or recurrent mutation.
Functional analysis of natural acrB polymorphisms.
Although only one nonsynonymous mutation was observed in acrB during the course of this study, a previous report suggested that even synonymous changes can alter the structure and hence the substrate specificity of the major mammalian multidrug efflux pump, MDR1 (P-gp) (16). Clones of acrB sequences in this study were prepared from noteworthy alleles and were tested for complementation in a ΔacrB background (AG102MB). Thus, we prepared clones from strain P1, which is identical to K-12 in acrB (pP1); strain P10, which harbors the nonsynonymous Q733R mutation background (pP10); and strain S1328, which represents a common silent SNP background containing 28 of the 69 polymorphisms observed in acrB, including several (22 of 28) in common with P10 (pS1328) (see Table S1 in the supplemental material). Interestingly, several additional synonymous and nonsynonymous mutations were generated during the cloning process (data not shown), an effect observed previously by us (10) that underscores the necessity of verifying cloned sequences of these large transporter proteins when overexpressed. However, since the physiological and phenotypic impacts of these “selected” secondary silent and nonsilent mutations remain unclear, a thorough DNA sequence screening was conducted to eliminate clones with these additional, otherwise unrelated mutations.
Regardless, we were able to distill this clone survey into useful species for MIC analyses using common pump substrates that included drugs, dyes, and natural bile acid detergents (Table 3). MICs for the strain background (AG102MB) with and without the cloning vector (pSportI) under IPTG induction conditions displayed high-level kanamycin (KAN) and AMP resistance, indicative of acrB inactivation and/or plasmid-mediated selection, respectively. These two data sets were otherwise similar but with some generally modest depression in resistance resulting from plasmid induction. We identified clones pP1 and pP10, which provided “clean” allele sequence backgrounds enabling us to compare native K-12 acrB complementation with the observed Q733R mutation. In this case, the mutation (with associated silent SNPs in the P10 background) caused some minor (2-fold) decreases in resistance to ciprofloxacin (CIP), ethidium bromide (EtBr), and taurocholic acid (TCA) and an increase in resistance to chloramphenicol (CHL) compared to the resistance observed with pP1 (native acrB). As expected, pP1 complementation generated severalfold increases in resistance to each of the substrates in the study (except for AMP and KAN) over that for the vector-only control (pSportI), suggesting our system was quite functional. Of the S1328 series, we were able to obtain a clone, pS1328, that provided only one additional silent mutation (GGT→GGC at position 2388) relative to the respective S1328 SNP background and likewise produced only 2-fold fluctuations from pP1 but with increases in resistance to cholic acid (CA), CIP, tetracycline (TET), erythromycin (ERY) (4-fold), and potentially tylosin (TYL) and a decrease in resistance to EtBr. Thus, our analysis suggested some modest contributions of silent SNPs or Q733R to drug resistance, but it remains to be determined whether the mutations significantly affect hitherto unknown natural or synthetic substrates or have physiological impacts.
TABLE 3.
Drug, dye, and detergent susceptibilities of the major efflux-deficient E. coli strain AG102MB complemented with acrB SNP allele clones
| Strain or clonea | MICb (μg/ml) of the following compoundc: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CHL | CA | CIP | CV | ERY | EtBr | FA | KAN | NOV | OLE | TCA | TET | TYL | |
| AG102MB | 16 | 2 | 4 | 0.063 | 0.5 | 4 | 4 | 16 | >1,024 | 4 | 256 | 8 | 1 | 128 |
| pSportI | >1,024 | 4 | 4 | 0.004 | 0.5 | 8 | 2 | 8 | >1,024 | 8 | 64 | 4 | 1 | 16 |
| pP1 | >1,024 | 16 | 8 | 0.5 | 4 | 128 | 128 | >512 | >1,024 | 512 | >1,024 | 32 | 8 | 1,024 |
| pP10 | >1,024 | 32 | 8 | 0.25 | 4 | 128 | 64 | >512 | >1,024 | 512 | >1,024 | 16 | 8 | 1,024 |
| pS1328 | >1,024 | 16 | 16 | 1 | 4 | 512 | 64 | >512 | >1,024 | 512 | >1,024 | 32 | 16 | >1,024 |
pSportI-based acrB clones expressed with 0.1 mM IPTG (final culture concentration) in AG102MB.
MIC values of all compounds except bile acids are reported in micrograms per milliliter. MIC values of bile acids (cholic acid and taurocholic acid) are reported in milligrams per milliliter.
Abbreviations: CA, cholic acid; CV, crystal violet; FA, fusidic acid; NOV, novobiocin; OLE, oleandomycin.
Expression profiling.
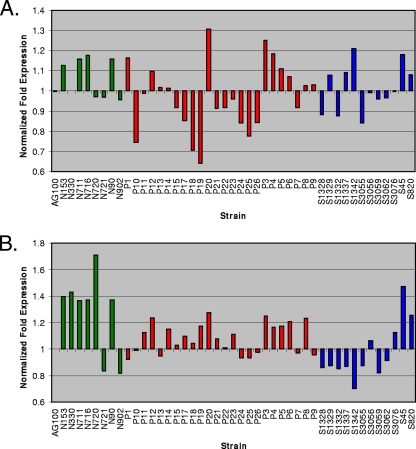
Previous studies have suggested a role for environment-induced hyperexpression of multidrug efflux pumps as an adaptive measure for bacterial drug resistance (4, 26, 33). As a corollary to our study, we measured expression profiles for each of the strains in log phase by real-time PCR employing AG100 as a benchmark (Fig. 4). After internal normalization of each sample to endogenous gyrB levels, the expression profiles revealed only modest fluctuations (<1.8-fold), with mostly inconsistent trends observed between acrA (Fig. 4A) and acrB (Fig. 4B), suggesting some potential independent regulation (representative profiles are shown). In addition, acrB appeared somewhat hyperexpressed in antibiotic-exposed pools in several cases (64% of bovine and 86% of catfish versus 31% of rodent isolate pools). The experiment was also conducted under stationary-phase conditions without any reproducible or noteworthy observations (data not shown).
FIG. 4.
Quantitative RT-PCR of log-phase acrA (A) and acrB (B) mRNA expression relative to expression in AG100, with internal normalization to gyrB levels.
DISCUSSION
The role of bacterial efflux pump nucleotide polymorphisms (SNP identification) in acquired drug resistance has not been addressed previously in the literature, presumably because of the sheer number and size of some pump genes. In this study, we have identified 69 SNPs in the 3,149-bp E. coli K-12 acrB gene sequence; approximately half of these 69 SNPs are conserved in isolates from disparate animal pools. These SNPs did not segregate in a biased way with regard to any structural significance in the final protein product. Comparatively, these observations were similar to those obtained with the cognate pump component, acrA. Interestingly, drug exposure (ceftiofur or oxytetracycline/Romet 30) seemed to produce certain SNPs that were unique to isolate source pools (N and P series), whereas we failed to identify SNPs from rodent isolates (“S series”) that were unique to this source pool. The significance of these findings is unclear. Thus, it is not known whether conserved gene-specific SNPs may have structural ramifications, but they may be potentially useful in future pursuits for source-tracking methodologies or strain history determinations.
Interestingly, conserved silent polymorphisms have recently been shown to affect the cotranslational membrane insertion (and functional structure) of the major mammalian efflux system, P-gp. This occurs through transitioning to potentially rare codons that cause tRNA species depletion, leading to ribosome stalling, thereby affecting cotranslational folding and the timing of membrane insertion (31). Such a mechanism has been implicated in altered substrate specificity and, hence, in alterations in therapeutic drug resistance (16). Our results using functional complementation with MICs as a metric were less straightforward; they were repeatable but may require further explanation with some structural analysis or quantitative measurement of efflux capacity. Alternatively, or in addition, the use of a larger panel of “substrates” may prove enlightening in identifying additional changes in MICs or efflux that are more quantitatively significant than those observed in this study. Unfortunately, these pursuits are beyond the scope of this study, since there is little rationale for drug inclusion, but once identified, these changes may also prove physiologically noteworthy.
It is also noteworthy that clonally distinct isolates produced identical nonsynonymous SNPs. With acrB, P7 and P10 exhibited a more basic (charged) amino acid substitution (Q733R), yet the two isolates display different pulsed-field gel electrophoresis (PFGE) and serotype profiles and slightly different drug resistances. Hence, the P7 serotype is O111:H+, whereas that of P10 is O15/O145:H11; P7 carries the stx1 gene and spectinomycin resistance, whereas P10 lacks stx1 and carries florfenicol resistance (5). Likewise with acrA, P15 and P18 are also clonally distinct, with different PFGE profiles, serotypes (O2:H30 versus O33:H17, respectively), resistance profiles (P15 is resistant to CHL and florfenicol), and virulence gene profiles (P15 carries F1845 fimbriae) (5), yet both conserve a nonsynonymous SNP producing the T104A mutation. The possibility that Q733R and T104A may represent old events in the history of the strain or may have unknown advantageous phenotypes may provide some interpretation of these sequence conservations.
Our results involving recombination breakpoints and the modular nature of transporter evolution may argue for an evolutionary scale of SNP acquisition for acrB. Interestingly, the predicted breakpoint at position 1563 divides the protein product into two topologically symmetrical halves (between transmembranes 6 and 7 [Fig. 1A]) that correspond to the primary predicted internal duplication event in RND transporter evolution (28). Furthermore, the breakpoint at position 1065 is conserved in both allele groups (Table 2) and corresponds to the transmembrane 1 through 2 coding regions, which include the large hydrophilic (periplasmic) domain characteristic of RND transporters (32). Given the modular nature of transporter evolution, this region may have been acquired independently of other transmembrane domains. Thus, these two breakpoints suggest an ancient origin for the hybrid SNP patterns observed in this study when taken in aggregate with the conserved nature of certain SNPs in disparate animal reservoirs. In contrast, the other potential breakpoints at either 1701, 2064, or 2232, which divide the protein in the large periplasmic loop 2-encoding domain, would argue for an adaptive functional role, given that certain substrate specificities are mapped in part to this region (30, 35). Thus, it remains interesting (from an adaptive perspective) that the only nonsynonymous mutations observed in this study for acrB and acrA map well within a loop region important in pump substrate specificity (30, 35) and within the structurally significant coiled-coil feature characteristic of membrane fusion proteins (12, 15), respectively (Fig. 1).
Previous research involving adaptive resistance mechanisms have focused on regulation-based responses affecting efflux pump expression (4, 26, 33). In our study, we failed to demonstrate striking changes in acrAB expression levels between the pools of isolates. This observation may not be all that surprising, considering the constitutive nature of acrAB-tolC expression in the E. coli background, although it has been reported that some hyperexpression of this system is typically induced upon entry into stationary phase. However, many other pump systems in the genome (with the exception of EmrAB-TolC) are expressed at low levels—if at all—under normal conditions. As a result, regulators of these systems can be responsive to drug induction or can be subject to mutations that cause their overexpression (19). In particular, systems providing resistance to non-AcrAB-TolC substrates may prove to be the most responsive under such conditions. Apart from the potential contributions of other pump systems, the study may have benefited from collecting and identifying strains a priori with constitutive hyperexpression of acrAB-tolC, since the study focused only on structural SNPs rather than on promoter regions or other nonencoding regulatory regions. This approach would naturally predispose any adaptive response involving further increases in resistance toward improved gene function.
Some of the issues discussed here suggest roles for multidrug efflux pumps apart from bacterial drug resistance. Certainly, these pumps are pervasive, with functional homologs in humans for anticancer and chemotherapy resistance (P-gp) and antifungal and antiprotozoal clinical resistance (27). Moreover, bacterial acrB RND-specific homologs exist in diverse genera, including Pseudomonas, Salmonella, Agrobacterium, Neisseria, Enterobacter, Burkholderia, Campylobacter, Vibrio, Francisella, Acinetobacter, and Serratia. Indeed, the association of this system with bacterial virulence and colonization is of particular relevance (23). Thus, tripartite AcrB-type systems may be required for Neisseria gonorrhoeae to colonize the genital tract (14) and for Salmonella to colonize the avian gut or invade macrophages (3). Moreover, recent observations made first by our group (7, 8) and subsequently by others (34) using transcriptomic analyses with knockouts of this pump system suggest global effects central to cell physiological systems. As a corollary, addressing the impact of gene-specific SNPssing the impact of gene-specific SNPs (synonymous and nonsynonymous) may require additional sensitive assay systems to assess cell fitness in particular niches, which may represent interesting future subjects of research. Using this perspective, the various nonsynonymous mutations arising (or “selected,” depending on one's perspective) during the cloning and overexpression of acrB in plasmid vectors may indeed have an as yet undefined functional or stabilizing role.
Supplementary Material
Acknowledgments
We thank Bhushan M. Jayarao (PSU) and Mohamed Nawaz (NCTR) for access to E. coli isolates, and we thank Roger Steele (NCTR) for isolating rodent strains. We also thank Tammy Barnaba (CFSAN) for performing some additional MIC determinations during the review and revision process.
The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant, D., and V. Moulton. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 3.Buckley, A. M., M. A. Webber, S. Cooles, L. P. Randall, R. M. La Ragione, M. J. Woodward, and L. J. Piddock. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8:847-856. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S., S. Cui, P. F. McDermott, S. Zhao, D. G. White, I. Paulsen, and J. Meng. 2007. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson, S. C., B. A. Straley, N. V. Hegde, A. A. Sawant, C. DebRoy, and B. M. Jayarao. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkins, C. A., and K. E. Beenken. 2005. Modeling the tripartite drug efflux pump archetype: structural and functional studies of the macromolecular constituents reveal more than their names imply. J. Chemother. 17:581-592. [DOI] [PubMed] [Google Scholar]
- 7.Elkins, C. A., T. Han, L. B. Mullis, and J. C. Fuscoe. 2008. Comparative transcriptomic analysis of AcrA function: the membrane fusion protein of the major tripartite multidrug efflux pump of Escherichia coli, abstr. K-015. Abstr. 108th Gen. Meet. Am. Soc. Microbiol., Boston, MA.
- 8.Elkins, C. A., T. Han, M. E. Munoz, L. B. Mullis, and J. C. Fuscoe. 2007. Functional transcriptomic analysis of the major tripartite multidrug efflux pumps AcrAB-TolC and EmrAB-TolC of Escherichia coli, abstr. K-034. Abstr. 107th Gen. Meet. Am. Soc. Microbiol., Toronto, Ontario, Canada.
- 9.Elkins, C. A., and L. B. Mullis. 2007. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob. Agents Chemother. 51:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. U. S. A. 101:9994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 14.Jerse, A. E., N. D. Sharma, A. N. Simms, E. T. Crow, L. A. Snyder, and W. M. Shafer. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 71:5576-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, J. M., and G. M. Church. 1999. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 287:695-715. [DOI] [PubMed] [Google Scholar]
- 16.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525-528. [DOI] [PubMed] [Google Scholar]
- 17.Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk, and S. D. Frost. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891-1901. [DOI] [PubMed] [Google Scholar]
- 18.Lacher, D. W., H. Steinsland, T. E. Blank, M. S. Donnenberg, and T. S. Whittam. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189:342-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 20.Mao, W., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 21.Nawaz, M., A. A. Khan, S. Khan, K. Sung, K. Kerdahi, and R. Steele. 2009. Molecular characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathog. Dis. 6:553-559. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishino, K., E. Nikaido, and A. Yamaguchi. 2009. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794:834-843. [DOI] [PubMed] [Google Scholar]
- 24.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, N. K., D. M. Dixon, S. M. Holland, and A. S. Fauci. 2008. The research agenda of the National Institute of Allergy and Infectious Diseases for antimicrobial resistance. J. Infect. Dis. 197:1087-1093. [DOI] [PubMed] [Google Scholar]
- 26.Poole, K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39:162-176. [DOI] [PubMed] [Google Scholar]
- 27.Saier, M. H., Jr., and I. T. Paulsen. 2001. Phylogeny of multidrug transporters. Semin. Cell Dev. Biol. 12:205-213. [DOI] [PubMed] [Google Scholar]
- 28.Saier, M. H., Jr., R. Tam, A. Reizer, and J. Reizer. 1994. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol. Microbiol. 11:841-847. [DOI] [PubMed] [Google Scholar]
- 29.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 30.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai, C. J., Z. E. Sauna, C. Kimchi-Sarfaty, S. V. Ambudkar, M. M. Gottesman, and R. Nussinov. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 383:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 33.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webber, M. A., A. M. Bailey, J. M. Blair, E. Morgan, M. P. Stevens, J. C. Hinton, A. Ivens, J. Wain, and L. J. Piddock. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.