Abstract
In Neisseria gonorrhoeae, the mosaic type of penA, which encodes penicillin-binding protein 2 (PBP 2), is associated with reduced susceptibility to oral cephalosporins. To investigate the relatedness of N. gonorrhoeae clinical isolates with reduced susceptibility, we sequenced the penA genes of 32 isolates. Five different amino acid sequence types of PBP 2 were identified, but all seemed to be derivatives of pattern X of PBP 2 (PBP 2-X). However, multilocus sequence typing of the isolates showed that the isolates belonged to six different sequence types. As PBP 2-X was identified in three different sequence types, horizontal transfer of the penA allele encoding PBP2-X was suggested. We demonstrated that the penA gene could be transferred from an isolate with reduced susceptibility to a sensitive isolate by natural transformation. Comparison of the sequence of the penA-flanking regions of 12 transformants with those of the donor and the recipient suggested that at least a 4-kb DNA segment, including the penA gene, was transferred. During horizontal transfer, some of the penA alleles also acquired variations due to point mutations and genetic exchange within the allele. Our results provide evidence that the capacity for natural transformation in N. gonorrhoeae plays a role in the spread of chromosomal antibiotic resistance genes and the generation of diversity in such genes.
Neisseria gonorrhoeae is one of the most common sexually transmissible infective agents. Humans are the only natural host for N. gonorrhoeae, and transmission is restricted to direct person-to-person sexual contact. As there is no vaccine for gonorrhea, the control of dissemination depends on timely identification and initiation of an appropriate antibiotic treatment for the infected person in order to prevent transmission.
N. gonorrhoeae strains that are resistant to various types of antibiotics have emerged, causing critical concern for public health around the world. Resistance to oral cephalosporins, such as cefixime, is emerging (2, 3, 10, 18), and approximately 30% of N. gonorrhoeae isolates in Japan now show reduced susceptibility to cefixime (20). The molecular mechanism of resistance has been elucidated as the formation of a mosaic structure of penA-encoded penicillin-binding protein 2 (PBP 2). The mosaic penA was generated by interspecies recombination with other neisserial species (3, 10), which is the same mechanism for chromosomally mediated penicillin resistance in N. gonorrhoeae (23). However, the precise junctions of recombination have not been fully elucidated.
penA-encoded PBP 2 proteins of N. gonorrhoeae are divided into several types on the basis of the amino acid sequence, and some of these types are associated with reduced susceptibility to cefixime (10, 14, 25, 27). Among these, the most common PBP 2 type is pattern X (PBP 2-X), implying the expansion of a single clone. According to the spread of isolates with reduced susceptibility to cefixime, the expansion of a single clone, which emerged at an early phase, is suggested (18). However, another possibility is that recombination of the penA gene occurred several times independently, followed by multiclonal expansion. Understanding of the mode of spread of antibiotic-resistant clones could help us construct a public health strategy for preventing the further spread of resistant clones.
To investigate the mode of dissemination of the newly emerged antibiotic-resistant N. gonorrhoeae isolates, we retrospectively characterized isolates with reduced susceptibility to cefixime (cefixime MIC ≥ 0.25 μg/ml, referred to hereafter as CefRs isolates), using penA sequencing and multilocus sequence typing (MLST) with seven housekeeping genes. We also examined whether the horizontal transfer of penA occurred in vitro, resulting in the one-step emergence of CefRs isolates from the susceptible isolate.
MATERIALS AND METHODS
Strains.
The Kanagawa Prefectural Institute of Public Health is a reference laboratory for N. gonorrhoeae in Kanagawa Prefecture, Japan, where the primary isolation of N. gonorrhoeae from clinical specimens collected at nine hospitals was carried out. The N. gonorrhoeae isolates were identified and stored as described previously (11, 28). A total of 32 N. gonorrhoeae clinical isolates with reduced susceptibility to cefixime comprising 3 to 7 CefRs isolates randomly selected from each year were examined (Table 1). N. gonorrhoeae isolates that belonged to sequence type (ST) 1901 (ST1901) were used for comparison of the sequences of the penA-flanking regions that we analyzed. Isolates NGON03-079, NGON03-092, NGON130-115, and NGON07-002 were collected at another hospital in Tokyo (Table 1). The MICs of cefixime and ciprofloxacin were determined by the agar dilution method (19).
TABLE 1.
Description of Neisseria gonorrhoeae isolates from Kanagawa Prefecture and Tokyo with reduced susceptibility to cefixime, 1998 to 2005
| Isolate | Yr of isolation | Hospital | Sexa | Specimenb | Cefixime MIC (μg/ml) | MLST ST | PBP 2 type |
|---|---|---|---|---|---|---|---|
| NG9806 | 1998 | Kanagawa, H1 | M | U | 0.25 | 7363 | X |
| NG9811 | 1998 | Kanagawa, H1 | F | VD | 0.25 | 7363 | X |
| NG9812 | 1998 | Kanagawa, H1 | F | VD | 0.25 | 7363 | X |
| NG9911 | 1999 | Kanagawa, H2 | M | UD | 0.25 | 7363 | X |
| NG9912 | 1999 | Kanagawa, H3 | M | U | 0.25 | 7363 | X |
| NG9913 | 1999 | Kanagawa, H3 | M | U | 0.25 | 7363 | X |
| NG9914 | 1999 | Tokyo, H9 | M | U | 0.25 | 7363 | X |
| NG0002 | 2000 | Kanagawa, H4 | F | VD | 0.25 | 1901 | X |
| NG0003 | 2000 | Kanagawa, H2 | M | UD | 0.25 | 7363 | X |
| NG0008 | 2000 | Kanagawa, H2 | M | UD | 0.25 | 7363 | X |
| NG0109 | 2001 | Kanagawa, H6 | M | UD | 0.25 | 7363 | X |
| NG0110 | 2001 | Kanagawa, H5 | M | UD | 0.25 | 1596 | X |
| NG0111 | 2001 | Tokyo, H9 | M | U | 0.25 | 1590 | XXXI |
| NG0201 | 2002 | Kanagawa, H7 | M | UD | 0.25 | 1596 | X |
| NG0204 | 2002 | Kanagawa, H7 | M | UD | 0.25 | 7363 | X |
| NG0205 | 2002 | Kanagawa, H2 | M | UD | 0.25 | 7363 | X |
| NG0206 | 2002 | Kanagawa, H5 | F | VD | 0.25 | 7363 | X |
| NG0207 | 2002 | Kanagawa, H5 | M | UD | 0.25 | 7363 | X |
| NG0303 | 2003 | Kanagawa, H3 | M | — | 0.25 | 7363 | X |
| NG0304 | 2003 | Tokyo, H9 | M | U | 1.0 | 7363 | XXX |
| NG0311 | 2003 | Tokyo, H9 | M | U | 0.5 | 7363 | XXX |
| NG0312 | 2003 | Tokyo, H9 | M | U | 0.5 | 7358 | XXVI |
| NG0401 | 2004 | Kanagawa, H7 | M | UD | 0.5 | 7363 | X |
| NG0404 | 2004 | Kanagawa, H8 | M | U | 0.5 | 7363 | X |
| NG0410 | 2004 | Tokyo, H9 | M | U | 0.5 | 7363 | X |
| NG0503 | 2005 | Kanagawa, H3 | M | U | 0.25 | 1901 | XXXII |
| NG0508 | 2005 | Kanagawa, H5 | M | — | 0.25 | 1596 | X |
| NG0509 | 2005 | Kanagawa, H3 | M | UD | 0.25 | 7363 | X |
| NG0511 | 2005 | Kanagawa, H3 | F | VD | 0.25 | 7363 | X |
| NG0512 | 2005 | Kanagawa, H3 | M | UD | 0.25 | 1588 | X |
| NG0513 | 2005 | Kanagawa, H3 | F | VD | 0.5 | 1901 | XXXII |
| NG0514 | 2005 | Kanagawa, H8 | M | U | 0.25 | 7363 | X |
| NG0202c | 2002 | Kanagawa, H7 | M | UD | <0.008 | 1901 | V |
| NG0402c | 2004 | Kanagawa, H5 | M | U | 0.031 | 1901 | V |
| NGON03-079c | 2003 | Tokyo, H10 | M | UD | 0.5 | 1901 | X |
| NGON03-092c | 2003 | Tokyo, H10 | M | UD | 0.25 | 1901 | X |
| NGON03-115c | 2003 | Tokyo, H10 | F | U | 0.5 | 1901 | X |
| NGON07-002c | 2007 | Tokyo, H10 | M | U | 0.25 | 1901 | X |
M, male; F, female.
U, urine; VD, vaginal discharge; UD, urethral discharge; —, no information.
ST1901 isolates used for analysis of penA-flanking region.
Sequencing of penA and the penA-flanking region.
To obtain genomic DNA, the clinical strains were suspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and boiled for 10 min. After the cell debris was removed by centrifugation, the supernatant was used directly as the template DNA for PCR. The penA gene was amplified and sequenced by using primers penA_F and penA_R (Table 2). The PCR mixtures were incubated for 2 min at 96°C, followed by 30 cycles of 10 s at 96°C, 10 s at 65°C, and 2 min at 72°C. Purification of the PCR products was done with an ExoSAP IT kit (GE Healthcare). Sequencing was carried out with the appropriate sequencing primers and an ABI BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems), followed by purification of the termination products. Both strands of the products were sequenced by use of an ABI 3130 xl sequencer. The translated amino acid sequences were compared with known PBP 2 amino acid sequences. Newly identified types were designated XXX to XXXII, as described by Ito et al. (10) and Whiley et al. (27).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | Primer use |
|---|---|---|
| penA_F | CGGGCAATACCTTTATGGTGGAAC | Amplification of penAa |
| penA_R | ACAACGGCGGCGGGGATATAAC | |
| penA_SF1 | CAAAGATAGAAGCAGCCTG | Sequencing of the penA region |
| penA_SF2 | GATATTGACGGCAAAGGTC | |
| penA_SF3 | CTTTGGATGTGCGCGGC | |
| penA_SR1 | GCCGTCGGTATATTCGC | |
| penA_SR2 | CCAAAGGGGTTAACTTGC | |
| penA_SR3 | TTCTCAACAAACCTGCAG | |
| penA_SR4 | CTTTGCCGTTTTGCGGGG | |
| penA_5′R | GCCATCAGGACGAAGCTAATCC | Amplification of the region upstream of penAb |
| mraW_F | GTGAGTGGAGCAGAAAGTTACCG | |
| mraW_S1 | CCGTTACTGGTCATCG | Sequencing of the PCR product from penA_5′R and mraW_F |
| mraW_S2 | TATCGGACCGGCAGTC | |
| mraW_S3 | CCTCGTGCAAATCCTG | |
| mraW_S4 | GGCGGTCAGAGAAGC | |
| penA_3′F | GCGGCAGCCTGAACATCTTGG | Amplification of the region downstream of penAb |
| dcaA_R | GGACACATCGGTAGCGGCTG | |
| murE_S1 | TTCAAGATCGGAAAAACG | Sequencing of PCR product from penA_3′F and |
| murE_S2 | TTGGCACAAAGCAAGG dcaA_R | |
| murE_S3 | TGCGCGGTTTCTTCC | |
| murE_S4 | TCGGACGGTTCAACG | |
| murE_S5 | GCAGGCTTTGTTAACTC | |
| dcaA_S1 | TCAATATCTTAACCGTATC | |
| dcaA_S2 | GCGTATCGGGCAATGG | |
| dcaA_S3 | CGGGAAGATTGCCGAC | |
| dcaA_S4 | GGGGTATTTGCTGACG | |
| dcaA_S5 | AGCTTGGCGAAGCAGG | |
| dcaA_S6 | CGGTTTGATGCATGTCG |
Amplification conditions were 96°C for 2 min and 30 cycles of 96°C for 10 s, 65°C for 10 s, and 72°C for 2 min.
Amplification conditions were 96°C for 2 min and 30 cycles of 96°C for 10 s, 63°C for 10 s, and 72°C for 2 min.
A neighbor-joining tree with 33 PBP 2 amino acid sequences was generated by using the MEGA program (version 4) (22, 26). The reliability of the inferred relatedness was evaluated by the use of bootstrap tests (1,000 replicates) (7).
Amplification of the penA-flanking DNA was done by using primer set penA_3′F and dcaA_R and primer set penA_5′R and mraW_F (Table 2). The PCR mixtures were incubated for 2 min at 96°C, followed by 30 cycles of 10 s at 96°C, 10 s at 63°C, and 2 min at 72°C. The primers listed in Table 2 were used to sequence each PCR product.
MLST.
PCR amplification and sequencing of the seven N. gonorrhoeae housekeeping genes (abcZ, adk, aroE, fumC, gdh, pdhC, and pgm) were undertaken by using a previously described protocol (12). All nucleotide sequences were determined directly from the purified PCR products. After end trimming of the data obtained and editing by using Sequencher software (gene codes), the allele numbers of the STs were assigned by querying the Neisseria MLST database (http://pubmlst.org./neisseria/) (13).
Pulsed-field gel electrophoresis (PFGE).
Agarose plugs into which DNA was embedded were prepared as described previously (10), and the samples were digested with SpeI. The SpeI-digested genomic DNA was analyzed on a 1% agarose gel with 0.5× Tris-boric acid-EDTA buffer at 14°C by using a CHEF Mapper apparatus (Bio-Rad). The run time was 19.5 h at 6 V/cm, and the initial and final switch times were 0.5 and 35 s, respectively. The gel was stained with ethidium bromide.
In vitro genetic exchange of the penA allele during cocultivation.
An in vitro interstrain genetic exchange experiment was performed with strain NG0003 (cefixime MIC, 0.25 μg/ml; ciprofloxacin MIC, 0.031 μg/ml) and strain NG0202 (cefixime MIC, 0.004 μg/ml; ciprofloxacin MIC, 8 μg/ml). NG0202 was selected from 58 isolates susceptible to cefixime (cefixime MIC ≤ 0.125 μg/ml) and on the basis of the ciprofloxacin MICs. The strains were grown on GC agar plates with 5% CO2 for 16 h and then suspended in GC broth. After adjustment of the optical density at 600 nm (OD600) of the culture to 0.02 with GC broth, suspensions of strains NG0003 and NG0202 (500 μl each) were mixed and statically incubated for 16 h. One hundred microliters of sample was placed onto GC agar plates containing both cefixime (0.031 μg/ml) and ciprofloxacin (2 μg/ml) (Cef+Cip GC agar plate) in duplicate. Neither NG0003 nor NG0202 is able to form colonies on this medium. The plates were incubated for 20 h at 37°C with 5% CO2, and the number of colonies on each plate was determined. The viable counts of NG0202 and NG0003 were determined on GC agar plates containing either cefixime (0.031 μg/ml) or ciprofloxacin (2 μg/ml). The experiment was repeated three times. The transformation frequency was estimated on the basis of the number of viable recipient NG0202 colonies that grew on the Cef+Cip agar plates compared to the number of NG0202 colonies that grew on Cip agar plates. The MICs of clones (n = 12) resistant to both cefixime and ciprofloxacin were determined; and MLST typing, PFGE, and sequence analysis of the penA-flanking region of the clones were performed.
Nucleotide sequence accession numbers.
The nucleotide sequences revealed in this study have been deposited in the DDBJ sequence library and assigned accession numbers AB511942 for penA-XXX, AB511943 for penA-XXXI, AB511944 for penA-XXXII, and AB511945 and AB511946 for the penA-flanking regions.
RESULTS
penA sequence variation.
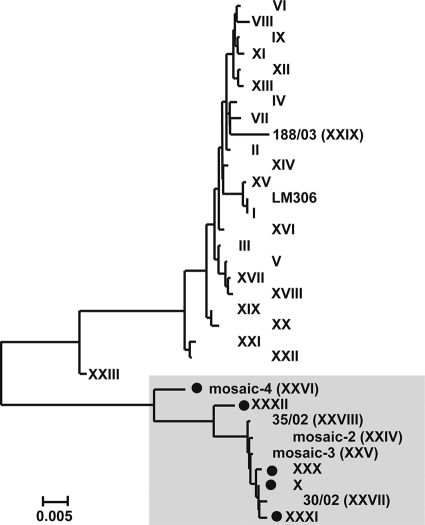
To examine the possibility of the expansion of a single clone with reduced susceptibility to cefixime, we sequenced the penA alleles of the CefRs isolates in our collection. Among 32 CefRs isolates obtained from 1998 to 2005, five PBP 2 types were revealed, including three newly identified types. PBP 2-X was the predominant type (26/32, 81.3%), which is consistent with the findings presented in previous reports (10, 27). PBP XXVI, originally designated mosaic 4 (25), was also found. Newly identified types PBP 2-XXX and PBP 2-XXXI had replacements of Ala by Val at positions 502 and 533, respectively, compared with the sequence of PBP 2-X. PBP 2-XXXII was identical to PBP 2-X from positions 1 to 548, but its C-terminal portion was identical to that of the PBP 2-I allele from a strain that is susceptible to cefixime, strain LM306, suggesting the creation of a new mosaic structure.
Using phylogenetic analysis, we demonstrated that the amino acids sequences of the PBP 2 alleles among the CefRs strains varied; however, the variation was restricted to a cluster, which was distinct from the other cluster formed by the PBP 2 types of Cef-susceptible isolates (Fig. 1), suggesting that penA of the CefRs isolates evolved from a single origin through a point mutation or the replacement of a short segment, such as that in PBP 2-XXXII.
FIG. 1.
Relationships of 33 PBP 2 types. A neighbor-joining tree was constructed from the PBP 2 amino acid sequences. The tree contains the LM306, PBP 2, and the PBP 2 types reported by Ito et al. (10), Whiley et al. (27), Takahata et al. (25), and Lindberg et al. (14) and in this study. The PBP 2 types that resulted in the reduced susceptibility of N. gonorrhoeae to cefixime are shaded in gray. Black dots indicate the PBP 2 types found in this study.
Multilocus sequence typing of CefRs isolates.
In order to examine whether the whole genomes of the CefRs strains were clonal, we applied an MLST strategy. Thirty-two CefRs isolates were divided into six different STs (Table 3), including three singleton STs. The predominant ST was newly assigned ST7363 (n = 23, 71.9%). ST1901 (n = 3) and ST1596 (n = 3) were the second most dominant STs among the CefRs isolates.
TABLE 3.
MLST types and penA alleles of isolates with reduced susceptibility to cefixime
| ST | No. of isolates | Allele at locusa: |
No. of isolates with penA allele: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| abcZ | adk | aroE | fumC | gdh | pdhC | pgm | X | XXVI | XXX | XXXI | XXXII | ||
| 7363 | 23 | 59 | 39 | 67 | 78 | 148 | 153 | 65 | 21 | 0 | 2 | 0 | 0 |
| 1596 | 3 | 59 | 39 | 67 | 78 | 148 | 71 | 65 | 3 | 0 | 0 | 0 | 0 |
| 1588 | 1 | 59 | 39 | 67 | 158 | 148 | 71 | 65 | 1 | 0 | 0 | 0 | 0 |
| 1590 | 1 | 126 | 39 | 67 | 78 | 149 | 153 | 65 | 0 | 0 | 0 | 1 | 0 |
| 7358 | 1 | 109 | 39 | 67 | 78 | 149 | 153 | 133 | 0 | 1 | 0 | 0 | 0 |
| 1901 | 3 | 109 | 39 | 170 | 111 | 148 | 153 | 65 | 1 | 0 | 0 | 0 | 2 |
Boldface data indicate alleles different from that of ST7363.
ST7363 and ST1588 differed from ST1596 only in the pdhC locus and the fumC locus, respectively, suggesting that ST7363, ST1588, and ST1596 are closely related to each other (Table 3). The MLST sequence type might alter during passages in vivo and in vitro due to a point mutation or an interstrain recombinational event. However, the other three STs, ST1901, ST1590, and ST7358, showed at least two differences from the other STs of the CefRs isolates. It is unlikely that this was because of allele exchange in all these isolates, indicating that the concept of the expansion of a single clone of CefRs could not completely explain the spread of CefRs.
Correlation of penA allele type with MLST typing.
If CefRs isolates emerged as different STs through independently generated mosaic structures of the penA allele, we would expect isolates with unique penA alleles in each ST. As shown in Table 3, the penA-X of the dominant PBP, PBP 2-X, was widely distributed in four different STs, ST7363, ST1596, ST1588, and ST1901, while unique penA alleles of PBP 2-XXX and PBP 2-XXXII, which were found in more than two isolates, were detected only in ST7358 and ST1901, respectively. From the results, we speculate that one of the possible reasons for this is that in some CefRs isolates the transfer of the penA-X allele occurs between different N. gonorrhoeae strains.
In vitro transfer of penA gene.
To explore the possibility that the penA-X allele spread between different N. gonorrhoeae strains, we tested whether penA-X could be transferred by the in vitro cocultivation of CefRs isolate (NG0003, ST7363) and a cefixime-susceptible (Cefs) strain (strain NG0202, ST1901). NG0003 is susceptible to ciprofloxacin, and NG0202 is resistant to ciprofloxacin.
When a portion (0.1 ml) of a 16-h static culture of strain NG0003 or strain NG0202 (0.71 × 108 and 1.01 × 108 CFU/ml, respectively) was plated on a Cef+Cip GC agar plate, no colonies appeared, indicating that no spontaneous antibiotic resistance mutations occurred (Table 4). When a mixture of NG0003 and NG0202 was plated after cocultivation for 16 h, we obtained colonies resistant to both drugs (4.33 × 103 CFU/ml) on Cef+Cip GC agar plates (Table 4). We randomly selected 12 colonies from these mutants. All clones were ST1901, and the PFGE profiles of all the resistant clones were identical to the PFGE profile of NG0202 (Fig. 2), suggesting that NG0202 received penA-X from NG0003 and became resistant to cefixime.
TABLE 4.
In vitro transfer of reduced susceptibility to cefixime
| Straina | No. of CFU on plates with: |
||
|---|---|---|---|
| Cefiximeb | Ciprofloxacinc | Cefixime and ciprofloxacind | |
| NG0202 (ST1901) | <10 | 0.71 × 108 | <10 |
| NG0003 (ST7363) | 1.01 × 108 | <10 | <10 |
| NG0202 + NG0003 | 1.03 × 108 | 0.2 × 108 | 4.33 × 103 |
Strain NG0202, strain NG0003, and a suspension of equal numbers of cells of both strains (OD600, 0.02) were incubated for 16 h.
Containing 0.031 μg/ml of cefixime.
Containing 2 μg/ml of ciprofloxacin.
Containing 0.031 μg/ml of cefixime and 2 μg/ml of ciprofloxacin.
FIG. 2.
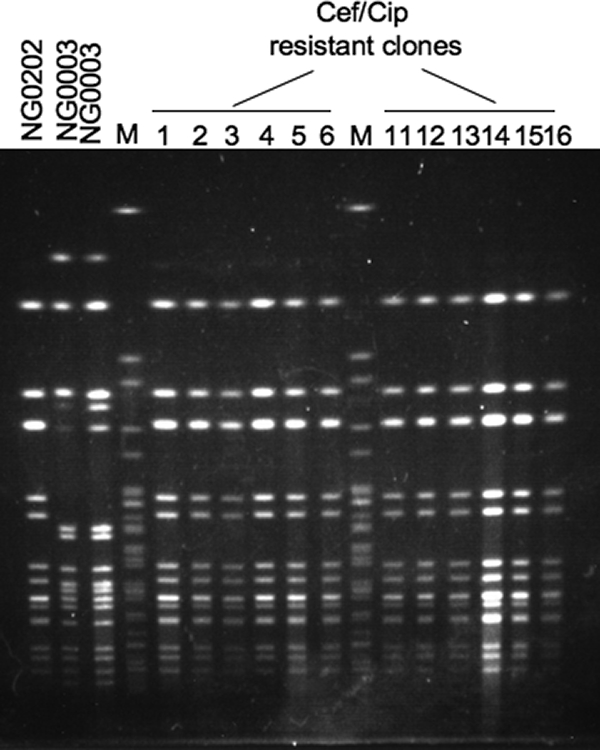
PFGE patterns of clones obtained by in vitro penA-X transfer. NG0202 (Cefs of ST1901) and NG0003 (CefRs of ST7363) were cocultivated overnight, and then colonies that were resistant to both cefixime (Cef) and ciprofloxacin (Cip) were identified by using GC agar plates containing 0.031 μg/ml of cefixime and 2 μg/ml of ciprofloxacin. SpeI-digested genomic DNA from 12 of the clones obtained was analyzed by PFGE. Lanes M, size marker consisting of SpeI-digested Salmonella enterica serovar Braendecup strain H9812 genomic DNA.
The transformation frequency was estimated to be 2.1 × 10−4 (Table 4). When DNase (200 μg/ml) was present in the cocultivated mixture, no colonies resistant to both drugs were obtained, suggesting that the transfer was dependent on naked DNA released from the donor strain in the broth.
Sequence comparison of penA alleles.
To confirm the transfer of the penA-X allele, we determined the nucleotide sequence of the penA allele (1,752 bp) in the double-resistant clones derived from NG0202, which originally possessed a penA-V allele. Sequence diversity between the penA-X and the penA-V alleles was identified at a total of 221 polymorphic sites after nucleotide position 294 of the penA gene, and the overall sequence identity was 87.3%. Eight of 12 clones had the same penA-X allele as NG0003. The clones with the other penA alleles, clones Tf-3, Tf-13, Tf-14, and Tf-15, had alleles highly similar to the alleles in penA-X (99.6 to 99.9%), and all sequence divergences were found between nucleotide positions 294 and 669 of penA (Fig. 3). We concluded that all clones analyzed acquired penA-X or its derivatives and that these were responsible for the reduced susceptibility to cefixime in the clones. We also showed that penA-X allelic diversity was generated in the Tf-3, Tf-13, Tf-14, and Tf-15 clones.
FIG. 3.
Comparison of sequences from nucleotides 241 to 720 among penA-X, penA-V, and minor variants of the transformants. Dashes indicate the same nucleotide as penA-X of strain NG0003 (shown on the first line). Shaded boxes in penA Tf-3 and NG0202 (positions 241 to 417) indicate regions where the sequences between them are identical. The underlined region indicates the possible junction site of Tf-3.
Junction site of recombination of penA-X.
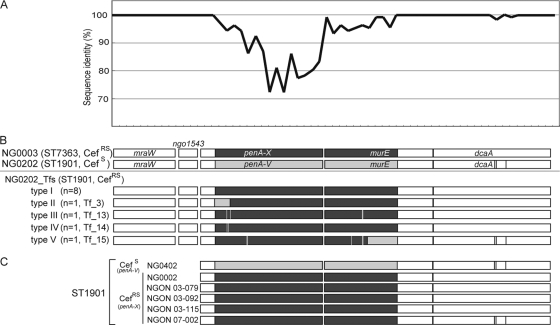
The 5′ portion of penA (positions 1 to 417) in clone Tf-3 was identical to that of penA-V in strain NG0202, while the 3′ portion after nucleotide position 456 was identical to that of penA-X (Fig. 3), implying that a recombination junction site was located between positions 417 and 456 in penA of Tf-3. To determine the junction sites of the other clones, we first sequenced the penA-flanking regions (6,299 bp) in strains NG0003 and NG0202 (Fig. 4A). The overall nucleotide sequence identity of the region between NG0003 and NG0202 was 95.7%, significantly less than the identity of the concatenated seven loci of ST7363 and ST1901 determined by MLST analysis (3 bp different in 3,284 bp; 99.9%). As shown in Fig. 4A, the sequence divergence accumulated in the penA locus and also in the 5′ part of murE. Only three polymorphic sites were identified in the dcaA gene (1,647 bp), at about nucleotide position 5500, outside the highly variable region (Fig. 4A and 4B).
FIG. 4.
Sequence diversity in penA-flanking regions (6,299 bp) among strain NG0003, strain NG0202, and the transformants generated by in vitro cocultivation. (A) Sequence identity of each 100 bp between NG0003 and NG0202. (B) Boxes indicate the five open reading frames in this region, mraW, NGO1543, penA, murE, and dcaA. Gray boxes, the highly variable region; dark gray boxes, sequences that are identical to the sequence of the penA-X-flanking region of NG0003; bright gray boxes, sequences identical to the sequence of the penA-V-flanking region of NG0202; fine vertical lines (white and black), polymorphic sites that match the nucleotide bases of NG0202. (C) Sequence diversity in the penA-murE-dcaA regions of additional clinical isolates of ST1901.
As shown in Fig. 4A and B, since the upstream region (positions 1 to 1590) was highly conserved and there were no polymorphic sites between strains NG0003 and NG0202, we could not determine the left junction site, other than that of Tf-3. As for the right junction site, we detected a possible junction site within the highly variable region in the penA-flanking region of Tf-15 (Fig. 4B). Although we could not determine the right junction site for penA recombination other than that in Tf-15, our analysis of the other 11 clones showed that the nucleotide sequence of dcaA was identical to that of NG0003, indicating that penA-X was replaced along with murE and dcaA.
Sequencing analysis of murE-dcaA region of ST1901 clinical isolates with PBP 2-X allele.
To investigate the horizontal transfer of penA, we analyzed a murE-dcaA region of additional an ST1901 Cefs isolate (n = 1) and ST1901 CefRs isolates (n = 5) (Table 1). As shown in Fig. 4C, the penA-murE-dcaA region of the ST1901 Cefs clinical isolate (NG0402) was identical to that of NG0202. The sequence of the penA-murE-dcaA region of the ST1901 CefRs strains NG0002, NGON03-079, NGON03-092, and NGON03-115 was identical to that of NG0003 and most of the clones (type I) obtained in the in vitro experiment. NGON07-002 had a murE sequence identical to that of NG0003, but the polymorphism sites in dcaA of NGON07-002 were the same as those of NG0202, implying that the recombination junction of NGON07-002 was within the region from positions 4100 to 5500. The results suggested that similar DNA transfer and recombination events involving penA-X might occur in vivo.
DISCUSSION
Reduced susceptibility to cefixime has been associated with the mosaic-type penA-X allele encoding PBP 2-X or its derivatives with minor differences (10, 25, 27). However, the genetic relatedness between CefRs N. gonorrhoeae isolates has not been completely elucidated. In the study described here, we applied MLST analysis to reveal the clonality of the CefRs N. gonorrhoeae isolates in our collection and showed that CefRs N. gonorrhoeae isolates belong to six different MLST types. One possible explanation for the wide distribution of CefRs N. gonorrhoeae is the introduction of penA from other species to these STs by interspecies recombination (3, 10). We found that the minor types of PBP 2, PBP 2-XXX and PBP 2-XXXII, were seen only in ST7363 and ST1901 strains, respectively. Although we should analyze more CefRs isolates, this may imply that the independent introduction of a DNA segment from a putative common ancestor has occurred, as proposed previously (24).
However, the putative original penA-X allele was the predominant allele among CefRs strains in this study as well as in other studies (10, 27). All nine types of CefRs-associated PBP 2 seem to be derived from the putative original penA-X (Fig. 1). Therefore, another possible explanation for the wide distribution of CefRs is that a putative original CefRs clone may emerge in a given lineage and clonally expand worldwide (14, 27). This is also suggested by our finding that ST7363 with the penA-X allele is predominant. During the spread of the CefRs clone, mutations may be introduced, resulting in the emergence of new variants of penA-X. Another possibility is that the observed predominance may reflect fitness. If the penA-X allele has an advantage in cell growth over other alleles, the result is the elimination of the other alleles, although there is no evidence for such a difference.
The horizontal transfer of the penA-X allele shown in the present study can explain the clonality of CefRs-associated PBP 2 even in isolates of different STs. We demonstrated the in vitro transfer of the penA-X allele from CefRs ST7363 to CefS ST1901. Our sequence analysis of the penA-flanking region in the clones that acquired penA-X (8 of 12) showed that penA and the downstream open reading frames for murE and dcaA were replaced. Furthermore, the sequences from the CefRs clinical isolates of ST1901 were also identical to those of the clones generated in vitro, supporting the possibility of the in vivo spread of the penA-flanking DNA segment. To our knowledge, this is the first case that suggests the interstrain transfer of a chromosomally encoded antibiotic resistance-conferring gene in N. gonorrhoeae by natural transformation in nature.
In addition to the horizontal transfer of penA, we observed the generation of penA allele diversity by the introduction of point mutations and the formation of a mosaic structure between a donor and a recipient in vitro. The penA alleles of one-third (4 of 12) of the transformants analyzed had minor differences from those of both the donor and the recipient. This is inconsistent with an observation mentioned by Spratt et al. (24). They could not detect any sequence variation during experimental transformation by using a PCR-amplified N. meningitidis penA gene. This discrepancy may be due to differences in the experimental procedures used, for example, a coculture assay versus transformation by use of a PCR product. However, we should examine more details about the natural transformation system, including the repair process, in N. gonorrhoeae. Nonetheless, the dynamic change observed in the allele during transformation may explain the diversity of the penA allele-derived CefRs clinical isolate. Determination of the mutation rate for the penA allele during in vitro passages and analysis of more CefRs isolates from various geographical areas will help improve our understanding of the diversity of the penA allele.
N. gonorrhoeae is a highly recombinogenic pathogen. DNA transformation contributes to the interspecies acquisition of chromosomally encoded antibiotic resistance (10, 23). DNA uptake in Neisseria is directly affected by piliation of the cells and the 10-bp-specific DNA uptake sequence (1, 9). After the DNA is internalized, it can be efficiently recombined with a homologous sequence on the recipient chromosome. As the efficiency of homologous recombination is correlated with sequence homology, intraspecies genetic exchange may be more efficient than interspecies exchange (8). If so, once N. gonorrhoeae acquires a genetic element from another bacterium that provides an advantage for N. gonorrhoeae survival in vivo, the acquired element would easily be spread among N. gonorrhoeae strains under selective pressure.
MLST is used for phylogenetic analysis for many other bacteria because the nucleotide sequence variation of housekeeping genes is likely to accumulate slowly and to be selectively neutral (4, 6, 16). However, the phylogeny of highly recombinogenic bacteria such as Neisseria species are difficult to study due to the exchange of DNA segments by natural transformation, resulting in the formation of nonclonal populations (21). Therefore, CefRs isolates also might exchange the allele(s) utilized in MLST analysis by a recombinational event. As the allele profiles of ST7363, ST1588, and ST1596 were very similar to each other, these STs might be expected to be genetically related (the ST1596 complex). If we can assume that the housekeeping genes are exchangeable between strains, CefRs isolates belonging to ST1596 complex might emerge by allele exchange, despite penA allele transfer. Other than the ST1596 complex, ST1901, which is one of the STs found in CefRs isolates with the penA-X allele, has three loci, abcZ, fumC, and aroE, different from those in ST7363 (Table 3). These loci are scattered on the N. gonorrhoeae chromosome (5). Because even the loci closest to each other, abcZ and fumC, are 140 kb apart on the N. gonorrhoeae chromosome (5), evolution from ST7363 to ST1901 (or the other direction) would require three independent genetic events. However, we cannot suggest that this scenario is completely exclusive, since N. gonorrhoeae has a high likelihood of acquiring DNA from other cells.
As N. gonorrhoeae is an obligate human pathogen, there is neither transmission to other animals nor an environmental reservoir. Genetic exchange between two different strains must take place when one strain meets another strain within an individual host. Recently, two independent groups showed evidence for N. gonorrhoeae mixed infections (15, 17). The spread of an antibiotic resistance gene demonstrated in this study could also occur during a mixed infection, probably in highly sexually active persons. It should be noted that the frequency of penA allele transfer was relatively high (approximately 2 cells per 104 recipients). As expected previously and also as demonstrated in this study, the high natural competence of N. gonorrhoeae plays an important role in the transfer of a mosaic penA allele among different types of N. gonorrhoeae strains. As a result, the prevalence of the allele would be increasing in the population, although it remains unclear whether the other determinants are spread like the penA allele. If it is assumed that the spread occurs frequently, we need to reinforce surveillance for asymptomatic mixed gonococcal infections to prevent the spread of resistance-conferring genes.
Acknowledgments
We thank H. Takahashi and H. Izumiya for helpful discussions. We thank H. Matsuoka for technical assistance.
This work was partly supported by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan (grants H21-Shinkou-Ippan-001 and H21-Shinkou-Ippan-012).
Footnotes
Published ahead of print on 22 December 2009.
REFERENCES
- 1.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 2.Akasaka, S., T. Muratani, Y. Yamada, H. Inatomi, K. Takahashi, and T. Matsumoto. 2001. Emergence of cephem- and aztreonam-high-resistant Neisseria gonorrhoeae that dose not produce beta-lactamase. J. Infect. Chemother. 7:49-50. [DOI] [PubMed] [Google Scholar]
- 3.Ameyama, S., S. Onodera, M. Takahata, S. Minami, N. Maki, K. Endo, H. Goto, H. Suzuki, and Y. Oishi. 2002. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46:3744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, J., K. A. Jolley, P. F. Sparling, N. J. Saunders, C. A. Hart, I. M. Feavers, and M. C. J. Maiden. 2007. Species status of Neisseria gonorrhoeae: evolutionary and epidemiological inferences from multilocus sequence typing. BMC Biol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, G. T., J. S. Yoo, H. B. Oh, Y. S. Lee, S. H. Cha, S. J. Kim, and C. K. Yoo. 2008. The complete genome sequence of Neisseria gonorrhoeae NCCP11945. J. Bacteriol. 190:6035-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, C., W. P. Hanage, and B. G. Spratt. 2007. Recombination and the nature of bacterial speciation. Science 315:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, M., T. Deguchi, K. Mizutani, M. Yasuda, S. Yokoi, S. Ito, Y. Takahashi, S. Ishihara, Y. Kawamura, and T. Ezaki. 2005. Emergence and spread of Neisseria gonorrhoeae clinical isolates harbouring mosaic-like structure of penicillin-binding protein 2 in central Japan. Antimicrob. Agents Chemother. 49:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda, W. M., and C. A. Gaydos. 2007. Neisseria, p. 601-620. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 12.Jolley, K. A. 2001. Multi-locus sequence typing, p. 173-186. In A. J. Pollard and M. C. Maiden (ed.) Meningococcal diseases: methods and protocols. Humana Press, Totowa, NJ.
- 13.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) database. BMC Bioinform. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindberg, R., H. Fredlund, R. Nicholas, and M. Unemo. 2007. Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrob. Agents Chemother. 51:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynn, F., M. M. Hobbs, J. M. Zenilman, F. M. T. F. Behets, K. Van Damme, A. Rasamindrakotroka, and M. C. Bash. 2005. Genetic typing of the porin of Neisseria gonorrhoeae from clinical noncultured samples for strain characterization and identification of mixed gonococcal infections. J. Clin. Microbiol. 43:368-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, I. M. C., and C. A. Ison. 2003. Detection of mixed infection of Neisseria gonorrhoeae. Sex. Transm. Infect. 79:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muratani, T., S. Akasaka, T. Kobayashi, Y. Yamada, H. Inatomi, K. Takahashi, and T. Matsumoto. 2001. Outbreak of cefzopran (penicillin, oral cephems, and aztreonam)-resistant Neisseria gonorrhoeae in Japan. Antimicrob. Agents Chemother. 45:3603-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing, 9th informational supplement M7-A4 (M100-S9). National Committee for Clinical Laboratory Standards, Wayne, PA.
- 20.Ochiai, S., H. Ishiko, M. Yasuda, and T. Deguchi. 2008. Rapid detection of the mosaic structure of the Neisseria gonorrhoeae penA gene, which is associated with decreased susceptibilities to oral cephalosporins. J. Clin. Microbiol. 46:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Rourke, M., and B. G. Spratt. 1994. Further evidence for the non-clonal population structure of Neisseria gonorrhoeae: extensive genetic diversity within isolates of the same electrophoretic type. Microbiology 140:1285-1290. [DOI] [PubMed] [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Spratt, B. G. 1988. Hybrid penicillin-binding proteins in penicillin-resistant gonococci. Nature 332:173-176. [DOI] [PubMed] [Google Scholar]
- 24.Spratt, B. G., L. D. Bowler, Q.-Y. Zhang, J. Zhou, and J. M. Smith. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115-125. [DOI] [PubMed] [Google Scholar]
- 25.Takahata, S., N. Senju, Y. Osaki, T. Yoshida, and T. Iida. 2006. Amino acid substitutions in mosaic penicillin-binding protein 2 associated with reduced susceptibility to cefixime in clinical isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 50:3638-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 27.Whiley, D. M., E. A. Limnios, S. Ray, T. P. Sloots, and J. W. Tapsall. 2007. Diversity of penA alterations and subtypes in Neisseria gonorrhoeae strains from Sydney, Australia, that are less susceptible to ceftriaxone. Antimicrob. Agents Chemother. 51:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamai, S., Y. Obara, T. Nikkawa, Y. Shimoda, and Y. Miyamoto. 1979. Preservation of Neisseria gonorrhoeae by the gelatin-disc method. Br. J. Vener. Dis. 55:90-93. [DOI] [PMC free article] [PubMed] [Google Scholar]